Abstract
Expression of programmed death 1 (PD-1) on CD8 T cells promotes T cell exhaustion during chronic antigen exposure. During acute infections, PD-1 is transiently expressed and has the potential to modulate CD8 T cell memory formation. Conserved Region C (CR-C), a promoter proximal cis-regulatory element that is critical to PD-1 expression in vitro, responds to NFATc1, FoxO1, and/or NF-κB signaling pathways. Here, a CR-C knockout mouse (CRC−) was established to determine its role on PD-1 expression and corresponding effects on T cell function in vivo. Deletion of CR-C decreased PD-1 expression on CD4 T cells and antigen-specific CD8 T cells during acute and chronic lymphocytic choriomeningitis virus (LCMV) challenges, but did not affect the ability to clear an infection. Following acute LCMV infection, memory CD8 T cells in the CRC− mouse were formed in greater numbers, were more functional, and were more effective at responding to a melanoma tumor than wild-type memory cells. These data implicate a critical role for CR-C in governing PD-1 expression, and a subsequent role in guiding CD8 T cell differentiation. The data suggest the possibility that titrating PD-1 expression during CD8 T cell activation could have important ramifications in vaccine development and clinical care.
INTRODUCTION
The immune-inhibitory receptor Programmed Death 1 (PD-1) is expressed on CD8 T cells upon activation (1–3). In chronic viral infections and in anti-cancer immune responses, PD-1 is highly expressed on antigen-specific T cells for the duration of the immune challenge (4–8). This high expression, combined with PD-1 binding to its ligands PD-L1 and PD-L2 (9, 10), results in CD8 T cell functional exhaustion, a cellular state characterized by reduced proliferation, cellular toxicity, and cytokine secretion (11, 12). Antibody blockade of the PD-1/PD-L interaction mediates reinvigoration of CD8 T cell function (8, 11). As such, this PD-1 immune checkpoint antibody blockade therapy is now used to treat patients with melanoma or non-small cell lung cancers (13–15). Understanding the molecular mechanisms that govern initial PD-1 induction may aid in the development of future therapies, as well as give an understanding of the context in which these therapies are applied.
A variety of factors tightly regulate Pdcd1, the gene that encodes PD-1, across multiple cell types and in response to different stimuli (reviewed in (16)). In T cells, TCR signaling induces Nuclear Factor of Activated T cells (NFAT)c1 (17) and Activator Protein-1 (18) binding to the Pdcd1 locus. TCR-mediated NFAT signaling is both necessary and sufficient to induce PD-1 expression in T cells. Other regulatory factors, including the transcription factors STAT3, STAT4 and IRF9, require TCR signaling in addition to their individual stimuli in order to augment expression of Pdcd1 (19–21).
In the mouse genome, conserved region C (CR-C) is located between 1009 bp and 1,301 bp upstream of the Pdcd1 transcriptional start site. This region is conserved across mammalian species and highly DNAse I hypersensitive (17). CR-C is a complex element that can respond to a variety of stimuli in a cell type specific manner. When bound by NFATc1 in response to TCR stimulation in CD8 T cells, CR-C is able to induce expression of a luciferase reporter in vitro (17, 19, 22). FoxO1, another transcriptional activator, also binds to CR-C and perpetuates PD-1 expression in CD8 T cells of mice that are chronically infected with lymphocytic choriomeningitis virus (LCMV) (23). In both T cells and macrophages exposed to acute activating factors, IRF9 binds to an interferon-sensitive response element in CR-C and promotes PD-1 expression (20, 21). Lastly, in murine macrophages activated through TLRs 2 or 4, CR-C binds NF-κB in a manner necessary for the transient induction of PD-1 in these cells (22).
CR-C also undergoes dynamic epigenetic modifications that are concordant with PD-1 expression. CpG dinucleotides within CR-C are highly methylated in naïve CD8 T cells. DNA methylation is associated with gene silencing (24). During the initial stages of an acute infection with LCMV, the CR-C region in antigen-specific CD8 T cells becomes demethylated as PD-1 is expressed, suggesting an increase in accessibility at the locus (25, 26). Additionally, CR-C chromatin gains the histone mark histone 3 lysine 27 acetylation (H3K27Ac) following T cell stimulation (27), a modification associated with active enhancers (28). Following resolution of an acute infection and loss of PD-1 expression, CR-C loses its active chromatin modifications and gains epigenetic marks associated with repressive chromatin structures, including H3K9me3, H3K27me3, and H4K20me3 (27). CR-C CpG loci also become remethylated at this stage. Thus, CR-C is a highly active and dynamic regulatory region, implicating it as a major control element of PD-1 expression.
PD-1 knockout mice exhibit altered immune cell development and function. Such mice displayed a higher frequency of thymocytes and early thymic emigrants (29, 30) and were more susceptible to autoimmune diseases (31, 32). Moreover, loss of PD-1 resulted in a much stronger memory response to an acute infection, in both number and effector function of cells produced (33). In chronic infections, PD-1 knockout CD8 T cells were more functionally active and induced fatal circulatory failure due to an over-active immune response (34). While these studies examined the complete loss of PD-1 on T cell responses, it is not known how Pdcd1 cis-regulatory elements alter PD-1 expression in vivo and influence T cell development or immune responses.
To derive a functional role for one critical element in vivo, mice carrying a genetic deletion of CR-C were generated (termed CRC− mice herein). T cells in CRC− mice appear to develop normally and there is no increase in susceptibility to autoimmunity. In cell culture, and in acute and chronic LCMV viral infection, CR-C deletion resulted in significant loss of PD-1 expression on both virus-specific CD8 T cells and CD4 T cells following activation. In CRC− mice bearing melanoma tumors, PD-1 expression was decreased on tumor-infiltrating T cells, as well as antigen-specific T-cells in the tumor draining lymph nodes. This resulted in a greater anti-tumor response and slower tumor growth. Although CRC− mice produced fewer antigen-specific CD8 T cells during primary infection, they displayed more effective memory responses in both quantity and quality of memory cells. Collectively, these data demonstrate that CR-C is critical to the expression of PD-1 in vivo and that alterations in PD-1 expression can mediate changes in the immune effector:memory axis.
