Abstract
Enlarged vestibular aqueduct (EVA) is one of the most common congenital inner ear malformations and accounts for 1–12% of sensorineural deafness in children and adolescents. Multiple genetic defects contribute to EVA; therefore, early molecular diagnosis is critical for EVA patients to ensure that the most effective treatment strategies are employed. This study explored a new genetic diagnosis method for EVA and applied it to clinic diagnoses of EVA patients. Using next-generation sequencing technology, we set up a multiple polymerase chain reaction enrichment system for target regions of EVA pathogenic genes (SLC26A4, FOXI1, and KCNJ10). Forty-six EVA samples were sequenced by this system. Variants were detected in 87.0% (40/46) of cases, including three novel variants (SLC26A4 c.923_929del, c.1002-8C>G, and FOXI1 c.519C>A). Biallelic potential pathogenic variants were detected in 27/46 patient samples, leading to a purported diagnostic rate of 59%. All results were verified by Sanger sequencing. Our target region capture system was validated to amplify and measure SLC26A4, FOXI1, and KCNJ10 in one reaction system. The result supplemented the mutation spectrum of EVA. Thus, this strategy is an economic, rapid, accurate, and reliable method with many useful applications in the clinical diagnosis of EVA patients.
Introduction
Enlarged vestibular aqueduct (EVA; MIM 600791) is an autosomal recessive genetic disease causing congenital inner ear malformation that accounts for 1–12% of sensorineural deafness in children and adolescents [1]. EVA can be divided into syndromic EVA (mostly Pendred syndrome [PDS]; MIM274600) and the more common nonsyndromic EVA (DFNB4; MIM 600791) depending on the presence of other inner ear malformations or diseases.
The SLC26A4 (DFNB4; MIM 605646) gene encodes pendrin, which is expressed in the inner ear and is responsible for EVA symptoms. In many EVA patients, SLC26A4 screening identified two disease-causing allele variants[2]. More than 300 variants have been identified in the SLC26A4 gene with EVA or PDS (www.healthcare.uiowa.edu/labs/pendredandbor), and each ethnic population has a different and diverse variant spectrum with their own specific mutation hot spots [3,4]. The function of SLC26A4 also has been explored in Pendrin knockout mice that recapitulate the pathology observed in humans: profound deafness and bulged endolymphatic spaces of the inner ear with striking [5]. Recent studies found that the SLC26A4 promoter contains a key transcriptional regulatory element that binds FOXI1 (MIM 601093), a transcriptional activator of the gene [6]. Additionally, double heterozygosity of SLC26A4 and KCNJ10 (MIM 602208) was identified in individuals with an EVA phenotype from two families, linking KCNJ10 variants with EVA [7].
Routine clinical examinations to diagnose EVA involve audiological tests (e.g., pure tone audiometry, acoustic immittance, auditory steady-state response, and auditory brainstem response) and temporal bone imaging (e.g., computed tomography and magnetic resonance imaging) to reveal expansile vestibular or endolymphatic sac. EVA manifests clinically as fluctuating or progressive sensorineural hearing loss, ranging from mild to profound deafness [8], so most patients are clinically diagnosed when their hearing is already poor. Therefore, early clinical genetic diagnoses of EVA patients are critical to clarify the molecular etiology and implement the appropriate disease control and prevention responses, such as avoiding head trauma, getting cold and noise stimulation. When patients with hearing loss were diagnosed of EVA, they can choose hearing aid or artificial cochlear implantation as soon as possible.
EVA molecular diagnoses have traditionally relied upon Sanger sequencing. More recently, variant detection systems using denaturing high-performance liquid chromatography (DHPLC) have been developed for PDS screening [9,10]. Array-based variant screenings also are a rapid and efficient technique to detect known variants. A recently developed genome-wide association approach to detect loci affecting PDS susceptibility used 597 genotyped sows with 62,163 single nucleotide polymorphisms (SNPs) [11]. We developed a microarray to detect 240 variants underlying syndromic and nonsyndromic sensorineural hearing loss, including 11 distinct variants in SLC26A4 [12]. However, all of these EVA genetic diagnosis strategies rely upon either full gene sequencing or test for common variants in only the SLC26A4 gene. These approaches are not optimal, as they do not allow for all EVA-associated genes to be simultaneously examined. Furthermore, these techniques are both time-consuming and costly, limiting their clinical application. Recent developments in next-generation sequencing (NGS) have widely expanded its use in scientific research and clinical fields, promoting the development of assays to rapidly and cost-effectively sequence all genes and noncoding regions of interest. NGS can provide more precise information about genetic causes of disease and recurrence risks, ultimately leading to better treatment [13,14]. Several NGS methods have been developed and clinically applied for hereditary hearing loss [14,15,16].
We developed a molecular diagnosis for EVA based on multiple polymerase chain reaction (PCR) targeted enrichment and NGS that includes three known EVA-associated genes. This genetic diagnostic can be extended to clinical practices and has the power to advance simple genetic screening into genetic diagnoses.
Materials and Methods
Subjects
A cohort of 46 sporadic Chinese probands diagnosed with EVA was recruited between 2014 and 2016 from the Otolaryngology Department of Xiangya Hospital, Central South University, including 32 males and 14 females, the average age was 6, range from 1 to 26 years old (Table 1). A detailed medical history was available for each proband. Every participant was examined thoroughly, including systemic and specialized physical examination, electric otoscopy and audiological assessment (pure tone audiometry, acoustic immittance, auditory steady-state response and auditory brainstem response). High Resolution Computed Tomography (HRCT) scanning of temporal bone and Magnetic Resonance Hydrography (MRH) examination of inner ear were performed on all the patients. Inclusion criteria of EVA patients were HRCT shows significant bone defect on posterior border of petrosum. The width of external opening of vestibular aqueduct is more than 1.5 millimeter, or the width of middle opening is over 2 millimeter. MRH shows expanded endolymphatic sac. Syndromic features were not detected. The controls consisted of 100 unrelated healthy Chinese volunteers with normal hearing and without another genetic disease. All patients and controls were ethnically Chinese. Written informed consent was obtained from all the participants or their parents (when participants were under 18 years old).and the research was approved by the Ethic Committee of the Xiangya Hospital of Central South University and is compliant with the Code of Ethics of the World Medical Association[17].
