ABSTRACT
This article discusses the historical perspective and the new findings of autophagy and ubiquitin-proteasome system cooperation during the post-fertilization sperm mitophagy, a process that eliminates potentially damaged paternal mitochondrial DNA from an early embryo. New insight into the mechanism that promotes clonal, maternal inheritance of mitochondrial genes may be helpful for managing mitochondrial disease and infertility in humans, as well as reproductive performance and production traits in agriculturally important domestic animals.
KEYWORDS: assisted reproductive therapy, autophagy, mitochondria, mitophagy, mtDNA, SQSTM1, ubiquitin-proteasome system, VCP
Clonal inheritance of mitochondria and mitochondrial DNA (mtDNA), sometimes referred to as the Mitochondrial Eve Paradigm, is well documented in mammals and other taxa. Interest in how such a uniparental inheritance pattern is propagated can be traced back to a 1940 Nature paper that explained it as a dispersion and dilution due to disproportionate number of mitochondria in an oocyte vs. spermatozoon. Indeed, there are about 100 mitochondria in an oocyte for each mitochondrion contributed by the fertilizing spermatozoon. Nevertheless, alternative explanations were sought, chief among them the notion that the fertilizing spermatozoon jettisons its tail with mitochondria, leaving it outside once the sperm head fuses with the oocyte. Textbooks and popular science have propagated such a theory as late as the 1990s, despite electron microscopy evidence of the sperm tail and sperm mitochondria being incorporated into the ooplasm in various species’ zygotes.
The boom of organelle-specific vital fluorescent probes in the 1990s armed fertilization biologists with new opportunities, including the ability to pre-label sperm mitochondria prior to in vitro fertilization and track their fate post-fertilization. We thus observed that in bovine, murine and rhesus monkey embryos, the initially straight sperm mitochondrial sheath (MS) became deformed, clumped and finally disassembled at the 2–4 cell stage of embryo development. As the initial alterations of the MS coincided with the progression of the first embryonic cell cycle (first embryo cleavage), we speculated that the process of what we now call sperm mitophagy could be related to cell cycle regulation by the ubiquitin-proteasome system (UPS) which, as it became known around that time, relies on precisely timed proteasomal degradation of cyclins at the metaphase-anaphase transition. Indeed, binding of ubiquitin to the sperm MS inside the ooplasm was documented and, later on, proteasomal inhibitors were shown to block sperm mitophagy in porcine zygotes. This species is particularly conducive to such research since therein, mitophagy occurs prior to the first embryo cleavage and proteasomal inhibitors thus do not interfere with its zygotic cell cycle. How the UPS, a cellar recycling and regulatory pathway that degrades proteins one molecule at a time, could dismantle an entire organelle remained to be shown.
In our early studies, we observed ultrastructural alterations of ooplasm-incorporated sperm mitochondria reminiscent of organelle autophagy, and treatment with inhibitors of the lysosome, the autophagic endpoint, indeed imposed some delay on the process. As the molecular machinery of autophagy became deciphered, sperm mitophagy was being revisited in new animal models including Drosophila and C. elegans, eventually revealing the link between the UPS and autophagy through ubiquitin-binding autophagy receptors such as SQSTM1, and the autophagy-related proteins GABARAP and LC3, but also suggesting that the mitophagy in the fertilized animal oocytes was somewhat different from canonical organelle autophagy described in somatic cells. With that knowledge, we uncovered that SQSTM1 and the protein dislocase VCP work in conjunction in the porcine zygote and that their co-inhibition delays sperm mitophagy from the one cell to 2–4 cell stage (Fig. 1), at which time point the inhibitory molecules wear out. We thus created heteroplasmic porcine embryos, along the lines of the Mitochondrial Steve moniker introduced by Bromham et al., in 2003. The ability of VCP to extract ubiquitinated proteins from mitochondrial membrane and serve them to the 26S proteasome explains why proteasomal inhibitors block post-fertilization sperm mitophagy. The recognition of the ubiquitinated mitochondrial shell by SQSTM1 shows how the cell disposes of the destabilized remnants of the organelle. Meanwhile, it is also possible that specific endonucleases may be involved in degrading the mtDNA cargo of such compromised sperm mitochondria. Altogether, sperm mitophagy may benefit embryo development by removing potentially harmful sperm mtDNA, which accumulates mtDNA mutations and strand breaks at a high rate, possibly due to harsh conditions rich in reactive oxygen species in both male and female reproductive tracts.