MATERIALS AND METHODS
Generation of CRC knockout mice
Homology targeting fragments (5′ and 3′, respectively) surrounding CR-C (chr1:95950331-95953209 and chr1:95949964-95950330) were inserted into the pM30 targeting plasmid (35) using SacII and MluI sites (upstream region) or Nhe1 and Kpn1 sites (downstream region). The pM30 plasmid was previously modified to contain an MluI site and a second LoxP site. A 400 bp fragment containing CR-C (chr1:95949964–95950330) was inserted between the LoxP sites using SalI. This yielded a plasmid-targeting construct that contained the CR-C fragment flanked by two LoxP sites, within the homology arms of the Pdcd1 locus. All insertion fragments were generated by PCR, and inserted using In-Fusion Dry-Down PCR Cloning Kit (Clontech). The Brigham and Women’s Hospital Transgenic Core Facility directed by Dr. Arlene Sharpe used the resulting pM30_PD-1_CRC plasmid to generate homologously recombined JM8.N4 ES cells (trans-NIH Knock-Out Mouse Project (KOMP) repository- www.komp.org), which are derived from the C57Bl/6 strain (36). These recombined cells were used to produce CRCfl/fl mice used in this study. CRCfl/fl founder mice were bred to B6-Tg (CAG-FLPe) mice to remove the neo cassette (37), and subsequently maintained on a C57Bl/6 background. To delete CR-C, mice were crossed with C57BL/6-Tg(Zp3-cre)93Knw/J mice to generate CRC− mice. Mice were genotyped using the following CR-C primers: 5′-CCTGAGCATGCCAGAAAGACA and 5′-CTAACACCAGGCTGGGGTAGACTC. Wild-type C57Bl/6 mice (Jackson laboratories) were used as controls for CRC− experiments, where indicated. Mice that were 6–8 weeks old were used for experiments unless otherwise noted. For establishing bone marrow chimeras, 107 bone marrow cells from CRC− or B6.SJL-Ptprca Pepcb/BoyJ (WT mice carrying the CD45.1 allele) were adoptively transferred into B6129S7-Rag1tm1Mom/J hosts (Jackson laboratories) that had been irradiated with a dose of 500 rads. Where indicated, B6.PL-Thy1a/CyJ mice (WT mice carrying the CD90.1 allele) were used as hosts in adoptive transfers. All mice were cared for in accordance with approved Emory University Institutional Animal Care and Use Committee protocols.
Experimental Autoimmune Encephalitis (EAE)
The EAE inducing peptide MOG35–55 (MEVGWYRSPFSRVVHLYRNGK) was synthesized in house on a Prelude Peptide Synthesizer as previously described (38). For EAE induction, 200 μg of the MOG35–55 peptide was emulsified in Complete Freund’s Adjuvant (CFA, desiccated M.Tb concentration at 5 mg/ml) and 150 μl of the emulsion was injected subcutaneously on days 0 and 7. On days 0 and 2, 300ng of pertussis toxin was injected intraperitoneally. EAE scoring was performed on the 5-point scale: 0=not sick, 0.5= distal limp tail, 1= proximal weak tail, 2= hind limb weakness, 3= complete hind limb paralysis, 4= inability to flip over, 5=death or moribund. Mice in EAE experiments were sacrificed if there was >25% body weight reduction for 72 hours.
Melanoma model
The B16–F10.gp33 melanoma cell line was kindly supplied by Dr. R. Ahmed with permission from Dr. H. Pircher (Universitätsklinikum Freiburg) (39). These cells were cultured with Dulbecco’s modified Eagles’s medium (DMEM) containing 10% fetal bovine serum (FBS), 1% glucose and 100 U/ml Penicillin/Streptomycin at 37 °C. Low passage number B16-F10.gp33 melanoma cells were harvested and then resuspended at 107 cells per ml in serum free DMEM media. 1x106 cells were injected into the study animals subcutaneously on the right flank. Tumor growth was measured daily using calipers. Tumor infiltrating lymphocytes were isolated from whole tumors and analyzed following purification by a Ficoll gradient.
Virus Infections and Plaque Assays
Viral stocks of LCMV strains Armstrong and Clone-13 were produced as described previously (40). Mice were injected with 2x105 pfu Armstrong intraperitoneally or with 2x106 pfu LCMV-Cl13 intravenously in the lateral tail vein. For day 28 chronic experiments, mice were injected intraperitoneally with 200 μg anti-CD4 antibody (clone GK1.5, BioXcell, West Lebanon, NH) at the day before and the day after viral infection to deplete CD4 T cells as described (41).
Vero cells (ATCC CCL-81) were cultured in DMEM containing 10% FBS and 100 U/ml Penicillin/Streptomycin at 37 °C and used for viral plaque assays as previously described (42). Briefly, 5x105 cells were plated in each well of a 6 well plate and allowed to rest for one day. Mice were bled by lancet from the submandibular vein. Serum samples were extracted from whole blood by centrifugation and serially diluted in 10-fold increments into DMEM +10%FBS. Diluted serum was adsorbed onto cells for one hour at 37°C. Samples were subsequently overlaid with 199 media containing 0.5% agarose and then incubated at 37°C. On Day 4, 199 media/agarose containing 0.002% Neutral Red were added to each well to stain infected/dead cells. Plaques were counted on day 5 and viral titers calculated.
Ex vivo T cell activation
Naïve CD8 T cells were isolated from spleens of healthy mice by magnetic activated cell sorting (MACS), using the CD8a+ T Cell Isolation Kit according to manufacturer’s instructions (Miltenyi Biotec, Auburn, CA). Anti-CD3 antibody (clone 145-2C11, Fisher Scientific) was adhered to a 24 well plate at 5 μg/ml in sterile PBS. The plate was sealed with parafilm and incubated at 37 °C for 2 hours to adhere antibodies. The plate was then placed at 4° C overnight. On the next day, the plate was washed twice with 500μl sterile PBS, and 0.5x106 primary CD8 T cells were incubated in each well with 10 μg/ml soluble anti-CD28 antibody (clone 37.51, Fisher Scientific) at 37 °C for the time points indicated. Cell samples were harvested and stained for flow or stored as a pellet at −80°C for RT-PCR. In some experiments a variable amount of anti-CD3/CD28 antibodies were used as indicated.
