Abstract
Understanding the biology and molecular pathogenesis of ovarian epithelial cancer (EOC) is key to developing improved diagnostic and prognostic indicators and effective therapies. Although research has traditionally focused on the hypothesis that high-grade serous carcinoma (HGSC) arises from the ovarian surface epithelium (OSE), recent studies suggest that additional sites of origin exist and a substantial proportion of cases may arise from precursor lesions located in the Fallopian tubal epithelium (FTE). In FTE cells, the transcription factor PAX8 is a marker of the secretory cell lineage and its expression is retained in 96% of EOC. We have recently reported that PAX8 is involved in the tumorigenic phenotype of ovarian cancer cells. In this study, to uncover genes and pathways downstream of PAX8 involved in ovarian carcinoma we have determined the molecular profiles of ovarian cancer cells and in parallel of Fallopian tube epithelial cells by means of a silencing approach followed by an RNA-seq analysis. Interestingly, we highlighted the involvement of pathways like WNT signaling, epithelial-mesenchymal transition, p53 and apoptosis. We believe that our analysis has led to the identification of candidate genes and pathways regulated by PAX8 that could be additional targets for the therapy of ovarian carcinoma.
Keywords: PAX8, ovarian cancer, fallopian tube secretory epithelial cells, RNA-seq, transcriptional networks
INTRODUCTION
Ovarian cancer, like other cancers, is a spectrum of diseases and not a single disease entity [1]. Malignant surface epithelial tumors (carcinomas) are the most common ovarian cancers, accounting for 90% of the cases. These tumors are divided in four major histotypes: serous, mucinous, endometrioid and clear cell. Of these, high-grade serous carcinoma (HGSC) is the most common and lethal histotype, associated with abnormalities of BRCA1, BRCA2 and p53 [2]. This cancer is extremely genetic unstable and heterogenous; it is rarely detected while confined to the ovary and in about 90% of the cases it is diagnosed when women have already intra-abdominal spread. Although research has traditionally focused on the hypothesis that HGSC arises from the ovarian surface epithelium (OSE), it has been recently suggested that it may arise from the Fallopian tube fimbria. In particular, the serous tubal intraepithelial carcinomas (STICs), which are identified in the distal end (fimbria) of the Fallopian tube and arise from p53 mutations, have been indicated as the primary lesions that evolve through subsequent oncogenic events into HGSC [3]. This hypothesis is further supported by genetically engineered murine models that mimic the transformation from Fallopian tubal secretory epithelial cells to HGSC [4].
The transcription factor PAX8 is a marker of the Fallopian tube secretory cell lineage and its expression is retained in 96% of the serous ovarian carcinomas, in 89% of the endometrioid and 100% of the clear cell carcinomas, whilst it is not detected in the mucinous carcinoma [5]. PAX8 is a member of the PAX (PAired boX) gene family, genes tightly regulated in both temporal and spatial expression patterns [6]. PAX8 is crucial for the organogenesis of the thyroid gland, kidney, nervous system and Mullerian system [7, 8]. In the adult, PAX8 is expressed in the thyroid gland, in the renal excretory system and in tissues derived from the Mullerian ducts [9, 10]. Our recent results demonstrated that PAX8 plays a critical role in cell cycle progression and cell survival of differentiated epithelial cells [11], reinforcing the involvement of this transcription factor in different biological processes. In tumors, PAX8 is involved in the progression of follicular thyroid carcinomas [12] and it is overexpressed in the majority of gliomas, Wilms tumors and well-differentiated pancreatic neuroendocrine tumors [10, 13, 14]. In the scenario of ovarian cancer, PAX8 is the currently available most important marker [15] being a useful IHC target for the diagnosis of Mullerian tumors [16]. Despite PAX8 is normally expressed in Fallopian tube secretory cells, namely the cell of origin of HGSC, and is mainly considered a marker of tumor origin, we have recently reported its pivotal function in the tumorigenic phenotype of ovarian cancer cells. In particular, we showed that PAX8 plays a critical role in the migration, invasion and tumorigenic ability of ovarian cancer cells. In our published study, PAX8 silencing strongly suppressed anchorage-independent growth in vitro and significantly inhibited tumorigenesis in vivo in a nude mouse xenograft model [17]. In addition, the Cancer Genome Atlas (TGCA) Project indicated PAX8 as a survival gene essential for the proliferation of ovarian cancer cells [5]. Overall, PAX8 belongs to a class of lineage-survival genes that are required for both normal development of specific tissues and for cancer cell proliferation/survival.
On the basis of such evidences, it is conceivable that PAX8 is intimately involved in the progression of ovarian carcinomas. However, its function in Fallopian tube secretory epithelial cells, as well as the entire molecular network that allows PAX8 to regulate cellular processes in these cells and in ovarian cancer cells, is still an unexplored field. To improve our knowledge of the complex role of this transcription factor, we thought that it would be beneficial to uncover the downstream gene network governed by this transcriptional factor both in Fallopian tubal epithelium and ovarian cancer cells. Hence, the aims of our study were (1) to compare the transcriptome of normal Fallopian tube secretory epithelial cells with that of ovarian cancer cells in order to uncover genes and pathways modified during the transformation process; (2) to investigate PAX8 downstream gene regulatory network in physiological and pathological conditions to identify genes and pathways regulated by PAX8 that could be additional targets for the therapy of ovarian carcinoma.
RESULTS
Identification of genes differentially expressed between FT194 and SKOV-3 cells
Primary questions in the field of ovarian cancer biology concern its developmental cell of origin. The epithelial cells covering the ovaries (OSE) have historically been considered the site of origin of all ovarian cancers but recent evidence suggests that high-grade serous carcinoma (HGSC) originates from the Fallopian tube (FT) epithelium or the tuboperitoneal junction rather than from the OSE [4, 18].
To identify genes differentially expressed in the Fallopian tubal epithelium and in ovarian carcinoma and to disclose some of the pathways that might contribute to the carcinoma formation, an RNA sequencing analysis was performed on FT194 Fallopian tube secretory epithelial cells and SKOV-3 ovarian adenocarcinoma cells and a total of 12628 genes were mapped to the reference human genome (Supplementary Table S1). Of these, 7451 genes were differentially expressed in the two cell types at significant level (FDR-adjusted p-value ≤ 0.05) (Supplementary Table S2): 3553 genes resulted more expressed in SKOV-3 cells than in FT194 cells and 3778 genes were found less expressed in SKOV-3 cells with respect to FT194 cells. Interestingly, 38 genes were exclusively expressed in SKOV-3 cells, while 82 genes were present only in FT194 cells (Supplementary Table S2).
To date, despite the identification of a number of key mutations in p53 and BRCA1/2 genes, the complexity of the molecular pathway(s) underlying epithelial ovarian cancer has not been yet fully elucidated. To focus our attention on genes showing the greatest expression differences between FT194 and SKOV-3 cells, we applied a 2 fold-change cutoff and we searched for enrichment of specific gene sets at the Molecular Signatures Database (MsigDB). In particular, we explored the hallmark gene sets that summarize and represent specific well-defined biological states or processes. As shown in Figure 1A, genes up-regulated ≥ 2 fold in SKOV-3 cells present a significant enrichment for pathways such as estrogen response, epithelial mesenchymal transition, response to UV DNA damage, angiogenesis and Wnt/β-catenin signaling. There are numerous evidences, both in vitro and in vivo studies, that estrogens might regulate ovarian carcinogenesis [19]. It is well known that hormones stimulate the proliferation of the cells, meaning that they might function as carcinogens. For example, in ovarian cancer cells 17β-estradiol (E2) increases the ROS and NO production that participates in cancer progression [20]. In addition, aberrant Wnt/β-catenin signaling is implicated in several cancers including epithelial ovarian cancer [21, 22] and it represents one of the pathway involved in epithelial-mesenchymal transition (EMT) responsible for cancer cell dissemination and metastasis formation [23]. Stressful conditions such as UV radiations, hypoxia, and alterations associated with changes in cell phenotype that include EMT as well as angiogenesis represent main processes that provide carcinogenesis. At the same time, genes down-regulated ≥ 2 fold in SKOV-3 cells (Figure 1B) are involved in pathways as TNF-α signaling, IFN-α and IFN-γ responses and inflammatory response. TNF-α, IFN-α and IFN-γ are pleiotropic cytokines with diverse physiological functions such as activation of macrophages, induction of immune response and apoptosis and inhibition of cell proliferation. It has been reported that in ovarian cancer the above-mentioned cytokines exert an anti-proliferative activity and induce apoptosis [24, 25]. Eventually, if the immune response might contribute to the tumorigenic process, it may be also helpful in preventing the tumor properties.
Figure 1. MsigDB analysis of the most relevant genes differentially expressed between FT194 and SKOV-3 cells.
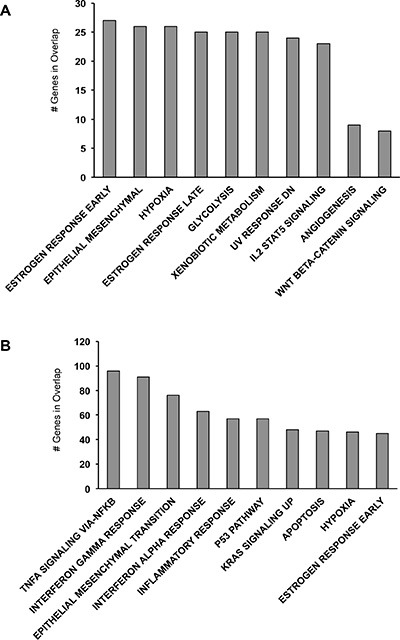
(A) MsigDB for genes upregulated ≥ 2 fold in SKOV-3 cells. (B) MsigDB for genes downregulated ≥ 2 fold in SKOV-3 cells.
Among the 38 genes exclusively expressed in SKOV-3 cells (Supplementary Table S2), ALDH1A1 [26], GABRA3 [27], FOLR1 [28], DPYSL5 [29], CGB8 [30], C8orf4 [31] and MAGEB2 [32] could be intriguing for the development and maintenance of the neoplastic phenotype. Similarly, among the 82 genes expressed only in FT194 cells (Supplementary Table S2) CDKN2A [33], MT1G [34], GPX7 [35], HCK [36], ZBTB16 [37] could be looked at as interesting genes being tumor suppressor epigenetically silenced in cancer cells.
PAX8 downstream target genes in FT194 and SKOV-3 cells
As already mentioned, in the Fallopian tube epithelium the transcription factor PAX8 is a marker of the secretory cell lineage. Our RNA sequencing analysis performed on FT194 Fallopian tube secretory epithelial cells and SKOV-3 ovarian adenocarcinoma cells confirmed that PAX8 is expressed at similar levels in these two cell types (Supplementary Table S2), supporting the hypothesis that PAX8 is not overexpressed in epithelial ovarian carcinoma but rather that its expression is conserved from the suggested cell of origin [38]. Recently, we have provided the first evidence of a clear involvement of PAX8 in the in vivo tumorigenesis of ovarian cancer cells [17], reinforcing the role of PAX genes in cancer through their effects on apoptosis resistance, tumor cell proliferation and migration, and repression of terminal differentiation.
