Abstract
The retinal pigment epithelium (RPE) is essential for retinoid recycling and phagocytosis of photoreceptors. Understanding of proteome changes that mediate oxidative stress-induced degeneration of RPE cells may provide further insight into the molecular mechanisms of retinal diseases. In the current study, comparative proteomics has been applied to investigate global changes of RPE proteins under oxidative stress.
Proteomic techniques including 2D SDS-PAGE, differential gel electrophoresis (DIGE) and tandem time-of-flight (TOF-TOF) mass spectrometry were used to identify early protein markers of oxidative stress in the RPE. Two biological models of RPE cells revealed several differentially-expressed proteins that are involved in key cellular processes such as energy metabolism, protein folding, redox homeostasis, cell differentiation, and retinoid metabolism. Our results provide a new perspective on early signaling molecules of redox imbalance in the RPE and putative therapeutic target proteins of RPE diseases caused by oxidative stress.
Keywords: Proteomics, Oxidative stress, Biomarker, Differential gel electrophoresis, Retinal pigment epithelium, Mass spectrometry
Introduction
The RPE, located between photoreceptors and choroid, is in a unique position to mediate the transport of nutrients, oxygen, and retinoids from blood to photoreceptors. For continued vision, the RPE is required for retinoid recycling and removal of molecular components that shed from the photoreceptor outer segment. RPE dysfunction leads to the depletion of nutrients and oxygen in photoreceptor cells, which initiates apoptosis and accumulation of retinoid byproducts that eventually block visual signaling [1,2]. Continuous exposure to light causes the RPE to consume a large amount of oxygen in order to complete the complex processes of nutrient transport, phagocytosis and the visual cycle. This oxidative environment in the RPE may contribute to the pathogenesis of retinal diseases. It is still not known why the initial retinal degeneration occurs and how the degenerative processes progress as a result of continued oxidative stress [3-5]. Adaptation to changes in oxidative environments is critical for the survival of retina and RPE cells. Clinical trials demonstrated a significant reduction toward retinal degeneration upon intake of antioxidants such as lutein, zeaxanthin, zinc, vitamin C, and vitamin E [6-7]. Oxidation of polyunsaturated fatty acids (PFA) and abundant photosensitizers in the RPE induce generation of reactive oxygen species (ROS) upon exposure to visible light [8,9]. Hydrogen peroxide (H2O2) is generated in the RPE during phagocytosis of the photoreceptor outer segment, and it has been used as a direct oxidative-inducing reagent to initiate cellular oxidative stress [10]. Understanding of molecular mechanisms that mediate oxidative stress-induced proteome changes in the RPE may provide insight into the pathogenesis of retinal degeneration. In the current study, comparative and differential proteomics have been applied to investigate global changes in the RPE proteome due to oxidative stress induced by H2O2. Two-dimensional fluorescent differential gel electrophoresis (2D-DIGE) is an advanced 2D technology. Protein samples were prelabeled with different fluorescent dyes and two different samples (control vs. treated) were run simultaneously on the same gel. By using 2D-electrophoresis and 2D-DIGE coupled with tandem time-of-flight mass spectrometry, unbiased system-wide analysis of proteome changes in oxidative stress was investigated in two different model systems. Identification of target proteins in the RPE under oxidative stress implies new insights into signaling mechanisms at the molecular level.
Material and Methods
The first sample preparation from bovine RPE cells
Fresh bovine eyes were obtained from a local abattoir (Brown Packing Company, Gaffney, SC) immediately after excision from the animal. The post-mortem stability, procedures for preparing RPE cells and general proteomic techniques have been described in detail previously [11-15]. Briefly, bovine eyes were opened 5 mm posterior to the limbus, and the vitreous and retina were removed. After washing with a phosphate-buffered saline (PBS), eye-cups were incubated in 0.25% trypsin in Dulbecco's minimum essential medium (DMEM; Gibco, Grand Island, NY) for 60 minutes at 37°C. RPE cells were collected under a dissecting microscope using a Pasteur pipette. After adding the culture medium (DMEM/F12) containing 10% fetal bovine serum (FBS), cells were centrifuged and resuspended in a culture medium, and plated into 6-well plates (Nunc). Second passage cells were used for experiments. After exposure to 200 μM H2O2 incubation (1 hr), followed by a 6 hr incubation, the medium was removed and cells were washed three times with serum-free DMEM. Cells were washed with ice-cold PBS and lysed with a buffer containing 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mg/mL leupeptin and 1 mM phenylmethylsulfonyl fluoride. Cells were collected by centrifugation.