Table 1. Details of Phenotype of the 46 patients.
| Patient number | Gender | Agea | Pre/Postlingual | Degree of hearing lossb | Evolution of hearing loss | IEM |
|---|---|---|---|---|---|---|
| 01 | Male | 6 | Prelingual | Profound | Progressive | EVA |
| 02 | Male | 7 | Prelingual | Profound | Fluctuating | EVA |
| 03 | Male | 2 | Prelingual | Profound | Stable | EVA |
| 04 | Male | 13 | Prelingual | Severe | Fluctuating | EVA |
| 05 | Female | 10 | Prelingual | Severe | Progressive | EVA |
| 06 | Female | 5 | Prelingual | Profound | Fluctuating | EVA |
| 07 | Female | 6 | Prelingual | Severe | Stable | EVA |
| 08 | Female | 11 | Postlingual | Profound | Fluctuating | EVA |
| 09 | Female | 2 | Prelingual | Profound | Stable | EVA |
| 10 | Female | 5 | Prelingual | Profound | Progressive | EVA |
| 11 | Female | 1 | Prelingual | Severe | Stable | EVA |
| 12 | Female | 15 | Postlingual | Severe | Progressive | EVA |
| 13 | Female | 1 | Prelingual | Profound | Stable | EVA |
| 14 | Female | 2 | Prelingual | Profound | Stable | EVA |
| 15 | Female | 2 | Prelingual | Profound | Stable | EVA |
| 16 | Female | 6 | Postlingual | Profound | Progressive | EVA |
| 17 | Female | 13 | Prelingual | Profound | Fluctuating | EVA |
| 18 | Female | 7 | Postlingual | Severe | Fluctuating | EVA |
| 19 | Female | 6 | Prelingual | Profound | Progressive | EVA |
| 20 | Female | 3 | Prelingual | Profound | Stable | EVA |
| 21 | Female | 2 | Prelingual | Severe | Stable | EVA |
| 22 | Female | 4 | Prelingual | Profound | Fluctuating | EVA |
| 23 | Female | 2 | Prelingual | Profound | Stable | EVA |
| 24 | Female | 1 | Prelingual | Profound | Stable | EVA |
| 25 | Female | 6 | Prelingual | Profound | Fluctuating | EVA |
| 26 | Female | 6 | Prelingual | Severe | Progressive | EVA |
| 27 | Female | 4 | Prelingual | Profound | Fluctuating | EVA |
| 28 | Female | 4 | Prelingual | Profound | Stable | EVA |
| 29 | Female | 4 | Prelingual | Profound | Stable | EVA |
| 30 | Female | 6 | Prelingual | Severe | Progressive | EVA |
| 31 | Female | 10 | Prelingual | Profound | Progressive | EVA |
| 32 | Female | 11 | Prelingual | Severe | Fluctuating | EVA |
| 33 | Female | 1 | Prelingual | Profound | Stable | EVA |
| 34 | Female | 3 | Prelingual | Profound | Stable | EVA |
| 35 | male | 13 | Prelingual | Profound | Progressive | EVA |
| 36 | male | 4 | Prelingual | Profound | Stable | EVA |
| 37 | Female | 15 | Prelingual | Severe | Fluctuating | EVA |
| 38 | male | 10 | Prelingual | Profound | Fluctuating | EVA |
| 39 | Female | 12 | Prelingual | Profound | Progressive | EVA |
| 40 | male | 2 | Prelingual | Profound | Stable | EVA |
| 41 | male | 26 | Postlingual | Severe | Fluctuating | EVA |
| 42 | Female | 4 | Prelingual | Profound | Stable | EVA |
| 43 | male | 5 | Prelingual | Profound | Stable | EVA |
| 44 | Female | 6 | Prelingual | Severe | Fluctuating | EVA |
| 45 | male | 6 | Prelingual | Profound | Progressive | EVA |
| 46 | male | 5 | Prelingual | Severe | Stable | EVA |
a Age (in years) at genetic consultation for study inclusion.
b Profound means hearing threshold consultation foeans hearing threshold 71-90dB, Moderate means hearing threshold 41-55dB, Mild means hearing threshold 26-40dB.
