ABSTRACT
The use of fluorescent proteins fused to other proteins has been very useful in revealing the location and function of many proteins. However, it is very important to show that the fusion of these reporter proteins does not impact the function of the protein of interest. Plants have 2 forms of the cap-binding protein that function in initiation of translation, eIF4E and a plant specific form, eIFiso4E. In an attempt to determine the cellular localization of eIFiso4E, fusions to GFP were made, but were found to not be competent to rescue the lethal phenotype of plants lacking eIF4E and eIFiso4E. This suggested that the GFP fusions at either the N- or C-terminus of eIFiso4E were not functional. Biochemical analysis of the fusions revealed that eIFiso4E•GFP fusions were not able to bind to m7GTP Sepharose indicating that they were not functional as cap-binding proteins. Analysis of eIF4E•GFP fusions, both in yeast and in vitro, showed that the N-terminal fusion may be functional, whereas the C-terminal fusion bound m7GTP Sepharose very poorly and functioned poorly in yeast. These results highlight the importance of verification both biochemically and in vivo that reporter fusions of proteins maintain activity and are stable in order to prevent observations that may result in artifacts.
KEYWORDS: cap-binding protein, eukaryotic initiation factor, fusion protein, GFP, translation
Introduction
Eukaryotic protein synthesis consists of 3 stages (initiation, elongation and termination) each involving numerous factors.1,2 In one of the early steps leading to the initiation of translation, the eukaryotic initiation factor (eIF) 4F cap-binding complex recognizes the 7-methylguanosine cap of the mRNA (mRNA).3 The eIF4F cap-binding complex is composed of eIF4E, which binds the mRNA cap structure, and eIF4G, often described as a scaffolding protein, which in turn binds the dead-box helicase eIF4A and the RNA binding protein eIF4B.1-3 The mRNA and associated eIF4 initiation factors are recruited to the 40S ribosome and associated eIFs to form the 48S initiation complex which subsequently joins the 60S ribosome to form the completed 80S complex to commence the elongation phase of translation.1,4
The cap-binding complex is conserved in mammals, plants and fungi and eIF4E is a target for the regulation of translation in various organisms through such mechanisms as 4E-binding proteins (4EBP)5 phosphorylation,6 and sumoylation.7 Plants do not appear to utilize these regulatory mechanisms for eIF4E.1 Additionally, plants have a second isoform of the cap-binding complex, eIFiso4F, composed of eIFiso4E and eIFiso4G.8 The distinct roles of the differing plant cap-binding protein isoforms have yet to be determined, but each is the preferred binding partner of its respective eIF4G subunit.9 All flowering plants have these 2 forms (eIF4E and eIFiso4E) of the cap-binding protein.8 In Arabidopsis thaliana a single gene encodes eIFiso4E and 3 genes are present which encode eIF4E proteins. Two of the eIF4E proteins (eIF4E1b, eIF4E1c) in Arabidopsis appear to be products of gene duplication specific to the Brassicaceae family and do not appear to be part of the canonical translation apparatus but may have a specialized function(s).10 Knockout mutants of either eIFiso4E or eIF4E are not lethal, but do lead to resistance to certain plant viruses11,12; however, double mutants lacking both eIF4E and eIFiso4E cannot be recovered.10,13 T-DNA insertions disrupting EIF4E1B or EIF4E1C do not appear to have any phenotype, nor has virus resistance been reported for these genes.14
To investigate the roles of plant cap-binding proteins, the use of green fluorescent protein (GFP) fusions have been employed. Martinez-Silva et al.15 published a study in which Arabidopsis wild type plants carried a 35S promoter-eIFiso4E-C-terminal fusion to GFP. There was no evidence to support substantial overexpression of the eIFiso4E GFP fusion protein or mRNA compared with the endogenous wild-type eIFiso4E. Although there was some evidence that the eIFiso4E-GFP fusion showed localized expression in some tissues, it was not clear if this was due to some other effect such as mislocalization and degradation. A C-terminal fusion to eIFiso4E was also utilized to monitor the interaction with the VPg of turnip mosaic virus in N. benthamiana.16 Subcellular localization of the eIFiso4E C-terminal fusion in infiltrated N. benthamiana leaves indicated localization of eIFiso4E was primarily with the ER and free GFP was found in the cytoplasm and nucleus.16 Neither of these reports15,16 showed that the fusion protein was functional in translation or was able to substitute in vivo and there was a background of wild type eIF4E and eIFiso4E present in both cases.