The second sample: human D407 RPE cell culture and protein labeling with fluorescent dye
D407 cells were maintained in Dulbecco's Modified Eagle Media (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM glutamine at 37oC and 5% CO2. When cells grown in a 6-well tissue culture plate reached 75-80 % confluence, they were treated with 200 μM H2O2 or PBS for 1 hour, followed by removal of H2O2, and then maintained in conditioning media for 6 hours. For 2D-DIGE, proteins were prelabeled with CyDye fluorescent saturation dye (CyDye DIGE labeling kit, GE Healthcare). The protein sample (5 μg) in a 9 μl cell lysis buffer was mixed with 1 μl of 2 mM tris(2-carboxyethyl)phosphine (TCEP) and incubated at 37°C for 1 hour in the dark. Then 2 μl of Cy5 was added to the control sample and Cy3 to the treated sample and the mixtures were incubated at 37°C for 30 minutes in the dark. The labeling reaction was stopped by adding an equal volume of 2X sample buffer (2 M thiourea, 7 M urea, 2% pH 3–10 pharmalyte for isoelectric focusing [IEF], 2% Dithiothreitol [DTT], 4% CHAPS). Labeled protein samples were then adjusted to 200 μl with a rehydration buffer (4% CHAPS, 8 M urea, 1% pharmalytes 3–10, 10 mM DTT) prior to isoelectric focusing (IEF).
2D-PAGE and 2D-DIGE
The samples were then subjected to electrophoresis process. IEF was performed with immobilized pH gradient (IPG) strips (11 cm ReadyStrip, pH 3–10, Biorad) at 20 °C with an Ettan IPGphor system (GE healthcare). For Coomassie staining, a total of 100 μg of proteins were dissolved in 200 μl of rehydration buffer (4% CHAPS, 8 M urea, 1% pharmalytes 3–10, 10 mM DTT) and IPG strips were rehydrated for 18 hours at 30 V. IEF focusing was performed at 500 V for 1 hour, 500 to 8000 V ramp for five hours, and 8000 v for 1 hour. After IEF, IPG strips were equilibrated for 15 minutes in a 10 ml equilibration buffer (50 mM Tris-HCl, 1.5 M, pH 8.8, 6 M urea, 30% glycerol [v/v], 2% SDS [w/v], trace amount of Bromophenol Blue, 50 mg DTT), followed by another 15 minutes incubation with a 10 ml equilibration buffer containing 450 mg iodoacetamide. Equilibrated strips were then placed on top of precast gradient gels (8–16% Tris-HCl, Criterion Precast Gel, Biorad), embedded in 0.5% agarose, and incased in a running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Proteins were separated at 100 volts for 2 hours. Coommassie blue stained gels were scanned with a transmission scanner and protein spot densities were measured quantitatively (Bio-Rad, PD Quest). For DIGE analysis, images were acquired on a multiwavelength scanner capable of resolving the three fluorescent colors. DIGE analysis included, i) spot detection, ii) background subtraction, iii) in-gel normalization, iv) gel artifact removal, v) gel-to-gel matching, vi) statistical analysis, as following manufacturer's instruction.
CyDye labeled proteins were visualized using a 532 nm laser and 580 nm emission filter for Cy3 images and a 633nm laser and 670nm filter for Cy5 images, respectively. Differentially expressed protein spots in Coomassie stained gels were excised from the gels and in-gel digested by trypsin. The proteins were then analyzed by MALDI-TOF and TOF-TOF mass spectrometry. For in-gel trypsin digestion, protein spots were excised from the gels and washed twice with 200 μl of 100 mM NH4HCO3/50% acetonitrile (MeCN) at 37°C for 45 minutes to remove the stain. Gel pieces were then dehydrated with 100 μl, 100% MeCN at room temperature for 5 minutes. MeCN was removed and gel pieces were dried in a speed vacuum. For protein digestion, the samples were incubated in 50 μl of 20 μg/ml trypsin (Promega) in 40 mM NH4HCO3/10 % MeCN at 37°C overnight. Peptides were extracted with an extraction buffer (50% MeCN, 0.1% trifluoroacetic acid [TFA]). To purify peptides, ziptip was used by MeCN (100%) activation, sample absorption, and elution using 70% MeCN/0.1% TFA.
Mass spectrometry analysis
Matrix-assisted-laser-desorption-ionization time-of-flight (MALDI-TOF) mass analysis was performed at the Mass spectrometry Center, University of South Carolina, using Bruker Ultraflex MALDI-TOF-TOF with Flex analysis 2.0 and Biotools 2.2 software. One μl of purified peptides was crystallized with an equal volume of a freshly prepared α-Cyano-4-hydroxy-cinnamic acid matrix solution (saturated, 0.5 ml/sample) in a 50% MeCN/0.1% TFA solution in a MALDI 100-well plate. Spectra were acquired manually with laser intensity set at 2400, with 200 shots per spectrum and a mass range of m/z 700 to m/z 3000. All spectra were calibrated using two trypsin auto digest ions (m/z=842.509, m/z=2211.104). To confirm protein identity, selected target proteins were further analyzed by MALDI-TOF-TOF. Peptide peaks from the target spot were submitted to Mascot (www.matrixscience.com) to obtain initial protein identification using the National Center for Biotechnology Information (NCBI) database. The criteria we based on our measurement on one missed trypsin cleavage, mass accuracy tolerance of 100 ppm, and methionine oxidation for matching the peptide mass values.
