Summary
Primary and motile cilia/flagella function as cellular antennae, receiving signals from the environment, and subsequently activating signaling pathways that are critical for cellular homeostasis and differentiation [1-3]. Recent work with the green alga Chlamydomonas and the nematode C. elegans demonstrated that ectosomes can be released from the cilium and can mediate the intercellular communication [4-9]. To better understand the function of flagellar ectosomes, we have compared their protein composition to that of the flagellar membrane from which they are derived. Ectosomes released from flagella have a unique protein composition, being enriched in a subset of flagellar membrane proteins, proteases, proteins from the endosomal sorting complex required for transport (ESCRT) [10-12], small GTPases, and ubiquitinated proteins. Live imaging showed that an ESCRT-related protein (PDCD6) was enriched in ectosomes released from flagella during gamete activation. We devised a sensitive and rapid assay to monitor ectosome release using luciferase fused to PDCD6 and a mutated ubiquitin. Ectosome release increased when cells underwent flagellar resorption. Knockdown of two ESCRT-related proteins, PDCD6 and VPS4, attenuated ectosome release during flagellar shortening and shortening was slowed. These data suggest that the ESCRT proteins mediate ectosome release and thereby influence flagellar shortening in Chlamydomonas. In addition, the prevalence of receptors such as agglutinin and ubiquitinated proteins in ciliary ectosomes suggests that they are involved in cell signaling and turnover of ciliary proteins.
Graphical abstract
Long et al. isolate flagellar ectosomes and demonstrate that flagellar ectosomes have a unique protein composition, especially being enriched in endosomal sorting complex required for transport (ESCRT) and ubiquitinated proteins. Knock down of ESCRT proteins attenuates the flagellar ectosome release and thereby influences flagellar shortening.
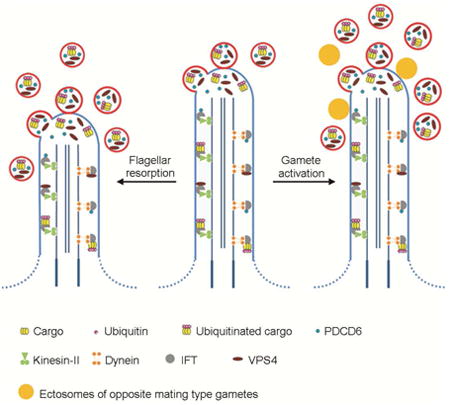
Results and Discussion
Purification of ectosomes from culture medium
Our previous work demonstrated that ectosomes are released from Chlamydomonas flagella and following cell division, these ectosomes contain a bioactive enzyme required to digest the mother cell wall [4]. In the present study we have focused on ectosomes released from growing cells (i.e. not just after division), which we can manipulate more easily in large quantities. Our first goal was to purify ciliary ectosomes and compare their composition to that of the flagellar membrane to determine whether their compositions were very different as would be expected if vesicle release were a specific process rather than a generalized sloughing off of the flagellar membrane.
We purified ectosomes from medium in which flagellated wild type cells had been maintained using differential centrifugation and Optiprep gradient centrifugation. The ectosomes appeared spherical by negative stain electron microscopy, with a diameter of approximately 100 nm (Figure 1A). When sedimented for thin section microscopy, some vesicles remained spherical, while others were collapsed forming double membrane sheets (Figure 1B). The size of isolated ectosomes determined by either method was variable and less than 500 nm, similar to the microvesicles or microparticles described in vertebrates [13, 14]. Immunoblot analysis showed that the ectosomes contained known flagellar membrane proteins such as the major flagellar membrane glycoprotein 1 (FMG1) and the polycystin-2 homologue, PKD2, and had little axonemal contamination as demonstrated using α-tubulin and radial spoke 1 (RSP1) antibodies (Figure 1C). Because Chlamydomonas is surrounded by a cell wall, the plasma membrane cannot release vesicles into the medium, ensuring that the sole source of the ectosomes in the medium was from the flagella. The purity of these Chlamydomonas flagellar ectosomes distinguishes the current studies from many other proteomic analyses of ectosomes, which also include exosomes derived from exocytosis of multivesicular bodies [15, 16].
Figure 1. Ectosomes have a unique protein composition.