On the basis of our previous data and in order to clarify PAX8 contribution to ovarian cancer through the identification of its downstream gene regulatory network, we have transiently knocked-down PAX8 expression in both FT194 and SKOV-3 cells. We chose to inhibit PAX8 expression also in the FT194 cell line because to date its role in the physiological contest of the Fallopian tube secretory cell has not been investigated. Moreover, we reasoned that genes affected by PAX8 silencing in both cell lines could be looked at as PAX8 targets with greater confidence.
Three independent silencing experiments were performed and analyzed by RNA-seq unraveling a total of 467 genes modulated by PAX8 expression in SKOV-3 and FT194 cells (Supplementary Table S3 and Supplementary Table S4). The reduced expression of PAX8 in the samples was confirmed by the RNA-seq findings, which showed an 80% decrease (FDR-adjusted p-value ≤ 0.05) (Supplementary Table S3 and Supplementary Table S4).
In SKOV-3 cells, 301 genes were significantly modulated by PAX8 silencing after correction for multiple testing (FDR-adjusted p-value ≤ 0.05) of which 214 were downregulated while 87 resulted upregulated (Supplementary Table S3). As shown in Table 1, among the most downregulated genes based on FDR there are TUBB2B, PPME1, ATP8A1, USP2, BLCAP, DIAPH1, FGF18 and KIF12.
Table 1. Differentially expressed genes that are significantly modulated after PAX8 silencing in SKOV– 3 cells.
| Gene Name | NCBI gene ID | FPKM1 | FPKM2 | Log2 fold change | FDR q value | Fold change | |
|---|---|---|---|---|---|---|---|
| 1 | ANKRD34B | 340120 | 0,517453 | 2,34955 | 2,18288 | 0,00400937 | 4,54059 |
| 2 | PAX8 | 7849 | 8,65929 | 30,224 | 1,80338 | 0,00400937 | 3,49037 |
| 3 | TUBB2B | 347733 | 0,483395 | 1,54683 | 1,67804 | 0,015208 | 3,19993 |
| 4 | PPME1 | 51400 | 11,285 | 34,0561 | 1,59351 | 0,00400937 | 3,01783 |
| 5 | LMLN | 89782 | 0,558739 | 1,61854 | 1,53445 | 0,00400937 | 2,89678 |
| 6 | H2AFY2 | 55506 | 6,68489 | 19,0865 | 1,51358 | 0,00400937 | 2,85518 |
| 7 | PHTF2 | 57157 | 3,23708 | 8,76508 | 1,43707 | 0,00400937 | 2,70770 |
| 8 | DCAF12L1 | 139170 | 1,66899 | 4,506 | 1,43287 | 0,00400937 | 2,69983 |
| 9 | GRIN2B | 2904 | 0,727982 | 1,94915 | 1,42087 | 0,0100329 | 2,67747 |
| 10 | EPRS | 2058 | 18,5003 | 47,5384 | 1,36154 | 0,00400937 | 2,56959 |
| 11 | PPAP2B | 8613 | 5,23887 | 13,3333 | 1,3477 | 0,00400937 | 2,54506 |
| 12 | ATP8A1 | 10396 | 0,686376 | 1,74582 | 1,34683 | 0,00400937 | 2,54353 |
| 13 | USP2 | 9099 | 0,582276 | 1,43193 | 1,29819 | 0,0127719 | 2,45920 |
| 14 | BLCAP | 10904 | 9,66632 | 23,4671 | 1,2796 | 0,00400937 | 2,42771 |
| 15 | FGF18 | 8817 | 2,82984 | 6,85593 | 1,27663 | 0,00400937 | 2,42272 |
| 16 | DIAPH1 | 1729 | 5,21853 | 12,5088 | 1,26123 | 0,00400937 | 2,39700 |
| 17 | KIF12 | 113220 | 2,32013 | 5,50046 | 1,24535 | 0,00400937 | 2,37076 |
| 18 | C10orf46 | 143384 | 14,0943 | 33,1881 | 1,23555 | 0,00400937 | 2,35471 |
| 19 | ANXA2P2 | 304 | 1,92906 | 4,516 | 1,22715 | 0,00400937 | 2,34104 |
| 20 | PSAP | 5660 | 201,624 | 467,609 | 1,21364 | 0,00400937 | 2,31922 |
| 21 | DIO2 | 1734 | 5,30475 | 1,69721 | – 1,64412 | 0,00400937 | 3,12557 |
| 22 | ROS1 | 6098 | 2,97713 | 1,06039 | – 1,48933 | 0,00400937 | 2,80759 |
| 23 | P4HA3 | 283208 | 1,91889 | 0,746284 | – 1,36248 | 0,00400937 | 2,57127 |
| 24 | PTPMT1 | 114971 | 14,4328 | 5,88494 | – 1,29426 | 0,0196014 | 2,45251 |
| 25 | MET | 4233 | 47,8695 | 20,5651 | – 1,21891 | 0,00400937 | 2,32771 |
| 26 | CCIN | 881 | 1,24831 | 0,566005 | – 1,14109 | 0,0361254 | 2,20548 |
| 27 | ARTN | 9048 | 5,77216 | 2,63816 | – 1,12958 | 0,00400937 | 2,18795 |
| 28 | LOC100507412 | 100507412 | 394,087 | 186,2 | – 1,08166 | 0,00400937 | 2,11647 |
| 29 | SERPINB5 | 5268 | 1,49223 | 0,722489 | – 1,04642 | 0,0340659 | 2,06540 |
| 30 | SERPINE1 | 5054 | 128,607 | 62,4787 | – 1,04153 | 0,00400937 | 2,05841 |
| 31 | DKK1 | 22943 | 85,9562 | 42,0833 | – 1,03035 | 0,00400937 | 2,04252 |
| 32 | SGK1 | 6446 | 18,4838 | 9,10546 | – 1,02146 | 0,00400937 | 2,02997 |
| 33 | FGF1 | 2246 | 1,67617 | 0,843698 | – 0,990371 | 0,00400937 | 1,98670 |
| 34 | BOD1 | 91272 | 27,4212 | 14,2379 | – 0,945548 | 0,00400937 | 1,92592 |
| 35 | CCND3 | 896 | 27,6423 | 14,3971 | – 0,941101 | 0,00400937 | 1,91999 |
| 36 | TGFB2 | 7042 | 13,4826 | 7,11507 | – 0,922151 | 0,00400937 | 1,89494 |
| 37 | CALU | 813 | 57,1015 | 30,1837 | – 0,919757 | 0,00400937 | 1,89180 |
| 38 | ANKRD1 | 27063 | 10,4157 | 5,56174 | – 0,905149 | 0,00400937 | 1,87274 |
| 39 | CGB8 | 94115 | 11,0552 | 5,93536 | – 0,897321 | 0,015208 | 1,86260 |
| 40 | C6orf120 | 387263 | 18,0817 | 9,71162 | – 0,896746 | 0,00400937 | 1,86186 |
Note: FPKM1 and FPKM2 indicate fragments per kilobase of exon per million mapped reads after and before PAX8 silencing, respectively. Genes from 1 to 20 are downregulated and genes from 21 to 40 are upregulated.
TUBB2B (tubulin beta 2B class IIb) may have important roles in tumor progression and chemoresistance [39]. PPME1 is a protein phosphatase 2A (PP2A)-specific methylesterase that mediates the demethylation and inactivation of PP2A. This protein is a human tumor suppressor that accounts for the majority of cellular serine/threonine phosphatase activity [40]. ATP8A1 is a member of P4-ATPase family with a role in the formation of membrane ruffles to promote cell migration [41]. BLPAP (Bladder cancer-associated protein) is a highly conserved protein among species displaying tissue-specific expression patterns, but also tissue-specific functions being both a tumor suppressor and a tumor promoting [42]. Recent data revealed that the blockade of DIAPH1-tubulin interaction might be a promising approach to inhibit one of the earliest steps in the metastatic cascade of colon cancer [43]. FGF18 is a pleiotropic growth factor involved in skeletal growth and development [44, 45]. Deregulated FGF signaling may affect some oncogenic mechanisms and properties in various tumor types [46]. KIF12 belongs to the kinesin superfamily of motor proteins that bind microtubules and mediate the intracellular transport of organelles and protein complexes [47].
Interestingly, in siPAX8-SKOV-3 cells among the most up-regulated genes based on FDR there are DKK1 and BOD-1 [48] (Table 1). DKK1 belongs to the Dickkopf gene family, which encodes secreted glycoproteins to control cell fate and neural patterning during embryonic development. It was first identified as an inhibitor of the Wnt signaling pathway. BOD-1 is a member of the FAM44 protein family and is highly conserved throughout metazoans. Depletion of BOD-1 in human cells causes severe biorientation defects [49].
In FT194 cells, 166 genes affected by PAX8 silencing reached genome-wide significance after correction for multiple testing (FDR-adjusted p-value ≤ 0.05) of which 119 were downregulated while 47 were upregulated (Supplementary Table S4). Among the most significantly downregulated genes there are: FGF18, CDH6, CHRD, ZBED2, CNTN4, ANXA2 (Table 2). Very interestingly, FGF18 results among the top 20 downregulated genes upon silencing of PAX8 in both SKOV-3 and in FT194 cells (Table 1 and Table 2).