Western blot analysis
We used western blot to confirm the identity of the proteins. Proteins extracted from bovine RPE cells or human RPE D407 cells were electrophoresed on a 12% SDS polyacrylamide gel. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (pore size 0.2 μm, Biorad). Nonspecific binding was blocked by incubation in 5% nonfat dry milk with 0.1% Tween20 in PBS at room temperature for 2 hours. The membrane was incubated with mouse monoclonal antibodies against 200 kDa+160 kDa neurofilament (Abcam, 1:2000 dilution) or 14-3-3 (Abcam, 1: 500 dilution) in PBS containing 5% nonfat milk and 0.1% Tween 20 overnight at 4°C with gentle agitation. Monoclonal anti RPE65 protein primary antibody (1:4000, Abcam) was used for RPE65 identification. After three washes with PBS/0.1% Tween 20, membranes were incubated with secondary antibody horseradish peroxidase coupled anti-mouse IgG (1:8000 dilution) for 1 hour at room temperature. The membranes were then washed three times with PBS/0.1% Tween 20 and blots were detected by using enhanced chemiluminescence (ECL) procedure (Pierce).
Normalizations were performed with polyclonal anti-actin antibody (Neomarkers).
Results
In our first biological model, bovine RPE cells were treated with 200 μM hydrogen Peroxide (H2O2). Proteins were extracted and separated by two-dimensional electrophoresis. Proteins were visualized by Coomasie blue staining. Proteins from PBS-treated bovine RPE cells were separated in parallel to serve as a control. Gel image overlay revealed that seventeen protein spots were up-regulated and seven proteins spots were down-regulated as a result of the H2O2 treatment (Fig. 1A). Quantitative analysis of each spot based on normalized volume and intensity is shown in Fig. 2A and Fig 2B. Differentially-expressed protein spots were excised and digested by trypsin followed by protein identification using MALDI-TOF and MALDI -TOF-TOF mass spectrometry.
Figure 1. Comparative proteome analysis of bovine RPE cells and human RPE D407 cells under oxidative stress.
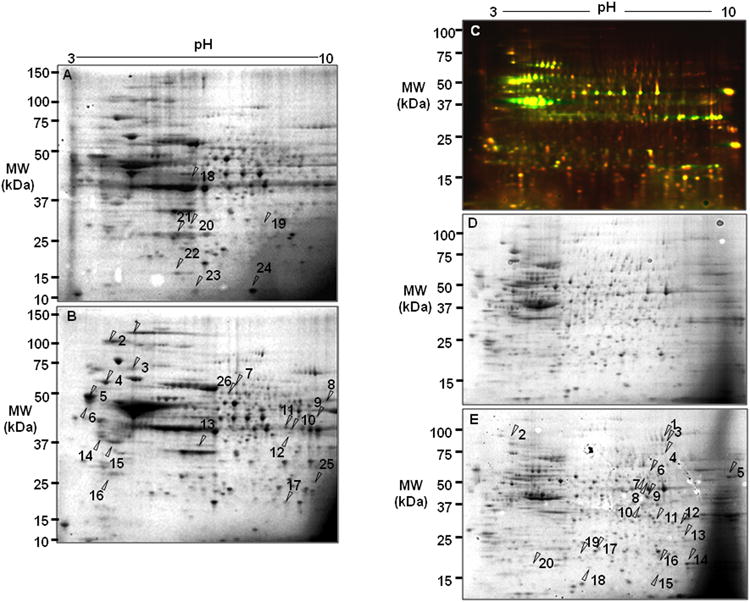
(A) 2D-SDS-PAGE analysis of bovine RPE proteins. RPE proteins (100 μg) were separated by electrophoresis and visualized by Coomassie blue staining. X axis represents isoelectric point (3-10) and Y axis shows molecular weight in kDa. Arrows indicate protein spots that were up-or down-regulated proteins under oxidative stress. (B) 2D-SDS-PAGE of control RPE cells with treated with PBS. (C) 2D-DIGE protein analysis of human RPE D407. Five μg of total proteins were prelabeled with Cy3 (untreated control, green, emission wavelength 580nm) and with Cy5 (oxidative stress, red, emission wavelength 670nm). Red spots indicate that proteins were up-regulated and green spots were down-regulated in H2O2 treated cells. (D) Coomassie blue stained gel (100 μg) from H2O2 treated RPE D407 cells. Arrows indicate protein spots that were up-regulated by the treatment. (E) Coomassie blue stained gel of control D407 cells.
Figure 2. Quantitative analysis of 2D-PAGE.