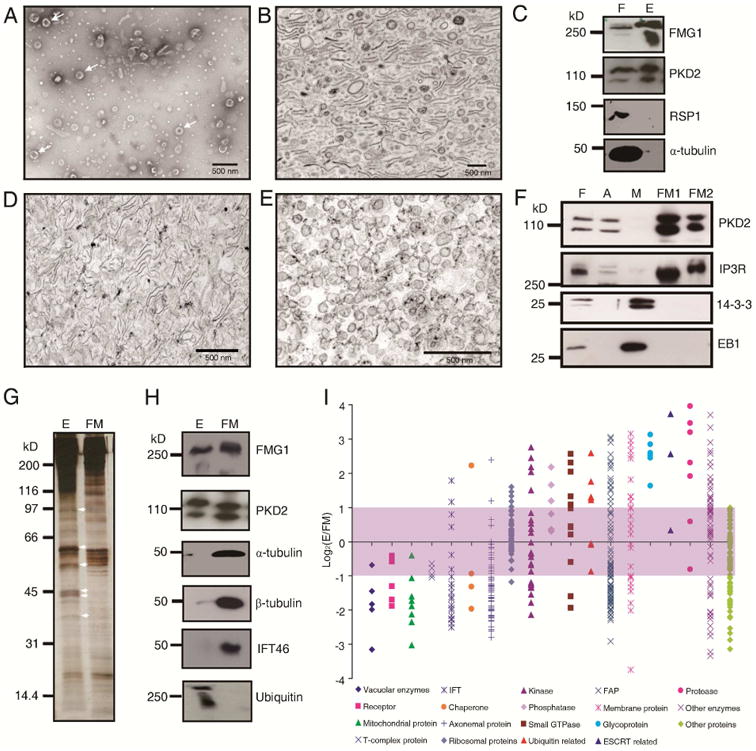
A) Negative stained isolated ectosomes (arrows) released from flagella of vegetative cells. B). Electron micrograph of thin sectioned, isolated ectosomes. C). Immunoblot of flagella (F) and ectosomes (E) with antibodies against flagellar membrane proteins (FMG1 and PKD2) and axonemal proteins (RSP1 and α-tubulin). D). Electron micrograph of flagellar membrane (FM1) isolated with 1% NP-40. E). Electron micrograph of flagellar membrane vesicles (FM2) recovered from the supernatant after removing the detergent. F).Western blot of 10 ug of protein from flagella (F), axoneme (A), matrix (M), FM1 and FM2 with antibodies against PKD2, IP3R (IP3 receptor), 14-3-3 and EB1. Membrane proteins are enriched in FM1 and FM2. G). Silver stained SDS-PAGE gel of proteins from ectosomes (E) versus flagellar membrane (FM). Arrows indicate the proteins enriched in ectosomes. H). Western blots of the same amount of protein of ectosomes (E) and flagellar membrane (FM) with antibodies recognizing the indicated proteins. I). Comparison of the proteins in ectosomes versus in the isolated flagellar membrane (FM1+FM2) using iTRAQ analysis. In total, 563 proteins with more than 3 peptides were categorized into 19 subgroups according to their known or putative function. Proteins with the Log2(E/FM) > 1 were considered as ectosome enriched, see in Table S1; -1 < Log2(E/FM) < 1 were considered to be equal in ectosomes and flagellar membranes; and Log2(E/FM) < -1 were considered to be enriched in the flagellar membrane, see in Table S2. Membrane proteins present in ectosome and flagellar membrane are shown in Table S3. Comparison of flagellar membranes isolated with detergent and freeze/thaw are shown in Figure S1.
Isolation of flagellar membrane
Flagellar ectosomes are derived from the flagellar membrane, so we compared the protein composition of the flagellar membrane to that of ectosomes to determine whether they were enriched in specific proteins. To this end, we first evaluated three methods of isolating flagellar membranes. After isolating flagella using the pH shock method, flagellar membranes were prepared using a freeze/thaw method, sonication followed by two-phase-partition, or detergent treatment. The freeze/thaw method was simple, without the interference with detergent; however, even after washing the axoneme pellet two times, sheets of flagellar membrane were still associated with the axonemes (Figure S1A), indicating this method did not harvest all the flagellar membrane. A two-phase-partition method, based on forming right-side-out membrane vesicles by sonication and partitioning these vesicles into a polyethylene glycol-rich phase, has been widely used in isolating the plasma membrane from the cell body of Chlamydomonas [17]. We adapted this method to isolate the flagellar membrane, but after flagella were broken down into small fragments by sonication, some of axonemal fragments were still associated with flagellar membrane (Figure S1B); therefore, this method was not suitable for isolating pure flagellar membrane.
