ABSTRACT
Protein insertional fusion and circular permutation are 2 promising protein engineering techniques for creating integrated functionalities and sequence diversity of a protein, respectively. Finding insertion locations for protein insertional fusion and new termini for circular permutation through a rational approach is not always straightforward, especially, for proteins without detailed structural knowledge. On the contrary, a combinatorial approach facilitates a comprehensive search to evaluate all potential insertion sites and new termini locations. Conventional methods used to create random insertional fusion libraries generate sub-optimal inter-domain linker length and composition between fused proteins. There are also methods available for construction of random circular permutation libraries. However, these methods too, impose many drawbacks, such as significant sequence modification at the new termini of circular permutants and additionally, require re-design of transposons for tailored expression of circular permutants. Furthermore, these conventional methods employ relatively inefficient blunt-end ligation during library construction. In this commentary, we present a concise overview and key findings of engineered Mu transposons, which have recently been developed in our group as a facile and efficient tool to alleviate limitations realized from conventional methods and to construct high quality libraries for random insertional fusion and random circular permutation.
KEYWORDS: combinatorial library, MuRCP transposon, MuST transposon, protein engineering, random circular permutation, random insertional fusion
Introduction
Transposons (or transposable elements) are mobile genetic elements that translocate from one genomic location to another in a random fashion. Depending on the intermediates formed during transposition, transposable elements are classified into 2 main groups: 1) Class I or retrotransposon, and 2) Class II or DNA transposon.1 Retrotransposons, which are mostly found in eukaryotic organisms, employ the “copy” mechanism: retrotransposons are reverse-transcribed to DNA before insertion of a new copy to another genome location.2 On the contrary, DNA transposons can be found in both prokaryotes and eukaryotes, and employ the “cut and paste” mechanism: DNA transposons use DNA directly as a transposition intermediate without forming RNA intermediates.3
DNA transposons can serve as in vitro molecular tools for various protein engineering applications due to their ability to integrate into various DNA sequences and thus generate extensive mutant libraries.4 In vitro transposition reactions have primarily been mediated by (1) bacterial transposons, such as Tn7,5 Tn3,6 Tn5,7 Tn552,8 Tn10 9 and IS911,10 (2) bacteriophage transposons, such as Mu,11 and (3) yeast transposons, such as Ty1.12 Transposons in the most simplistic form, called mini-transposons, have also been developed to facilitate in vitro transposition reactions.13,14 The minimal elements required for in vitro transposition include the terminal inverted repeat nucleotides within transposons (i.e. transposase recognition site), transposase (i.e., enzyme), the target host DNA, and a reaction buffer.
A bacteriophage Mu transposon is one of the most useful transposable elements in nature due to its high integration efficiency and non-specific target site selection.14 Accordingly, the in vitro Mu transposition reaction has been studied extensively.15 The Mu transposon has 22 bp-long terminal inverted repeats, which is a recognition sequence for MuA transposase.15 Random integration of Mu transposon into target DNA occurs through the following 3 steps; 1) the MuA transposase binds to the symmetrical sequence of the Mu transposon and forms a transposome assembly15; 2) the transposome assembly assists in Mu transposon's self-cleavage at cleavage site (i.e. TA/CA)16; 3) the Mu transposon is integrated into the target DNA with precise 5 bp duplication.15
Most protein engineering approaches involve mutations in the primary sequence of proteins for the improvement of desired properties.17,18 However, introducing mutations often compromised intrinsic properties of proteins, such as enzyme activity.19 In contrast, protein insertional fusion and circular permutation are among other protein engineering approaches, which involve no change in the primary sequence except for its arrangement and linear order, respectively.20,21 When a rational design is not available, especially for the protein without detailed structural knowledge, a combinatorial protein engineering tools is extremely beneficial. This commentary presents our recent development on combinatorial approaches for the construction of insertional fusion libraries and circular permutation libraries using engineered Mu transposons.22,23
Transposons for random insertional fusion of protein domains
Insertional protein fusion is an advanced engineering approach to generate novel proteins with integrated functions. Three critical parameters that can determine the success of functional integration between fused proteins are the insertion location within the host protein,24,25 the inter-domain linker length26,27 and the inter-domain linker composition.26,28 Inter-domain linker length and composition should be carefully designed to avoid a steric hindrance between the 2 fused domains and to facilitate maximum inter-domain interactions.29 Among various methods for generating protein insertional fusions, a combinatorial approach is especially beneficial because robust guidelines for selection of insertion sites are unavailable. Construction of a random insertional fusion library covering all possible insertion sites within the host protein domain, followed by high-throughput screening of the resulting variants is an effective strategy for identifying insertional fusions with desired functional outcomes.