Table 2. Differentially expressed genes that are significantly modulated after PAX8 silencing in FT194 cells.
| Gene Name | NCBI gene ID | FPKM1 | FPKM2 | Log2 fold change | FDR q value | Fold change | |
|---|---|---|---|---|---|---|---|
| 1 | CDH6 | 1004 | 0,904512 | 5,66947 | 2,648 | 0,00642248 | 6,26798 |
| 2 | SNORA26 | 677810 | 12,5059 | 73,4428 | 2,55401 | 0,00642248 | 5,87264 |
| 3 | FGF18 | 8817 | 2,9921 | 14,7197 | 2,29852 | 0,00642248 | 4,91953 |
| 4 | CHRD | 8646 | 1,14353 | 5,39776 | 2,23887 | 0,00642248 | 4,72027 |
| 5 | KLHL14 | 57565 | 0,218892 | 0,999955 | 2,19164 | 0,00642248 | 4,56824 |
| 6 | EPHB1 | 2047 | 0,17794 | 0,77373 | 2,12044 | 0,0162802 | 4,34827 |
| 7 | ZBED2 | 79413 | 0,98563 | 4,13057 | 2,06722 | 0,00642248 | 4,19078 |
| 8 | TLL2 | 7093 | 0,625627 | 2,60957 | 2,06043 | 0,00642248 | 4,17111 |
| 9 | PAX8 | 7849 | 9,23106 | 33,8269 | 1,8736 | 0,00642248 | 3,66446 |
| 10 | BAALC | 79870 | 0,360111 | 1,28504 | 1,8353 | 0,0333357 | 3,56846 |
| 11 | MAL | 4118 | 7,14709 | 25,0391 | 1,80876 | 0,00642248 | 3,50341 |
| 12 | DCDC2 | 51473 | 4,33336 | 14,9979 | 1,7912 | 0,00642248 | 3,46103 |
| 13 | LOC643201 | 643201 | 1,32389 | 4,51658 | 1,77045 | 0,00642248 | 3,41160 |
| 14 | ADAMTS14 | 140766 | 0,900768 | 3,0108 | 1,74092 | 0,00642248 | 3,34248 |
| 15 | CNTN4 | 152330 | 0,800182 | 2,67321 | 1,74017 | 0,00642248 | 3,34075 |
| 16 | ANXA2 | 302 | 572,227 | 1838,55 | 1,68391 | 0,00642248 | 3,21298 |
| 17 | FAT2 | 2196 | 0,373409 | 1,18513 | 1,66622 | 0,00642248 | 3,17382 |
| 18 | ANXA2P2 | 304 | 1,12435 | 3,55187 | 1,65948 | 0,0115711 | 3,15903 |
| 19 | ROR1 | 4919 | 7,26579 | 22,3551 | 1,62141 | 0,00642248 | 3,07676 |
| 20 | LMLN | 89782 | 0,291647 | 0,876636 | 1,58775 | 0,0115711 | 3,00580 |
| 21 | SERPINB2 | 5055 | 4,52656 | 1,12888 | – 2,00352 | 0,00642248 | 4,00977 |
| 22 | FPR1 | 2357 | 18,91 | 7,18704 | – 1,39568 | 0,00642248 | 2,63113 |
| 23 | PAPPA | 5069 | 4,39506 | 1,94254 | – 1,17794 | 0,00642248 | 2,26253 |
| 24 | PTGS2 | 5743 | 14,4361 | 6,46952 | – 1,15795 | 0,00642248 | 2,23140 |
| 25 | NRK | 203447 | 2,78453 | 1,25142 | – 1,15387 | 0,00642248 | 2,22510 |
| 26 | ST6GALNAC5 | 81849 | 10,3815 | 4,81544 | – 1,10827 | 0,00642248 | 2,15587 |
| 27 | THBD | 7056 | 7,82686 | 3,66541 | – 1,09446 | 0,00642248 | 2,13533 |
| 28 | CCL20 | 6364 | 60,3995 | 30,2135 | – 0,999342 | 0,0162802 | 1,99909 |
| 29 | RHOB | 388 | 44,0161 | 22,2603 | – 0,983562 | 0,00642248 | 1,97734 |
| 30 | F3 | 2152 | 132,89 | 67,4915 | – 0,977451 | 0,00642248 | 1,96898 |
| 31 | TGFB2 | 7042 | 53,4068 | 27,4917 | – 0,958027 | 0,00642248 | 1,94265 |
| 32 | ZCCHC2 | 54877 | 9,21038 | 4,7504 | – 0,955212 | 0,00642248 | 1,93886 |
| 33 | DCN | 1634 | 20,2125 | 10,5292 | – 0,940855 | 0,00642248 | 1,91967 |
| 34 | MX2 | 4600 | 66,0911 | 34,8487 | – 0,923353 | 0,00642248 | 1,89652 |
| 35 | RSAD2 | 91543 | 72,3042 | 38,7711 | – 0,899096 | 0,00642248 | 1,86490 |
| 36 | MSRB3 | 253827 | 35,8124 | 19,2352 | – 0,896709 | 0,00642248 | 1,86181 |
| 37 | MET | 4233 | 54,6992 | 30,1665 | – 0,858572 | 0,00642248 | 1,81324 |
| 38 | MX1 | 4599 | 264,614 | 148,11 | – 0,837216 | 0,00642248 | 1,78660 |
| 39 | CYP1B1 | 1545 | 64,4002 | 36,0841 | – 0,835702 | 0,00642248 | 1,78473 |
| 40 | NCOA7 | 135112 | 27,7387 | 15,6627 | – 0,824561 | 0,00642248 | 1,77100 |
Note: FPKM1 and FPKM2 indicate fragments per kilobase of exon per million mapped reads after and before PAX8 silencing, respectively. Genes from 1 to 20 were downregulated and genes from 21 to 40 were upregulated.
CDH6, a member of the cadherin superfamily, is required for the kidney development as well as ganglia formation [50, 51]. Increased expression of this gene may be associated with tumor growth and metastasis [52]. CHRD functions as a dorsalizing factor for early vertebrate embryonic tissues by binding to ventralizing TGF-beta family bone morphogenetic proteins (BMPs) and sequestering them in latent complexes [53]. ZBED2 (Zinc Finger, BED-Type Containing 2) is a member of family factors involved in the regulation of various functions in vertebrates [54]. CNTN4 is a glycosylphosphatidylinositol (GPI)-anchored neuronal membrane protein responsible for the formation of axon connections in the developing nervous system [55]. Loss of heterozygosity (LOH) and gene sequencing analysis investigate the possibility of CNTN4 to function as tumor suppressor gene in ovarian cancer [56]. ANXA2 is a calcium-binding cytoskeletal protein aberrantly expressed in a wide spectrum of cancers and in EOC may promote cell proliferation [57].
Additionally, in siPAX8-FT194 cells among the top 20 significantly upregulated genes there are: SERPINB2, FPR1, PAPPA, CCL20, RHOB, DCN (Table 2).
SERPINB2, also named Plasminogen Activator Inhibitor, Type II (PAI-2) is upregulated by numerous inflammatory conditions [58]; in cancer disease its expression is often an indicator of positive prognosis as described in ovarian cancer [59, 60]. FPR1 is a G protein-coupled receptor that promotes growth, angiogenesis and invasion in glioblastoma tumor [61]. FPR1 as the other two FPRs play a pivotal role in inflammatory response, tissue repair, tumor growth, physiological and pathological angiogenesis [62]. PAPPA (pregnancy-associated plasma protein A) regulates mitotic progression through modulating the IGF1 signaling pathway [63]. CCL20 is a small cytokine constitutively produced by Fallopian tube epithelial cells and able to function as endogenous anti-viral microbicide of female reproductive tract [64]. In ovarian cancer, CCL20 is one of the primary chemokine induced via NF-kB pathway [65]. RHOB, is a member of small GTPases belonging to the Ras protein superfamily, might have a suppressive activity in cancer progression, in fact its loss occurs frequently in ovary carcinogenesis and progression. Moreover, ectopic expression of RhoB into nude mice is highly effective in suppressing tumor growth of ovarian cancer xenografts [66]. DCN is a small leucine-rich proteoglycan, component of connective tissue responsible for the matrix assembly. Also this factor might function as a tumor suppressor in ovarian cancer, because its expression is lost during epithelial transformation [67].
PAX8 regulates the expression of shared and exclusive sets of genes in SKOV-3 and FT194 cells
As illustrated in the above paragraph, following PAX8 silencing 214 downregulated and 87 upregulated genes were identified in siPAX8-SKOV-3 cells while 119 downregulated and 47 up-regulated were defined in siPAX8-FT194 cells (FDR-adjusted p-value ≤ 0.05) (Supplementary Table S3 and Supplementary Table S4). A Venn diagram was constructed to identify shared and distinct sets of genes regulated by PAX8 in the two cell types. As shown in Figure 2A, 47 genes are commonly downregulated in siPAX8-SKOV-3 and siPAX8-FT194 cells, whereas the genes downregulated only in one of the samples are 167 and 72 (SKOV-3 and FT194, respectively). Analogously, 15 genes are commonly upregulated in siPAX8-SKOV-3 and siPAX8-FT194 cells while 72 and 32 genes are exclusively upregulated in SKOV-3 and FT194 cells, respectively (Figure 2B). The complete lists of the genes classified according to the Venn diagram are shown in Table 3.
Figure 2. The Venn diagram of genes modulated upon PAX8 knockdown in SKOV-3 and FT194 cells.
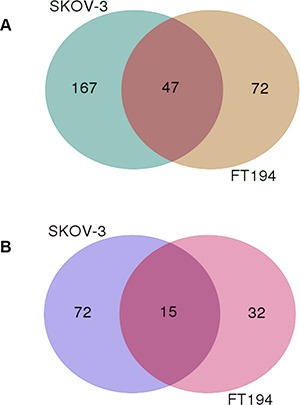
(A) Overlap and differences of downregulated genes following PAX8 silencing between SKOV-3 and FT194 cells. (B) Overlap and differences of upregulated genes following PAX8 silencing between SKOV-3 and FT194 cells.
Table 3A. List of shared and exclusive genes downregulated upon PAX8 silencing in SKOV-3 and FT194 cells.