Detergent extraction appeared more satisfactory (Figure S1C): when isolated flagella were treated with 1% NP-40 for 30 minutes twice at room temperature, the resulting axonemes were devoid of membrane sheets or vesicles (Figure S1D). After sedimenting the axonemes, the supernatant was centrifuged at 228,000 g and the resultant pellet, flagellar membrane fraction 1 (FM1), contained flagella membrane, much of which did not appear vesicular (Figure 1D). The 228,000 g supernatant still contained flagellar membrane proteins, so to recover these proteins, the detergent was removed and following sedimentation, we obtained a pellet, flagellar membrane fraction 2 (FM2), containing many small (<100 nm) spherical vesicles (Figure 1E). FM1 and FM2 contained approximately equal concentrations of the flagellar membrane proteins PKD2 and the IP3 receptor. Neither fraction contained flagellar matrix proteins such as 14-3-3 and EB1 (Figure 1F). These data suggested that both fractions contain pure flagellar membrane, so we combined FM1 and FM2 to produce the flagellar membrane (FM) fraction. We also compared the protein composition of flagellar membrane isolated with detergent versus isolated with freeze/thaw (Figure S1E to S1K): both samples contained little radial spoke 1 protein, no matrix proteins, and were enriched in flagellar membrane proteins. The detergent method released more FMG-1 and IP3 receptor from axonemes than did the freeze/thaw method, suggesting the former is more efficient in harvesting the flagellar membrane.
Comparison of proteins in ectosome and in flagellar membrane
We first used SDS-PAGE to compare the protein composition of ectosomes to that of flagellar membrane. At least 6 protein bands were more prominent in ectosomes than in flagellar membrane (Figure 1G). When the two fractions were probed with flagellar antibodies, flagellar membrane proteins FMG-1 and PKD2 were detected in both ectosomes and flagellar membranes; however, α-tubulin, β-tubulin and IFT46 were much more prevalent in the flagellar membrane fraction. The association of tubulin with flagellar membranes has been reported previously [18]. On the other hand, high molecular weight ubiquitinated proteins were only detected in the ectosome fraction (Figure 1H).
We used Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) to better compare the proteins in ectosomes and flagellar membranes. We identified 563 proteins, and compared their abundance in ectosomes (E) versus flagellar membrane (FM). The quantitative proteomic data paralleled the western blot data: the ratio of IFT46 in E versus FM was 0.26 according to iTRAQ analysis, compared to 0.17 seen on immunoblots.
The proteins were categorized into 19 subgroups according to known or putative functions (Figure 1I). 140 proteins were concentrated in the ectosome fraction, i.e., the value of Log2(E/FM) was greater than 1 (Table S1). This fraction includes 13 membrane proteins, 7 small GTPase, 6 glycoproteins, 6 proteases, 6 kinases, 5 ubiquitin system proteins, 4 phosphatases, 2 IFT related proteins, 2 chaperones, 2 axonemal proteins, 2 ESCRT proteins, 19 putative flagellar associated proteins, 22 other enzymes and 37 uncharacterized proteins. We also found 173 proteins that were dominant in the flagellar membrane fraction, i.e., the value of Log2(E/FM) was less than -1 (Table S2). Almost all of the axonemal proteins, IFT proteins and mitochondrial proteins were enriched in this fraction. The iTRAQ data confirmed that the ectosomes released from flagella have a vastly different protein composition from the flagellar membrane, suggesting that ectosome formation involves the active accumulation of specific proteins for release into the medium, rather than a general sloughing off of flagellar membrane.
In total, 64 known or predicted membrane proteins were identified in this iTRAQ analysis. Among them, 13 proteins including flagellar membrane glycoprotein 1B, fibrocystin-like protein, flagellar membrane protein AGG2, a calcium-transporting ATPase (FAP39), glycosyl transferase GTR11 and FAP212 were enriched in ectosomes (Table S3). On the other hand, other membrane proteins, including the IP3 receptor, PKD2, and an ABC transporter, were more prevalent in the flagellar membrane fraction (Table S3). The different distribution of membrane proteins in these two fractions suggests that there is a sorting mechanism for flagellar membrane proteins targeted to ectosomes.
Eight proteases were enriched in ectosomes including the vegetative lytic enzyme (VLE) (Table 1), which digests the mother cell wall following cell division and which was shown to localize on ectosomes [4, 19]. The amount of this protein in ectosomes isolated from non-dividing cells was remarkably lower than the amount obtained from ectosomes isolated following cell division (Figure S2) in keeping with its critical role in hatching of daughter cells following cell division [4].
Table 1.
Quantitative analysis of proteases, small GTPase, ubiquitin and ESCRT-related proteins present in ectosomes (E) versus flagellar membranes (FM). See also Figure S2.