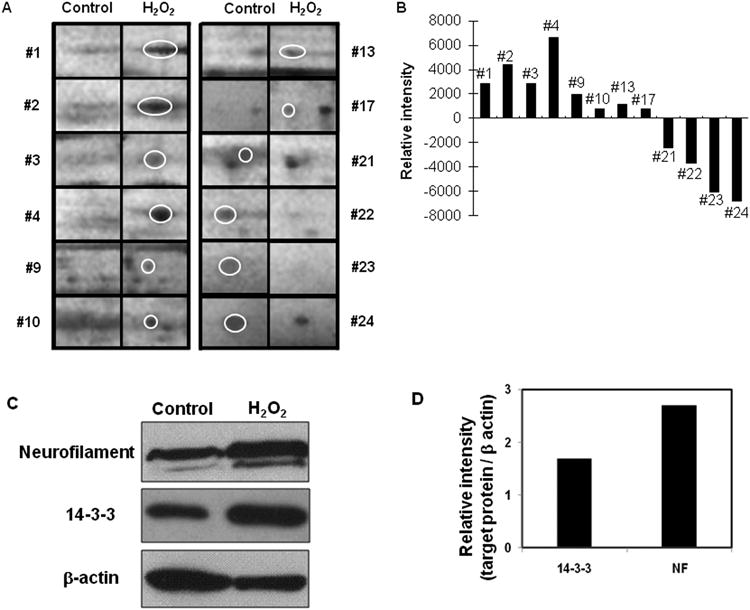
(A) Magnified protein spots in oxidative stress are shown with the control. (B) Each protein spots were analyzed based on volume and intensity. (C) Western blot analysis of 14-3-3 and neurofilament in control (lane 1) and H2O2 treated (lane 2) bovine RPE cells. (D) Quantification of western blot shows that 14-3-3 and neurofilament in oxidative stress were up-regulated by 1.8 fold and 2.8 fold compared to control.
Protein name, predicted function, probability score, sequence coverage, calculated molecular weight, pI, and related diseases are shown in Table 1. Among these, retinoid metabolism related proteins, including RPE65, an all-trans-retinyl ester isomerohydrolase in the visual cycle, and retinol binding protein are of interest considering perturbed retinoid metabolism under oxidative stress in the RPE. Apoptotic (annexin V), redox (GST, DADH), and DNA repair (RAD23) proteins were up-regulated and aging related (prohibitin), respiratory (COX) proteins were down-regulated in bovine RPE cells.
Table 1. Up- or down-regulated proteins in bovine RPE cells under oxidative stress.
| Up-down/Spot# | Protein identified | Predicted Function | Scorea | Coverage | Mass | pI | Human diseaseb |
|---|---|---|---|---|---|---|---|
| Up N1 | Neurofilament triplet H protein | Cytoskeletal protein | 208 | 30 | 112168 | 8.02 | ALS |
| Up N2 | Neurofilament triplet M protein | 115 | 12 | 103074 | 4.91 | Alzheimer's | |
| Up N3 | peripherin | 146 | 21 | 63566 | 5.42 | RP | |
| Up N4 | Neurofilament triplet L protein | 329 | 45 | 62665 | 4.59 | ALS | |
| Up N14 | Glial fibrillary acidic protein | 237 | 71 | 49538 | 5.36 | Alexander D | |
|
| |||||||
| Up N16 | 14-3-3 zeta | Cell signaling | 71 | 45 | 28272 | 4.75 | schizophrenia |
|
| |||||||
| Up N5, 6 | Calreticulin | Chaperon | 212/104 | 51/29 | 46524 | 4.31 | Alzheimer's |
|
| |||||||
| Up N7 | TNFR-associated protein 1 | Inflammation | 101 | 32 | 79331 | 6.66 | Autoimmune disease |
|
| |||||||
| Up N13 | Dimethylarginine Dimethylamino hydrolase 1 | NO synthesis | 104 | 33 | 30573 | 5.71 | Alzheimer's |
|
| |||||||
| Up N15 | Annexin V | apoptosis | 194 | 56 | 35992 | 4.86 | aging |
|
| |||||||
| Up N8 | Pyruvate kinase M1 | Metabolic enzyme | 167 | 52 | 58994 | 7.96 | cancer |
| Up N11, 12 | Pyruvate dehydrogenase E1 | 206 | 40 | 43835 | 8.17 | - | |
|
| |||||||
| Up N9 | UV excision repair protein RAD23 | DNA repair | 68 | 21 | 33511 | 4.7 | - |
|
| |||||||
| Up N10 | (2-5) oligo (A) synthetase | RNA synthesis | 67 | 33 | 58071 | 9.23 | - |
| Up N17 | HNRPM protein | 72 | 25 | 58716 | 8.94 | - | |
|
| |||||||
| Up N25 | Glutathione S-transferase Pi | Redox enzyme | 71 | 28 | 23826 | 6.89 | Lung/prostate cancer |
|
| |||||||
| Up N26 | RPE65 | Visual cycle | 161 | 33 | 61590 | 6 | LCA, RP |
|
| |||||||
| Down N18 | Tripeptidyl-peptidase 1 | Protein degradation | 92 | 22 | 61770 | 6.28 | Lipofuscinosis (NCL) |
|
| |||||||
| Down N19 | DNA replication factor | Cell cycle | 73 | 33 | 73163 | 9.76 | - |
|
| |||||||
| Down N20 | Endothelial differentiation-related factor | Cell differentiation | 72 | 57 | 16359 | 9.95 | - |
|
| |||||||
| Down N21 | Prohibitin | Cell proliferation | 100 | 63 | 29843 | 5.57 | aging |
|
| |||||||
| Down N22 | Retinol binding protein (RBP) | Retinoid metabolism | 82 | 69 | 15473 | 5.89 | recess RPE degeneration |
|
| |||||||
| Down N23 | Cytochrome c oxidase VIc | Cell respiratory | 55 | 82 | 8473 | 10.28 | - |
|
| |||||||
| Down N24 | Platelet-derived growth factor R like | Cell signaling | 72 | 27 | 57143 | 9.36 | - |
Probability-based MASCOT MOWSE score is -10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 55 are significant (p<0.05).