When attempting to generate various tagged versions of eIFiso4E we discovered that tagged versions could not rescue the lethal phenotype in plants lacking both eIFiso4E and eIF4E10,17 suggesting that the tagged versions of eIFiso4E are not functional in vivo. In this study, biochemical characterization of the fusion proteins finds that neither N- nor C-terminally GFP tagged eIFiso4E is able to bind m7GTP-Sepharose and appears to be biochemically non-functional, consistent with the inability to rescue the lethal phenotype. Further analysis of the other cap-binding protein in plants, eIF4E shows that a C-terminal GFP tagged version of eIF4E, like eIFiso4E, was non-functional in yeast and bound m7GTP-Sepharose poorly, but N-terminal GFP tagged eIF4E may be functional in yeast. There was significant degradation of the N-terminal GFP tagged eIF4E fusion protein in yeast which may be responsible for the apparent rescue in yeast. E. coli expressed N-terminal GFP tagged eIF4E bound m7GTP-Sepharose similarly to untagged eIF4E suggesting that the ability to bind m7G caps is not compromised.
Results and discussion
In planta expression of GFP tagged eIFiso4E
To investigate the localization and function of eIFiso4E, C- or N-terminally GFP-tagged eIFiso4E was transformed into an A. thaliana background homozygous for the loss of eIFiso4E18 and heterozygous in the cum1 mutant allele for eIF4E.19 Progeny were screened for the null background of eIF4E (cum1) to ensure that only the exogenous version of the eIFiso4E protein was present in planta. Although transgenic unmodified eIFiso4E readily rescued the lethal phenotype of the null eIF4e/eIFiso4E, plants containing N- or C-terminal tagged eIFiso4E were not recovered (see Table 1). This result suggests that N- and C-terminally tagged versions of eIFiso4E are not functional in planta.
Table 1.
Screen for plants with tagged eIFiso4E.
| Transgene Line | Plants Screened | Lines retaining cum1 mutation |
|---|---|---|
| eIFiso4E (untagged transgene) | 100 | 3 homozygous 8 heterozygous |
| N-terminal GFP•eIFiso4E | 203 | 0 |
| C-terminal GFP•eIFiso4E | 129 | 0 |
| C-terminal 3x FLAG•eIFiso4E | 120 | 0 |
Biochemical characterization of eIFiso4E fusion proteins
To determine the reason for the lack of rescue of the lethal phenotype by eIFiso4E fusions, a biochemical characterization was undertaken. Constructs were prepared similarly to Martinez-Silva et al.15 with the 16 amino acid linker for the C-terminal fusion and no spacer for the N-terminal fusion and expressed in E. coli. In both cases the correct size fusion proteins were expressed (see Fig. 1) and the extracts demonstrated fluorescence from the GFP tag; however, neither the N- nor C-terminal fusion of GFP to eIFiso4E were able to bind m7GTP-Sepharose to any significant degree and no protein was recovered upon elution with m7GTP (see Fig. 1B, C). This is in contrast to Martinez-Silva et al.15 in which they were able to demonstrate modest binding of a small amount of the fusion protein from plant extracts, but most of the plant protein bound to m7GTP-Sepharose appeared to be wild type eIFiso4E, not the fusion protein. The expressed N- and C-terminal GFP•eIFiso4E fusions described here appear to not be functional as cap-binding proteins based on the inability to bind m7GTP-Sepharose in vitro and the inability to recover plants containing a tagged version of eIFiso4E in the absence of another functional cap-binding protein (eIF4E). It is likely that the fusion proteins in plants do not bind well to m7G of mRNAs.
Figure 1.