| Functional groups | Protein | Uniprot Accession No. | Peptides | E/FM |
|---|---|---|---|---|
| Proteases | Prohead core scaffold and protease | A8J7I6_CHLRE | 6 | 0.57 |
| Subtilase-like serine protease, VLE | A9ZNH0_CHLRE | 15 | 1.50 | |
| Metalloproteinase of VMP family, MMP6 | A8JBG9_CHLRE | 4 | 1.50 | |
| Matrix metalloproteinase-like protein | A8IEN8_CHLRE | 3 | 3.75 | |
| Matrix metalloproteinase-like protein, MMP13 | A8J363_CHLRE | 6 | 4.93 | |
| Matrix metalloproteinase, MMP1 | A8JII3_CHLRE | 18 | 9.19 | |
| Type I metacaspase | A8J698_CHLRE | 2 | 11.06 | |
| Matrix metalloproteinase, MMP3 | A8IZV1_CHLRE | 14 | 15.49 | |
| Zinc-dependent metalloprotease | A8J840_CHLRE | 8 | 17.27 | |
|
| ||||
| Small GTPase | Small rab-related GTPase, IFT27 | A8HN58_CHLRE | 8 | 0.26 |
| Ras GTPase-like protein, IFT22 | A8HME3_CHLRE | 11 | 0.33 | |
| small Ran-like small GTPase, RAN1 | A8IRX5_CHLRE | 3 | 0.66 | |
| Small rab-related GTPase, RABG1 | A8ILX2_CHLRE | 5 | 1.15 | |
| Small rab-related GTPase, RABB1 | A8J195_CHLRE | 7 | 1.34 | |
| ARF-like GTPase, ARL13 | A8INQ0_CHLRE | 4 | 1.37 | |
| Small rab-related GTPase, RABH1 | A8HN42_CHLRE | 3 | 1.37 | |
| Small rab-related GTPase, RABA1 | A8IRT2_CHLRE | 5 | 1.96 | |
| Small rab-related GTPase, RABF1 | A8J6A0_CHLRE | 7 | 1.99 | |
| Small rab-related GTPase, RAB23 | A8HX77_CHLRE | 3 | 2.01 | |
| Small rab-related GTPase, RABE1 | A8JHI5_CHLRE | 2 | 2.06 | |
| Small ARF-related GTPase, ARFA1A | A8IL29_CHLRE | 11 | 2.11 | |
| Small Rab GDP dissociation inhibitor protein, GDIC1 | A8J146_CHLRE | 4 | 2.50 | |
| ARF-like GTPase, ARL3 | A8ISN6_CHLRE | 5 | 4.10 | |
| Small GTPase superfamily, Rab family, YptC1 | A8JHQ7_CHLRE | 10 | 4.96 | |
| Small Rab GAP/TBC protein | A8JCA4_CHLRE | 2 | 5.89 | |
|
| ||||
| Ubiquitin related | Ubiquitin-protein ligase, UBC7 | A8IYE1_CHLRE | 11 | 0.55 |
| Cullin, CUL3 | A8IW43_CHLRE | 2 | 0.94 | |
| Ubiquitin-conjugating enzyme E2 36-like | A8HQ77_CHLRE | 2 | 0.97 | |
| Predicted E3 ubiquitin ligase | A8J9K5_CHLRE | 4 | 2.34 | |
| Predicted E3 ubiquitin ligase | A8IE95_CHLRE | 3 | 2.46 | |
| Ubiquitin-activating enzyme E1, UBA1 | A8J1C1_CHLRE | 22 | 2.47 | |
| Ubiquitin-conjugating enzyme E2, UBCX | Q8VZX3_CHLRE | 2 | 3.36 | |
| Ubiquitin | A8IZZ4_CHLRE | 8 | 6.01 | |
|
| ||||
| ESCRT related | AAA-ATPase of VPS4/SKD1 family, VPS4 | A8IAJ1_CHLRE | 4 | 1.26 |
| Programmed cell death protein 6-interacting protein | A8JG06_CHLRE | 4 | 5.92 | |
| Calclium binding protein, PDCD6 | A8IXR0_CHLRE | 5 | 13.23 | |
Several other classes of proteins were enriched in the ectosomes. For example, seven small GTPases, including ARF like GTPase (ARL3), Rab family protein YptC1 and a small Rab GAP/TBC protein (A8JCA4) were abundant in ectosomes (Table 1). In addition, the ubiquitin conjugation system including the ubiquitin-activating enzyme E1 (UBA1), a ubiquitin-conjugating enzyme E2 (UBCX), two E3 ubiquitin ligases and ubiquitin were enriched in ectosomes. Notably, the amount of ubiquitin was 6 times higher in ectosomes than in the bulk flagellar membrane. Much, or all, of this ubiquitin was conjugated to proteins (Figure 1H). We also found that two ESCRT related proteins, the programmed cell death protein 6 (PDCD6), also named ALG-2 [20], and its binding protein PDCD6-interacting protein (PDCD6IP, also named Alix) were enriched in ectosomes (Table 1). The latter was also found in isolated ciliary transition zones [21].