| Name | Total | Elements |
|---|---|---|
| SKOV3-FT194 | 47 | NUP35 ANXA2 HDGFRP3 STX3 DUSP11 AP1G1 ENPP4 PHLDA3 PSAP ENPP1 EPRS RHPN2 C10orf46 GPR63 CDK2AP1 H2AFY2 PPME1 LMLN KITLG CDH6aFAM107B C1orf186 BLCAP TEX30 LONRF1 DIAPH1 FGF18a TCN2 ROR1 AKR1B1 TGOLN2 KAZN PHTF2 ARL4C MAL ADCY9 PPAP2B PAX8a ARL5A MAL2 MRE11A HTATSF1 GPR56 PTGIS ANXA2P2 ABCC4 SLC35B4 |
| SKOV3 | 167 | ACTB TSPAN1 RAB12 OSBPL11 L1CAM OSBPL10 LRRC20 RRAD ANKRD34B RNF38 HES2 SMC5 FAM3C PRKAA2 KCNH3 LMAN1 ST3GAL2 AFF1 TAF4B IFNGR1 FHDC1 CGNL1 CA13 SYPL1 MKNK2 SLC39A10 AMMECR1 ZNF114 GAS2L3 CERK BAG4 SASH1 C6orf228 TBC1D13 C7orf29,LRRC61 TUBB2B GTF2E1 ABCC3 PLCH1 CLDND1 MBP CYB5R4 SLC39A14 AKR1B10 CELSR2 PDZK1 FOXN3 PIK3R1 GRIN2B CIAPIN1 CORO1C RNFT1 UPP1 SHROOM2 FAM45A,FAM45B ANKRD33B ZNF702P ID4 PPM1B PER1 SLC44A1 CDH16 PCDH20 ADAM10 RNF130 CBS KIF12aKLHL15 ZNF28 ATP8B2 EGFR MIR4723,TMEM199 DICER1 DPP4 ITGB3 DENND1B SGK2 BASP1P1 USP18 RAB3D DCUN1D4 SNTB1 PGGT1B CNKSR3 CHST15 MCAM TYW1 GCH1 TRIM2 C12orf23 FNDC5 IGFN1 USP2 TERF1 SORT1 CRYAB GMCL1 WNT7Aa MXRA5 AGL RNF144B ASRGL1 MPHOSPH6 QPRT AATF LIN7C NXT2 C2orf72 RRM2 BCAT1 SLC7A11 ADAMTS9 TMSB4X TPK1 KPNA6 ATP8A1 CHST2 THBS1 CYP4F11 RNF145 ADAMTS5 FLJ26245 SYTL2 UBE2H DCAF12L1 RAB11FIP2 ABI2 PTPRB FIGN PAFAH1B2 TCF12 PMAIP1 WDR44 DCUN1D3 SLC30A6 EPHA4 NEBL DSC2 CNOT6 ZNF611 MYO10 MAP2 KCNS3 SLC26A2 HDHD2 SLAIN1 PLXNC1 MGST1 NUPL1 COL12A1 KCTD5 RCBTB2 CANT1 MTPN FN1 UBE2D1 ZCCHC24 TMTC2 TRIM24 FAM174B C10orf26 TMCO7 GUCY1B3 ARHGEF1 ANKRD52 RC3H2 ZNF618 |
| FT194 | 72 | KLHL13 ZNF185 PPARGC1A WASF1 PLAU KLHL14 TLL2 CHRDa CLGN NGFR ARHGEF37 FLG GAS7 TGFBR3 RGS20 ILDR2 SPON1 MMD CD24 AP1M2 TNFSF4 THY1 ADAMTS14 ZBED2a PLCB4 ADAMTS1 LOC643201 NOV MPP7 SHISA2 ST3GAL1 SOX17 GDF6 ANK3 RASGEF1B CNTN4 C13orf15 SULF2 ADORA1 MEGF9 KLRG2 ITGB8 DANCR,SNORA26 SLC6A6 BAALC CTHRC1 G0S2 TMEM117 AIF1L GJB2 PRKAG2 FAT2 MST1 DCDC2 OXTR SDC2 INHBB KIAA1456 BTBD11 CA2 NID2 SLC47A1 CDH5 PDE1A COL13A1 IGFBP5 TMEM170B CTGF EPHB1 GPRC5C DKK1 CDKN1C |
Genes that were validated using qRT-PCR.
Table 3B. List of shared and exclusive genes upregulated upon PAX8 silencing in SKOV-3 and FT194 cells.
| Name | Total | Elements |
|---|---|---|
| SKOV3-FT194 | 15 | ANKRD1 F3 G3BP2 TGFB2 MAPK1IP1L TOMM20 MBTPS1 DNMT3B PTPN1 MSRB3 MET RBPJ TCEB3 WDR1 STX12 |
| SKOV3 | 72 | DIO2b ZCCHC3 NTN4 NGRN TRAPPC2 CALU MOK DCBLD1 PTPMT1 FGF1 C1D AHRR,PDCD6 LOC100507412,RN45S PCSK7 CPA4 CIRBP SERPINE1 C6orf120 PODXL CCDC80 DOCK10 PYGO2 P4HA3 MCFD2 DSEL MYPN LPXN CCND3 CHFR MAP2K4 GRWD1 CCBE1 UBE2G1,ZZEF1 BOD1b NCF2 ODZ2 ERRFI1 CCIN SERPINB5 ATG12 CGB8 FLRT2 AIM1 ROS1 C7orf58 ALS2CL FRMD6 KRT5 SSFA2 PIP4K2A SOCS7 NT5E ESYT2 SMAP1 ARTN SCRN1 DKK1 EFEMP1 RFK SGK1 CCDC68 CRK FOXD1 PGM2L1 PCDH10 SEMA7A ZFAND3 IFFO2 CCNC RCAN1 KHNYN POLR3F |
| FT194 | 32 | CMPK2 RAD54L2 DDX58 IFIT1 SP110 PTPN13 DCN PAPPAb RHOB CCL20 GBP1 SERPINB2b OAS3 PDZD2 IFIH1 NCOA7 OAS2 RSAD2 PTGS2 MX1 ZCCHC2 CYP1B1 DHX58 DDX60L HMGA2 FARP2 MX2 NRK FPR1 THBD ST6GALNAC5 THBS2 |
Genes that were validated using qRT-PCR.
We reasoned that genes modulated upon PAX8 silencing in both cell lines could be looked at as putative PAX8 targets with greater confidence. Hence, we focused our attention on the intersections between SKOV-3 and FT194 cells and we observed that they include several interesting genes such as: PSAP, FGF18, CDH6, ROR1, RBPJ and DNMT3B whose role in cell proliferation, cell survival and tumorigenic process has already been reported. In particular, PSAP (prosaposin) is a pleiotropic growth factor able to prevent cell death or apoptosis and to promote cell survival [68]; FGF18 controls migration, invasion and tumorigenicity of ovarian cancer cells [69]; CDH6 is a new TGF-beta target modulated as a mesenchymal marker in EMT [52]; ROR1, a receptor tyrosine kinase orphan receptor 1, is involved in migration, invasion and EMT in ovarian cancer cells [70]; RBPJ (recombination signal-binding protein Jk) is a key transcription factor in the Notch signaling pathway and its inhibition reduces cell growth [71]; DNMT3B encodes a DNA methyltransferase involved in de novo DNA methylation whose role in cancer development is not clear but several reports suggest a role as tumor suppressor [72].
At the same time, we believe that genes modulated upon PAX8 silencing exclusively in FT194 or in SKOV-3 cells should be considered relevant because they could reflect the continuous process from the precancerous to the cancerous condition. In addition, the tumorigenic process might itself promote the expression of some genes making them available for PAX8 transcriptional regulation. For example, WNT7A known to function as tumor-promoting in ovarian cancer [73] is present at very low level in FT194 cells but becomes highly expressed in SKOV-3 cells (Supplementary Table S1) where is significantly downregulated upon PAX8 silencing (Supplementary Table S3 and Table 3A). Similarly, ASRGL1 (asparaginase like 1) is highly expressed in ovarian carcinoma and confers a selective growth advantage [74]; accordingly, our data show that ASRGL1 is preferentially expressed in SKOV-3 cells where it is downregulated following PAX8 silencing (Table 3A).
We validated the data obtained by RNA-seq by means of qRT-PCR for 13 genes, including PAX8. For these genes, we confirmed the different expression between the two cell types (Figure 3A) and in OVCAR-3, PEA1 and PEO14 ovarian cancer cell lines (Supplementary Figure S1). The same genes were also validated upon silencing of PAX8 in FT194 and SKOV-3 cells (Figure 3B and 3C). In particular, we confirmed that KIF12, DIO2 and WNT7A are expressed preferentially in SKOV-3 cells (Figure 3A) and in these cells are modulated upon silencing of PAX8 (Figure 3B). At difference, FGF18, CDH6, ANXA2 and ROR1 are expressed and modulated in both cell lines (Figure 3A, 3B, 3C). CHRD, PAPPA and SERPINB2 are expressed almost exclusively in FT194 cells (Figure 3A) and are modulated upon silencing of PAX8 (Figure 3C). ZBED2 and BOD1 are expressed in both cell lines but following PAX8 silencing ZBED2 is modulated at significant level only in FT194 cells, while BOD1 only in SKOV-3 cells. To strengthen our observations we validated the same genes also in PEA1 cells after PAX8 silencing (Supplementary Figure S2).
Figure 3. Validation of representative genes by qRT-PCR analysis.
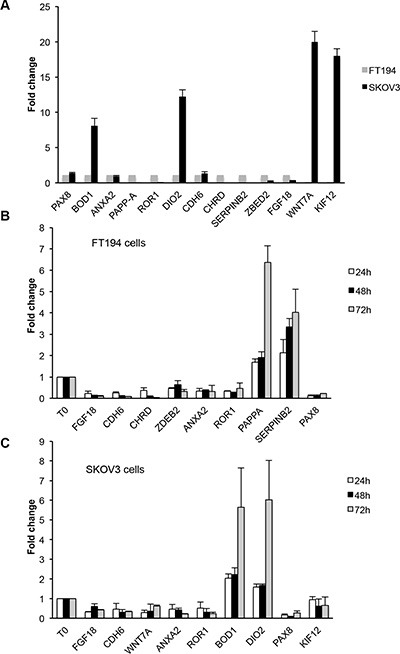
(A) Expression levels of 13 genes measured on total RNA prepared from SKOV-3 and FT194 cells. The values are means ± SD of three independent experiments in duplicate, normalized by the expression of IP08 and expressed as fold change with respect to FT194 cells. (B and C) Expression levels of some representative genes measured on total RNA prepared from FT194 and SKOV-3 cells transiently transfected with PAX8 siRNA or scramble siRNA 24 h (white bars), 48 h (black bars) and 72 h (grey bars) after transfection. The values are means ± SD of three independent experiments in duplicate, normalized by the expression of IP08 and expressed as fold change with respect to the cells transfected with the scramble siRNA, whose value was set at 1.0. p-value was calculated by t-test 0.001 ≤ p ≤ 0.1.
Subsequently, we analyzed the 5′-flanking regions of the genes commonly regulated in the two cell lines in order to recognize DNA binding motifs for PAX8 matrices using the PASTAA method (http://trap.molgen.mpg.de) [75] which utilizes the prediction of binding affinities of a transcription factor to regulatory regions. The genes containing PAX8 DNA binding motifs were ranked according to the prediction of binding affinity of their 5′-flanking region to the PAX8 binding sites (Supplementary Table S5). To unambiguously determine whether PAX8 directly binds to the above mentioned regulative genomic sequences, we performed a computational analysis for some representative genes chosen among those commonly regulated in the FT194 and SKOV3 cells, using the MatInspector Software (Genomatix). We searched for PAX8 binding sites in a region of about 2 Kb in their 5′-flanking region and we found several PAX8 consensus sequences. To confirm the predictions of the MatInspector analysis, we carried out chromatin immunoprecipitation (ChIP) assays on FT194 cells using a polyclonal antibody against PAX8. The ChIP results indicate that in vivo PAX8 is able to bind the regulatory regions of all selected genes (Figure 4).
Figure 4. Direct binding of PAX8 on the regulatory regions of putative target genes.
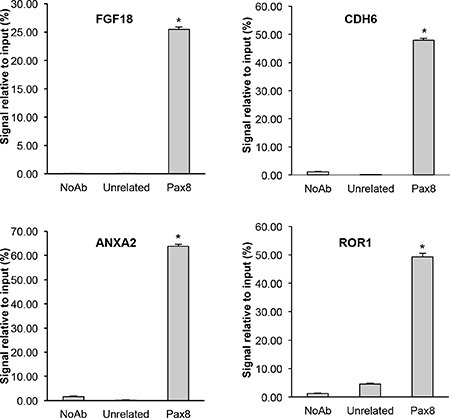
Chromatin immunoprecipitation assays were performed to determine the binding of PAX8 to the 5′-flanking region of representative genes. Chromatin was subjected to quantitative real-time PCR analysis using appropriate primers (see Materials and Methods). Error bars indicate s.d. between two experiments performed in duplicate (p < 0,001).