The conventional methods for constructing a combinatorial protein fusion library are based-on random DNA cut by endonucleases.30,31 Unfortunately, these methods generate uncontrollable tandem duplication and deletion around insertion sites within the host DNA. Upon translation, the tandem duplication of the host DNA generates uncontrollable linker length between fused proteins. In addition, uncontrolled truncation deteriorates the integrity of the primary sequence of a host protein, affecting its other intrinsic properties. Also, this method involves the final step of blunt-end ligation between guest and host genes, which is less efficient than sticky-end ligation, lowering library construction efficiency.32,33 Alternatively, exploiting the inherent nature of transposon for its random insertion into host DNA sequences has gained interest for the construction of domain insertion library. In these alternative methods, transposons are designed to include flanking guest DNA sequences along with other genetic components required for transposition. However, no mechanism is implemented for removal of transposon elements (including 22-bp recognition sequence) after transposition, and the remaining nucleotides encode suboptimal inter-domain linkers upon translation.34-36 In another method, control of inter-domain linker length and composition is made possible by the removal of an inserted transposon from a host DNA sequence after transposition.26 However, similar to the endonuclease-based methods, this transposon-based method depends on the blunt-end ligation between host and guest DNAs.
To alleviate all limitations imposed by the conventional endonuclease- and transposon-based methods, we developed an engineered Mu transposon, MuST, for the construction of random insertion library, which employ (1) the sticky-end ligation between guest and host DNAs and (2) provide optimal control over inter-domain linker length and composition.22
A schematic of the construction of random domain insertion libraries using the MuST transposon is shown in Fig. 1A. Here, the method is illustrated with model systems bcx (Bacillus circulans Xylanase) and PUC19 containing lacZα as guest and host genes, respectively. The library was constructed in 3 phases; (1) the MuST transposon was randomly inserted into the host plasmid by transposition; 2) the entire MuST transposon was then removed from the plasmid, producing random single-cut plasmids; 3) the random single-cut host DNA was subsequently sticky-end ligated with guest DNA.
Figure 1.
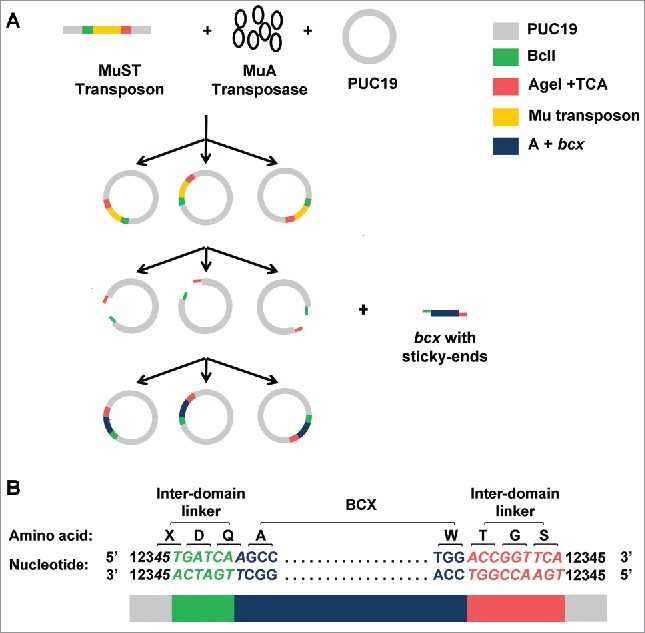
(A) A schematic of random protein domain insertion is illustrated with bcx and PUC19 containing lacZα as model guest and host genes, respectively. Random transposition of the MuST transposon into the host plasmid PUC19 is mediated by MuA transposase. After random transposition, the MuST transposon is removed by double digestion using BclI and AgeI restriction enzymes. The double-digested PUC19 is then sticky-end ligated with a guest DNA. Additional 5′ adenine is contained upstream of the bcx for in-frame connection between BCX and the desired inter-domain linker residues. (B) A sequence schematic of bcx randomly inserted into PUC19. The BclI (TGATCA) site is shown in green. The AgeI (ACCGGT) + TCA site is shown in red. Numbers (i.e., 12345) represent nucleotide sequences derived from the host DNA. Nucleotide sequences encoding linkers (i.e., 45TGATCAA and ACCGGTTCA) are shown in italic. The first and last codons of BCX (i.e., GCC and TGG, respectively) are shown in plain font. X represents one of 15 amino acids (i.e., Ala, Arg, Asn, Asp, Cys, Gly, His, Ile, Leu, Phe, Pro, Ser (twice), Thr, Tyr, and Val) encoded by a codon containing 3′ thymine. Colored blocks shown at the bottom indicate the following: gray for nucleotide sequences derived from PUC19, green for the BclI site, dark blue for additional 5′ adenine + bcx, and red for the AgeI + TCA site. (Reproduced with permission from Fig 1 in Shah V. (2013), Random domain insertion using an engineered transposon. Anal Biochem., 432, 97–102).