Release of ectosome associated PDCD6 and ubiquitinated proteins from flagella
To confirm the proteomic data and set up a quick assay to monitor the amount of ectosomes release, we selected two ectosome-enriched proteins for further analysis: the ESCRT-related calcium binding protein PDCD6 and ubiquitin. We expressed a PDCD6∷luciferase fusion protein driven by a constitutive promoter, PsaD, in Chlamydomonas [22]. As expected, luciferase activity was detected in cell bodies and flagella of transformants (Figure S3A), suggesting that PDCD6 was found in flagella prior to being released into medium. To determine whether the fusion proteins were released to the medium, transformants that expressed the fusion proteins were moved to fresh medium, and ectosomes were isolated from the supernatant after 0, 1, 2 and 4 hours. Luciferase activity in the medium increased continuously from 0 to 4 h (Figure 2A). We also compared the PDCD6∷luciferase activity in ectosomes versus flagella (Figure S3B), the ratio was increased from 15 to 553. All these data demonstrate that PDCD6 containing ectosomes accumulate in the medium over time.
Figure 2. Ectosomes are released when cells are shifted to fresh medium and during gamete activation.
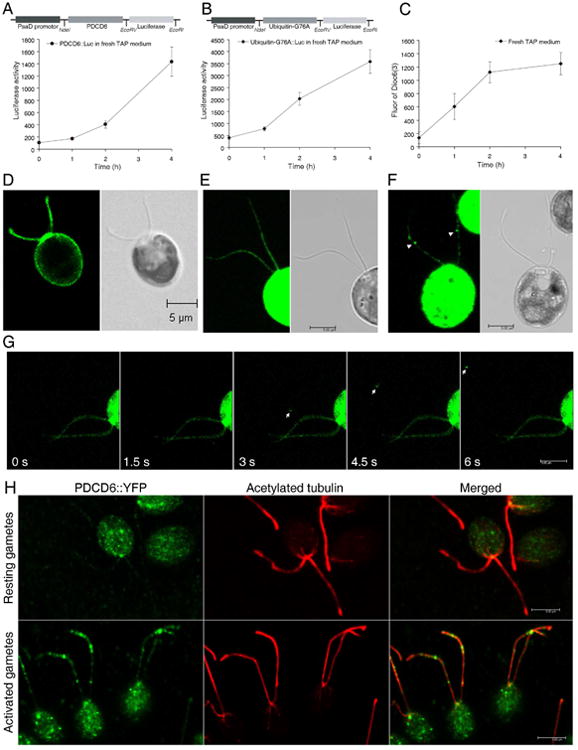
PDCD6 (A) and mutated ubiquitin (B) were fused with luciferase and expressed in Chlamydomonas cells. Cells were placed in fresh medium, ectosomes were isolated from the medium and their luciferase activity was measured. The luciferase activity of PDCD6 in both cell bodies and flagella is shown in Figure S3A. The activity of PDCD6∷luciferase in ectosomes from medium was normalized with the luciferase activity in flagella (Figure S3B). As negative control, IFT46∷luciferase was not detected in flagellar ectosomes (Figure S3C). C). CC1009 cells labeled with Dioc6(3) were transferred to fresh medium, and the fluorescence in the medium was measured periodically. Bar graphs represent mean ± SEM. D). The flagella and plasma membrane of CC1009 cells were labeled with the lipid dye Dioc6(3). Scale bars: 5μm. E). PDCD6∷YFP was localized uniformly in flagella of gametes that had not been activated. Western blot showed that PDCD6∷YFP was expressed in transformants (Figure S3D). F). PDCD6∷YFP enriched ectosomes (arrowheads) were formed on flagella during gamete activation. G). Live-imaging of PDCD6∷YFP enriched ectosomes (arrows) released from a flagellum during gamete activation. The source video, Movie S1 can be found in the Supplemental Information. H). Immunostaining showing PDCD6∷YFP was localized mainly in cell body in resting gametes (upper panel) and in ectosomes associated with flagella of gametes activated by adding the ectosomes isolated from the medium of the opposite mating type gametes (lower panel).
We also fused luciferase to a mutated form of ubiquitin in which the terminal glycine of ubiquitin was changed to alanine to avoid cleavage of the ubiquitin∷luciferase fusion in Chlamydomonas cells. As seen with PDCD6∷luciferase, after shifting the cells expressing ubiquitin (G76A)∷luciferase to fresh medium, the luciferase activity in ectosomes in the medium increased for 4 hours (Figure 2B). On the other hand, the activity of a control protein not found in ectosomes, IFT46∷luciferase, was detected only in flagella and not in the medium (Figure S3C). These results confirmed the iTRAQ data showing that PDCD6 and ubiquitinated proteins are shuttled into ectosomes that are released into the medium.
As a second means to monitor ectosome release, the flagella of Chlamydomonas were labeled with a lipid dye, Dioc6(3), and the appearance of lipid-bound dye in the medium was measured. As reported by Wright [23], the flagellar membrane, as well as the plasma membrane, is labeled with this dye (Figure 2D). When the labeled cells were shifted to fresh medium, the dye fluorescence in medium increased continuously for 4 hours (Figure 2C). These data suggest that flagellar lipids formed the ectosomes that were shed into the medium. The lipid, PDCD6∷luciferase and ubiquitin∷luciferase all were located in ectosomes, showing similar, increasing patterns when cells were moved to fresh medium. Fluorescence of lipid dye and the luciferase activity of these two fusion proteins, therefore, can be used to quickly assay the release of ectosomes from flagella and thereby explore the novel functions of flagellar ectosome release.