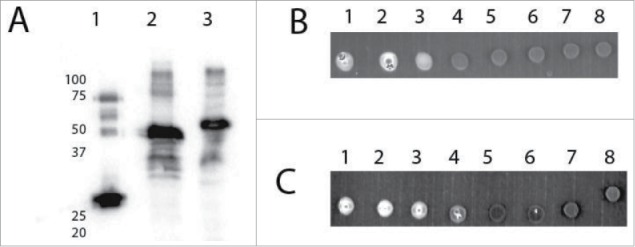
Expression of eIFiso4E with C and N-terminal GFP. Panel A, Western of E. coli expressed N- and C-terminal GFP•eIFiso4E. E. coli extracts were separated by SDS PAGE, transferred to PVDF and probed with Arabidopsis eIFiso4E antibody (1/1000); Lane 1, 0.005 µg purified eIFiso4E Lane 2, 5 µl N-terminal GFP•eIFiso4E E. coli extract; Lane 3, 5 µl C-terminal GFP•eIFiso4E E. coli extract. Panel B, Fluorescence of C-terminal GFP•eIFiso4E, 5 µl of each indicated fraction was spotted on Parafilm: Lane 1, E. coli extract loaded to m7GTP-Sepharose; Lane 2, m7GTP-Sepharose run through; Lane 3, m7GTP-Sepharose wash 1; Lane 4, m7GTP-Sepharose wash 2; Lane 5, m7GTP-Sepharose wash 3; Lane 6, m7GTP-Sepharose eluant 1; Lane 7, m7GTP-Sepharose eluant 2; Lane 8, elution buffer only. Panel C, Fluorescence of N-terminal GFP•eIFiso4E, 5 µl of each indicated fraction was spotted on Parafilm: Lane 1, N-terminal GFP•eIFiso4E E. coli extract loaded to m7GTP-Sepharose; Lane 2, m7GTP-Sepharose run through; Lane 3, m7GTP-Sepharose wash 1; Lane 4, m7GTP-Sepharose wash 2; Lane 5, m7GTP-Sepharose wash 3; Lane 6, m7GTP-Sepharose eluant 1; Lane 7, m7GTP-Sepharose eluant 2; Lane 8, elution buffer only. Fluorescence was detected using a BioRad ChemDoc on the fluorescein setting.
Are eIF4E-GFP fusions functional in yeast?
To further investigate this phenomenon and to see if it extends to the other plant cap-binding protein, eIF4E, we utilized the more tractable yeast system. It is known that A. thaliana eIF4E, but not eIFiso4E, is able to substitute for yeast eIF4E.20,21 The 16 amino acid linker region described in Martinez-Silva et al.15 was used to make a C-terminal GFP fusion to eIF4E while an N-terminal direct fusion of GFP to eIF4E was also produced for expression in yeast. A yeast strain with the genomic copy of eIF4E deleted and carrying eIF4E (CDC33/GAL1:URA) on a plasmid10,22 was transformed with plasmids (TRP) containing A. thaliana eIF4E, N-terminal GFP•eIF4E or C-terminal GFP•eIF4E. An exogenous yeast eIF4E copy on a plasmid with a different nutritional marker (HIS) was used as a control. All the yeast strains were cured of the CDC33/GAL1:URA plasmid by treatment with 5-Fluoroorotic acid (5-FOA) and loss of the plasmid was confirmed by PCR (see Supplemental Fig. 2) and the lack of yeast eIF4E confirmed by western analysis (see Fig. 3A). The yeast strains were grown in both minimal media for their respective marker (HIS or TRP) and also on rich media (YPD). As shown in Figure 2, the untagged version of A. thaliana eIF4E was able to substitute as expected, as was the N-terminal GFP•eIF4E plasmid; however the C-terminal GFP•eIF4E appears to be less robust at 30°C and does not grow at 37°C. This suggests that the C-terminal GFP fusion to eIF4E functions poorly in yeast.
Figure 3.