The engineered MuST transposon was derived from the commercially available Mu transposon – with a built-in chloramphenicol resistance gene as a selection marker – by appendages of BclI (TGATCA) and Agel+TCA (ACCGGTTCA) at the 5′ and 3′ ends, respectively. Four criteria were considered for the selection of these restriction sites. First, the included BclI and AgeI sites encode inter-domain linkers with optimal linker length and composition: that is, Asp-Gln (second reading frame) and Thr-Gly-Ser (first reading frame) at the N- and C-termini of a guest protein, respectively (Fig. 1B).22 Second, placement of the restriction sites at the ends required minimal mutations outside or on the near ends of the MuA transposase recognition binding sites. The minimal sequence change was desired to maintain the transposition efficiency as compared to the wild-type Mu transposon.37 Note that the cleavage sites (TG/CA) within the transposon must be conserved as it is critical for transposition.16 Third, the restriction enzyme sites facilitated sticky-end ligation between the host and the guest DNAs. Fourth, the restriction enzyme sites were absent in the selected host and guest DNA fragments (e.g. PUC19 and bcx, respectively).
In some cases, other restriction enzyme sites than BclI and AgeI need to be included in engineered transposons to fulfill the first and fourth criteria For example, BclI and AgeI might not be unique with respect to other host and guest DNAs. Also, the inter-domain linker composition derived from BclI and AgeI might not be preferable for certain fusions. A number of MuST transposon variants containing different restriction sites in place of BclI and/or AgeI can be realized (Table 1). Inclusion of a certain restriction enzyme site (e.g., TfiI) requires even fewer mutations than BclI on the near ends of the Mu transposon recognition sites. While transposition efficiencies of the MuST transposon variants have yet to be examined, these options would help to generate different inter-domain linker compositions. It is important to note that regardless of selected restriction sites, our method has a feature that allows change in reading frames of an inserted protein with respect to a host protein. For example, the BclI site was placed in the second reading frame by placing an additional adenine at the upstream of the 5′ end of bcx (Fig. 1B). It should also be noted that inter-domain linker length can readily be increased by addition of nucleotides/codons at the termini of guest DNA, before ligation to a random single-cut host DNA fragment.
Table 1.
Different options of restriction enzyme sites for designing MuST transposon variants.
| Restriction enzyme site at the 5′end | Restriction enzyme site at the 3′end | Nucleotides encoding Inter-domain linker | Inter-domain linker composition | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T+TfiI | 4 | 5 | T | G | A | A | T | C | N | X-Glu-Ser | |||||||
| 4 | 5 | A | C | T | T | A | G | N | |||||||||
| BsrGI | 4 | 5 | T | G | T | A | C | A | N | X-Val-His/Gln | |||||||
| 4 | 5 | A | C | A | T | G | T | N | |||||||||
| TG+Hind III | 4 | 5 | T | G | A | A | G | C | T | T | N | N | X-Glu-Ala-Y | ||||
| 4 | 5 | A | C | T | T | C | G | A | A | N | N | ||||||
| TGAAG +EagI | 4 | 5 | T | G | A | A | G | C | G | G | C | C | G | N | N | X-Glu-Ala-Ala-Z | |
| 4 | 5 | A | C | T | T | C | G | C | C | G | G | C | N | N | |||
| BssHII +CTTCA | N | G | C | G | C | G | C | C | T | T | C | A | Gly/Ser/Arg/Cys-Ala-Pro-Ser | ||||
| N | C | G | C | G | C | G | G | A | A | G | T | ||||||
| BtsI+TCA | G | C | A | G | T | G | T | C | A | Ala-Val-Ser | |||||||
| C | G | T | C | A | C | A | G | T | |||||||||
| NgoMIV +TCA | G | C | C | G | G | C | T | C | A | Ala-Gly-Ser | |||||||
| C | G | G | C | C | G | A | G | T | |||||||||
Note. The numbers represent nucleotide sequences derived from a host DNA. N represents an additional nucleotide placed either upstream of the 5′ end or downstream of the 3′end of the guest DNA to ensure inter-domain linker in second and first reading frames, at the N and C terminus of the guest protein, respectively. The mutated nucleotides introduced in the MuST transposon variants compared to the wild-type Mu transposon are shown with underlines. X represents one of 15 amino acids (i.e., Ala, Arg, Asn, Asp, Cys, Gly, His, Ile, Leu, Phe, Pro, Ser (twice), Thr, Tyr, and Val) encoded by codons containing 3′ thymine. Y represents one of 6 possible amino acids (i.e. Phe, Leu, Ser, Tyr, Cys, Trp) encoded by codons containing 5′ thymine. Z represents one of 5 possible amino acids (i.e., Val, Ala, Asp, Glu, Gly) encoded by codons containing 5′ guanine.
The MuST transposon was constructed by PCR. Due to the presence of the 22 bp symmetrical recognition sequence at the termini of Mu transposon, both forward and reverse primers can anneal at either ends during the annealing step. The mixed annealing can consequently result in a mixture of 4 different constructs in the final PCR product. From the sequencing results the desired construct (i.e., BclI at the 5′ and AgeI+TCA at the 3′ end) was identified and used as a template for the subsequent PCR for amplification of the MuST transposon.