All together, our results show that the expression profiles affected by PAX8 silencing might reflect an ongoing transformation process reinforcing the involvement of this transcription factor in ovarian cancer.
Pathways regulated by PAX8 in ovarian cancer
The pathways that PAX8 may regulate in ovarian cancer and in Fallopian tube secretory cells are still undefined. To categorize PAX8 associated pathways that are represented in our PAX8-silenced SKOV-3 and FT194 cells, we classified all the dysregulated genes (301 for SKOV-3 and 166 for FT194) using the Gene annotations co-occurrence discovery web-based tool (GeneCodis; http://genecodis.dacya.ucm.es/). Several significant GO categories appeared enriched including signal transduction, cell adhesion, blood coagulation, positive regulation of cell migration, angiogenesis and cell differentiation (Figure 5A and 5B). At the same time, the most affected pathways upon silencing of PAX8 comprise Wnt signaling, cadherin signaling, integrin signaling and TGF-beta signaling (Figure 5C and 5D).
Figure 5. Biological processes and pathways altered in siPAX8 cells.
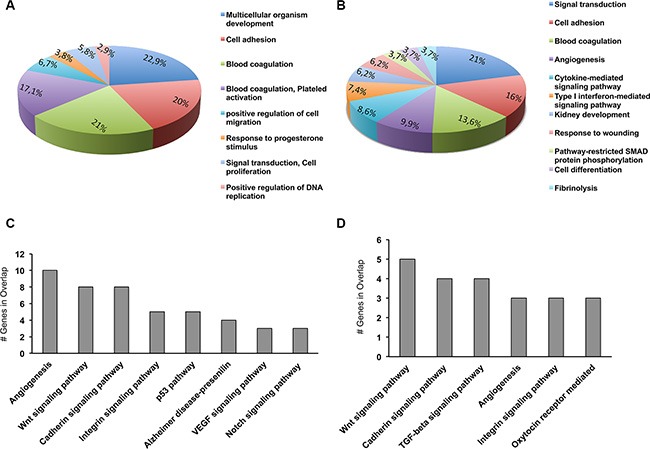
Gene ontology (GO) and Panther pathway anlysis have been performed using GeneCodis (http://genecodis.dacya.ucm.es/). (A) GO categories enriched for genes modulated upon PAX8 silencing in SKOV-3 cells. (B) GO categories enriched for genes modulated upon PAX8 silencing in FT194 cells. (C) Panther pathways enriched for genes modulated upon PAX8 silencing in SKOV-3 cells. (D) Panther pathways enriched for genes modulated upon PAX8 silencing in FT194 cells.
To further investigate the functional associations of downregulated and upregulated genes following PAX8 knockdown we performed an MsigDB analysis. Interestingly, we observed that for the significantly upregulated genes (87 for SKOV-3 and 47 for FT194) the most affected pathways in both cells lines are interferon response, TNFa signaling, inflammatory response, apoptosis, UV response and epithelial mesenchymal transition (Figure 6). In the same way, for the downregulated genes (214 for SKOV-3 and 119 for FT194) the most affected pathways in both cells lines are epithelial mesenchymal transition, UV response, Kras signaling, estrogen response and p53 pathway (Figure 6). In agreement with our previous studies, the pathways perturbed upon PAX8 silencing such as IFN-g and TGF-a signaling, EMT, apoptosis, hypoxia as well as UV response strengthen the new role of PAX8 in the regulation of cell survival, proliferation and in the maintenance of oncogenic properties [11, 17].
Figure 6. Pathway analysis of differentially expressed genes after PAX8 silencing.
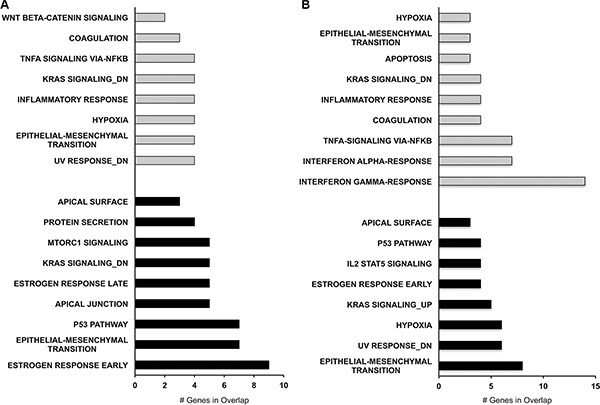
MsigDB software was used to identify the pathways most affected by the gene dysregulation. (A) MsigDB for SKOV-3 downregulated (black bars) and upregulated genes (gray bars). (B) MsigDB for FT194 downregulated (black bars) and upregulated genes (gray bars).
In conclusion, we think that the majority of the genes affected by PAX8 silencing are associated with important biological cellular processes and we believe that our analysis provides a solid basis for the identification of relevant molecules involved in ovarian cancer.
DISCUSSION
Recent studies suggest that a substantial proportion of cases of ovarian high-grade serous carcinoma may arise from precursor lesions located in the Fallopian tubal epithelium (FTE). In our study, by means of RNA-seq analysis we investigated the expression of genes modified during the transformation process from Fallopian tube secretory epithelial cells to HGSC cells. Moreover, genes and pathways downstream the transcription factor PAX8 have been analyzed in both cell types. Our goal was to identify new targets for diagnostic and/or therapeutic approaches for HGSC that is the third most common cause of death among gynecologic malignancies worldwide. Actually, improved screening strategies for HGSC diagnosis in early stages, as well as effective treatments are greatly needed.
We highlighted that ≈ 60% of the genes differentially expressed between the two cell types is modulated at a significant level (FDR-adjusted p-value ≤ 0.05). Applying a 2 fold-change cutoff and performing a Molecular Signatures Database analysis, we determined in SKOV-3 cells a significant enrichment of estrogen response, EMT, angiogenesis and Wnt/β catenin pathways. At the same time, in FT194 cells we observed an enrichment of pathways like TNF-α, IFN-α, IFN-γ and inflammatory responses. Furthermore, we consider intriguing those genes that turned out to be preferentially, or in some cases exclusively, expressed in FT194 or SKOV-3 cells. There is no doubt that the future challenge will be to examine the potential role of genes differentially expressed between the two cell types as specific and sensitive biomarkers. In fact, several ovarian cancer screening studies have established that acombinatorial biomarker strategy is more reliable, sensitive and specific than using a single protein biomarker [76]. A recent study identified a 5-marker panel for early detection of ovarian cancer that includes five serum biomarkers, namely macrophage-stimulating protein alpha, tissue inhibitor of metalloproteinases-4, platelet-derived growth factor receptor alpha (PDGF-R alpha), osteoprotegerin, and CA-125 [77]. Preliminary insights that we obtained using ProteINSIDE, a novel web service [78], suggest that among the genes highly expressed in SKOV-3 cells there are some that encode for secreted proteins like CGB5 [79], CGB8 [79], FOLR1 [28], SPP1 [80], IGFBP3 [81], NFASC [82] emphasizing that our study may indeed provide a solid basis for the identification of new biomarkers. Of interest, among the top genes highly expressed in SKOV-3 cells there is PRAME that in a wide variety of human malignancies correlates with poor clinical outcome [83]. It has been reported that PRAME proteins are associated to self-renewal cell maintenance [84] and are currently considered as potential target to hamper cancer cell proliferation [85].
In 2003, a gene expression study carried out in ovarian carcinoma [86] reported the transcription factor PAX8, normally absent in ovarian surface epithelial cells, among the most highly expressed genes. The recent carcinogenesis model proposes the secretory cells of the Fallopian tubal mucosa as the cell of origin for the majority of EOC. In agreement with this hypothesis, PAX8 would not be overexpressed in epithelial ovarian carcinoma but rather its expression would be conserved from the cell of origin [38]. To date, the function of PAX8 in Fallopian tube epithelial secretory cells has not been clarified. It has been demonstrated a continually regulated cycle of growth, differentiation, death and renewal in the epithelium of the mammalian oviduct [87] and in this context PAX8 could exert its role. However, more studies are needed to shed light on the function of this transcription factor in this cell type.
We have recently reported that PAX8 is involved in the tumorigenic phenotype of ovarian cancer cells [17]. In the present study, we intend to clarify PAX8 contribution to ovarian cancer through the identification of its downstream gene regulatory network. MsigDB analysis revealed that in both FT194 and SKOV-3 cells the most affected pathways are interferon response, TNFa signaling, inflammatory response, apoptosis, UV response and epithelial mesenchymal transition (considering the upregulated genes) and epithelial mesenchymal transition, UV response, estrogen response and p53 pathway (considering the downregulated genes). Interestingly, our RNA-seq analysis identifies, among PAX8 potential targets, genes that have been reported in the literature having a role in ovarian cancer. One of such genes is FGF18 whose expression is significantly reduced after PAX8 knockdown. It has been demonstrated that FGF18 regulates both tumor cells and tumor microenvironment to facilitate the progression of serous ovarian cancer, enhancing angiogenesis and tumor-associated macrophage infiltration [69, 88]. In addition, FGF18 was identified as the gene possessing the strongest prognostic value in segment 5q31-5q35.3 that was found amplified on microdissected HGSC samples [89]. Furthermore, a recent study provides a platform for the identification of blood-based biomarkers (“Secretome”) for high-grade, advanced-stage serous ovarian tumors and identifies two new markers, FGF18 and GPR172 [90]. Here, we hypothesize that one way in which PAX8 confers a proliferative advantage to ovarian cancer cells is through the regulation of procancer factors, like FGF18, in a pathological microenvironment.
Abnormal activation of the WNT/b-catenin signaling pathway has been associated with ovarian carcinomas. It has been reported that WNT7A is abundantly expressed in ovarian carcinoma and is able to control cell division, adhesion and motility [73]. In addition, WNT7A knockdown cancer cells show a significantly decreased growth rate and invasion ability in a xenografts model [73]. Intriguingly, the phenotype of WNT7A knockdown xenografts resembles that described for the xenograft model of the siPAX8-SKOV-3 cells [17] and this study shows that WNT7A is abundantly expressed only in SKOV-3 cells where is a downstream target of PAX8.
In addition, the expression of some genes regulated by PAX8 in Fallopian tubes secretory cells may be lost during the neoplastic transformation. For example, our data show that chordin (CHRD), a BMP extracellular regulator that behaves as suppressor of tumorigenesis in ovarian carcinoma cells, is abundantly expressed and regulated by PAX8 in FT194 cells, while is almost absent in SKOV-3 cells.