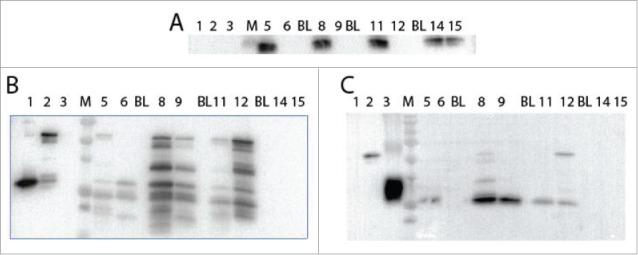
Western blot analysis of expression of N and C-terminal GFP fusions with eIF4E in yeast. Yeast strains with yeast eIF4E carried on a GAL1:URA plasmid were transformed with plasmids containing either A. thaliana eIF4E (TRP), N-terminal GFP•eIF4E (TRP), C-terminal GFP/eIF4E (TRP) or yeast eIF4E (CDC33; HIS). Cells were grown to ∼0.8 A600, recovered by centrifugation and the protein extracted, 8 µl of lysate was loaded in each well. Lane 1, purified A. thaliana eIF4E (0.1 µg); Lane 2, purified N-terminal GFP•eIF4E (0.1 µg); Lane 3, purified GFP (0.3 µg); M, BioRad Precision Plus Protein Standards (250, 150, 100, 75, 50, 37, 25, 20, 15 kDa); Lane 5, A. thaliana eIF4E/CDC33; Lane, 6, A. thaliana eIF4E; BL, blank lane; Lane 8, N-terminal GFP•eIF4E/CDC33; Lane 9, N-terminal GFP•eIF4E; BL, blank lane; Lane 11, C-terminal GFP•eIF4E/CDC33; Lane 12, C-terminal GFP•eIF4E; BL, blank lane; Lane 14, yeast eIF4E (HIS)/CDC33; Lane 15, yeast eIF4E (HIS). Panel A, probed with antibody to yeast eIF4E (1/2000); Panel B, probed with antibody to A. thaliana eIF4E (1/1000); Panel C, probed with antibody to GFP (1/10,000).
Figure 2.
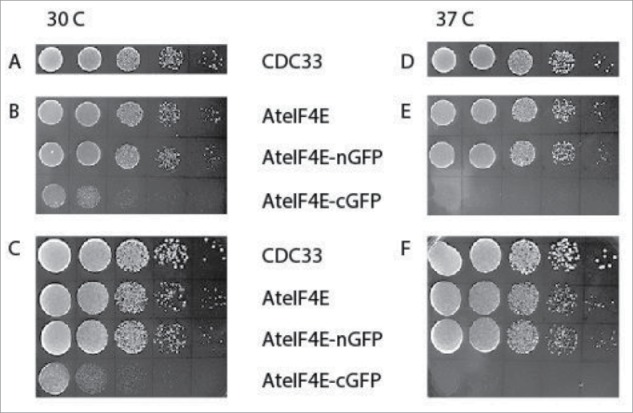
A) Yeast strains with yeast eIF4E carried on a GAL1:URA plasmid were transformed with plasmids containing either A. thaliana eIF4E (TRP), N-terminal GFP/eIF4E (TRP), C-terminal GFP/eIF4E (TRP) or yeast eIF4E (CDC33; HIS). Yeast were cured of the GAL1:URA plasmid using 5FOA and only had the remaining plasmid (TRP or HIS) for survival. Panels A and D. CDC 33 (HIS) grown on minimal media minus HIS; Panels B and E. A. thaliana eIF4E, N and C-terminal GFP/eIF4e fusions grown on minimal media minus TRP; Panels C and F. All plasmids grown on YPD rich media. Samples were grown to ∼1 OD600 (except C-terminal GFP/eIF4E which was ∼0.36) and diluted to 0.3 OD600 and 10-fold serial dilutions were made. For each dilution 10 µl was spotted onto a YPD plate and grown at 30 or 37°C.
The status of the fusion protein expression in yeast was investigated by western blot using both eIF4E and GFP antibodies. The fusion protein for GFP•eIF4E is present for both the N and C-terminal fusions; however, there appears to be significant presence of degradation products detected by eIF4E antibody (Fig. 3B) and similar evidence of degradation products was observed with GFP antibody. This suggests that the GFP fusion protein is unstable and that a degradation product which retains eIF4E activity may be responsible for the apparent ability to rescue deletion of the yeast eIF4E rather than the fusion protein itself. Therefore, it was necessary to determine if eIF4E-GFP fusion proteins retain any biochemical functionality.