Although this method of constructing the MuST transposon is quite straightforward, its success is heavily dependent on obtaining the desired construct from the mixture of the 4 possible constructs. A better method is proposed to construct the MuST transposon. Briefly, the Mu transposon can be split into 2 fragments of unequal size by single digestion of a natively present unique restriction site (e.g. NcoI: CCATTG). Splitting the Mu transposon in this manner separates the inverted repeats into both ends of the transposon. BclI and AgeI restriction sites can then be attached at the 5' end of the first fragment and the 3' end of the second fragment, respectively, during 2 separate PCR. In the last step, both fragments can be annealed by overlap extension PCR. This technique can be utilized with much higher efficiency to place any restriction site at the 5′ and 3′ ends of the MuST transposon.
Sequencing of library members suggested that a guest gene was randomly inserted with 5 bp precise duplication into a host gene. Also, the ratio of having correctly oriented to inversely oriented insertions of a guest DNA was about 1:1, though slight variation was also observed from run to run.22,23
Transposons for random circular permutation of proteins
Circular permutation is a protein engineering approach that connects the original N- and C- termini of proteins through a linker and, instead, introduces new termini elsewhere in the sequence.38 As more than 50% of single domain proteins have their N- and C- termini proximal,39 circular permutation could be an effective tool to generate diversity in the linear order of amino acid sequences. Proteins with distant N- and C- termini could also be linked with a relatively long linker, further highlighting the wide applicability of circular permutation. Circular permutation has proven effective for improving proteolytic resistance,40 enzyme activity41 and stability of proteins. 42
As insertion location is an important factor for the functional outcome of insertional protein fusion, the success of circular permutation is dependent on the location of new termini. The new termini should be carefully selected to avoid structural disturbance of protein's key domains (e.g.,, active sites) upon generation of new open ends. A combinatorial strategy is often advantageous when it is difficult to rationally locate the new termini.
Similar to protein insertional fusion, the conventional methods for constructing random circular permutation of proteins rely on endonucleases and thus suffer from limitations associated with tandem duplication and deletion, and blunt-end ligation. As an alternative, a transposon-based method was developed.43 Although this method alleviates the limitation of uncontrolled duplication and deletion, and provides controlled, precise 5 bp duplication, it however creates other significant drawbacks due to the lack of a transposon removal mechanism. For example, a 20 amino acid-long fragment, which was derived from the recognition site of the transposon, became attached to the new termini of circularly permuted proteins. This 20 amino acid appendage may exert a negative impact on protein properties, such as stability.44,45 In this method, the expression level of a random circular permutation library was determined by a part of the transposon (i.e, built-in components for the expression of protein), which remained intact during circular permutation. As such, the expression level of random circular permutants can only be changed by redesigning the whole transposon and reconstructing the library. This limited tunability may be a critical obstacle in library construction and screening.
In order to mitigates limitations imposed by the conventional methods, we engineered a variation of the Mu transposon, MuRCP (Mu transposon for random circular permutation), for random circular permutation.23 The design of MuRCP transposon involves attachment of 2 restriction sites at the 5′ and 3′ ends of the Mu transposon, respectively, which not only facilitate sticky-end ligation in library construction, but also reduces modifications at the new termini of random circular permutants. Additionally, the MuRCP transposon requires no permanent built-in elements for expression; instead, expression can be externally modulated by the selection of an appropriate cloning vector.
A schematic for construction of a random circular permutation library using bcx as a model gene and the MuRCP transposon is shown in Fig. 2. The method consists of 5 steps; (1) the transposition of the MuRCP transposon into the host vector was carried out in the presence of MuA transposase; (2) MuRCP inserted itself randomly throughout the host vector; (3) the library DNA was digested with SpeI restriction enzyme, followed by isolation of randomly inserted MuRCP into the target gene; 4) the target gene with randomly inserted MuRCP was then circularized by self-ligation via the SpeI site; and 5) the inserted MuRCP transposon was removed to create randomly circularly permuted target genes, which were subsequently, sticky-end ligated to the expression vector. The sequencing results confirmed the randomness and singular insertion of MuRCP transposon into a target gene. Contrary to the MuST transposon, inversely oriented insertion of the MuRCP transposon into a target gene represents the desired insertion.
Figure 2.