It is worth noting that it has recently been demonstrated that Fallopian tube secretory cell expansion and the ratio between secretory/ciliated cells (S/C ratio) are linked to pelvic serous neoplasia [91] and in this scenario the cancer cells of the serous tubal intraepithelial carcinoma (STIC) bearing a “p53 signature” invade onto the ovary and implant on peritoneal surfaces [92–94]. During this process, the secretory cells retain the expression of PAX8 that possibly continues to exert its transcriptional activity on its physiological targets and in addition may also function on new targets that become available after the tumorigenic hits (BRCA mutations, p53 signature etc).
Our final suggestion is that the relevance of PAX8 in ovarian carcinoma lies in the downstream network(s) regulated by this transcription factor that contribute to ovarian carcinoma pathogenesis. In this respect, we believe that our study has led to the identification of genes and pathways regulated by PAX8 that could be valuable for future studies to uncover the molecular mechanisms leading to EOC.
MATERIALS AND METHODS
Cell culture and RNA interference
The human ovarian carcinoma cell line SKOV-3 was obtained from the CEINGE Cell Culture Facility (Naples, Italy) and was grown in RPMI-1640 medium (Euroclone) containing 10% fetal bovine serum (Euroclone). The immortalized Fallopian tube secretory epithelial cell line FT194 was kindly provided by Dr. R. Drapkin (Boston, USA) and was maintained in DME-F12 medium (Euroclone) containing 2% Ultroser G serum (PALL). The OVCAR-3 cell line was obtained from ATCC and was maintained in RPMI-1640 medium (Euroclone) containing 20% fetal bovine serum (Euroclone) and 0.01 mg/ml bovine insulin. PEA1 and PEO14 cells were purchased from Sigma-Aldrich and mantained in RPMI-1640 medium (Euroclone) containing 10% fetal bovine serum (Euroclone), 2 mM glutamine and 2 mM sodium pyruvate (Gibco, Life Technologies).
For RNA interference, SKOV-3, FT194 cells and PEA1 were plated at 2 × 105 cells/60-mm tissue culture dish 24 h prior to transfection and were transfected in replicates with 5 nM PAX8 siRNA (Ambion, Life Techonologies, siRNA ID s15403) or siRNANon-Targeting (Ambion, Life Technologies, siRNA ID 4390843) as scramble, using the Lipofectamine RNAiMAX transfection reagent (Invitrogen) following the manufacturer's protocol. Cells were harvested 24 h after transfection and the total RNA was prepared. Human Fallopian tubes RNA was from Origene (CR559726).
RNA extraction, qRT–PCR, RNA-seq and data mining
For both qRT-PCR and RNA-seq experiments, total RNA was extracted using the RNeasy Mini kit (Qiagen). For qRT-PCR, the cDNA was synthesized using the iScript cDNA Synthesis kit (BIORAD, Hercules, CA). Real time RT–PCR analysis was performed using the IQ™ SYBR Green PCR Master Mix (BIORAD) in a CFX96 Real-Time PCR Detection System (BIORAD) with gene-specific oligonucleotides (Supplementary Table S6).
For RNA-seq, 2 μg of total RNA extracted 24 h after transfection were sent to the Genomics4Life Company (University of Salerno, Italy). Independent silencing experiments were performed from which biological replicates of each condition (siPAX8 or control) were processed for the RNA-seq analysis.
The extracted RNA samples were sequenced using the Illumina HiSeq 1500 platform, TruSeq Stranded Total RNA, 100 bp paired-end reads at the Genomics4Life Company (University of Salerno, Italy).
Analysis was performed using the EPIGEN project sequence facility, RAP (RNA-Seq Analysis Pipeline) [95], available on the following website https://bioinformatics.cineca.it/. Sequence quality was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and NGS QC Toolkit [96], using default parameters (PHRED quality score: 20 and for percentage of read length of given quality: 70). Hence, paired-end reads were mapped to the reference human genome build (hg19/GRCh37) using Tophat 2.0.12 [97] with default parameters. The resulting alignment files are provided to Cufflinks [98] to generate a transcriptome assembly and to estimate the expression level expressed as units of FPKM (Fragment mapped per kilobase of exons per million mapped reads). Differential expression analyses were performed with Cuffdiff2 [99] using default parameters. An alpha level of 0.05 was used for all statistical tests. Gene expression data have been submitted to the Gene Expression Omnibus (GEO), Accession number GSE79572.
Chromatin immunoprecipitation assay
ChIP was performed as previously described [100]. Precleared chromatin from FT194 cells was incubated with 3 μg of affinity-purified rabbit polyclonal anti-PAX8 antibody (Thermo Scientific, PA1-112) or polyclonal anti-TAZ antibody as unrelated (Santa Cruz Biotechnology, sc-17130) and rotated at 4°C for 16 h. Thereafter, the immunoprecipitated DNAs were amplified by quantitative real-time PCR with the following primers:
FGF18 for. 5′-gtgggtagccagtcaagagg-3′; rev. 5′-ctccccaagaacgcagttag-3′;
ANXA2 for. 5′-Gctaaacggctgcaagaaac-3′; rev. 5′-Cgtagcaggcagtcctgag-3′;
CDH6 for. 5′-Atccaacagtggctgactcc-3′; rev. 5′-tctggaaagttgccgaagtt-3′;
ROR1 for. 5′-CAGATCACAGCTGCCTTCAC-3′; rev. 5′-ATTTCACATTCATCGCGACA-3′.
Pathway analysis
Gene ontology (GO) and Panther pathway analysis have been performed using the GeneCodis tool (http://genecodis.cnb.csic.es) previously described in references [101–103].
Genes showing a two-fold change in expression were analyzed at Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb) using the “Hallmark” gene set.
SUPPLEMENTARY FIGURES AND TABLES
ACKNOWLEDGMENTS AND FUNDING
We greatly thank L. Nitsch and M. De Felice for helpful discussions and suggestions.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no known conflicts of interest in this work.
GRANT SUPPORT
This work was supported by a grant from the Italian Ministry of Economy and Finance to the CNR for the Project FaReBio di Qualità, by the grant Medical Research in Italy (MERIT) RBNE08YFN3_001 and by the Epigenomic Flagship Project-Epigen of the CNR.
O.A. is a recipient of a doctoral fellowship from the Ph.D. of Computational Biology and Bioinformatics, University of Naples Federico II, Naples, Italy.
A.A.S. is a recipient of a doctoral fellowship from the Ph.D. of Molecular Medicine and Medical Biotechnology, University of Naples Federico II, Naples, Italy.
REFERENCES
- 1.Saad AF, Hu W, Sood AK. Microenvironment and pathogenesis of epithelial ovarian cancer. Horm Cancer. 2010;1:277–290. doi: 10.1007/s12672-010-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw PA, McLaughlin JR, Zweemer RP, Narod SA, Risch H, Verheijen RH, Ryan A, Menko FH, Kenemans P, Jacobs IJ. Histopathologic features of genetically determined ovarian cancer. Int J Gynecol Pathol. 2002;21:407–411. doi: 10.1097/00004347-200210000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Nik NN, Vang R, Shih Ie M, Kurman RJ. Origin and pathogenesis of pelvic (ovarian, tubal, and primary peritoneal) serous carcinoma. Annu Rev Pathol. 2014;9:27–45. doi: 10.1146/annurev-pathol-020712-163949. [DOI] [PubMed] [Google Scholar]
- 4.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, Chen JY, Ohman AW, Stepule CD, Kwak S, Karst AM, Hirsch MS, Setlur SR, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24:751–765. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- 7.Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plachov D, Chowdhury K, Walther C, Simon D, Guenet JL, Gruss P. Pax8, a murine paired box gene expressed in the developing excretory system and thyroid gland. Development. 1990;110:643–651. doi: 10.1242/dev.110.2.643. [DOI] [PubMed] [Google Scholar]
- 9.Ozcan A, Liles N, Coffey D, Shen SS, Truong LD. PAX2 and PAX8 expression in primary and metastatic mullerian epithelial tumors: a comprehensive comparison. Am J Surg Pathol. 2011;35:1837–1847. doi: 10.1097/PAS.0b013e31822d787c. [DOI] [PubMed] [Google Scholar]
- 10.Poleev A, Fickenscher H, Mundlos S, Winterpacht A, Zabel B, Fidler A, Gruss P, Plachov D. PAX8, a human paired box gene: isolation and expression in developing thyroid, kidney and Wilms' tumors. Development. 1992;116:611–623. doi: 10.1242/dev.116.3.611. [DOI] [PubMed] [Google Scholar]
- 11.Di Palma T, Filippone MG, Pierantoni GM, Fusco A, Soddu S, Zannini M. Pax8 has a critical role in epithelial cell survival and proliferation. Cell Death Dis. 2013;4:e729. doi: 10.1038/cddis.2013.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raman P, Koenig RJ. Pax-8-PPAR-gamma fusion protein in thyroid carcinoma. Nat Rev Endocrinol. 2014;10:616–623. doi: 10.1038/nrendo.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangoi AR, Ohgami RS, Pai RK, Beck AH, McKenney JK, Pai RK. PAX8 expression reliably distinguishes pancreatic well-differentiated neuroendocrine tumors from ileal and pulmonary well-differentiated neuroendocrine tumors and pancreatic acinar cell carcinoma. Mod Pathol. 2011;24:412–424. doi: 10.1038/modpathol.2010.176. [DOI] [PubMed] [Google Scholar]
- 14.Chen YJ, Campbell HG, Wiles AK, Eccles MR, Reddel RR, Braithwaite AW, Royds JA. PAX8 regulates telomerase reverse transcriptase and telomerase RNA component in glioma. Cancer Res. 2008;68:5724–5732. doi: 10.1158/0008-5472.CAN-08-0058. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Ma H, Pan Y, Xiao W, Li J, Yu J, He J. PAX2 and PAX8 reliably distinguishes ovarian serous tumors from mucinous tumors. Appl Immunohistochem Mol Morphol. 2015;23:280–287. doi: 10.1097/PAI.0000000000000065. [DOI] [PubMed] [Google Scholar]