Flow cytometry and Antibodies
Cells were stained for flow cytometry in FACS buffer (PBS, 1% BSA, and 1 mM EDTA) and antibodies at 4°C for 30 minutes, and then fixed in 1% paraformaldehyde for 30 minutes. Antigen-specific CD8 T cells were gated by CD8+CD44+CD62L-Tetramer+ (including gp33, gp276 and np396 tetramers). Activated CD4 T cells were gated on CD4+CD44+CD62L− markers. Antibodies used included CD4 PerCP-Cy5.5 (clone RM4.5, Tonbo Biosciences, San Diego, CA), CD8 FITC (clone 53–6.7, Tonbo Biosciences), CD44 APC-Cy7 (clone IM7, BioLegend, San Diego, CA), CD62L Alexa Fluor 700 (clone MEL-14, BioLegend, San Diego, CA), CD69 PE-Cy7 (Clone H1.2F3, BioLegend, San Diego, CA) CD127 BV510 (Clone SB/199, BD Horizon), PD-1 PE (clone RMP1–30, BioLegend, San Diego, CA). gp33 var C41M (KAVYNFATM), gp276 (SGVENPGGYCL), and np396 (FQPQNGQFI) biotinylated monomers on H-2Db were obtained from the NIH Tetramer Core facility at Emory University and tetramerized to streptavidin-APC (Prozyme). Flow cytometry was performed on a BD LSR II and analyzed using FlowJo 9.6.4 software. Intracellular cytokine staining was performed using the Fixation and Permeabilization Solution Kit with BD GolgiPlug (Becton, Dickinson and company) according to the manufacturer’s protocols.
Real-time PCR
Total RNA was isolated with the RNeasy Mini Prep kit (Qiagen, Germantown, MD). cDNA was prepared from 1 μg total RNA with SuperScript II reverse transcriptase (Invitrogen). mRNA levels were quantified in technical duplicates by real time PCR. mRNA levels were calculated relative to expression of 18s rRNA. Primers used for real-time PCR include the following: PD-1 forward 5′-GCTGAAGGCTCCTCCTTCTGACAT; PD-1 reverse, 5′-AGATATCCCAGCCCCTCGCCC; IL-2 forward, 5′-ACCCACTTCAAGCTCCACTTCA; IL-2 reverse, 5′-TGGCCTGCTTGGGCAAGTAAA; 18s forward, 5′-GTAACCCGTTGAACCCCATT; 18s reverse, 5′-CCATCCAATCGGTAGTAGCCG.
Bisulfite Sequencing
Genomic DNA was isolated from antigen specific CD8 T cells of mice infected with LCMV Armstrong 5 days post infection. DNA was bisulfite converted using the EpiTect bisulfite kit (Qiagen). Bisulfite-converted DNA was amplified through PCR and then cloned using the TOPO TA cloning kit (Life Technologies). Twelve colonies from each sample were analyzed by DNA sequencing. Primers used for cloning and sequencing 5′-GGTGGGTTTTTATTTTTTAGGGATTGAGG and 5′-CTAAAACTAAAACCAAACTCTTATCCC.
Statistical Analyses
Student’s T-tests were carried out to assess the significance of differences in results where indicated. Two-way ANOVA was used for comparisons in ex vivo stimulations and viral time-courses, as well as comparing tumor growth across time. Wilcoxon rank sum test was used to compare survival rates. Fisher’s exact test was used to compare DNA methylation across different populations in bisulfite conversion experiments. Statistical analyses were performed using data combined from independent groups of mice.
RESULTS
Deletion of CR-C does not alter thymocyte maturation
To characterize the effects of the CR-C region on the expression of PD-1 and the development of immune responses, mice containing a floxed CR-C conditional allele (CRCfl) were generated (Fig 1A). These mice were crossed to Cre-zp3 transgenic mice, resulting in germ-line deletion of the locus and generation of a mouse strain lacking CR-C. These mice were termed CRC−. Using a PCR based assay, appropriate deletion of CR-C was observed in total splenic CD8 T cells from CRC− but not in WT mice (Figure 1B).
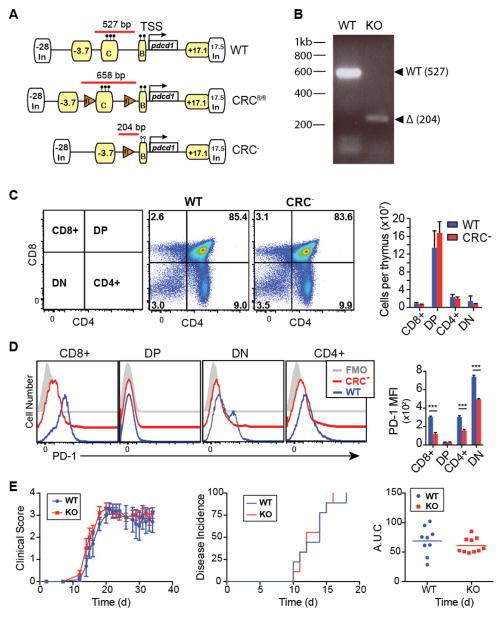
Figure 1. Deletion of CR-C does not affect immune development.
A) Schematic showing the Pdcd1 region in CRCfl/fl and CRC− mice compared to wild-type B6. Known regulatory elements are labeled in yellow (17, 19). Chromatin insulator sites are labeled with white boxes. B) Representative genotyping gel for Cre− and Cre+ CRCfl/fl, WT, or CRC− mice. PCR amplicons and relative sizes for genotyping are shown using red bars in (a). C) Representative flow plots and characterization of frequencies of different T cell populations collected from the thymi of naïve, 6-week old WT or CRC− mice. Absolute numbers of cells within each thymus were calculated using frequencies of each population out of a sample of 2 x106 cells from each thymus, multiplied by the total number of cells per thymus. N=6. D) Representative histograms and quantification of PD-1 expression on each population of immature thymocytes from (C). E) Average clinical score of EAE in WT or CRC− mice (left), time of onset to disease symptoms per mouse (middle), and total disease severity as measured by area under the curve (right). Graphs indicate mean plus standard deviation. Two groups of 5 mice per genotype were analyzed. Panels C and D were analyzed for significance by Student’s T test. Statistical significance for Panel E was determined using a Mann-Whitney test. *** P<0.001.