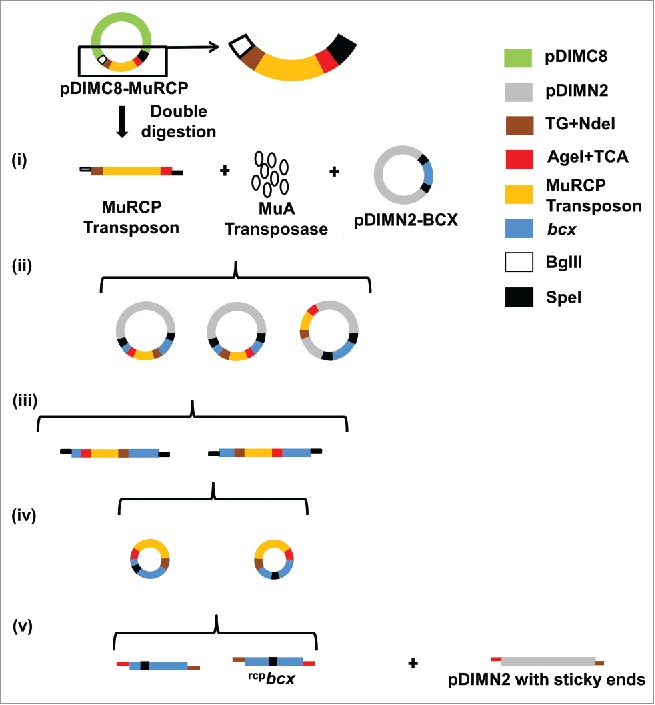
Schematic of random circular permutation of bcx as a model target gene using the MuRCP transposon. The MuRCP transposon was produced from double digestion of pDIMC8–MuRCP using BglII and SpeI restriction enzymes. (i) The MuRCP transposon was mixed with MuA transposase and a host vector containing the target gene (e.g. pDMIN2–BCX). (ii) The MuRCP transposon was randomly inserted into the pDIMN2-BCX. (iii) The target gene into which the MuRCP transposon was randomly inserted was isolated by a single digestion using an SpeI restriction site. (iv) The bcx gene with the randomly inserted MuRCP transposon was circularized. (v) The circularized bcx gene with the randomly inserted MuRCP transposon was double digested by NdeI and AgeI restriction enzymes to remove the MuRCP transposon and to prepare randomly circularly permuted bcx genes (referred to as rcpbcx) with sticky-ends for ligation to a host vector, pDIMN2. (Reproduced with permission from Fig 1 in Pierre B. (2015), Random circular permutation using a transposon. Anal. Biochem., 474, 16–24).
Selection criteria for the MuRCP restriction enzyme sites remain similar to the MuST transposon, specifically; (1) the restriction sites were unique to the host vector and the target gene; (2) the number of mutations made outside or on the near ends of the MuA transposase recognition binding sites were kept at minimum to maintain the transposition efficiency; (3) the restriction sites facilitated sticky-end ligation between an expression vector and randomly circularly permutated genes; (4) upon translation, the minimal number of amino acids were derived from the restriction enzyme sites at the new N- and C- termini of circular permutants. Consistent with these 4 criteria, the TG+NdeI (TGCATATG) restriction site at the 5′ end of the MuRCP transposon was selected due to the built-in start codon (i.e. shown with underline), reducing the number of attached amino acids to 2, Met-His or Met-Gln, at the N-terminus of the random circular permutants (Fig. 3A,B). For the 3′ end, the suitable restriction enzyme site was AgeI+TCA (ACCGGTTCA), which upon translation resulted in 3 amino acid attachment, Leu-Asn-Arg or Met-Asn-Arg or Val-Asn-Arg at the C-terminus of the random circular permutants (Fig 3A). A stop codon, immediately after Arg, was derived from genetically attached nucleotides on the host vector. The alternative restriction enzyme site at the 3′ end was AflII+TCA (CTTAAGTCA), which has a built-in stop codon (shown in bold), further reducing the number of attached amino acids to 2, Leu-Thr or Met-Thr or Val-Thr (Fig. 3B). It should, however, be noted that the transposition efficiency of the MuRCP transposons was lower by >∼6-fold than the MuST transposon.23 This differential transposon efficiency might be due to the difference in the number of necessary nucleotide mutations in order to incorporate the restriction sites at the ends of transposons. For the MuRCP transposon, inclusion of the NdeI site at the 5′ end requires 4 nucleotide mutations as compared to 2 nucleotide mutations for the BclI site included in the MuST trasnsposon. The alternative design of the MuRCP transposon with AflII site at the 3′ end requires 6 additional mutations, making the transposition efficiency even lower. This result supports the importance of conserving the near ends of transposon recognition sites for transposition efficiency.37 Overall, enzyme restriction sites need to be selected based on the trade-off between the transposition efficiency and the reduced number of attached amino acids at the termini.
Figure 3.