Biochemical characterization of GFP•eIF4E fusion proteins
Constructs for recombinant expression of the N- and C-terminal GFP fusions to eIF4E in E. coli were prepared. The first biochemical attribute tested was the capability of the fusion protein to display GFP fluorescence; both N- and C-terminal GFP fused to eIF4E appeared fluorescent in extracts and the correct sized fusion protein was expressed as detected by both eIF4E and GFP antibodies (Fig. 4A,B). The next attribute was the capability bind to m7GTP-Sepharose. Unlike the N-terminal eIFiso4E-GFP fusion, the N-terminal GFP•eIF4E construct bound to m7GTP-Sepharose resin similarly to untagged eIF4E and was eluted with m7GTP (see Fig. 4C). However, like the C-terminal eIFiso4E GFP fusion, the C-terminal GFP•eIF4E fusion protein did not bind well to the m7GTP-Sepharose column and was primarily in the column flow through (although a small amount of protein could be recovered). These results show that the C-terminal GFP•eIF4E fusion protein binds poorly to m7GTP-Sepharose which could compromise its ability to function as a cap-binding protein in vivo, as reflected by its poor ability to complement yeast lacking eIF4E.
Figure 4.
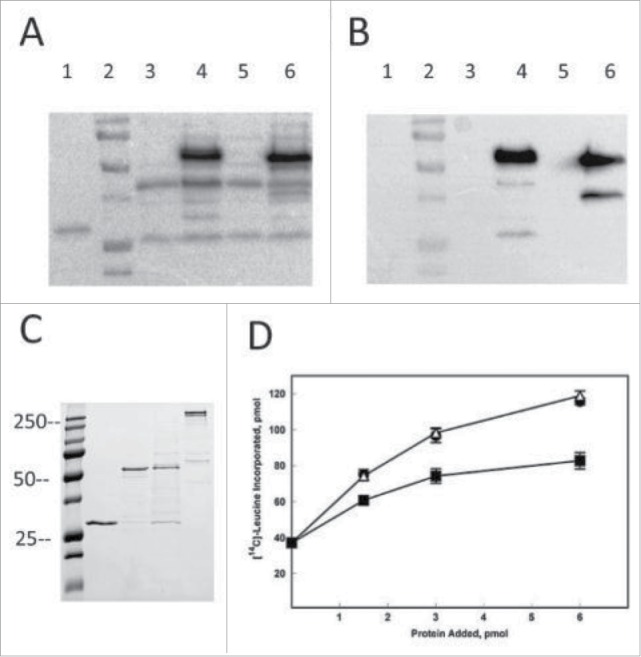
Expression of Arabidopsis eIF4E N and C-terminal GFP fusion proteins. Lane 1, purified Arabidopsis eIF4E (0.2 µg); Lane 2, BioRad Precision Plus Protein Standards (100, 75, 50, 37, 25, 20 kDa); Lane 3, uninduced C-terminal GFP fusion protein; Lane 4, induced C-terminal GFP fusion protein; Lane 5, uninduced N-terminal GFP fusion protein; Lane 6, induced N-terminal GFP fusion protein. Cell lysates (15 µl) were loaded on a 12% SDS-PAGE and transferred to PVDF for western analysis. Panel A, western blot probed with affinity purified rabbit polyclonal antibody to Arabidopsis eIF4E (1/500). Panel B), western blot probed with mouse monoclonal antibody to GFP (Invitrogen A11121, 1/1000). Panel C) SDS-PAGE of purified proteins. Lane 1, purified Arabidopsis eIF4E (0.9 µg); Lane 2, purified Arabidopsis N-terminal GFP/eIF4E fusion (0.9 µg); Lane 3, purified Arabidopsis C-terminal GFP/eIF4E fusion (0.9 µg); Lane 4, purified Arabidopsis eIF4G (0.9 µg)10 The gel was stained with Coomassie Brilliant blue. Panel D) In vitro assay of eIF4E, N-terminal GFP•eIF4E and C-terminal GFP•eIF4E with eIF4G. Equal molar amounts of A. thaliana eIF4G and the respective eIF4E were mixed and added as indicated. The reaction mixture contained 5 pmol of barley α-amylase mRNA and 15 µl of a wheat germ S30 extract that had been partially depleted of eIF4F and eIFiso4F by passage over a m7GTP-Sepharose column as previously described.10 eIF4G/eIF4E (•); eIF4G/ N-terminal GFP•eIF4E (Δ); eIF4G/C-terminal GFP•eIF4E (▪). Experiments were done in triplicate and averaged. The incorporation of [14C]-Leucine in the absence of any added factor was ∼37 pmol.