Related diseases based on Retnet database (http://www.sph.uth.tmc.edu/retnet/disease.htm) and IHOP (information hyperlinked over proteins, http://www.ihop-net.org/UniPub/iHOP/).
Identified proteins were confirmed by western blot. Representative 14-3-3 protein that binds specifically to phosphoserine/phosphothreonine-containing proteins was up-regulated under oxidative stress shown by quantitative analysis (Fig. 2C) [17,18]. Neurofilament protein, a major intermediate filament protein involved in neurodegenerative diseases [18,19] was up-regulated 2.8 fold (Fig. 2C). RPE65, a putative isomerohydrolase of all-trans-retinyl ester and retinoid binding protein in the RPE, is cleaved to 45 kD and 20 kD upon oxidative stress in vitro [15]. Prohibitin, a tumor suppressor and mitochondrial chaperone, was shown to be down-regulated in the bovine RPE in oxidative stress (Table 1) was also down-regulated in mild oxidative stress in vitro [20].
Next, we used human RPE D407 to gain a better understanding of early molecular events induced by oxidative stress in immortalized RPE cells. D407 cells were treated with 200 μM H2O2 for 1 hour, then H2O2 was removed. Cells were incubated for additional 6 hours in conditioning media. No significant cytotoxicity was observed under these conditions. Total proteins were labeled with Cy3 (PBS-treated control) or Cy5 (H2O2-treated cells) followed by co-separation of the two samples on the same gel, which enables for accurate spot matching and interpretation [16]. DIGE image analysis (Fig. 1C) showed that 18 proteins were up-regulated under oxidative stress (red spots). Green spots represent down-regulated proteins. The DIGE gel served as an analytical gel to detect minor proteins with lower concentrations in the RPE. However, to have enough protein amounts to identify by mass spectrometry, separate preparative gels were loaded with 20-fold more total protein and stained with Coomassie blue to allow for subsequent spot picking and mass spectrometric identification. The preparative gels were matched with the analytical gel based on image overlay and visual inspection (Fig. 1C, 1D, 1E). Identified proteins, accession numbers, and molecular weight and pI values are listed in Table 2.
Table 2. Up-regulated proteins in human RPE D407 cells under oxidative stress.
| Spot number | Protein identified | Predicted functiona | MOWSE Score | % Coverage | Mass | pI | Human disease |
|---|---|---|---|---|---|---|---|
| D1 | EF2 | Protein synthesis | 214 | 39 | 96246 | 6.41 | Alzheimer's |
| D9 | Elongation factor Tu | 168 | 61 | 49852 | 7.26 | - | |
|
| |||||||
| D2 | Hsp 90 alpha | Chaperone | 61 | 20 | 85006 | 4.94 | lens apoptosis |
| D17 | Hsp beta 1 | 94 | 42 | 22826 | 5.98 | ||
|
| |||||||
| D3 | Ribonucleoside-diphosphate reductase | DNA synthesis | 78 | 27 | 90925 | 6.76 | Lung cancer cell line |
| D7 | Inosine-5′-monophosphate dehydrogenase 2 (IMPDH2) | 122 | 49 | 56226 | 6.44 | IMPDH1 with RP | |
|
| |||||||
| D4 | DDX3X | RNA transcription | 61 | 28 | 73597 | 6.73 | Breast cancer |
|
| |||||||
| D5 | Plasminogen activator inhibitor 1 (SERBP1) | RNA binding | 66 | 25 | 44995 | 8.66 | Ovarian cancer |
|
| |||||||
| D6 | ATP dependent RNA helicase DDX1 | RNA metabolism | 112 | 31 | 83349 | 6.81 | Neuroblastoma |
|
| |||||||
| D8 | Lamin-A/C (d) | Nuclear envelope | 148 | 40 | 74380 | 6.57 | Lipodystrophy, cardiomyopathy, aging |
|
| |||||||
| D10 | BUB3 | Cell cycle | 77 | 36 | 37587 | 6.36 | Bladder/gastric/breast cancer |
|
| |||||||
| D11 | RAN | GTP binding protein | 72 | 44 | 24579 | 7.01 | Kennedy's disease |
| D13 | Guanine binding protein B2L1 | 88 | 46 | 35511 | 7.6 | - | |
|
| |||||||
| D12 | Voltage-dependent anion-selective channel protein VDAC2 | Channel protein | 84 | 48 | 32060 | 7.49 | Alzheimer's, Down syndrome |
|
| |||||||
| D14 | Peroxiredoxin-1 | Antioxidant enzyme | 191 | 73 | 22324 | 8.27 | Parkinson's |
| D16 | Peroxiredoxin-1 | 191 | 73 | 22324 | 8.27 | ||
| D19 | Thioredoxin-dependent peroxide reductase (PRDX3) | 56 | 21 | 28017 | 7.67 | - | |
| D20 | Peroxiredoxin 2 | 74 | 50 | 15243 | 5.82 | Parkinson's | |
|
| |||||||
| D18 | Phosphomevalonate kinase | Metabolic enzyme | 149 | 66 | 22152 | 5.56 | - |
Chaperone (Hsp), RNA metabolic (DDX), and antioxidant (peroxiredoxins) proteins were up-regulated in human RPE D407 cells under oxidative stress.