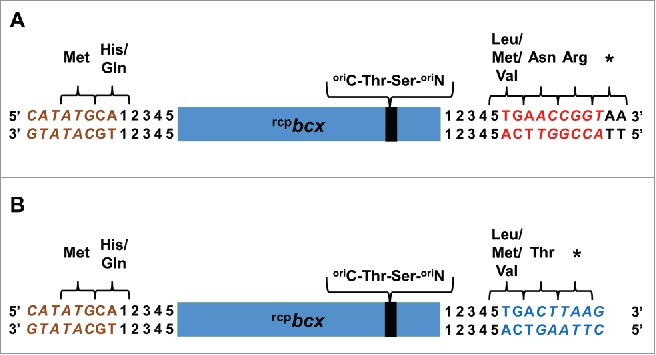
Comparison of the terminal amino acids of random circular permutants of a target protein (e.g., BCX) created using the MuRCP transposon containing (A) AgeI (ACCGGT)+TCA and (B) AflII (CTTAAG)+TCA sites. Complements of the TG + NdeI (CATATG), AgeI + TCA, AflII + TCA sites are shown in brown, red and blue, respectively, and the 3 restriction enzyme sites are shown in italic font. Nucleotides derived from a cloning vector, pDIMN2, are shown in plain black uppercase letters (i.e., AA). Numbers (i.e., 12345) represent nucleotide sequences derived from bcx. The rcpbcx represents randomly circularly permuted bcx gene. The original N- and C- termini of BCX are represented as oriN- and oriC-, respectively. (Reproduced with permission from Fig S3 in Pierre B. (2015), Random circular permutation using a transposon. Anal. Biochem., 474, 16–24).
The MuRCP was constructed by replacing the BclI with TG+NdeI restriction site from the MuST transposon by PCR. Also, to avoid sequence uncertainty from PCR due to the presence of symmetrical ends, 2 cloning sites, BglII and SpeI, were genetically appended upstream of TG+NdeI and downstream of AgeI+TCA, respectively, and sticky-end ligated to a cloning vector, pDIMC8, to construct pDIMC8-MuRCP. Whenever necessary, the MuRCP transposon was produced by replicating pDIMC8-MuRCP in E. coli., followed by DNA extraction and double digestion with BglII and SpeI.
The host plasmid (e.g. pDIMN2) containing the insert target gene (e.g., bcx) for transposition was prepared using NdeI and EcoRI as cloning sites. The SpeI restriction site (ACTAGT) was attached between the cloning sites and the 2 ends of the bcx to separate the bcx containing MuRCP transposon after transposition (Fig. 2(iii)), and subsequently, to circularize the isolated DNA fragment through self-ligation (Fig. 2(iv)). The SpeI site would encode a backbone linker, Thr-Ser, upon translation of circularly permuted bcx (Fig. 3A,B). A longer linker can readily be introduced by including additional codons between the Spe1 sites and the target gene. Any restriction site other than SpeI may be selected and incorporated at both ends of the target gene, which would code a different peptidyl linker.23
Conclusion
We developed engineered transposons, MuST and MuRCP, as combinatorial tools for protein domain insertion and circular permutation, respectively. Collective features of these transposons address many limitations realized from conventional endonucleases- and transposon-based methods. However, design of engineered transposons should be carefully done due to possible compromise of transposition efficiency.
Disclosure of potential conflicts of interest
The authors have filed patents for both transposon based methods. No other potential conflicts of interest were disclosed.
Funding
The authors are grateful for support from the National Science Foundation (CBET-1134247).
References
- [1].Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al.. A unified classification system for eukaryotic transposable elements. Nat Rev Genet 2007; 8:973-82; PMID:17984973; http://dx.doi.org/ 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- [2].Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 1990; 9:3353-62; PMID:1698615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Munoz-Lopez M, Garcia-Perez JL. DNA transposons: nature and applications in genomics. Curr Genomics 2010; 11:115-128; PMID:20885819; http://dx.doi.org/ 10.2174/138920210790886871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Reznikoff WS. Tn5 transposition: a molecular tool for studying protein structure-function. Biochem Soc Trans 2006; 34:320-3; PMID:16545104; http://dx.doi.org/ 10.1042/BST0340320 [DOI] [PubMed] [Google Scholar]
- [5].Bainton RJ, Kubo KM, Feng JN, Craig NL. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell 1993; 72:931-43; PMID:8384534; http://dx.doi.org/ 10.1016/0092-8674(93)90581-A [DOI] [PubMed] [Google Scholar]
- [6].Maekawa T, Yanagihara K, Ohtsubo E. A cell-free system of Tn3 transposition and transposition immunity. Genes Cells 1996; 1:1007-16; PMID:9077463; http://dx.doi.org/ 10.1046/j.1365-2443.1996.d01-216.x [DOI] [PubMed] [Google Scholar]
- [7].Reznikoff WS, Goryshin IY, Jendrisak JJ. Tn5 as a molecular genetics tool: In vitro transposition and the coupling of in vitro technologies with in vivo transposition. Methods Mol Biol 2004; 260:83-96; PMID:15020804 [DOI] [PubMed] [Google Scholar]