To begin the characterization of these mice and to determine if deletion of CR-C affected T cell development, thymocytes from naïve mice were counted and analyzed by flow cytometry (Figure 1C). The numbers of double negative (DN), double positive (DP), CD4+, and CD8+ thymocyte populations were unaffected by the deletion CR-C, indicating that normal T cell development occurred. This differs significantly from the phenotype seen in PD-1−/− mice, which have increased frequency of both DN and DP thymocytes due to disrupted positive and negative selection processes mediated by PD-1 (29, 30). However, changes in PD-1 expression on developing thymocytes were observed in CRC− mice. PD-1 expression on DN cells (in which PD-1 is usually highly expressed on the DN4 population (43)), as well as on both CD4 and CD8 single positive immature thymocyte populations was significantly reduced in CR-C knockout cells (Figure 1D), indicating that CR-C was necessary for full and normal thymic PD-1 expression. The residual levels of PD-1 on these cell populations may be sufficient to facilitate the positive- and negative-selection associated anergy that is mediated by PD-1 (29, 30) and account for the differences in T cell development in these mice compared to PD-1−/− mice.
PD-1 expression on thymocytes is necessary for development of central and peripheral tolerance (30, 32). To test if the observed differences in PD-1 expression in the CRC− mice led to a breakdown of tolerance despite comparable numbers of developing thymocytes, mice were tested for susceptibility to developing immunity to the self-antigen MOG using experimental autoimmune encephalitis (EAE). The clinical scores of EAE in CRC− mice were not significantly greater than in the WT (Figure 1E). This indicates that the CRC− mice are not pre-disposed to autoimmunity or a breakdown of immune tolerance, despite lower thymic expression of PD-1. Thus, given the normal numbers of immature thymocyte populations and no increased susceptibility to a model of autoimmune disease, CRC− mice appear to have normal immune systems.
CR-C is required for ex vivo expression of PD-1
In promoter reporter assays, CR-C was shown to be a critical regulator of Pdcd1 expression (17, 19), suggesting that it is necessary for activating expression in responses to TCR stimulation. To test if CR-C was necessary for ex vivo TCR-mediated expression of Pdcd1, magnetically enriched primary splenic CRC− or WT CD8 T cells (Figure 2A) were stimulated with anti-CD3/CD28 beads in culture for 24 h. In WT mice, ex vivo induction of Pdcd1 mRNA peaked at 24 hours after activation (Figure 2B). In contrast, CRC− CD8 T cells failed to fully induce Pdcd1, increasing mRNA expression only 3 fold over baseline, compared to over 15 fold seen in the WT cells. Importantly, both wild-type and knockout populations induced very high and similar levels of IL-2 mRNA (Figure 2B, right), indicating that comparable activation of these cells occurred, and the lower PD-1 levels were not due to a failure to integrate TCR signaling. To correlate surface expression with mRNA levels, CD8 T cells were also analyzed by flow cytometry on days 1, 2, and 4 after activation. Surface expression of PD-1 peaked on WT cells within 24 hours and subsequently declined over time to day 4 (Figure 2C). In CRC− cells, PD-1 levels were significantly lower than in WT cells at all times after Day 0. Using CD69 as a marker for T cell activation, both WT and knockout populations showed comparable levels of stimulation (Figure 2D), indicating that failure to express PD-1 was not due to failure to stimulate T cells in the CRC− cultures.
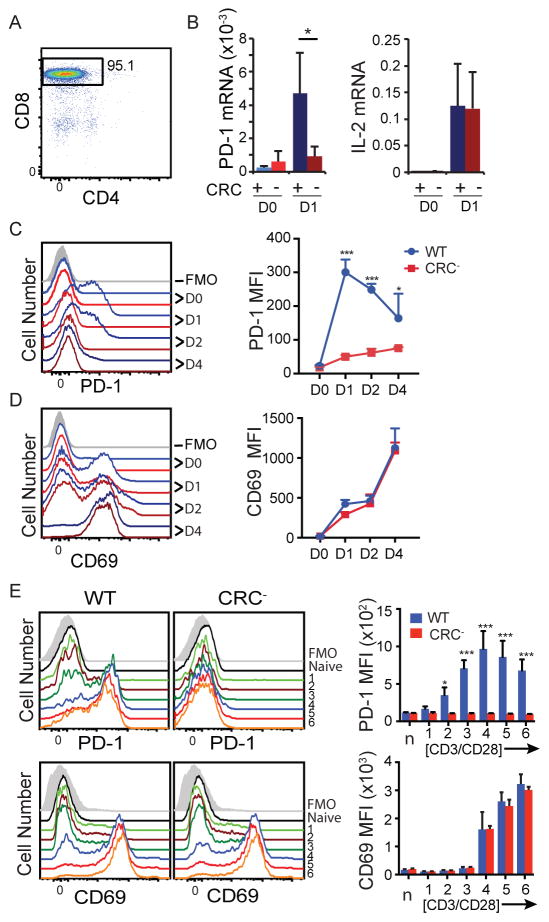
Figure 2. CRC is necessary for PD-1 expression during ex vivo stimulation.