Discussions
Recently, pioneering proteomic profiling studies revealed the protein expressions in human RPE [21], drusen [22] and lipofuscin [23]. Other studies compared native differentiated to cultured dedifferentiated RPE cells [24,25]. Proteomics proved useful in delineating changes in RPE associated with the progressive stages of AMD [26,27,29], and diabetes [28]. Also, proteomic tools were used to study changes in the vitreous humor associated with diabetic retinopathy [30] and retinal proteins in a glaucoma model [31]. Since oxidative stress is implicated in the etiology of several RPE diseases such as AMD, identification of oxidative stress molecular mediators or early signaling molecules is a crucial step for understanding these diseases and discovering novel therapeutic approaches. However, proteomic limitations, including minor proteins with low concentration, hydrophobic proteins, reproducibility, and time and labor demanding processes, exist for a better understanding of proteome changes in different biological systems.
2-D DIGE is an advanced proteomic technique that labels minor protein samples with fluorescent dyes before 2-D electrophoresis. It enables accurate analysis of differences in protein concentrations between samples. Thus, DIGE method reduces experimental variations and technical errors. It is possible to separate up to three different samples within the same 2-D gel using three different fluorescent molecules. However, there are limitations of DIGE methods also, including covalent modifications on cysteine or lysine. We confirmed fluorescent dye labeled proteins with traditional Coomassie blue staining to show its sensitivity and reproducibility.
We used two different biological models, primary bovine RPE cells and the human RPE cell line D407, to uncover differential biosignatures under oxidative stress in the RPE. We employed a combination of proteomic technologies, including sensitive fluorescent labeling and TOF-TOF mass spectrometry analysis with high mass accuracy and low tolerance in the range of 50 ppm (0.05 Dalton). Identified novel proteins were confirmed by quantitative western blot. We observed expression changes of cellular signaling related molecules, including intermediate filament structure, retinoid metabolism, energy metabolism, and antioxidant proteins in bovine RPE cells. Several intermediate filament proteins, including neurofilament H, M, L proteins and glial fibrillary acidic protein, were up-regulated in bovine RPE cells. Intermediate filament proteins are cytoskeletal components that form fibrils with an average diameter of 10 nm. Neurofilament H, M, and L proteins are specifically expressed in neurons. Glial fibrillary acidic proteins are biomarker protein in glial cells [32,33]. Previous studies showed that environmental changes, such as culture conditions, could induce dedifferentiation of RPE cells and give rise to mesenchymal-like cells [34,35].
Proteome changes related to energy metabolism were also observed under oxidative stress in bovine RPE cells. Pyruvate dehydrogenase E1 transforms pyruvate into acetyl-CoA that is used in the citric acid cycle to generate ATP. Pyruvate kinase M1 transfers a phosphate group from phosphoenolpyruvate (PEP) to adenosine diphosphate (ADP), to synthesize ATP and pyruvate. Up-regulation of both enzymes may indicate higher energy consumption to compensate oxidative stress-induced faster turnover of proteins in the RPE.
In the immortalized human RPE, several molecular chaperones were altered as a result of H2O2 treatment. Hsp 90α and Hsp β1 exhibit general protective chaperone properties such as preventing unspecific aggregation of non-native proteins [36]. Calreticulin, up-regulated in bovine RPE cells, is a multifunctional protein that binds to Ca2+ and misfolded proteins to export them from the endoplasmic reticulum (ER) to the Golgi apparatus [37]. Elimination of misfolded proteins in the ER affects cellular homeostasis and survival, so chaperone function of heat shock proteins is indispensible under stress conditions.
Retinoid metabolism in primary bovine RPE cells is altered under oxidative stress conditions. Photosensitivity and the steady state of retinoid concentrations are controlled by regeneration of 11-cis-retinoid, which is called the visual cycle. RPE65, a peripheral membrane protein of RPE cells, is thought to be an all-trans-retinyl ester isomerohydrolase. Indeed, we showed that RPE65 in bovine RPE is up-regulated under oxidative stress. We recently showed that RPE65 is cleaved into a 45 kDa and 20 kDa truncation forms under oxidative stress [15]. This data indicates that oxidative stress can influence the visual cycle by up-regulation of RPE65 and cleavage of this protein. Further studies of whether this truncated form might be a biomarker of oxidative stress are currently under investigation. It seems that immortalized human RPE cells suppress RPE65 expressions as a basic regulatory mechanism for the visual cycle that is not essential in this stable cell line for survival. Prohibitin, a mitochondrial chaperone involved in oxidative stress and aging, was found to be down-regulated under oxidative stress in primary bovine RPE cells.