- [8].Leschziner AE, Griffin TJT, Grindley ND. Tn552 transposase catalyzes concerted strand transfer in vitro. Proc Natl Acad Sci U S A 1998; 95:7345-50; PMID:9636151; http://dx.doi.org/ 10.1073/pnas.95.13.7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chalmers RM, Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J Biol Chem 1994; 269:8029-35; PMID:8132525 [PubMed] [Google Scholar]
- [10].Polard P, Ton-Hoang B, Haren L, Bétermier M, Walczak R, Chandler M. IS911-mediated transpositional recombination in vitro. J Mol Biol 1996; 264:68-81; PMID:8950268; http://dx.doi.org/ 10.1006/jmbi.1996.0624 [DOI] [PubMed] [Google Scholar]
- [11].Poussu E. Mu in vitro transposition applications in protein engineering [dissertation]. [Helsinki]: University of Helsinki; 2007. 64 p. [Google Scholar]
- [12].Devine SE, Boeke JD. Efficient integration of artificial transposons into plasmid targets in vitro: a useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res 1994; 22:3765-72; PMID:7937090; http://dx.doi.org/ 10.1093/nar/22.18.3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].de Lorenzo VC, Herrero M, Sánchez JM, Timmis KN. Mini-transposons in microbial ecology and environmental biotechnology. FEMS Microbiol Ecol 1998; 27:211-24; http://dx.doi.org/ 10.1111/j.1574-6941.1998.tb00538.x [DOI] [Google Scholar]
- [14].Haapa S, Taira S, Heikkinen E, Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res 1999; 27:2777-84; PMID:10373596; http://dx.doi.org/ 10.1093/nar/27.13.2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem 1992; 61:1011-51; PMID:1323232; http://dx.doi.org/ 10.1146/annurev.bi.61.070192.005051 [DOI] [PubMed] [Google Scholar]
- [16].Goldhaber-Gordon I, Early MH, Baker TA. The terminal nucleotide of the Mu genome controls catalysis of DNA strand transfer. Proc Natl Acad Sci U S A 2003; 100:7509-14; PMID:12796508; http://dx.doi.org/ 10.1073/pnas.0832468100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Steipe B. Evolutionary approaches to protein engineering. Curr Top Microbiol Immunol 1999; 243:55-86; PMID:10453638 [DOI] [PubMed] [Google Scholar]
- [18].Turanli-Yildiz B, Alkim C, Cakar ZP. Protein engineering methods and applications. INTECH Open Access Publisher, 2012. (Kaumaya P, editor. Protein Engineering.) http://dx.doi.org/ 10.5772/1286 [DOI] [Google Scholar]
- [19].Caetano-Anolles G, Mittenthal J. Exploring the interplay of stability and function in protein evolution: new methods further elucidate why protein stability is necessarily so tenuous and stability-increasing mutations compromise biological function. Bioessays 2010; 32:655-8; PMID:20658703; http://dx.doi.org/ 10.1002/bies.201000038 [DOI] [PubMed] [Google Scholar]
- [20].Yu Y, Lutz S. Circular permutation: a different way to engineer enzyme structure and function. Trends Biotechnol 2011; 29:18-25; PMID:21087800; http://dx.doi.org/ 10.1016/j.tibtech.2010.10.004 [DOI] [PubMed] [Google Scholar]
- [21].Doi N, Yanagawa H. Insertional gene fusion technology. FEBS Lett 1999; 457:1-4; PMID:10486551; http://dx.doi.org/ 10.1016/S0014-5793(99)00991-6 [DOI] [PubMed] [Google Scholar]
- [22].Shah V, Pierre B, Kim JR. Facile construction of a random protein domain insertion library using an engineered transposon. Anal Biochem 2013; 432:97-102; PMID:23026779; http://dx.doi.org/ 10.1016/j.ab.2012.09.030 [DOI] [PubMed] [Google Scholar]
- [23].Pierre B, ¸ Shah V, Xiao J, Kim JR. Construction of a random circular permutation library using an engineered transposon. Anal Biochem 2015; 474:16-24; PMID:25576953; http://dx.doi.org/ 10.1016/j.ab.2014.12.011 [DOI] [PubMed] [Google Scholar]
- [24].Berrondo M, Ostermeier M, Gray JJ. Structure prediction of domain insertion proteins from structures of individual domains. Structure 2008; 16:513-27; PMID:18400174; http://dx.doi.org/ 10.1016/j.str.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schuyler AD, Carlson HA, Feldman EL. Computational methods for predicting sites of functionally important dynamics. J Phys Chem B 2009; 113:6613-22; PMID:19378962; http://dx.doi.org/ 10.1021/jp808736c [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Edwards WR, Busse K, Allemann RK, Jones DD. Linking the functions of unrelated proteins using a novel directed evolution domain insertion method. Nucleic Acids Res 2008; 36:e78; PMID:18559359; http://dx.doi.org/ 10.1093/nar/gkn363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 2013; 65:1357-69; PMID:23026637; http://dx.doi.org/ 10.1016/j.addr.2012.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gokhale RS, Khosla C. Role of linkers in communication between protein modules. Curr Opin Chem Biol 2000; 4:22-7; PMID:10679375; http://dx.doi.org/ 10.1016/S1367-5931(99)00046-0 [DOI] [PubMed] [Google Scholar]