A) Representative enrichment by MACS of CD8 T cells from total splenocytes used for ex vivo experiments. B–D) Enriched CD8 T cells were stimulated with CD3/CD28 antibodies. B) mRNA was harvested at 0 and 24 hours, and analyzed by real-time PCR. Values for Pdcd1 and IL2 mRNA are graphed as a percentage of 18s rRNA. C and D) Cells were harvested at 0, 24, 48, and 96 hours after stimulation as above and stained for flow cytometry. Representative plots (left) and MFI (right) at each time point are shown for PD-1 (C) and CD69 (D). Data are graphed as mean plus standard deviation, and represent six independent experiments. E) Naïve CD8 T cells were enriched by MACS and stimulated with the following doses of anti-CD3/CD28 antibodies: 0.04/0.08 μg/ml (1), 0.2/0.4 μg/ml (2), 1/2 μg/ml (3), 2.5/5 μg/ml (4), 5/10 μg/ml (5), and 10/20 μg/ml (6). Cells were harvested at 24 h after stimulation and stained for flow cytometry. Representative flow plots for both WT and CRC− cells are shown on the left, and average plus standard deviation from three independent replicates is graphed to the right. Panel B used a Student’s T test and Panels C, D, and E used a two-way ANOVA to determine statistical significance. * P< 0.05; *** P<0.001
To determine if increased stimulation of CRC− cells could invoke a WT level of PD-1 induction, CRC− and WT cells were incubated with various doses of anti-CD3/CD28 antibodies for 24 h (Figure 2E). The results showed that despite a 4 fold higher concentration of stimulant, CRC− cells did not show an appreciable increase in PD-1 expression at this time point. Together, these data indicate that CR-C is necessary in culture to induce PD-1 expression through TCR stimulation, and redundant mechanisms such as those mediated by Pdcd1’s distal enhancer regions, which also bind NFATc1 (19), are insufficient to induce PD-1 expression under these experimental conditions.
CR-C is necessary for normal PD-1 expression in acute infection
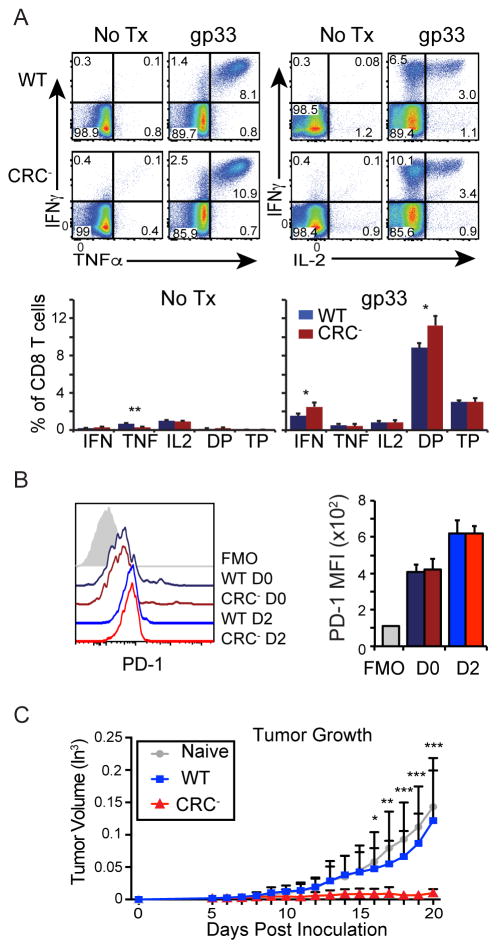
To assess the function of CR-C in vivo, CRC− mice were infected with LCMV Armstrong, which produces an acute infection that is typically cleared within eight days (12). Viral load and the frequency of LCMV specific CD8 T cells were similar between CRC− and WT mice (Figures 3A and B). However, Pdcd1 mRNA expression was reduced by greater than 50% on CD8 T cells from acutely infected CRC− mice compared to WT mice at day 6 after infection (Figure 3C). Irrespective of the strain, PD-1 mRNA expression in naïve CD8 T cells were similar (Figure 3C). Flow cytometry analysis of CD8 T cells, using the gating strategy shown in Figure 3D, showed a nearly 50% loss of PD-1 expression on virus-specific CD8 T cells at both day 4 (Figure 3E) and day 6 (Figure 3F). Differences in PD-1 expression were not due to changes in levels of T cells activation, as CD69 and KLRG1 expression profiles at these time points were similar between CRC− and WT mice (Figure 3E and F). These data argue that CR-C is necessary for maximal PD-1 expression following acute activation of T cells in vivo.
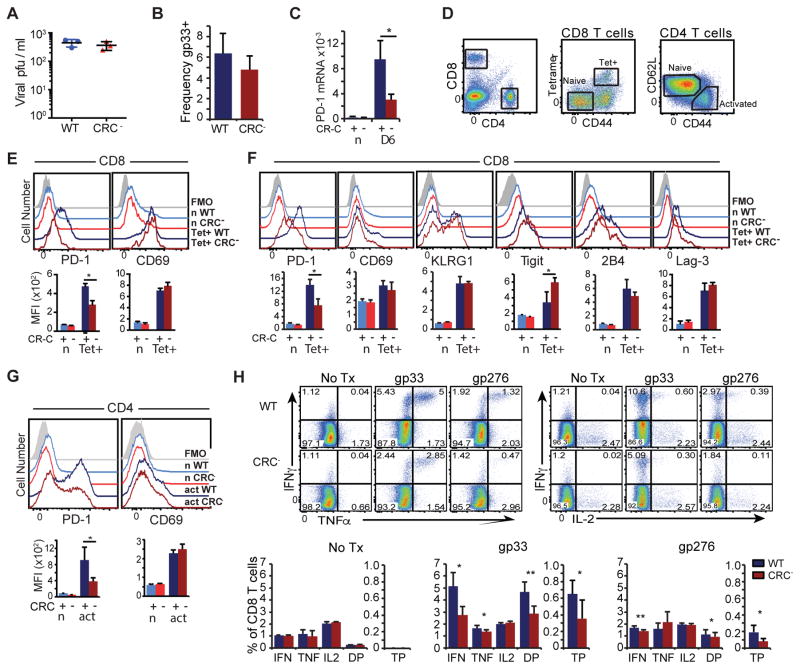
Figure 3. Acute viral infection induces PD-1 through CR-C.