IEM: inner ear malformation
Design of Captured Target Genome Regions and Multiple PCR Enrichment System
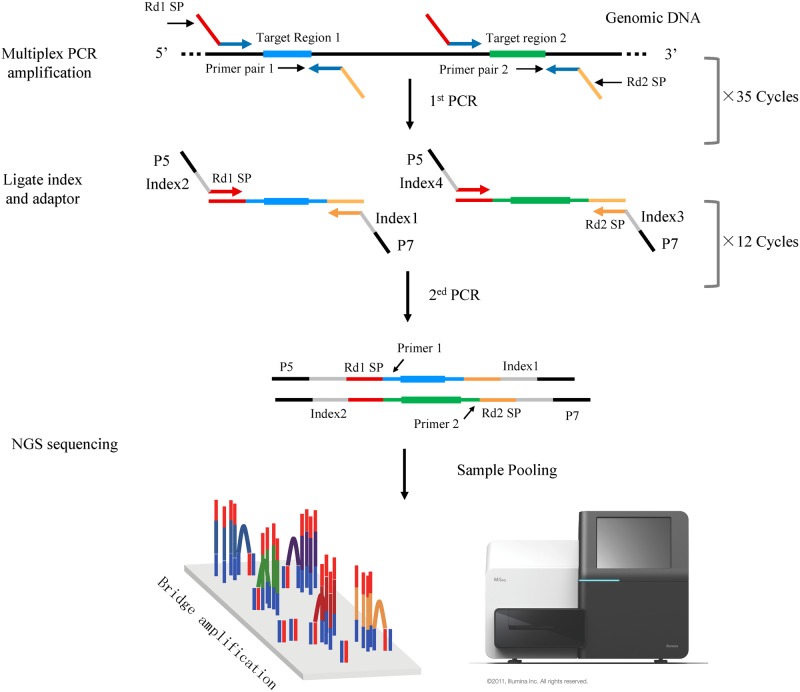
Genomic DNA was extracted from peripheral blood using standard phenol-chloroform protocols and stored at -20°C. After purification and quality testing, multiple PCR enrichment was performed in accordance with the special reaction conditions developed in this study. The target genome regions of the three candidate genes (SLC26A4, FOXI1, and KCNJ10) were designed to include their promoter regions (~500 bp), 5`untranslated region (5`UTR), coding regions, splice sites (~8 bp), and 3' untranslated region (3' UTR) (Table 2). Thirty-nine primer pairs were designed using FastTarget Primer (V5.0.1) software developmented by Genesky, with the most stringent conditions (no SNPs in primer annealing region, amplicon length between 230–315 bp, GC content between 30 and 80%). These primers were synthesized and assigned into four multiplex PCR panels to amplify all the target regions of the three genes. The first round enrichment amplification reactions were carried out on a ABI 2720 Thermal Cycler (Life Technologies Corporation, USA) with following cycling program: 95°C for 2 min; 11 cycles of 94°C for 20 s, 63°C-0.5°C per cycle for 40 s, 72°C for 1mins; 24 cycles of 94°C for 20 s, 65°C for 30 s, 72°C for 1 mins; 72°C for 2 min. In the second round, four multiple PCR reaction products from the first round were mixed, and a pair of universal primer with an added index sequence was used to amplify for distinguishing different samples. The EVA sequencing library was constructed after the two rounds of amplification (Fig 1).
Table 2. EVA-associated genes and their PCR target regions.
| Gene Name | NM_Accession | Exon Number | Gene Length (bp) | mRNA Length (bp) | Target regions length |
|---|---|---|---|---|---|
| FOXI1 | NM_012188.4 | 2 | 3813 | 2296 | 619|coding+5'UTR,1677(563)|coding+3'UR |
| KCNJ10 | NM_002241.4 | 2 | 32795 | 5306 | 5066(1141)|coding+3'UTR,240|5'UTR |
| SLC26A4 | NM_000441.1 | 21 | 57175 | 4930 | 221|5'UTR,167|coding+5'UTR,140,111,185,165,153,83,148,114,78,96,107,70,93,96,231,55,146,84,2387(24)|coding+3'UTR |
Fig 1. Multiple PCR Target Enrichment and Next-Generation Sequencing.
Next-Generation Sequencing
The PCR production of each sample was labeled with 8bp index, all the libraries of each sample were pooled. After Cluster Generation and hybridization of sequencing primer, base incorporation was carried out on MiSeq Benchtop Sequencer (Illumina, Inc, San Diego, CA) in one single lane following the manufacturer's standard cluster generation and sequencing protocols, for 608 cycles of sequencing per read to generate paired-end reads including 300bp at each end and 8 bp of the index tag. The average effective sequencing depth of every sample was300× and sequencing depth of all bases was above 20×.
Variants Analysis
Sequencing reads were aligned to hg19 using the Burrows–Wheeler Aligner (BWA) [18]. SNV calling was performed using both GATK and Varscan programs [19, 20], and the called SNV data were then combined. The Annovar program was used for SNV annotation [21]. The functional effect of non-synonymous SNVs was assessed by the PolyPhen-2, SIFT, and MutationTaster[22,23,24]. Non-synonymous SNVs with SIFT score of <0.05, Polyphen-2 score of >0.85 or MutationTaster score of >0.85 were considered as significant of not being benign. To sort potentially deleterious variants from benign polymorphisms, perl scripts were used to filter the SNVs against those of dbSNP135. Any SNV recorded in dbSNP135 and with a minor allele frequency of ≥1% in Chinese from 1000 genome database was considered as benign polymorphisms and therefore removed for subsequent analysis. We also test all the variants for their allele frequency in the Exac exome variant database (http://exac.broadinstitute.org/) to further support the novel variants being pathogenic.
Sanger Sequencing
Variants selected and suspected to be pathogenic were confirmed by Sanger sequencing. Parental samples were used for segregation analysis of the sequence variants identified in the index proband via Sanger sequencing. In addition, 100 controls were sequenced for the variants detected to evaluate the population-wide incidence of the novel variants. Data were analyzed using DNASTAR software program (DNASTAR, Inc., Madison, Wisconsin, US).
Results
All probands were diagnosed as DFNB4. Possibly pathogenic gene variants were found in 40 of 46 cases (87%). Thirty-eight cases carried SLC26A4 variants and two cases carried FOXI1 variants. KCNJ10 gene variants were not detected. By analyzing variant results of all the available DNA of patients’ parents, we found that 27 cases conformed to cosegregation principles, including 19 compound heterozygous, two homozygous variants and six heterozygous (Table 3), leading to a purported diagnostic rate of 59%.