- [29].George RA, Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng 2002; 15:871-9; PMID:12538906; http://dx.doi.org/ 10.1093/protein/15.11.871 [DOI] [PubMed] [Google Scholar]
- [30].Wright CM, Wright RC, Eshleman JR, Ostermeier M. A protein therapeutic modality founded on molecular regulation. Proc Natl Acad Sci U S A 2011; 108:16206-11; PMID:21930952; http://dx.doi.org/ 10.1073/pnas.1102803108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci U S A 2005; 102:11224-9; PMID:16061816; http://dx.doi.org/ 10.1073/pnas.0502673102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sgaramella V, Ehrlich SD. Use of the T4 polynucleotide ligase in the joining of flush-ended DNA segments generated by restriction endonucleases. Eur J Biochem 1978; 86:531-7; PMID:350585; http://dx.doi.org/ 10.1111/j.1432-1033.1978.tb12336.x [DOI] [PubMed] [Google Scholar]
- [33].Loukianov E, Loukianova T, Periasamy M. Efficient cloning method that selects the recombinant clones. Biotechniques 1997; 23:292-5; PMID:9266085 [DOI] [PubMed] [Google Scholar]
- [34].Mealer R, Butler H, Hughes T. Functional fusion proteins by random transposon-based GFP insertion. Methods Cell Biol 2008; 85:23-44; PMID:18155457; http://dx.doi.org/ 10.1016/S0091-679X(08)85002-9 [DOI] [PubMed] [Google Scholar]
- [35].Gregory JA, Becker EC, Jung J, Tuwatananurak I, Pogliano K. Transposon assisted gene insertion technology (TAGIT): a tool for generating fluorescent fusion proteins. PLoS One 2010; 5:e8731; PMID:20090956; http://dx.doi.org/ 10.1371/journal.pone.0008731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Osawa M, Erickson HP. Probing the domain structure of FtsZ by random truncation and insertion of GFP. Microbiology 2005; 151:4033-43; PMID:16339948; http://dx.doi.org/ 10.1099/mic.0.28219-0 [DOI] [PubMed] [Google Scholar]
- [37].Goldhaber-Gordon I, Early MH, Baker TA. MuA transposase separates DNA sequence recognition from catalysis. Biochem 2003; 42:14633-42; PMID:14661976; http://dx.doi.org/ 10.1021/bi035360o [DOI] [PubMed] [Google Scholar]
- [38].Goldenberg DP, Creighton TE. Circular and circularly permuted forms of bovine pancreatic trypsin inhibitor. J Mol Biol 1983; 165:407-13; PMID:6188846; http://dx.doi.org/ 10.1016/S0022-2836(83)80265-4 [DOI] [PubMed] [Google Scholar]
- [39].Krishna MM, Englander SW. The N-terminal to C-terminal motif in protein folding and function. Proc Natl Acad Sci U S A 2005; 102:1053-8; PMID:15657118; http://dx.doi.org/ 10.1073/pnas.0409114102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Whitehead TA, Bergeron LM, Clark DS. Tying up the loose ends: circular permutation decreases the proteolytic susceptibility of recombinant proteins. Protein Eng Des Sel 2009; 22:607-13; PMID:19622546; http://dx.doi.org/ 10.1093/protein/gzp034 [DOI] [PubMed] [Google Scholar]
- [41].Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J Am Chem Soc 2005; 127:13466-7; PMID:16190688; http://dx.doi.org/ 10.1021/ja053932h [DOI] [PubMed] [Google Scholar]
- [42].Chen WT, Chen T, Cheng CS, Huang WY, Wang X, Yin HS. Circular permutation of chicken interleukin-1 β enhances its thermostability. Chem Commun (Camb) 2014; 50:4248-50; PMID:24634912; http://dx.doi.org/ 10.1039/c3cc48313d [DOI] [PubMed] [Google Scholar]
- [43].Mehta MM, Liu S, Silberg JJ. A transposase strategy for creating libraries of circularly permuted proteins. Nucleic Acids Res 2012; 40:e71; PMID:22319214; http://dx.doi.org/ 10.1093/nar/gks060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Goda S, Takano K, Yamagata Y, Katakura Y, Yutani K. Effect of extra N-terminal residues on the stability and folding of human lysozyme expressed in Pichia pastoris. Protein Eng 2000; 13:299-307; PMID:10810162; http://dx.doi.org/ 10.1093/protein/13.4.299 [DOI] [PubMed] [Google Scholar]
- [45].Liu L, Zhang G, Zhang Z, Wang S, Chen H. Terminal amino acids disturb xylanase thermostability and activity. J Biol Chem 2011; 286:44710-5; PMID:22072708; http://dx.doi.org/ 10.1074/jbc.M111.269753 [DOI] [PMC free article] [PubMed] [Google Scholar]