To image the release of ectosomes in real time, PDCD6 was fused to YFP and expressed in wild type Chlamydomonas. A fusion protein of the appropriate molecular weight 43 kD was detected with a GFP antibody in transformants 18, 26 and 35 (Figure S3D). The fusion protein was distributed uniformly throughout the flagella of gametes (Figure 2E). For visualization of ectosome release we took advantage of the increase in the release of flagellar ectosomes stimulated by the mating reaction in Chlamydomonas [9]. Gamete formation is induced in Chlamydomonas by starving the cells for nitrogen. When gametes of + and − mating types are mixed, their flagella adhere via mating type specific agglutinin molecules, activating the gametes and inducing an increase in the formation of flagellar ectosomes. Flagellar ectosomes derived from gametes of one mating type can, likewise, stimulate ectosome formation when added to gametes of the opposite mating type. When unlabeled ectosomes of mating type CC-124 (−) gametes were added to mating type + gametes expressing PDCD6∷YFP, release of flagellar ectosomes was observed in real time (Figure 2F, 2G, Movie S1), one ectosome formed from a flagellum and floated around in the medium. Together with the immunostaining data showing newly formed ectosomes containing PDCD6∷YFP associated with the flagellar membrane (Figure 2H), these data demonstrate that PDCD6 is an ectosomal protein.
Ectosome release is involved in flagellar resorption
When flagella shorten, for example, before division or when flagella are induced to resorb with NaPPi, some axonemal proteins are transported by IFT to the cell body, where they can be re-used to build new flagella [1, 24]. The fate of flagellar membrane lipid and membrane proteins, however, is unclear. To address this issue, we labeled the flagella of WT Chlamydomonas cells with Dioc6(3), induced flagellar shortening by the addition of NaPPi, and measured the fluorescence of dye released into the medium. We observed that the intensity of fluorescence increased for 90 minutes when the flagella had shortened from 10.31±1.36 μm to 3.16±1.64 μm (Figure 3A). The fluorescence increase only occurred in flagellated wild type cells, but not in the flagella-less mutant ift88 also treated with NaPPi [25] (Figure S4). These data suggest that the lipid of flagellar membrane was released to medium during flagellar resorption.
Figure 3. Ectosome release is involved in flagellar disassembly.
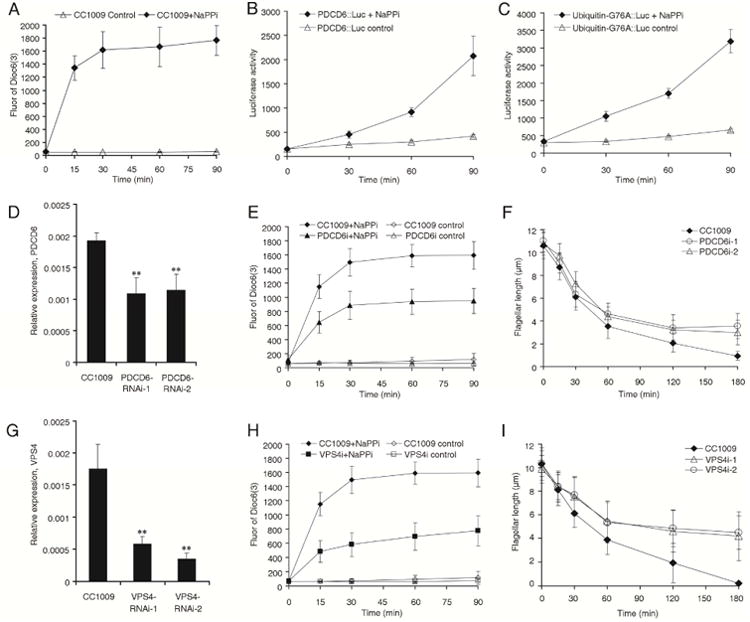
Flagella, labeled with lipid dye Dioc6(3) (A), were induced to shorten by NaPPi, and the fluorescence of Dioc6(3) in medium was measured. The flagella from the cells expressing PDCD6∷luciferase (B) or mutated ubiquitin∷luciferase (C) were induced to shorten by NaPPi. Ectosomes were isolated from the medium and their luciferase activity was measured. The fluorescence of dye in the supernatant or the luciferase activity in ectosomes increased during flagellar shortening. Figure S4 shows that ectosomes were mainly released from flagella during flagellar shortening. D). The PDCD6 mRNA level in the knockdown strains and WT (CC-1009) cells was quantified using real-time PCR. Differences between groups were analyzed by one-way ANOVA followed by Tukey post hoc test, using GraphPad Prism. A P-value of less than 0.05 was considered to be statistically significant, **p<0.01. E). Dioc6(3)-labeled flagella from PDCD6 knock down and WT cells were induced to shorten by adding NaPPi and the fluorescence in the supernatant was measured. F). Comparison of the kinetics of flagellar shortening in PDCD6 knock down cells and WT cells. G). The VPS4 mRNA level in the knockdown strains and WT (CC-1009) cells was quantified using real-time PCR. **p<0.01. H). Dioc6(3)-labeled flagella from VPS4 knock down and WT cells were induced to shorten by adding NaPPi, the fluorescence in the supernatant was measured. I). Comparison of the kinetics of flagellar shortening in VPS4 knock down cells and WT cells. Bar graphs represent mean ± SEM.