Table 3. SNVs detected in 46 EVA probands.
| Patient number | SLC26A4 | FOXI1 | KCNJ10 | Genotype | Inheritance statusa | |
|---|---|---|---|---|---|---|
| Mat | Pat | |||||
| 01 | c.919-2A>G | wt | wt | HE | N/A | N/A |
| 02 | wt | c.519C>A | wt | HE | wt | wt |
| 03 | c.1343C>T;c.2168A>G | wt | wt | Comp HE | c.1343C>T | c.2168A>G |
| 04 | c.919-2A>G;c.1975G>C | wt | wt | Doub HE | N/A | N/A |
| 05 | c.2T>C;c.269C>T | wt | wt | Doub HE | wt | c.269C>T |
| 06 | c.1229C>T | wt | wt | HOM | N/A | N/A |
| 07 | wt | wt | wt | - | N/A | N/A |
| 08 | c.754T>C;c.919-2A>G | wt | wt | Comp HE | c.754T>C | c.919-2A>G |
| 09 | c.919-2A>G;c.1229C>T | wt | wt | Comp HE | c.919-2A>G | c.1229C>T |
| 10 | wt | wt | wt | - | N/A | N/A |
| 11 | wt | c.716C>T | wt | HE | wt | c.716C>T |
| 12 | c.2168A>G | wt | wt | HE | wt | c.2168A>G |
| 13 | c.919-2A>G | wt | wt | HE | c.919-2A>G | wt |
| 14 | c.919-2A>G | wt | wt | HE | c.919-2A>G | wt |
| 15 | c.109G>T;c.1079C>T | wt | wt | Doub HE | wt | c.109G>T |
| 16 | c.919-2A>G | wt | wt | HOM | c.919-2A>G | c.919-2A>G |
| 17 | c.919-2A>G | wt | wt | HOM | c.919-2A>G | c.919-2A>G |
| 18 | c.919-2A>G;c.2000T>C | wt | wt | Comp HE | c.2000T>C | c.919-2A>G |
| 19 | c.919-2A>G;c.2000T>C | wt | wt | Comp HE | c.2000T>C | c.919-2A>G |
| 20 | c.754T>C;c.919-2A>G | wt | wt | Comp HE | c.754T>C | c.919-2A>G |
| 21 | c.1174A>T;c.1716T>A | wt | wt | Doub HE | N/A | N/A |
| 22 | c.422T>C;c.1229C>T | wt | wt | Comp HE | c.1229C>T | c.422T>C |
| 23 | c.1174A>T;c.1229C>T | wt | wt | Comp HE | c.1174A>T | c.1229C>T |
| 24 | wt | wt | wt | - | N/A | N/A |
| 25 | wt | wt | wt | - | N/A | N/A |
| 26 | c.589G>A;c.919-2A>G | wt | wt | Comp HE | c.589G>A | c.919-2A>G |
| 27 | c.919-2A>G;c.1547dupC | wt | wt | Comp HE | c.1547dupC | c.919-2A>G |
| 28 | c.919-2A>G;c.1547dupC | wt | wt | Comp HE | c.1547dupC | c.919-2A>G |
| 29 | c.1173C>A;c.1229C>T | wt | wt | Doub HE | N/A | N/A |
| 30 | c.919-2A>G;c.1786C>T | wt | wt | Comp HE | c.919-2A>G | c.1786C>T |
| 31 | c.754T>C;c.919-2A>G | wt | wt | Comp HE | c.754T>C | c.919-2A>G |
| 32 | c.754T>C | wt | wt | HE | c.754T>C | c.919-2A>G |
| 33 | wt | wt | wt | - | N/A | N/A |
| 34 | c.2162C>T | wt | wt | HE | wt | c.2162C>T |
| 35 | c.919-2A>G; c.923_929del | wt | wt | Comp HE | c.919-2A>G | c.923_929del |
| 36 | c.754T>C;c.1229C>T | wt | wt | Comp HE | c.754T>C | c.1229C>T |
| 37 | c.1229C>T;c.2168A>G | wt | wt | Comp HE | c.1229C>T | c.2168A>G |
| 38 | c.1229C>T;c.2168A>G | wt | wt | Comp HE | c.1229C>T | c.2168A>G |
| 39 | c.919-2A>G;c.1694dupA | wt | wt | Doub HE | N/A | N/A |
| 40 | c.919-2A>G | wt | wt | HE | N/A | N/A |
| 41 | c.919-2A>G | wt | wt | HE | N/A | N/A |
| 42 | c.919-2A>G;c.2168A>G | wt | wt | Comp HE | c.919-2A>G | c.2168A>G |
| 43 | c.919-2A>G | wt | wt | HE | N/A | N/A |
| 44 | wt | wt | wt | - | N/A | N/A |
| 45 | c.919-2A>G;c.1002-8C>G | wt | wt | Doub HE | N/A | N/A |
| 46 | c.919-2A>G;c.1229C>T | wt | wt | Comp HE | c.1229C>T | c.919-2A>G |
a: Whether the variant was inherited from the parents or not;
Mat:maternal; Pat:paternal; HE: heterozygous; HOM: homozygous; Comp HE: compound heterozygous; Doub HE: double heterozygote, which need further analyses to conform that the two heterozygous mutations are located in one allele or two alleles respectively. wt: wild-type; N/A: data was not available.
We identified a total of 24 potentially pathogenic variants in these three genes (Table 4), including three novel variants (SLC26A4 c.923_929del, c.1002-8C>G and FOXI1 c.519C>A), which were absent in 100 control subjects and not reported in the dbSNP, 1000 Genomes Project database and the Exac exome variant database. To see if these mutations were de novo, we also sequenced their parental DNA, and found that the variant FOXI1 c.519C>A was not inherited from the parents. All the three novel variants were uploaded to the Leiden Open Variation Database (http://www.lovd.nl/3.0/home).