Analysis of the ability of the N-terminal fusion of GFP•eIF4E to interact with eIF4G and promote translation initiation was performed using in vitro translation assays. As shown in Figure 4D, the N-terminal fusion of GFP•eIF4E was able to support initiation of translation to the same extent as untagged eIF4E; however, the small amount of C-terminal fusion of GFP•eIF4E that did bind m7GTP-Sepharose and was recovered by elution with m7GTP was less robust in translation. These results taken together suggest that the N-terminal GFP fusion to eIF4E may be able to rescue the lethal phenotype in planta as it functions both biochemically and in yeast; whereas it is likely that the C-terminal fusion to eIF4E would likely not be able to rescue the lethal phenotype. Interestingly, we previously reported that a N-terminal extension of eIF4E1c (a non-canonical form of eIF4E) precluded function in yeast10 suggesting that each isoform of eIF4E may have different tolerances for tags and should be extensively tested for function both biochemically and in vivo prior to localization studies.
Conclusions
Although tagged proteins can be very useful and provide insights into localization and other aspects of protein interactions in the cell, they should be used with caution and tested for proper biological function as fusion proteins. It is not surprising that tagging a protein with something similar in size (cap binding proteins are about the same size as GFP, ∼24 kDa) could have profound effects on its ability to form complexes or interact with other molecules. In the case of cap-binding proteins, the protein must be able to form a complex with its binding partner, eIF4G for eIF4E or eIFiso4G for eIFiso4E, and be able to interact with the m7GpppG cap of mRNAs. It appears that the N-terminal GFP•eIF4E fusions may be able to function in planta based on the biochemical analysis and apparent ability to substitute for yeast eIF4E in vivo, although the fusion protein appears degraded in yeast and the free eIF4E protein may be responsible for the apparent substitution. It is not clear why the GFP•eIF4E fusion proteins are functional (N-terminal) to partly functional (C-terminal) and eIFiso4E fusions are not functional at all; however it likely has to do with either folding issues of the cap-binding pocket or interaction with other components of the translational machinery that influence cap-binding that are significantly compromised in the eIFiso4E fusion protein. It is also possible that various fusions and linkers may have influences that vary, so testing several configurations may be necessary.
Thus “tails” can be useful, but they should be used with caution and fusion proteins should always be shown to have biological function similar to the native proteins prior to use to avoid observing interactions or properties that may be meaningless and generate artifacts.
Materials and methods
E. coli and yeast expression clones
Clones were generated by overlap PCR of smGFP23 with codon optimized versions of A. thaliana eIF4E10 or eIFiso4E. N-terminal direct GFP fusions and C-terminal GFP fusions utilizing a 16 amino acid linker region15 were cloned into pCR-Blunt-II-TOPO (Invitrogen) and subcloned into pET22b (Novagen/EMD Millipore) for expression. GFP fusions for yeast expression were cloned similarly but with subcloning into pG1.1.10
Protein expression
GFP fusions to eIFiso4E or eIF4E were expressed and purified as previously described.10,24
In Vitro Translation Assays. Assays were performed as previously described with in vitro transcribed capped barley α-amylase mRNA and a wheat germ S30 extract that had been partially depleted of eIF4F and eIFiso4F by passage over a m7GTP-Sepharose column as previously described.10 Assays contained the indicated amounts of A. thaliana eIF4E or eIF4E-GFP fusion proteins.