- 16.Heidarpour M, Tavanafar Z. Diagnostic utility of PAX8 in differentiation of mullerian from non-mullerian tumors. Adv Biomed Res. 2014;3:96. doi: 10.4103/2277-9175.129366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Palma T, Lucci V, de Cristofaro T, Filippone MG, Zannini M. A role for PAX8 in the tumorigenic phenotype of ovarian cancer cells. BMC Cancer. 2014;14:292. doi: 10.1186/1471-2407-14-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci U S A. 2011;108:7547–7552. doi: 10.1073/pnas.1017300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modugno F, Laskey R, Smith AL, Andersen CL, Haluska P, Oesterreich S. Hormone response in ovarian cancer: time to reconsider as a clinical target? Endocr Relat Cancer. 2012;19:R255–279. doi: 10.1530/ERC-12-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maleki J, Nourbakhsh M, Shabani M, Korani M, Nourazarian SM, Ostadali Dahaghi MR, Moghadasi MH. 17beta-Estradiol Stimulates Generation of Reactive Species Oxygen and Nitric Oxide in Ovarian Adenocarcinoma Cells (OVCAR 3) Iran J Cancer Prev. 2015;8:e2332. doi: 10.17795/ijcp2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herr P, Hausmann G, Basler K. WNT secretion and signalling in human disease. Trends Mol Med. 2012;18:483–493. doi: 10.1016/j.molmed.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Gatcliffe TA, Monk BJ, Planutis K, Holcombe RF. Wnt signaling in ovarian tumorigenesis. Int J Gynecol Cancer. 2008;18:954–962. doi: 10.1111/j.1525-1438.2007.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arend RC, Londono-Joshi AI, Straughn JM, Jr, Buchsbaum DJ. The Wnt/beta-catenin pathway in ovarian cancer: a review. Gynecol Oncol. 2013;131:772–779. doi: 10.1016/j.ygyno.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Yoo HJ, Byun HJ, Kim BR, Lee KH, Park SY, Rho SB. DAPk1 inhibits NF-kappaB activation through TNF-alpha and INF-gamma-induced apoptosis. Cell Signal. 2012;24:1471–1477. doi: 10.1016/j.cellsig.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Wall L, Burke F, Smyth JF, Balkwill F. The anti-proliferative activity of interferon-gamma on ovarian cancer: in vitro and in vivo. Gynecol Oncol. 2003;88:S149–151. doi: 10.1006/gyno.2002.6707. [DOI] [PubMed] [Google Scholar]
- 26.Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, Matei D. beta-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34:2297–2308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Li YH, Guo FJ, Wang JJ, Sun RL, Hu JY, Li GC. Gamma-aminobutyric acid promotes human hepatocellular carcinoma growth through overexpressed gamma-aminobutyric acid A receptor alpha 3 subunit. World J Gastroenterol. 2008;14:7175–7182. doi: 10.3748/wjg.14.7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurosaki A, Hasegawa K, Kato T, Abe K, Hanaoka T, Miyara A, O'shannessy DJ, Somers EB, Yasuda M, Sekino T, Fujiwara K. Serum folate receptor alpha (FRA) as a biomarker for ovarian cancer: Implications for diagnosis, prognosis and predicting its local tumor expression. Int J Cancer. 2016;138:1994–2002. doi: 10.1002/ijc.29937. [DOI] [PubMed] [Google Scholar]
- 29.Brot S, Malleval C, Benetollo C, Auger C, Meyronet D, Rogemond V, Honnorat J, Moradi-Ameli M. Identification of a new CRMP5 isoform present in the nucleus of cancer cells and enhancing their proliferation. Exp Cell Res. 2013;319:588–599. doi: 10.1016/j.yexcr.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Giovangrandi Y, Parfait B, Asheuer M, Olivi M, Lidereau R, Vidaud M, Bieche I. Analysis of the human CGB/LHB gene cluster in breast tumors by real-time quantitative RT-PCR assays. Cancer Lett. 2001;168:93–100. doi: 10.1016/s0304-3835(01)00496-7. [DOI] [PubMed] [Google Scholar]
- 31.Xu HT, Liu Y, Liu SL, Miao Y, Li QC, Wang EH. TC-1 (C8orf4) expression is correlated with differentiation in ovarian carcinomas and might distinguish metastatic ovarian from metastatic colorectal carcinomas. Virchows Arch. 2013;462:281–287. doi: 10.1007/s00428-013-1375-7. [DOI] [PubMed] [Google Scholar]
- 32.Peche LY, Ladelfa MF, Toledo MF, Mano M, Laiseca JE, Schneider C, Monte M. Human MageB2 Protein Expression Enhances E2F Transcriptional Activity, Cell Proliferation, and Resistance to Ribotoxic Stress. J Biol Chem. 2015;290:29652–29662. doi: 10.1074/jbc.M115.671982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felix AS, Sherman ME, Hewitt SM, Gunja MZ, Yang HP, Cora RL, Boudreau V, Ylaya K, Lissowska J, Brinton LA, Wentzensen N. Cell-cycle protein expression in a population-based study of ovarian and endometrial cancers. Front Oncol. 2015;5:25. doi: 10.3389/fonc.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu J, Lv H, Guan H, Ma X, Ji M, He N, Shi B, Hou P. Metallothionein 1G functions as a tumor suppressor in thyroid cancer through modulating the PI3K/Akt signaling pathway. BMC Cancer. 2013;13:462. doi: 10.1186/1471-2407-13-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peppelenbosch MP, Spaander MC, Bruno MJ. Glutathione peroxidase 7 prevents cancer in the oesophagus. Gut. 2014;63:537–538. doi: 10.1136/gutjnl-2013-304906. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Zhao H, Li J, Liu H, Wang F, Wei Y, Su J, Zhang D, Liu T, Zhang Y. The identification of specific methylation patterns across different cancers. PLoS One. 2015;10:e0120361. doi: 10.1371/journal.pone.0120361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh CL, Botta G, Gao S, Li T, Van Allen EM, Treacy DJ, Cai C, He HH, Sweeney CJ, Brown M, Balk SP, Nelson PS, Garraway LA, et al. PLZF, a tumor suppressor genetically lost in metastatic castration-resistant prostate cancer, is a mediator of resistance to androgen deprivation therapy. Cancer Res. 2015;75:1944–1948. doi: 10.1158/0008-5472.CAN-14-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'shannessy DJ, Jackson SM, Twine NC, Hoffman BE, Dezso Z, Agoulnik SI, Somers EB. Gene expression analyses support fallopian tube epithelium as the cell of origin of epithelial ovarian cancer. Int J Mol Sci. 2013;14:13687–13703. doi: 10.3390/ijms140713687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi R, Pan C, Zhou Y, Liu Y, Jin G, Liu F. Identification of the metastasis potential and its associated genes in melanoma multinucleated giant cells using the PHA-ECM830 fusion method. Oncol Rep. 2016;35:211–218. doi: 10.3892/or.2015.4376. [DOI] [PubMed] [Google Scholar]
- 40.Wandzioch E, Pusey M, Werda A, Bail S, Bhaskar A, Nestor M, Yang JJ, Rice LM. PME-1 modulates protein phosphatase 2A activity to promote the malignant phenotype of endometrial cancer cells. Cancer Res. 2014;74:4295–4305. doi: 10.1158/0008-5472.CAN-13-3130. [DOI] [PubMed] [Google Scholar]
- 41.Dong W, Yao C, Teng X, Chai J, Yang X, Li B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016;37:2973–85. doi: 10.1007/s13277-015-3452-9. [DOI] [PubMed] [Google Scholar]
- 42.Gromova I, Gromov P, Kroman N, Wielenga VT, Simon R, Sauter G, Moreira JM. Immunoexpression analysis and prognostic value of BLCAP in breast cancer. PLoS One. 2012;7:e45967. doi: 10.1371/journal.pone.0045967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin YN, Bhuwania R, Gromova K, Failla AV, Lange T, Riecken K, Linder S, Kneussel M, Izbicki JR, Windhorst S. Drosophila homologue of Diaphanous 1 (DIAPH1) controls the metastatic potential of colon cancer cells by regulating microtubule-dependent adhesion. Oncotarget. 2015;6:18577–18589. doi: 10.18632/oncotarget.4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franco-Montoya ML, Boucherat O, Thibault C, Chailley-Heu B, Incitti R, Delacourt C, Bourbon JR. Profiling target genes of FGF18 in the postnatal mouse lung: possible relevance for alveolar development. Physiol Genomics. 2011;43:1226–1240. doi: 10.1152/physiolgenomics.00034.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haque T, Nakada S, Hamdy RC. A review of FGF18: Its expression, signaling pathways and possible functions during embryogenesis and post-natal development. Histol Histopathol. 2007;22:97–105. doi: 10.14670/HH-22.97. [DOI] [PubMed] [Google Scholar]
- 46.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 47.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Porter IM, Schleicher K, Porter M, Swedlow JR. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat Commun. 2013;4:2677. doi: 10.1038/ncomms3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porter IM, McClelland SE, Khoudoli GA, Hunter CJ, Andersen JS, McAinsh AD, Blow JJ, Swedlow JR. Bod1, a novel kinetochore protein required for chromosome biorientation. J Cell Biol. 2007;179:187–197. doi: 10.1083/jcb.200704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q, Dalman MR, Sarmah S, Chen S, Chen Y, Hurlbut AK, Spencer MA, Pancoe L, Marrs JA. Cell adhesion molecule cadherin-6 function in zebrafish cranial and lateral line ganglia development. Dev Dyn. 2011;240:1716–1726. doi: 10.1002/dvdy.22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubota F, Murakami T, Mogi K, Yorifuji H. Cadherin-6 is required for zebrafish nephrogenesis during early development. Int J Dev Biol. 2007;51:123–129. doi: 10.1387/ijdb.062200fk. [DOI] [PubMed] [Google Scholar]
- 52.Sancisi V, Gandolfi G, Ragazzi M, Nicoli D, Tamagnini I, Piana S, Ciarrocchi A. Cadherin 6 is a new RUNX2 target in TGF-beta signalling pathway. PLoS One. 2013;8:e75489. doi: 10.1371/journal.pone.0075489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zakin L, De Robertis EM. Extracellular regulation of BMP signaling. Curr Biol. 2010;20:R89–92. doi: 10.1016/j.cub.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayward A, Ghazal A, Andersson G, Andersson L, Jern P. ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One. 2013;8:e59940. doi: 10.1371/journal.pone.0059940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansford LM, Smith SA, Haber M, Norris MD, Cheung B, Marshall GM. Cloning and characterization of the human neural cell adhesion molecule, CNTN4 (alias BIG-2) Cytogenet Genome Res. 2003;101:17–23. doi: 10.1159/000073412. [DOI] [PubMed] [Google Scholar]
- 56.Manderson EN, Birch AH, Shen Z, Mes-Masson AM, Provencher D, Tonin PN. Molecular genetic analysis of a cell adhesion molecule with homology to L1CAM, contactin 6, and contactin 4 candidate chromosome 3p26pter tumor suppressor genes in ovarian cancer. Int J Gynecol Cancer. 2009;19:513–525. doi: 10.1111/IGC.0b013e3181a3cd38. [DOI] [PubMed] [Google Scholar]