A) Frequency of LCMV gp33-specific CD8 T cells as a percentage of total CD8 T cells from WT (Blue) or CRC− (Red) mice day 6 PI with LCMV Armstrong. B) Viral titers in the serum of LCMV Armstrong infected animals at day 4. C) PD-1 mRNA as a % of 18s rRNA from virus-specific CD8 T cells collected from WT (blue) and CR-C (red) mice infected with LCMV Armstrong at days 0 and 6. D) Gating strategy showing naïve (n) and tetramer+ (Tet+) populations of CD8 T cells, or naïve (n) and activated (act) populations of CD4 T cells. Representative flow cytometry histograms and quantification of MFIs from CD8 T cells collected from WT or CRC− mice at day 4 (E) or day 6 (F). Flow analysis of CD4 T cells at Day 6 PI is shown in (G). H) CD8 T cells from day 6 LCMV Armstrong infected mice were stimulated with no peptide, gp33, or gp276 ex vivo in the presence of brefeldin A for 5 hours. Frequency of IFNγ, TNFα, and IL-2 single positive cells, cells expressing any two cytokines (DP), and triple positive (TP) cells as a proportion of all CD8 T cells were graphed below. For panels A, B, C, D, F, and G, four groups of 3 or more mice/genotype were used. For panels E and H, two groups of 3 mice per genotype were used. Graphs represent mean plus standard deviation for individual representative experiments. Student’s T test was used to determine significance with * P< 0.05; ** P<0.01.
Loss of PD-1 expression has been shown to result in compensatory increased expression of other immune-inhibitory receptors (44). To determine if CR-C deletion and its effects on PD-1 expression affected other inhibitory pathways, surface expression of other immune-inhibitory receptors (Tigit, 2B4, and Lag-3) was analyzed. While surface levels of 2B4 and Lag-3 showed no significant changes in WT compared to the CRC− virus-specific T cells, expression of Tigit was increased in the CRC− animals (Figure 3F). It is unlikely that deletion of a regulatory element in the Pdcd1 locus would directly affect expression of Tigit. Instead, this change agrees with previous reports, suggesting that an intricate feedback network may regulate expression of some immune-inhibitory receptors (44).
As PD-1 is also significantly induced on CD4 T cells during acute infection (45, 46), changes in PD-1 expression were also examined on these cells on day 6 after acute LCMV infection. Similar to CD8 T cells, CRC− CD4 T cells showed a greater than 50% loss of PD-1 expression compared to WT (Figure 3G). This indicates that CR-C is also necessary in CD4 T cells to induce maximal induction of PD-1 in an acute viral infection. As in CD8 T cells, CRC− and WT CD4 T cells showed similar levels in CD69 expression (Figure 3G, right), indicating comparable activation. Thus, CR-C is important for induction of PD-1 in both CD4 and CD8 T cells.
CR-C-induced PD-1 expression is necessary for maximal effector T cell formation
To determine if there were functional consequences resulting from lower early PD-1 expression, the ability of cells from LCMV Armstrong infected CRC− mice to secrete cytokines was measured by intracellular cytokine staining (ICCS). At day 6 post infection, a lower frequency of CD8 T cells from CRC− mice secreted either IFNγ or TNFα in response to LCMV peptides gp33 or gp276 compared to WT (Figure 3H). Although, the frequency of cells secreting IL-2 was comparable between strains, the frequency of cells secreting at least two cytokines or all three cytokines in response to gp33 or gp276 stimulation was decreased in cells from CRC− animals (Figure 3H). Thus, concurrent with lower PD-1 expression, there were fewer poly-functional virus-specific effector CD8 T cells at day 6 in CRC− mice than in WT. This suggests that optimal cytokine responses are governed in part by CR-C mediated PD-1 expression.
Changes to PD-1 expression in CRC mice are cell intrinsic
Changes in PD-1 expression in CRC− mice were observed on CD4 and CD8 T cells during an immune response, and frequency of some effector cell populations was altered. However, because PD-1 was deleted in all cells, this raises the possibility that the loss of expression and these subsequent changes may be the result of variations in the cellular environment. To determine if the changes to PD-1 expression observed on CRC− CD4 or CD8 T cells were cell intrinsic, mixed bone marrow chimeras using CD45.1 WT and CD45.2 CRC− bone marrow were established in sublethally-irradiated RAG1−/− hosts. CD8 T cells carrying the WT allele (CD45.1) or CRC− CD8 T cells (CD45.2) were analyzed from the spleens of established chimeric mice 6 days after infection with LCMV Armstrong (Figure 4A). Total WT and CRC− CD4 and CD8 T cells were represented in equal numbers, indicating that there was no competitive advantage/disadvantage or survival to the mutant T cells (Figure 4A, left). Fewer virus-specific CRC− cells were found within each chimera compared to WT virus-specific cells (Figure 4A, right), indicating that these cells are less capable of proliferating or responding to virus than their WT counterparts within the same inflammatory environment. Furthermore, in agreement with Figure 3, surface expression of PD-1 on CRC− virus-specific CD8 T cells was approximately 50% lower compared to WT cells within the same chimeric animals (Figure 4B). As both WT and CRC− cells were derived from the same environment, cellular stimulation was identical, as indicated by similar up-regulation of the activation marker, KLRG1 (Figure 4B). Moreover, within the chimeras CRC− CD4 T cells expressed less PD-1 on their surface than WT CD4 T cells (Figure 4C). The activation marker CD69 showed a slight decrease in expression in CRC− cells (Figure 4B and 4C). However, because CD69 is expressed fully at much earlier time points (as shown in Figure 3D and 3E), the biological significance of this is not known. Cumulatively, these data demonstrate that CR-C is intrinsically necessary for full PD-1 expression in an acute inflammatory environment.
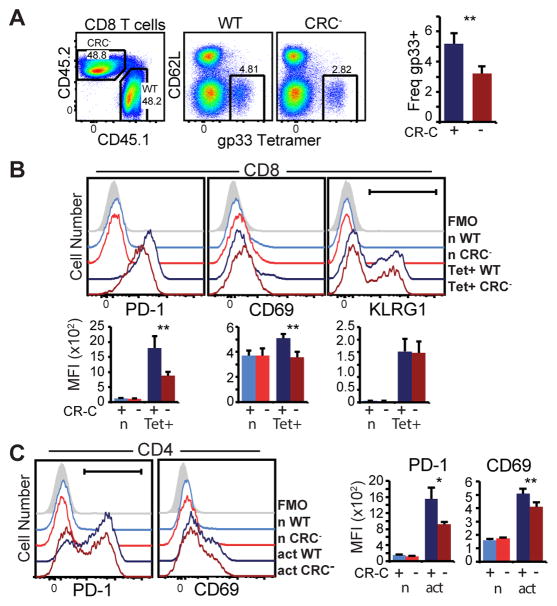
Figure 4. Changes to PD-1 expression in CR-C mice are cell intrinsic.