SDS PAGE and western blotting of yeast extracts
Yeast extracts were prepared as described.25 SDS PAGE was performed using 12.5% acrylamide gels in a Bio-Rad Mini-PROTEAN chamber (Hercules, CA) with Tris-glycine SDS running buffer (25 mM Tris; 192 mM glycine; 0.1% SDS). Proteins were transferred to Immobilon-P membrane (EMD Millipore, Darmstadt, Germany) for 30 minutes using the BioRad Turbo Blot (Bio-Rad, Hercules, CA). The membrane was blocked at room temperature for 2 hours by HNAT buffer (10 mM Hepes/KOH pH 7.6, 150 mM NaCl, 0.2% BSA, 0.2% Tween 20) containing 5% non-fat dried milk (boiled and filtered). The membrane was probed with rabbit antisera raised to recombinant A. thaliana eIFiso4E or eIF4E (1:3000 dilution) or monoclonal antibody to GFP (Thermo Fisher A11121, 1/1000) at 4°C overnight and washed 3-times with HNAT buffer. Goat anti-rabbit secondary antibody coupled to horse radish peroxidase was obtained from Kirkegaarde Perry Laboratories (Gaithersburg, MD) and used at a 1:20,000 dilution for 1 hour at room temperature in 5% milk in HNAT. The blots were washed with HNAT 3 times and visualized with Super Signal West Pico Chemiluminescent substrate (Thermo Fisher, Waltham, MA) in ChemiDocTM System (Bio-Rad, Hercules, CA).
Cloning of eIFiso4E fusions for in planta analysis
Genomic clones of AteIFiso4E were made for analysis in plants. A genomic interval containing the upstream sequence including the 5′UTR (4829 nucleotides) of At5g35620 (AteIFiso4E; 1386 nucleotides) and downstream sequence including the 3′UTR (1282 nucleotides) was amplified by PCR and cloned into the gateway shuttle vector pG125 via an In-Fusion Cloning Kit (Clontech). Overlap extension PCR was then used to make the N-terminal GFP•eIFiso4E, C-terminal GFP•eIFiso4E or C-terminal 3x Flag•eIFiso4E fusions. The tagged eIFiso4E sequence interval and oligonucleotides used for the cloning are listed in supplemental Figure S1 and Table S1. The eIFiso4E constructs were subcloned into the binary Gateway expression vector pMDC99 (hygromycin resistance) by LR recombination (ThermoFisher). Arabidopsis plants expressing only exogenous AteIFiso4E were generated using agrobacterium mediated transformation of AteIFiso4E-pMDC99 plasmids into plants carrying a homozygous T-DNA insertion in AteIFiso4E (AT5G3562018) and a heterozygous point mutation of AteIF4E1 (AT4G18040, cum119). Resulting progeny were screened for transformants on 1xMS agar plates containing hygromycin (50mg/L). The surviving plants were genotyped for the homozygous null background of AteIF4E1 (cum1).
Yeast complementation assays. Yeast complementation assays using untagged and GFP tagged versions of eIF4E were performed as previously described.10
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank W. Allen Miller for valuable comments on the manuscript.
Funding
This work was supported by grants from the National Science Foundation (MCB1052530 and Arabidopsis 2010 S-0000335) to KSB.
References
- [1].Browning KS, Bailey-Serres J. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 2015; 13:e0176; PMID:26019692; http://dx.doi.org/ 10.1199/tab.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 2014; 83:779-812; PMID:24499181; http://dx.doi.org/ 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- [3].Valasek LS. ‘Ribozoomin’–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 2012; 13:305-30; PMID:22708493; http://dx.doi.org/ 10.2174/138920312801619385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 2012; 4:55-70; http://dx.doi.org/ 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hernandez G, Proud CG, Preiss T, Parsyan A. On the diversification of the translation apparatus across eukaryotes. Comp Funct Genomics 2012; 2012:256848; PMID:22666084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mnks Proud CG., eIF4E phosphorylation and cancer. Biochim Biophys Acta 2015; 1849:766-73; PMID:25450520; http://dx.doi.org/ 10.1016/j.bbagrm.2014.10.003 [DOI] [PubMed] [Google Scholar]