Finally, antioxidant proteins were up-regulated in RPE D407 cells. Peroxiredoxins are a ubiquitous family of antioxidant enzymes that can be regulated by changes of redox potential in the cell. Thioredoxin-dependent peroxide reductase is an antioxidant enzyme that provides protection mechanism against oxidative stress. It reduces H2O2 by hydride provided by thioredoxin, thioredoxin reductase, and NADPH [38]. Up-regulation of antioxidants can regulate redox homeostasis in RPE cells and thus protect cells from oxidative stress-induced damage.
Both biological models using primary bovine RPE cells and immortalized D407 cells have advantages and limitations. We observed differences in protein expressions correlated with oxidative stress in two biological models used in our study, which can be attributed, at least partially, to the inherent differences in two different RPE cell types. It has been reported that cytoskeletal remodeling and cell survival factors are differentially expressed in primary culture and immortalized human RPE cells [25]. Dedifferentiated, immortalized human RPE cells results in down-regulation of specific proteins associated with retinoid metabolism. At the same time, they induce the differential expressions related with cytoskeletal organization, cell shape, migration, and proliferation. Thus, we speculate that two different proteome list in two different systems such as primary bovine vs. immortalized human RPE cells is due to different cell type and not due to species difference or detection system.
Oxidative stress alone is the major risk factor of retinal degeneration compared to genetic risk factors [39-41]. Furthermore, oxidative stress may influence the expression of genetic risk factor, such as complement factor H (CFH) [42]. Cellular processes induced by oxidative stress in RPE cells and the molecular mechanisms under the oxidative stress that contribute to retinal degeneration are not clear at this point. In this study, we revealed several expression changes in the RPE proteome that is induced by H2O2 treatment. These changes can aid in understanding early molecular signaling events of oxidative stress in the RPE as biosignatures. Targeting these early signaling proteins could be a useful therapeutic strategy for the treatment of degenerative diseases of the retina and the RPE.
Acknowledgments
The authors thank Dr. Elizabeth Hager, Dr. Steven Bailey, and Matthew Durocher for their critical reading and suggestions. This study was supported by the Centenary Award from the University of South Carolina; a start-up fund from the Department of Ophthalmology, University of South Carolina; the New Investigator Award from the International Foundation; the Century II Equipment fund, a start-up package, and the Research Excellent Fund from Michigan Technological University.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 2.Dorey CK, Wu G, Ebenstein D, Garsd A, Weiter JJ. Cell loss in the aging retina. Relationship to lipofuscin accumulation and macular degeneration. Invest Ophthalmol Vis Sci. 1989;30:1691–1699. [PubMed] [Google Scholar]
- 3.Cai J, Nelson KC, Wu M, Sternberg P, Jr, Jones DP. Oxidative damage and protection of the RPE. Prog Retin Eye Res. 2000;19:205–221. doi: 10.1016/s1350-9462(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 4.Winkler BS, Boulton ME, Gottsch JD, Sternberg P. Oxidative damage and age-related macular degeneration. Mol Vis. 1999;5:32–42. [PMC free article] [PubMed] [Google Scholar]
- 5.Liang FO, Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res. 2003;76:397–403. doi: 10.1016/s0014-4835(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 6.Chong EW, Wong TY, Kreis AJ, Simpson JA, Guymer RH. Dietary antioxidants and primary prevention of age related macular degeneration: systematic review and meta-analysis. BMJ. 2007;335:7623–7755. doi: 10.1136/bmj.39350.500428.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parisi V, Tedeschi M, Gallinaro G, Varano M, et al. CARMIS Study Group. Carotenoids and Antioxidants in Age-Related Maculopathy Italian Study Multifocal Electroretinogram Modifications after 1 Year. Ophthalmology. 2008;115:324–333. doi: 10.1016/j.ophtha.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 8.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 9.Gaillard ER, Atherton SJ, Eldred G, Dillon J. Photophysical studies on human retinal lipofuscin. Photochem Photobiol. 1995;61:448–453. doi: 10.1111/j.1751-1097.1995.tb02343.x. [DOI] [PubMed] [Google Scholar]
- 10.Ballinger SW, Van Houten B, Jin GF, Conklin CA, Godley BF. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;686:765–772. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- 11.Jahng WJ, David C, Nesnas N, Nakanishi K, Rando RR. A cleavable affinity biotinylating agent reveals a retinoid binding role for RPE65. Biochemistry. 2003;42:6159–68. doi: 10.1021/bi034002i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. Epub 2009 Jan 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon YH, Jeong KH, Shim MJ, Koh JY. High vulnerability of GABA-immunoreactive neurons to kainate in rat retinal cultures: correlation with the kainate-stimulated cobalt uptake. Brain Res. 1999;27:33–41. doi: 10.1016/s0006-8993(98)01336-5. [DOI] [PubMed] [Google Scholar]