Table 4. Summary of variants.
| Gene | Sample Number | Exons/ Introns | Variants Type | Variants | Amino Acid Change | References |
|---|---|---|---|---|---|---|
| FOXI1 | 1 | exon1 | Missense | c.519C>A | p.H173Q | de novo |
| FOXI1 | 1 | exon2 | Missense | c.716C>T | p.P239L | [25] |
| SLC26A4 | 1 | exon2 | Missense | c.2T>C | p.M1T | [26] |
| SLC26A4 | 1 | exon2 | Missense | c.109G>T | p.E37X | [27] |
| SLC26A4 | 1 | exon3 | Missense | c.269C>T | p.S90L | [3] |
| SLC26A4 | 1 | exon5 | Missense | c.422T>C | p.F141S | [28] |
| SLC26A4 | 1 | exon5 | Missense | c.589G>A: | p.G197R | [27] |
| SLC26A4 | 5 | exon6 | Missense | c.754T>C | p.S252P | [3] |
| SLC26A4 | 24 | IVS7 | Splicing | c.919-2A>G | - | [29] |
| SLC26A4 | 1 | exon8 | Deletion | c.923_929del | p.308_310del | Novel |
| SLC26A4 | 1 | IVS8 | Splicing | c.1002-8C>G | - | Novel |
| SLC26A4 | 1 | exon9 | Missense | c.1079C>T | p.A360V | [30] |
| SLC26A4 | 1 | exon10 | Missense | c.1173C>A: | p.S391R | [31] |
| SLC26A4 | 2 | exon10 | Missense | c.1174A>T | p.N392Y | [3] |
| SLC26A4 | 9 | exon10 | Missense | c.1229C>T | p.T410M | [32] |
| SLC26A4 | 1 | exon12 | Missense | c.1343C>T | p.S448L | [33] |
| SLC26A4 | 2 | exon14 | Insertion | c.1547dupC | p.F515fs | [3] |
| SLC26A4 | 1 | exon15 | Insertion | c.1694dupA | p.F565fs | [34,35,36] |
| SLC26A4 | 1 | exon16 | Missense | c.1716T>A | p.F572L | [37] |
| SLC26A4 | 1 | exon16 | Missense | c.1786C>T | p.Q596X | [38] |
| SLC26A4 | 1 | exon17 | Missense | c.1975G>C | p.V659L | [39] |
| SLC26A4 | 2 | exon17 | Missense | c.2000T>C | p.F667S | [40] |
| SLC26A4 | 1 | exon19 | Missense | c.2162C>T | p.T721M | [41] |
| SLC26A4 | 5 | exon19 | Missense | c.2168A>G | p.H723R | [4] |
Novel refers to variants were absent in 100 control subjects and not reported in the dbSNP, 1000 Genomes Project database and the Exac exome variant database.
de novo refers to variants were absent in the parents, 100 control subjects and not reported in the dbSNP, 1000 Genomes Project database and the Exac exome variant database.
Of these variants, 22 were SLC26A4 variants and two were FOXI1 variants. Twenty-four variants included 19 missense, two insertions, one deletion, and two splicing variants. In the SLC26A4 gene, 19 compound heterozygous variants (50%), nine heterozygous variants (23.7%), and three homozygous variants (7.9%) were detected. Besides, we detected seven double heterozygote (18.4%), including five patients whose parents’ DNA was not available and two patients(05 and 15) carried two heterozygote, one of which derived from the paternal transmission, however, the other heterozygote was not inherited from the parents. We cannot be sure that the two heterozygous mutations of the seven patients are located in one allele or two alleles respectively, which needs further analyses. Both FOXI1 gene variants were heterozygous.
Sanger sequencing completely verified the NGS results, indicating that the NGS accuracy rate was 100% in our study.
Discussion
EVA is an autosomal recessive hereditary disease with obvious genetic heterogeneity that complicates investigations into its molecular mechanism. Currently studies suggest that an SLC26A4 biallelic variant (compound heterozygous or homozygous) was the main cause of EVA and PDS. EVA patients carrying SLC26A4 biallelic variants usually can be verified by videography diagnosis [27]. In our research, 50% of cases had compound heterozygous variants, 23.7% had heterozygous variants, and 7.9% had homozygous variants, consistent with previous studies. EVA patients carrying SLC26A4 monoallelic variants might only be carriers. However, there were a considerable number of EVA patients who carried SLC26A4 monoallelic variants or variants not detected in SLC26A4. Some arguments support that probably there are other undetected mutations harboring in the promoter region or in a potential splice site of intron of the second SLC26A4 allele, which was not searched in the present study, or there might be a digenic pattern of inheritance with the implication of a second gene [6]. In addition, some researchers suggested that the interaction between genetic and environmental factors may play a role in the pathogenic process of EVA [42].
To date, three genes have been associated with EVA: SLC26A4, FOXI1, and KCNJ10. Variants in SLC26A4 reportedly account for about 50% of PDS and nonsyndromic EVA cases [43], while FOXI1 and KCNJ10 account for only <1% of all cases [44]. Our result indicates that 82% of patients had SLC26A4 variants, further confirming that SLC26A4 is the most common pathogenic gene of EVA. At the time of writing, more than three hundred SLC26A4 variants have been reported (http://www.hgmd.cf.ac.uk/ac/index.php). In this study, we identified two novel mutations of SLC26A4 were not reported in the NCBI dbSNP, 1000 Genomes Project database and the Exac exome variant database, of which, a deletion (c.923_929del TAATTGC) was predicted to cause frameshift and produce truncated proteins by premature stops. The truncated region caused by the variation was located in a highly conserved region among mammals and located in the predicted SulP(high affinity sulphate transporter 1) domain, which were predicted to be disease causing by MutationTaster. In addition, a splice site change (c.1002-8C>G) of SLC26A4 was reported for the first time in this study. The variation was predicted to cause aberrant splicing and considered as pathogenic by MutationTaster. It was confirmed that SLC26A4 c.1002–4 C>G was contributed to PDS by mRNA studies revealing the splice mutation resulted in a putative truncated protein [45]. Interestingly, the novel variation (c.1002-8C>G) discovered in our study adjacent to the reported mutation (c.1002–4 C>G), which is possible impairs the same functional region of SLC26A4 lead to EVA by causing a frameshift and introduction of a premature stop codon. Functional analyses are suggested to be completed.