- [7].Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. J Biol Chem 2010; 285:18139-43; PMID:20421305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Patrick RM, Browning KS. The eIF4F and eIFiso4F complexes of plants: an evolutionary perspective. Comp Funct Genomics 2012; 2012:287814; PMID:22611336; http://dx.doi.org/ 10.1155/2012/287814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mayberry LK, Allen ML, Nitka KR, Campbell L, Murphy PA, Browning KS. Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F: molecular specificity of subunit binding. J Biol Chem 2011; 286:42566-74; PMID:21965660; http://dx.doi.org/ 10.1074/jbc.M111.280099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Patrick RM, Mayberry LK, Choy G, Woodard LE, Liu JS, White A, Mullen RA, Tanavin TM, Latz CA, Browning KS. Two Arabidopsis thaliana loci encode novel eIF4E isoforms that are functionally distinct from the conserved plant eIF4E. Plant Physiol 2014; 164:1820-30; PMID:24501003; http://dx.doi.org/ 10.1104/pp.113.227785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jiang J, Laliberte JF. The genome-linked protein VPg of plant viruses-a protein with many partners. Curr Opin Virol 2011; 1:347-54; PMID:22440836; http://dx.doi.org/ 10.1016/j.coviro.2011.09.010 [DOI] [PubMed] [Google Scholar]
- [12].Wang A, Krishnaswamy S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol Plant Pathol 2012; 13:795-803; PMID:22379950; http://dx.doi.org/ 10.1111/j.1364-3703.2012.00791.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Callot C, Gallois JL Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal Behav 2014; 9:e27940; PMID:24492391; http://dx.doi.org/17316629 10.4161/445 psb.27940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nicaise V, Gallois JL, Chafiai F, Allen LM, Schurdi-Levraud V, Browning KS, Candresse T, Caranta C, Le Gall O, German-Retana S. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett 2007; 581:1041-6; PMID:17316629; http://dx.doi.org/ 10.1016/j.febslet.2007.02.007 [DOI] [PubMed] [Google Scholar]
- [15].Martinez-Silva AV, Aguirre-Martinez C, Flores-Tinoco CE, Alejandri-Ramirez ND, Dinkova TD. Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in arabidopsis thaliana seedlings. PLoS ONE 2012; 7:e31606; PMID:22363683; http://dx.doi.org/ 10.1371/journal.pone.0031606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Beauchemin C, Boutet N, Laliberte JF. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso4E In Planta. J Virol 2007; 81:775-82; PMID:17079311; http://dx.doi.org/ 10.1128/JVI.01277-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Callot C, Gallois JL. Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal Behav 2014; 9:e27940; PMID:24492391; http://dx.doi.org/ 10.4161/psb.27940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 2002; 32:927-34; PMID:12492835; http://dx.doi.org/ 10.1046/j.1365-313X.2002.01481.x [DOI] [PubMed] [Google Scholar]
- [19].Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 2004; 78:6102-11; PMID:15163703; http://dx.doi.org/ 10.1128/JVI.78.12.6102-6111.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rodriguez CM, Freire MA, Camilleri C, Robaglia C. The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J 1998; 13:465-73; PMID:9680993; http://dx.doi.org/ 10.1046/j.1365-313X.1998.00047.x [DOI] [PubMed] [Google Scholar]
- [21].German-Retana S, Walter J, Doublet B, Roudet-Tavert G, Nicaise V, Lecampion C, Houvenaghel MC, Robaglia C, Michon T, Le Gall O. Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J Virol 2008; 82:7601-12; PMID:18480444; http://dx.doi.org/ 10.1128/JVI.00209-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Altmann M, Müller PP, Pelletier J, Sonenberg N, Trachsel H. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J Biol Chem 1989; 264:12145-7; PMID:2663851 [PubMed] [Google Scholar]
- [23].Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 1998; 36:521-8; PMID:9484447; http://dx.doi.org/ 10.1023/A:1005991617182 [DOI] [PubMed] [Google Scholar]
- [24].Mayberry LK, Dennis MD, Allen ML, Nitka KA, Murphy PA, Campbell L, Browning KS. Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol 2007; 430:397-408; PMID:17913646; http://dx.doi.org/ 10.1016/S0076-6879(07)30015-3 [DOI] [PubMed] [Google Scholar]
- [25].Eudes A, Baidoo EE, Yang F, Burd H, Hadi MZ, Collins FW, Keasling JD, Loqué D. Production of tranilast [N-(3′,4′-dimethoxycinnamoyl)-anthranilic acid] and its analogs in yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 2011; 89:989-1000; PMID:20972784; http://dx.doi.org/ 10.1007/s00253-010-2939-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