To determine whether flagellar proteins are also released to the medium during flagellar resorption, cells expressing PDCD6∷luciferase and mutated ubiquitin∷luciferase were induced to shorten their flagella by adding NaPPi to the medium, ectosomes were isolated at times after inducing flagellar shortening, and luciferase activity of the ectosomes was measured. We observed that luciferase activity increased dramatically in the ectosomes for 90 minutes when the flagella had shortened to less than half length (Figure 3B, 3C). These data suggest that some flagellar proteins are also released in ectosomes during flagellar resorption.
To determine whether ectosome release played a role in flagellar shortening, we knocked down the ectosome-enriched PDCD6 and another ESCRT related protein, VPS4, using amiRNA technology [26]. Real time PCR showed that the amount of PDCD6 mRNA in the knock down cells was less than 60% of that in wild type cells (Figure 3D). The length of flagella in the knock down cells was similar to wild type cells: flagellar lengths for the two PDCD6i strains were 10.78±1μm and 11.03±0.99μm, for VPS4i strains they were 9.88±1.18μm and 10.36±1.11μm, while the flagellar length of wild type CC1009 cells was 10.31±1.36 μm. To measure the amount of ectosomes released, the flagella of PDCD6 knock down cells and wild type cells were labeled with Dioc6(3), then the flagella were induced to shorten by adding NaPPi. Without NaPPi, both PDCD6 knock down cells and wild type cells showed little ectosome release as judged by fluorescence increase. When NaPPi was added, ectosome release increased in both PDCD6 knock down and wild type cells, but the amount of release was about half as much in PDCD6 knock down cells as in wild type cells (Figure 3E). This was also true for another ESCRT protein, VPS4: when the VPS4 mRNA level was knocked down to less than 30% (Figure 3G), ectosome release was slowed compared to wild type cells during flagellar disassembly (Figure 3H). These data suggest that the ESCRT-mediated ectosome release plays a role in the shortening of flagella although an indirect effect of ESCRT knockdown on flagellar resorption cannot be ruled out since the ESCRT machinery plays diverse roles in membrane trafficking and cytokinesis [11, 12, 27, 28].
In keeping with the reduced amount of ectosome release in the knockdown cells, we observed that the rate of flagellar disassembly was slower in the knockdown cells than in wild type cells. The original length of flagella of knock down cells and wild type cells were similar, but after 180 minutes treatment, the flagellar length of wild type cells was less than 1 μm, whereas the flagellar length of PDCD6 knock down cells was 3.57±1.1 μm or 3±1.08 μm (Figure 3F). The effect was more dramatic when the VPS4 was knocked down. When the flagella of wild type cells resorbed completely, the flagella of VSP4 knockdown cells were still 4.18±2.07 μm or 4.48±1.41 μm long (Figure 3I). These data indicate that the release of ectosomes may be involved in flagellar resorption and the ESCRT complex may participate in this process.
Based on the well characterized involvement of ESCRT proteins and ubiquitination pathway in viral budding [29, 30], we propose that in the flagella ubiquitinated proteins are clustered by a mechanism involving Alix and PDCD6 [31, 32], which can form a ternary complex with ESCRT-I [33]. These ubiquitinated proteins are then concentrated in membrane buds that are released into the medium by a mechanism involving ESCRT-III proteins and VPS4 [28, 34]. The VPS4 that depolymerizes the ESCRT-III filaments remains in the flagella and therefore was not concentrated in the ectosomes (Table 1). This might be a common mechanism involved in the turnover of membrane proteins in all cilia.
Supplementary Material
Highlights.
Ectosomes released from the flagella have a unique protein composition
ESCRT proteins mediate ectosome release and thereby influence flagellar shortening
Ectosome participates in trafficking and turnover of ciliary membrane proteins
Acknowledgments
This work was supported in part by the National Nature Science Foundation of China (Grant 31171287 to Huang K. and 31401154 to Long H.) and National Institutes of Health of USA grant GM014642 to JLR. Thanks to Fang Zhou of the Institute of Hydrobiology, C.A.S. for helping with live cell imaging.