Though FOXI1 and KCNJ10 have been confirmed to be related to EVA, most research devoted to these genes has failed to find specific pathogenic variants through large sample screening studies [46,47]. Two FOXI1 gene variants were detected in our work and one KCNJ10 variant (c.812G>A) was detected in another study [48]. FOXI1 can activate and regulate the transcription of SLC26A4 gene by combined with two binding sites, FBS1 and FBS2, in promoter region of SLC26A4. A missense mutation (c.519C>A) of FOXI1 was reported for the first time in this study, which was absent in the parents, 100 control subjects and not reported in the dbSNP database. This variant lies within the conserved forkhead DNA-binding domain. The significance of variant located in this domain has been substantiated that compromise FOXI1 transactivation ability of SLC26A4 expression and are causally related to disease phenotype in EVA patients [6]. Therefore, it is possible that the mutation discovered in our study impairs its ability to activate SLC26A4 transcription. Additionally, the missense variant was predicted as disease causing by MutationTaster, a potential detrimental effect at the EVA phenotype is still possible to hypothesize. Functional study will be performed for verification in the future.
Several methods have been traditionally used for deafness gene detection (e.g., Sanger sequencing, restriction enzyme fingerprinting-single strand conformation polymorphism analysis, restriction fragment length polymorphism, DHPLC, gene chip, and mass spectrometry). While each of these technologies has its advantages, they also tend to be time-consuming, tedious, costly, and overall not suitable for large-scale detection in clinical applications. EVA displays high genetic heterogeneity with a genetic diagnosis involving multiple known and unknown loci. Thus, good diagnoses require simultaneous high-throughput detection of multiple gene variants. Since its introduction in 2005, NGS has revolutionized genomic research by providing more cost-effective, faster, and more high-throughput sequencing than traditional technologies [49, 50]. Three main NGS platforms currently exist: Illumina/Solexa, Roche/454, and Life Technologies/SOLiD [51]. In this study, the target region capture system used multiple PCR enrichment with special reaction conditions in a PCR-based non-hybridization gene enrichment scheme. Multiple PCR enrichment technology can run 140–200 multiplex PCRs of 150–450 bp simultaneously. This system is easy to use and allows simple NGS library preparation. The capture range is small and amplified segments overlap to optimize cost and uniform coverage. This method is customizable for application in unique research cases. The NGS-based targeted sequencing method developed in this study could directly achieve nearly complete coverage of all coding regions of the three EVA genes. Furthermore, our results manifested that this sequencing technology is highly sensitive and specific in detecting sequence variants in these EVA genes. We propose that this NGS-based screening strategy is an effective alternative method to identify the multiple genetic causes of EVA that will improve the molecular diagnosis of EVA patients in clinical applications.
Acknowledgments
We gratefully thank all subjects for their participation in this study.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was partly supported by the National Basic Research Program of China (973 Program) (2014CB943003), the National Nature Science Foundation of China (81470705 and 81301172), the Special Scientific Research Fund for Public Welfare Industry of the Ministry of Health in China (201302001), the Natural Science Foundation of Hunan Province, China (14JJ7009), and the Science and Technology Project of Hunan Province, China (S2012F1023). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Genesky Diagnostics Inc provided support in the form of salaries for author Deyuan Liu, but did not have any additional role in the research funding, study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of Deyuan Liu are articulated in the ‘author contributions’ section.
References
- 1.Arcand P, Desrosiers M, Dube J, Abela A. The large vestibular aqueduct syndrome and sensorineural hearing loss in the pediatric population. J Otolaryngol. 1991;20(4):247–50. Epub 1991/08/01. [PubMed] [Google Scholar]
- 2.Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, et al. A mutation in PDS causes non-syndromic recessive deafness. Nat Genet. 1998;18(3):215–7. Epub 1998/03/21. 10.1038/ng0398-215 [DOI] [PubMed] [Google Scholar]
- 3.Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40(4):242–8. Epub 2003/04/05. 10.1136/jmg.40.4.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Hauwe P, Everett LA, Coucke P, Scott DA, Kraft ML, Ris-Stalpers C, et al. Two frequent missense mutations in Pendred syndrome. Hum Mol Genet. 1998;7(7):1099–104. Epub 1998/06/09. [DOI] [PubMed] [Google Scholar]
- 5.Everett LA, Belyantseva IA, Noben-Trauth K, Cantos R, Chen A, Thakkar SI, et al. Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum Mol Genet. 2001;10(2):153–61. Epub 2001/01/12. [DOI] [PubMed] [Google Scholar]