- 14.Hyun HJ, Sohn JH, Ha DW, Ahn YH, Koh JY, Yoon YH. Depletion of intracellular zinc and copper with TPEN results in apoptosis of cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2001;42:460–5. [PubMed] [Google Scholar]
- 15.Chung H, Lee H, Lamoke F, Hrushesky WJ, Wood PA, Jahng WJ. Neuroprotective role of erythropoietin by antiapoptosis in the retina. J Neurosci Res. 2009;87:2365–74. doi: 10.1002/jnr.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 17.Julien JP, Mushynski WE. Neurofilaments in health and disease. Prog Nucleic Acid Res Mol Biol. 1998;61:1–23. doi: 10.1016/s0079-6603(08)60823-5. [DOI] [PubMed] [Google Scholar]
- 18.Wilker E, Yaffe MB. 14-3-3 proteins---a focus on cancer and human diseases. J Mol Cell Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 19.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 20.Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West KA, Yan L, Shadrach K, Sun J, Hasan A, Miyagi M, Crabb JS, Hollyfield JG, Marmorstein AD, Crabb JW. Protein database, human retinal pigment epithelium. Mol Cell Proteomics. 2003;2:37–49. doi: 10.1074/mcp.d200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2000;12:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schütt F, Völcker HE, Dithmar S. N-acetylcysteine improves lysosomal function and enhances the degradation of photoreceptor outer segments in cultured RPE cells. Klin Monatsbl Augenheilkd. 2007;224:580–584. doi: 10.1055/s-2007-963339. [DOI] [PubMed] [Google Scholar]
- 24.Alge CS, Suppmann S, Priglinger SG, Neubauer AS, May CA, Hauck S, Welge-Lussen U, Ueffing M, Kampik A. Comparative proteome analysis of native differentiated and cultured dedifferentiated human RPE cells. Invest Ophthalmol Vis Sci. 2003;44:3629–3641. doi: 10.1167/iovs.02-1225. [DOI] [PubMed] [Google Scholar]
- 25.Alge CS, Hauck SM, Priglinger SG, Kampik A, Ueffing M. Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival. J Proteome Res. 2006;5:862–878. doi: 10.1021/pr050420t. [DOI] [PubMed] [Google Scholar]
- 26.Nordgaard CL, Berg KM, Kapphahn RJ, Reilly C, Feng X, Olsen TW, Ferrington DA. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:815–822. doi: 10.1167/iovs.05-0976. [DOI] [PubMed] [Google Scholar]
- 27.An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman EP, Strunnikova N, Csaky K, Hathout Y. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res. 2006;5:2599–2610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- 28.Decanini A, Karunadharma PR, Nordgaard CL, Feng X, Olsen TW, Ferrington DA. Human retinal pigment epithelium proteome changes in early diabetes. Diabetologia. 2007;51:1051–1061. doi: 10.1007/s00125-008-0991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T, Kim SJ, Kim K, Kang UB, Lee C, Park KS, Yu HG, Kim Y. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics. 2007;7:4203–4215. doi: 10.1002/pmic.200700745. [DOI] [PubMed] [Google Scholar]
- 31.Tezel G, Yang X, Cai J. Proteomic identification of oxidatively modified retinal proteins in a chronic pressure-induced rat model of glaucoma. Invest Ophthalmol Vis Sci. 2005;46:3177–3187. doi: 10.1167/iovs.05-0208. [DOI] [PubMed] [Google Scholar]
- 32.Pitz S, Moll R. Intermediate-filament expression in ocular tissue. Prog Retin Eye Res. 2002;21:241–262. doi: 10.1016/s1350-9462(01)00025-8. [DOI] [PubMed] [Google Scholar]
- 33.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 34.Casaroli-Marano RP, Pagan R, Vilaró S. Epithelial-mesenchymal transition in proliferative vitreoretinopathy: intermediate filament protein expression in retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1999;40:2062–2072. [PubMed] [Google Scholar]
- 35.Alizadeh M, Wada M, Gelfman CM, Handa JT, Hjelmel LM. Downregulation of differentiation specific gene expression by oxidative stress in ARPE-19 cells. Invest Ophthalmol Vis Sci. 2001;42:2706–2713. [PubMed] [Google Scholar]
- 36.Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410:439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 37.Michalak M, Mariani P, Opas M. Calreticulin, a multifunctional Ca2+ binding chaperone of the endoplasmic reticulum. Biochem Cell Biol. 1998;76:779–785. doi: 10.1139/bcb-76-5-779. [DOI] [PubMed] [Google Scholar]
- 38.Chae HZ, Chung SJ, Rhee SG. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 39.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards AO, Ritter R, Abel ШKJ, Manning A, Panhuysen C, Farrer LA. Complement Factor H Polymorphism and Age-Related Macular Degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z, Lauer TW, Sick A, Hackett SF, Campochiaro PA. Oxidative stress modulates complement factor H expression in retinal pigmented epithelial cells by acetylation of FOXO3. J Biol Chem. 2007;282:22414–22425. doi: 10.1074/jbc.M702321200. [DOI] [PubMed] [Google Scholar]