Bone marrow chimeras were established in RAG−/ − mice irradiated with 500 rads. 107 WT (CD45.1) or CRC− (CF45.2) bone marrow cells from the femurs of healthy mice were adoptively transferred into irradiated hosts. 6 weeks after transfer, mice were bled to ensure chimerism and then infected with LCMV Armstrong. Splenocytes from infected animals were analyzed at day 6 post infection. A) Flow cytometry plots showing WT (CD45.2) and CRC− (CD45.1) CD8 T cells in chimeric animals at day 6, and gp33 tetramer-specific cells within each population. The frequency of virus-specific cells in WT (CRC+) and CRC- populations within each chimera are shown to the right. B) Flow cytometry histograms (top) and average MFI (bottom) in naïve (CD44+ CD62L−) or virus-specific CD8 T cells within WT (CD45.1) or CRC− (CD45.2) populations. C) Flow cytometry histograms for CD4 cells from the above animals. Two groups of 3–4 host mice with independent donors of each genotype were used for these experiments. Mean and standard deviation of a representative experiment is graphed. Paired Student’s T test was used to calculate significance. * P< 0.05; ** P<0.01
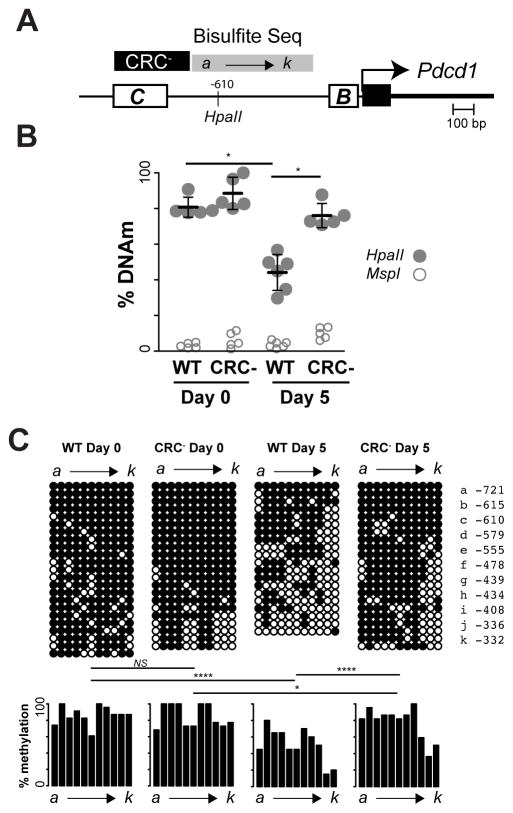
CR-C coordinates downstream epigenetic changes
CR-B is a promoter proximal element that binds AP-1 in response to T cell activation. The methylation of cytosines upstream of CR-B (−332 to -721 bp from the promoter) and those within CR-C itself are highly correlative with PD-1 expression (25, 26). As CR-C is required for TCR-mediated induction, this raises the question as to whether CR-C is required for subsequent alterations in DNA methylation at other regions, such as the sequences upstream of CR-B or whether these epigenetic changes could occur in the absence of CR-C. To address this, a DNA methylation-sensitive restriction enzyme digest assay within the above region was performed. Using the relative frequency of uncut (methylated) to cut (unmethylated) DNA from T cells of naïve mice, 80–100% methylation was observed at the queried CpG site (Figure 5B). As expected, the antigen-specific CD8 T cells from WT mice lost over 50% methylation at this site by day 5 post infection with LCMV Armstrong. In contrast, cells from the CRC− mouse showed no significant loss of DNA methylation.
Figure 5. CR-C coordinates epigenetic changes at the PD-1 locus.
A) Schematic of bisulfite sequencing primers relative to the CR-C and PD-1 promoter. The region deleted in the CRC− mice is also indicated. B) % of DNA methylation at a CpG site (−610 from the promoter) within the bisulfite sequencing region, as identified by its inability to be cleaved by the methylation sensitive restriction enzyme HpaII at days 0 and 5 after LCMV Armstrong infection. C) Clonal bisulfite sequencing from virus-specific CD8 T cells at days 0 and 5 after LCMV Armstrong in WT or CRC− mice. Each lettered column represents a specific CpG locus as designated in the side bar. Each row represents an allele. Black circles indicate methylated CpGs, whereas white circles are unmethylated. Frequency of methylation and statistical differences between groups are shown below. Panel B used two groups of 2–3 mice for each experiment and Panel C used two mice from independent experiments. Error bars in (B) represent standard deviation. The Fishers-Exact test was used to determine significance. * P< 0.05; *** P<0.001
When multiple CpGs within the region were analyzed by clonal bisulfite sequencing, a large loss of overall methylation across the entire region was observed by day 5 in cells from WT mice (Figure 5C). Although some demethylation occurred at the locus in the CRC− mouse, cells from these mice retained significantly more methylation after LCMV Armstrong challenge compared to the WT mouse. These data suggest that the loss of DNA methylation in this region is coordinated by the actions of CR-C.