- 6.Yang T, Vidarsson H, Rodrigo-Blomqvist S, Rosengren SS, Enerback S, Smith RJ. Transcriptional control of SLC26A4 is involved in Pendred syndrome and nonsyndromic enlargement of vestibular aqueduct (DFNB4). Am J Hum Genet. 2007;80(6):1055–63. Epub 2007/05/16. 10.1086/518314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T, Gurrola JG 2nd, Wu H, Chiu SM, Wangemann P, Snyder PM, et al. Mutations of KCNJ10 together with mutations of SLC26A4 cause digenic nonsyndromic hearing loss associated with enlarged vestibular aqueduct syndrome. Am J Hum Genet. 2009;84(5):651–7. Epub 2009/05/12. 10.1016/j.ajhg.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saliba I, Gingras-Charland ME, St-Cyr K, Decarie JC. Coronal CT scan measurements and hearing evolution in enlarged vestibular aqueduct syndrome. Int J Pediatr Otorhinolaryngol. 2012;76(4):492–9. Epub 2012/01/28. 10.1016/j.ijporl.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 9.Men M, Xue J, Jiang L, Wang H, Pan Q, Feng Y. Novel multiplex primer extension and denaturing high-performance liquid chromatography for genotyping of the deafness gene mutations. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011;36(11):1079–84. Epub 2011/12/16. 10.3969/j.issn.1672-7347.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 10.Dai P, Han D, Cao J, Zhai S, Kang D, Liu X, et al. [Genotypic analysis of familial dilated vestibular aqueduct syndrome]. Lin Chuang Er Bi Yan Hou Ke Za Zhi. 2006;20(4):147–50. Epub 2006/05/23. [PubMed] [Google Scholar]
- 11.Preissler R, Tetens J, Reiners K, Looft H, Kemper N. A genome-wide association study to detect genetic variation for postpartum dysgalactia syndrome in five commercial pig breeding lines. Anim Genet. 2013;44(5):502–8. Epub 2013/06/08. 10.1111/age.12047 [DOI] [PubMed] [Google Scholar]
- 12.Wu H, Feng Y, Jiang L, Pan Q, Liu Y, Liu C, et al. Application of a New Genetic Deafness Microarray for Detecting Mutations in the Deaf in China. PLoS One. 2016;11(3):e0151909 Epub 2016/03/29. 10.1371/journal.pone.0151909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi BY, Park G, Gim J, Kim AR, Kim BJ, Kim HS, et al. Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS One. 2013;8(8):e68692 Epub 2013/08/31. 10.1371/journal.pone.0068692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huijun Y, Yu L. [Application of next generation sequencing in gene identification and genetic diagnosis of hereditary hearing loss]. Yi Chuan. 2014;36(11):1112–20. Epub 2015/01/09. [DOI] [PubMed] [Google Scholar]
- 15.Yan D, Tekin M, Blanton SH, Liu XZ. Next-generation sequencing in genetic hearing loss. Genet Test Mol Biomarkers. 2013;17(8):581–7. Epub 2013/06/07. 10.1089/gtmb.2012.0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tekin D, Yan D, Bademci G, Feng Y, Guo S, Foster J 2nd, et al. A next-generation sequencing gene panel (MiamiOtoGenes) for comprehensive analysis of deafness genes. Hear Res. 2016;333:179–84. Epub 2016/02/07. 10.1016/j.heares.2016.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dale O, Salo M. The Helsinki Declaration, research guidelines and regulations: present and future editorial aspects. Acta Anaesthesiol Scand. 1996;40(7):771–2. Epub 1996/08/01. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. Epub 2010/01/19. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2014;20(9):1297–303. Epub 2010/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. Epub 2012/02/04. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164 Epub 2010/07/06. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. Epub 2003/06/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. Epub 2010/04/01. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–6. Epub 2010/08/03. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 25.Cirello V, Bazzini C, Vezzoli V, Muzza M, Rodighiero S, Castorina P, et al. Molecular and functional studies of 4 candidate loci in Pendred syndrome and nonsyndromic hearing loss. Mol Cell Endocrinol. 2012;351(2):342–50. Epub 2012/01/31. 10.1016/j.mce.2012.01.013 [DOI] [PubMed] [Google Scholar]
- 26.Prasad S, Kolln KA, Cucci RA, Trembath RC, Van Camp G, Smith RJ. Pendred syndrome and DFNB4-mutation screening of SLC26A4 by denaturing high-performance liquid chromatography and the identification of eleven novel mutations. Am J Med Genet A. 2004;124A(1):1–9. Epub 2003/12/18. 10.1002/ajmg.a.20272 [DOI] [PubMed] [Google Scholar]
- 27.Wang QJ, Zhao YL, Rao SQ, Guo YF, Yuan H, Zong L, et al. A distinct spectrum of SLC26A4 mutations in patients with enlarged vestibular aqueduct in China. Clin Genet. 2007;72(3):245–54. Epub 2007/08/28. 10.1111/j.1399-0004.2007.00862.x [DOI] [PubMed] [Google Scholar]
- 28.Chen K, Zong L, Liu M, Wang X, Zhou W, Zhan Y, et al. Developing regional genetic counseling for southern Chinese with nonsyndromic hearing impairment: a unique mutational spectrum. J Transl Med. 2014;12:64 Epub 2014/03/13. 10.1186/1479-5876-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coucke PJ, Van Hauwe P, Everett LA, Demirhan O, Kabakkaya Y, Dietrich NL, et al. Identification of two different mutations in the PDS gene in an inbred family with Pendred syndrome. J Med Genet. 1999;36(6):475–7. Epub 2000/06/30. [PMC free article] [PubMed] [Google Scholar]
- 30.Lai CC, Chiu CY, Shiao AS, Tso YC, Wu YC, Tu TY, et al. Analysis of the SLC26A4 gene in patients with Pendred syndrome in Taiwan. Metabolism. 2007;56(9):1279–84. Epub 2007/08/19. 10.1016/j.metabol.2007.05.013 [DOI] [PubMed] [Google Scholar]
- 31.Albert S, Blons H, Jonard L, Feldmann D, Chauvin P, Loundon N, et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur J Hum Genet. 2006;14(6):773–9. Epub 2006/03/30. [DOI] [PubMed] [Google Scholar]