- 57.Deng Y, Chen C, Hua M, Xi Q, Liu R, Yang S, Liu J, Zhong J, Tang M, Lu S, Tang C, Wang Y. Annexin A2 plays a critical role in epithelial ovarian cancer. Arch Gynecol Obstet. 2015;292:175–182. doi: 10.1007/s00404-014-3598-5. [DOI] [PubMed] [Google Scholar]
- 58.Schroder WA, Major L, Suhrbier A. The role of SerpinB2 in immunity. Crit Rev Immunol. 2011;31:15–30. doi: 10.1615/critrevimmunol.v31.i1.20. [DOI] [PubMed] [Google Scholar]
- 59.Croucher DR, Saunders DN, Lobov S, Ranson M. Revisiting the biological roles of PAI2 (SERPINB2) in cancer. Nat Rev Cancer. 2008;8:535–545. doi: 10.1038/nrc2400. [DOI] [PubMed] [Google Scholar]
- 60.Chambers SK, Ivins CM, Carcangiu ML. Expression of plasminogen activator inhibitor-2 in epithelial ovarian cancer: a favorable prognostic factor related to the actions of CSF-1. Int J Cancer. 1997;74:571–575. doi: 10.1002/(sici)1097-0215(19971219)74:6<571::aid-ijc2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 61.Liu M, Zhao J, Chen K, Bian X, Wang C, Shi Y, Wang JM. G protein-coupled receptor FPR1 as a pharmacologic target in inflammation and human glioblastoma. Int Immunopharmacol. 2012;14:283–288. doi: 10.1016/j.intimp.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prevete N, Liotti F, Marone G, Melillo RM, de Paulis A. Formyl peptide receptors at the interface of inflammation, angiogenesis and tumor growth. Pharmacol Res. 2015;102:184–191. doi: 10.1016/j.phrs.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 63.Loddo M, Andryszkiewicz J, Rodriguez-Acebes S, Stoeber K, Jones A, Dafou D, Apostolidou S, Wollenschlaeger A, Widschwendter M, Sainsbury R, Tudzarova S, Williams GH. Pregnancy-associated plasma protein A regulates mitosis and is epigenetically silenced in breast cancer. J Pathol. 2014;233:344–356. doi: 10.1002/path.4393. [DOI] [PubMed] [Google Scholar]
- 64.Ghosh M, Shen Z, Schaefer TM, Fahey JV, Gupta P, Wira CR. CCL20/MIP3alpha is a novel anti-HIV-1 molecule of the human female reproductive tract. Am J Reprod Immunol. 2009;62:60–71. doi: 10.1111/j.1600-0897.2009.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Son DS, Kabir SM, Dong Y, Lee E, Adunyah SE. Characteristics of chemokine signatures elicited by EGF and TNF in ovarian cancer cells. J Inflamm (Lond) 2013;10:25. doi: 10.1186/1476-9255-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Couderc B, Pradines A, Rafii A, Golzio M, Deviers A, Allal C, Berg D, Penary M, Teissie J, Favre G. In vivo restoration of RhoB expression leads to ovarian tumor regression. Cancer Gene Ther. 2008;15:456–464. doi: 10.1038/cgt.2008.12. [DOI] [PubMed] [Google Scholar]
- 67.Nash MA, Deavers MT, Freedman RS. The expression of decorin in human ovarian tumors. Clin Cancer Res. 2002;8:1754–1760. [PubMed] [Google Scholar]
- 68.Wu Y, Sun L, Zou W, Xu J, Liu H, Wang W, Yun X, Gu J. Prosaposin, a regulator of estrogen receptor alpha, promotes breast cancer growth. Cancer Sci. 2012;103:1820–1825. doi: 10.1111/j.1349-7006.2012.02374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei W, Mok SC, Oliva E, Kim SH, Mohapatra G, Birrer MJ. FGF18 as a prognostic and therapeutic biomarker in ovarian cancer. J Clin Invest. 2013;123:4435–4448. doi: 10.1172/JCI70625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan H, He Q, Gong G, Wang Y, Li J, Wang J, Zhu D, Wu X. miR-382 inhibits migration and invasion by targeting ROR1 through regulating EMT in ovarian cancer. Int J Oncol. 2016;48:181–190. doi: 10.3892/ijo.2015.3241. [DOI] [PubMed] [Google Scholar]
- 71.Lv Q, Shen R, Wang J. RBPJ inhibition impairs the growth of lung cancer. Tumour Biol. 2015;36:3751–3756. doi: 10.1007/s13277-014-3015-5. [DOI] [PubMed] [Google Scholar]
- 72.Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, Reddy S, Bell GW, Jaenisch R. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011;108:18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshioka S, King ML, Ran S, Okuda H, MacLean JA, 2nd, McAsey ME, Sugino N, Brard L, Watabe K, Hayashi K. WNT7A regulates tumor growth and progression in ovarian cancer through the WNT/beta-catenin pathway. Mol Cancer Res. 2012;10:469–482. doi: 10.1158/1541-7786.MCR-11-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weidle UH, Evtimova V, Alberti S, Guerra E, Fersis N, Kaul S. Cell growth stimulation by CRASH, an asparaginase-like protein overexpressed in human tumors and metastatic breast cancers. Anticancer Res. 2009;29:951–963. [PubMed] [Google Scholar]
- 75.Roider HG, Manke T, O'Keeffe S, Vingron M, Haas SA. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nolen BM, Lokshin AE. Biomarker testing for ovarian cancer: clinical utility of multiplex assays. Mol Diagn Ther. 2013;17:139–146. doi: 10.1007/s40291-013-0027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang W, Huang R, Duan C, Fu L, Xi Y, Yang Y, Yang WM, Yang D, Yang DH, Huang RP. Identification of five serum protein markers for detection of ovarian cancer by antibody arrays. PLoS One. 2013;8:e76795. doi: 10.1371/journal.pone.0076795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaspric N, Picard B, Reichstadt M, Tournayre J, Bonnet M. ProteINSIDE to Easily Investigate Proteomics Data from Ruminants: Application to Mine Proteome of Adipose and Muscle Tissues in Bovine Foetuses. PLoS One. 2015;10:e0128086. doi: 10.1371/journal.pone.0128086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szczerba A, Sliwa A, Kubiczak M, Nowak-Markwitz E, Jankowska A. Human chorionic gonadotropin beta subunit affects the expression of apoptosis-regulating factors in ovarian cancer. Oncol Rep. 2016;35:538–545. doi: 10.3892/or.2015.4386. [DOI] [PubMed] [Google Scholar]
- 80.Lim W, Jeong W, Kim J, Ka H, Bazer FW, Han JY, Song G. Differential expression of secreted phosphoprotein 1 in response to estradiol-17beta and in ovarian tumors in chickens. Biochem Biophys Res Commun. 2012;422:494–500. doi: 10.1016/j.bbrc.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 81.Douglas JB, Silverman DT, Pollak MN, Tao Y, Soliman AS, Stolzenberg-Solomon RZ. Serum IGF-I, IGF-II, IGFBP-3, and IGF-I/IGFBP-3 molar ratio and risk of pancreatic cancer in the prostate, lung, colorectal, and ovarian cancer screening trial. Cancer Epidemiol Biomarkers Prev. 2010;19:2298–2306. doi: 10.1158/1055-9965.EPI-10-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He LH, Ma Q, Shi YH, Ge J, Zhao HM, Li SF, Tong ZS. CHL1 is involved in human breast tumorigenesis and progression. Biochem Biophys Res Commun. 2013;438:433–438. doi: 10.1016/j.bbrc.2013.07.093. [DOI] [PubMed] [Google Scholar]
- 83.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 84.Tagliaferri D, De Angelis MT, Russo NA, Marotta M, Ceccarelli M, Del Vecchio L, De Felice M, Falco G. Retinoic Acid Specifically Enhances Embryonic Stem Cell Metastate Marked by Zscan4. PLoS One. 2016;11:e0147683. doi: 10.1371/journal.pone.0147683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kewitz S, Staege MS. Knock-down of PRAME increases retinoic acid signaling and cytotoxic drug sensitivity of Hodgkin lymphoma cells. PLoS One. 2013;8:e55897. doi: 10.1371/journal.pone.0055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, Skomedal H, Tu IP, Hernandez-Boussard T, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowen NJ, Logani S, Dickerson EB, Kapa LB, Akhtar M, Benigno BB, McDonald JF. Emerging roles for PAX8 in ovarian cancer and endosalpingeal development. Gynecol Oncol. 2007;104:331–337. doi: 10.1016/j.ygyno.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 88.El-Gendi S, Abdelzaher E, Mostafa MF, Sheasha GA. FGF18 as a potential biomarker in serous and mucinous ovarian tumors. Tumour Biol. 2016;37:3173–83. doi: 10.1007/s13277-015-4129-0. [DOI] [PubMed] [Google Scholar]
- 89.Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, Welch WR, Berkowitz RS, Mok SC. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 90.Vathipadiekal V, Wang V, Wei W, Waldron L, Drapkin R, Gillette M, Skates S, Birrer M. Creation of a Human Secretome: A Novel Composite Library of Human Secreted Proteins: Validation Using Ovarian Cancer Gene Expression Data and a Virtual Secretome Array. Clin Cancer Res. 2015;21:4960–4969. doi: 10.1158/1078-0432.CCR-14-3173. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, Li L, Wang Y, Tang SN, Zheng W. Fallopian tube secretory cell expansion: a sensitive biomarker for ovarian serous carcinogenesis. Am J Transl Res. 2015;7:2082–2090. [PMC free article] [PubMed] [Google Scholar]
- 92.Quartuccio SM, Karthikeyan S, Eddie SL, Lantvit DD, E Oh, Modi DA, Wei JJ, Burdette JE. Mutant p53 expression in fallopian tube epithelium drives cell migration. Int J Cancer. 2015;137:1528–1538. doi: 10.1002/ijc.29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Munakata S, Yamamoto T. Incidence of serous tubal intraepithelial carcinoma (STIC) by algorithm classification in serous ovarian tumor associated with PAX8 expression in tubal epithelia: a study of single institution in Japan. Int J Gynecol Pathol. 2015;34:9–18. doi: 10.1097/PGP.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medeiros F, Muto MG, Lee Y, Elvin JA, Callahan MJ, Feltmate C, Garber JE, Cramer DW, Crum CP. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am J Surg Pathol. 2006;30:230–236. doi: 10.1097/01.pas.0000180854.28831.77. [DOI] [PubMed] [Google Scholar]
- 95.D'Antonio M, D'Onorio De Meo P, Pallocca M, Picardi E, D'Erchia AM, Calogero RA, Castrignano T, Pesole G. RAP: RNA-Seq Analysis Pipeline, a new cloud-based NGS web application. BMC Genomics. 2015;16:S3. doi: 10.1186/1471-2164-16-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Di Palma T, Conti A, de Cristofaro T, Scala S, Nitsch L, Zannini M. Identification of novel Pax8 targets in FRTL-5 thyroid cells by gene silencing and expression microarray analysis. PLoS One. 2011;6:e25162. doi: 10.1371/journal.pone.0025162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