Footnotes
Author Contributions: H.L. designed and performed the experiments in proteomic analysis and live cell imaging, and also analyzed the data and wrote the paper. F.Z. performed the experiments in knock-down and luciferase assay. N.X. performed the experiments in screening transformants. G.L. participated in expression the fusion protein of IFT46∷luciferase. D.D. performed the EM experiments and contributed critical discussion of the experiments and the data, and also editing the manuscript. J.R designed the experiments, stimulated and monitored this study. K. H designed the experiments, performed the flagellar membrane and ectosome isolation for original proteomic work, analyzed the data, and wrote the paper. All of the authors discussed the results and read and contributed to the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan J, Snell WJ. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev Cell. 2005;9:431–438. doi: 10.1016/j.devcel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Nachury MV. How do cilia organize signalling cascades? Philos Trans R Soc Lond B Biol Sci. 2014;5:369. doi: 10.1098/rstb.2013.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood CR, Rosenbaum JL. Ciliary ectosomes: transmissions from the cell's antenna. Trends Cell Biol. 2015;5:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Barr MM. Ciliary Extracellular Vesicles: Txt Msg Organelles. Cell Mol Neurobiol. 2016;36:449–457. doi: 10.1007/s10571-016-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Kaletsky R, Silva M, Williams A, Haas LA, Androwski RJ, Landis JN, Patrick C, Rashid A, Santiago-Martinez D, et al. Cell-specific transcriptional profiling of ciliated sensory neurons reveals regulators of behavior and extracellular vesicle biogenesis. Curr Biol. 2015;25:3232–3238. doi: 10.1016/j.cub.2015.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;4:e05242. doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat Cell Biol. 2012;14:38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 12.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 16.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11:709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 17.Page MD, Kropat J, Hamel PP, Merchant SS. Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell. 2009;21:928–943. doi: 10.1105/tpc.108.064907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens RE, Oleszko-Szuts S, Good MJ. Evidence that tubulin forms an integral membrane skeleton in molluscan gill cilia. J Cell Sci. 1987;88:527–535. doi: 10.1242/jcs.88.4.527. [DOI] [PubMed] [Google Scholar]
- 19.Kubo T, Kaida S, Abe J, Saito T, Fukuzawa H, Matsuda Y. The Chlamydomonas hatching enzyme, sporangin, is expressed in specific phases of the cell cycle and is localized to the flagella of daughter cells within the sporangial cell wall. Plant Cell Physiol. 2009;50:572–583. doi: 10.1093/pcp/pcp016. [DOI] [PubMed] [Google Scholar]
- 20.Maki M, Suzuki H, Shibata H. Structure and function of ALG-2, a penta-EF-hand calcium-dependent adaptor protein. Sci China Life Sci. 2011;54:770–779. doi: 10.1007/s11427-011-4204-8. [DOI] [PubMed] [Google Scholar]
- 21.Diener DR, Lupetti P, Rosenbaum JL. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr Biol. 2015;25:379–384. doi: 10.1016/j.cub.2014.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer N, Rochaix JD. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;265:888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- 23.Wright R. Fluorescence labeling of flagellar membranes. Methods Cell Biol. 1995;47:413–418. doi: 10.1016/s0091-679x(08)60839-0. [DOI] [PubMed] [Google Scholar]
- 24.Coyne B, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. II. Re-utilization of flagellar proteins. J Cell Biol. 1970;47:777–781. doi: 10.1083/jcb.47.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Deng X, Shao N, Wang G, Huang K. Rapid construction and screening of artificial microRNA systems in Chlamydomonas reinhardtii. Plant J. 2014;79:1052–1064. doi: 10.1111/tpj.12606. [DOI] [PubMed] [Google Scholar]
- 27.Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 2011;12:1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- 28.Adell MA, Migliano SM, Teis D. ESCRT-III and Vps4: a dynamic multipurpose tool for membrane budding and scission. FEBS J. 2016:13688. doi: 10.1111/febs.13688. [DOI] [PubMed] [Google Scholar]
- 29.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 32.Hurley JH, Odorizzi G. Get on the exosome bus with ALIX. Nat Cell Biol. 2012;14:654–655. doi: 10.1038/ncb2530. [DOI] [PubMed] [Google Scholar]
- 33.Okumura M, Katsuyama AM, Shibata H, Maki M. VPS37 isoforms differentially modulate the ternary complex formation of ALIX, ALG-2, and ESCRT-I. Biosci Biotechnol Biochem. 2013;77:1715–1721. doi: 10.1271/bbb.130280. [DOI] [PubMed] [Google Scholar]
- 34.Adell MA, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