- 32.Coyle B, Reardon W, Herbrick JA, Tsui LC, Gausden E, Lee J, et al. Molecular analysis of the PDS gene in Pendred syndrome. Hum Mol Genet. 1998;7(7):1105–12. Epub 1998/06/09. [DOI] [PubMed] [Google Scholar]
- 33.Wu CC, Yeh TH, Chen PJ, Hsu CJ. Prevalent SLC26A4 mutations in patients with enlarged vestibular aqueduct and/or Mondini dysplasia: a unique spectrum of mutations in Taiwan, including a frequent founder mutation. Laryngoscope. 2005;115(6):1060–4. Epub 2005/06/04. [DOI] [PubMed] [Google Scholar]
- 34.Chen DY, Chen XW, Jin X, Zuo J, Wei CG, Cao KL, et al. [Screening of SLC26A4 (PDS) gene mutation in cochlear implant recipients with inner ear malformation]. Zhonghua Yi Xue Za Zhi. 2007;87(40):2820–4. Epub 2008/01/03. [PubMed] [Google Scholar]
- 35.Jiang L, Feng Y, Chen H, He C, Mei L. [An investigation of SLC26A4 gene mutation in nonsydromic hearing impairment in Hunan province of China]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2010;24(13):587–91. Epub 2010/09/17. [PubMed] [Google Scholar]
- 36.Lim L, Subramaniam S, LiQing X, Khor CC, Goh D, Berne YI. Clinical, audiometric, radiologic, and genetic profiles of Southeast Asian children with hearing loss due to enlarged vestibular aqueduct. Otol Neurotol. 2011;32(9):1464–7. Epub 2011/10/12. [DOI] [PubMed] [Google Scholar]
- 37.Zhao J, Wu LQ, Feng Y, Pan Q, Zhao K, Li HY, et al. [Mutational screening of the SLC26A4 gene in patients with nonsyndromic hearing loss by denaturing high-performance liquid chromatography]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26(1):21–5. Epub 2009/02/10. 10.3760/cma.j.issn.1003-9406.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 38.Wu CC, Lu YC, Chen PJ, Yeh PL, Su YN, Hwu WL, et al. Phenotypic analyses and mutation screening of the SLC26A4 and FOXI1 genes in 101 Taiwanese families with bilateral nonsyndromic enlarged vestibular aqueduct (DFNB4) or Pendred syndrome. Audiol Neurootol. 2010;15(1):57–66. Epub 2009/08/04. 10.1159/000231567 [DOI] [PubMed] [Google Scholar]
- 39.Hu H, Wu L, Feng Y, Pan Q, Long Z, Li J, et al. Molecular analysis of hearing loss associated with enlarged vestibular aqueduct in the mainland Chinese: a unique SLC26A4 mutation spectrum. J Hum Genet. 2007;52(6):492–7. Epub 2007/04/20. [DOI] [PubMed] [Google Scholar]
- 40.Chen K, Wang X, Sun L, Jiang H. Screening of SLC26A4, FOXI1, KCNJ10, and GJB2 in bilateral deafness patients with inner ear malformation. Otolaryngol Head Neck Surg. 2012;146(6):972–8. Epub 2012/03/14. 10.1177/0194599812439670 [DOI] [PubMed] [Google Scholar]
- 41.Usami S, Abe S, Weston MD, Shinkawa H, Van Camp G, Kimberling WJ. Non-syndromic hearing loss associated with enlarged vestibular aqueduct is caused by PDS mutations. Hum Genet. 1999;104(2):188–92. Epub 1999/04/06. [DOI] [PubMed] [Google Scholar]
- 42.Bogazzi F, Russo D, Raggi F, Ultimieri F, Berrettini S, Forli F, et al. Mutations in the SLC26A4 (pendrin) gene in patients with sensorineural deafness and enlarged vestibular aqueduct. J Endocrinol Invest. 2004;27(5):430–5. Epub 2004/07/29. [DOI] [PubMed] [Google Scholar]
- 43.Anwar S, Riazuddin S, Ahmed ZM, Tasneem S, Ateeq ul J, Khan SY, et al. SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred's syndrome in Pakistanis. J Hum Genet. 2009;54(5):266–70. Epub 2009/03/17. 10.1038/jhg.2009.21 [DOI] [PubMed] [Google Scholar]
- 44.Choi BY, Madeo AC, King KA, Zalewski CK, Pryor SP, Muskett JA, et al. Segregation of enlarged vestibular aqueducts in families with non-diagnostic SLC26A4 genotypes. J Med Genet. 2009;46(12):856–61. Epub 2009/07/07. 10.1136/jmg.2009.067892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massa G, Jaenen N, de Varebeke SJ, Peeters N, Wuyts W. Solitary thyroid nodule as presenting symptom of Pendred syndrome caused by a novel splice-site mutation in intron 8 of the SLC26A4 gene. Eur J Pediatr. 2003;162(10):674–7. Epub 2003/08/16. [DOI] [PubMed] [Google Scholar]
- 46.Jonard L, Niasme-Grare M, Bonnet C, Feldmann D, Rouillon I, Loundon N, et al. Screening of SLC26A4, FOXI1 and KCNJ10 genes in unilateral hearing impairment with ipsilateral enlarged vestibular aqueduct. Int J Pediatr Otorhinolaryngol. 2010;74(9):1049–53. Epub 2010/07/14. 10.1016/j.ijporl.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 47.Landa P, Differ AM, Rajput K, Jenkins L, Bitner-Glindzicz M. Lack of significant association between mutations of KCNJ10 or FOXI1 and SLC26A4 mutations in Pendred syndrome/enlarged vestibular aqueducts. BMC Med Genet. 2013;14:85 Epub 2013/08/24. 10.1186/1471-2350-14-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Yuan Y, Huang S, Huang B, Cheng J, Kang D, et al. KCNJ10 may not be a contributor to nonsyndromic enlargement of vestibular aqueduct (NSEVA) in Chinese subjects. PLoS One. 2014;9(11):e108134 Epub 2014/11/06. 10.1371/journal.pone.0108134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim KM, Park JH, Bhattacharya D, Yoon HS. Applications of next-generation sequencing to unravelling the evolutionary history of algae. Int J Syst Evol Microbiol. 2014;64(Pt 2):333–45. Epub 2014/02/08. 10.1099/ijs.0.054221-0 [DOI] [PubMed] [Google Scholar]
- 50.Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5(1):16–8. Epub 2008/01/01. 10.1038/nmeth1156 [DOI] [PubMed] [Google Scholar]
- 51.Xuan J, Yu Y, Qing T, Guo L, Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340(2):284–95. Epub 2012/11/24. 10.1016/j.canlet.2012.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.