Figure 1.
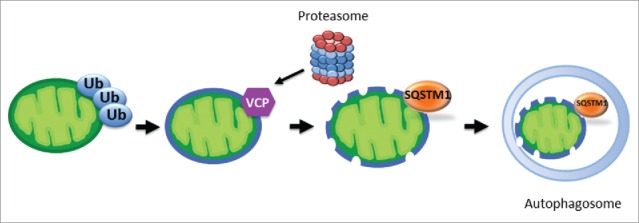
Proposed model of sperm mitophagy in the porcine zygote relies on a combined action of SQSTM1-dependent autophagy and VCP-mediated presentation of ubiquitinated (Ub) sperm mitochondrial proteins to the 26S proteasome. Sperm mitochondria become ubiquitinated before fertilization, and some of the ubiquitinated mitochondrial membrane proteins are extracted by VCP after fertilization. Concurrently, the ooplasmic SQSTM1 binds to other ubiquitinated mitochondrial proteins, targeting the destabilized organelle for sequestration within an autophagosome.
This work is significant both for human medicine, and for livestock fitness and productivity. The few documented cases of paternal heteroplasmy suggest that in humans, it could be associated with mitochondrial disease. Although the notion of paternal mtDNA leakage in human and chimpanzee populations has been around since the 1990s, patients with mitochondrial disease are not routinely, or even occasionally, screened for it. What is the true incidence of it in human populations and if it is prevalent, is it the root cause of certain mitochondrial diseases? Could this be managed in human assisted reproductive therapy (ART), wherein the prevalent ICSI-sperm injection method might delay mitophagy by introducing a spermatozoon with intact membranes (they are removed as the spermatozoon enters the oocyte during the natural fertilization process)? How about the practice of oocyte rejuvenation by mitochondrial donation in female infertility patients of advanced reproductive age?
If paternal leakage occurs in livestock species other than interspecific crosses, could it affect mitochondrion-rich musculature and by translation fitness and carcass quality which is paramount to profitability of beef and pork producers? Similarly, somatic cell nuclear transfer (SCNT), now used to replicate valuable genomes of sires with outstanding production traits, could be hindered by donor cell-derived heteroplasmy. Could we attach autophagy receptors to mitochondrial membranes of those donor cells? Such experiments could answer questions about the mechanisms that guide sperm mitophagy and make it species specific, only occurring in intraspecific crosses in mammals. Besides transgenic approaches, we are now pursuing studies in a cell-free system consisting of demembranated spermatozoa coincubated with mammalian oocyte extracts, which mimic the recognition of sperm mitochondria by SQSTM1 and VCP seen post-fertilization. Such a cell-free system can expose large numbers of spermatozoa to ooplasm from hundreds of oocytes, whereas even in a polyspermic animal zygote generated by in vitro fertilization, only 2–5 sperm MS can be observed in the fertilized ooplasm.
Altogether, the historical perspective and the new findings of autophagy and UPS cooperation deepen our understanding of sperm mitophagy in mammals, which may be helpful for managing mitochondrial disease and infertility in humans, as well as reproductive performance and production traits in agriculturally important domestic animals.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work discussed here was funded by National Institute of Food and Agriculture within the US. Department of Agriculture, National Institutes of Health Office of Research Infrastructure Programs, and by the Food for the 21st Century program at the University of Missouri.