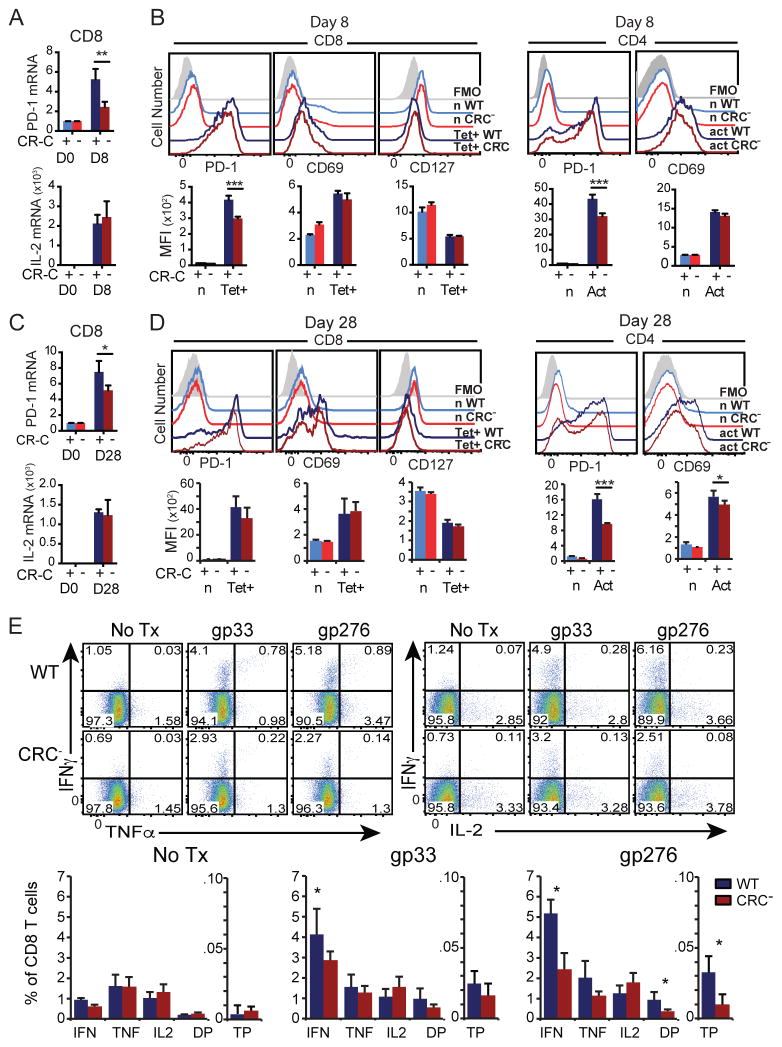
CR-C deletion decreases PD-1 expression in chronic infection
The expression and function of PD-1 as an immune-inhibitory receptor is exemplified during chronic infections (11, 41). To determine if CR-C is involved in PD-1 regulation during chronic antigenic stimulation, WT or CRC− mice were infected with LCMV Clone-13 under conditions that lead to chronic infection. At day 8 post infection, virus-specific CD8 T cells from CRC− mice displayed a decrease of Pdcd1 mRNA (Figure 6A). IL-2 mRNA expression was unchanged in CRC− mice (Figure 6A, bottom), indicating that cellular activation and effector function were not affected by the CR-C deletion.
Figure 6. CR-C is not necessary for PD-1 expression in chronic viral infection.
WT or CRC− mice were infected with LCMV Cl-13 for 8 (A–B) or 28 (C–E) days, and spleens were harvested for analysis. A and C) mRNA from MACS-purified CD8 T cells, graphed as a percentage of 18s rRNA. B and D) Representative flow cytometry histograms (top) and MFI quantifications (below) for naïve (n) or tetramer-specific (Tet+) CD8 T cells (left), or naïve and activated (act) CD4 T cells (right). E) Total splenic cells were stimulated with no peptide, gp33, or gp276 ex vivo in the presence of brefeldin A for 5 hours, and then stained by ICCS for flow cytometry. Frequency and estimated total number of cells per spleen of IFNγ, TNFα, and IL-2 single positive cells, cells expressing any two cytokines (DP), and triple positive (TP) cells out of all CD8 T cells are graphed below. Two groups of 4 mice per genotype were used for these experiments. Graphs show mean of a single group plus standard deviation for each genotype. Student’s T tests were used to assess statistical significance. * P< 0.05; ** P<0.01; *** P<0.001
PD-1 surface expression on virus-specific tetramer+ CD8 T cells during chronic infection was also analyzed using flow cytometry. As with mRNA levels, PD-1 surface levels were decreased in CRC− mice at day 8-post infection (Figure 6B, left). Similarly PD-1 expression was also decreased on CD4 T cells at this same time point (6B, right). The loss of PD-1 expression on CD8 T cells in chronically infected CRC− mice was less severe than that seen during acute infection. Again, as in acute infection, there was no overall change in cell activation to account for the differences in PD-1 expression as measured by up-regulation of CD69 or loss of the IL-7 receptor CD127 on CD8 T cells or induction of CD69 on CD4 T cells (Figure 6B).
PD-1 expression was also detected on CD8 and CD4 T cells of chronically infected animals at day 28 post infection with Clone-13 virus. Similarly to day 8, PD-1 mRNA was decreased at day 28, although to a lesser degree (Figure 6C). However, unlike at day 8, CRC− CD8 T cells at day 28 did not show a significant loss of PD-1 surface expression, although PD-1 expression did trend lower in correlation with changes in mRNA (p=0.06) (Figure 6D, left). This may indicate that other regulatory elements, such as the −3.7 and +17.1 distal enhancer regions (19) are sufficient to induce or maintain PD-1 on CD8 T cells at late time points in chronic infection. Collectively, these data indicate that CR-C is necessary for early PD-1 expression during acute infection and early chronic infection prior to the establishment of T cell exhaustion, but is not necessary for high PD-1 expression seen on exhausted CD8 T cells during prolonged antigen exposure. In contrast, activated CD4 T cells showed a decrease in PD-1 expression at day 28 after chronic infection (Figure 6D, right), indicating that CR-C continues to play a role in PD-1 expression on certain cell types even during prolonged antigen exposure.
The frequency of cytokine secreting effector CD8 T cells was also measured at day 28 after chronic Clone-13 infection. In both genotypes, there were fewer CD8 T cells in the chronic infection capable of secreting cytokines in response to gp33 or gp276 than seen in the acute infection (Fig 3H compared with 6E). This is most evident in the populations of polyfunctional double-positive or triple-positive cells responding to each peptide, which were depleted (Figure 6E compared to 3H)). These results are indicative of CD8 T cell exhaustion in both mutant and WT mice. However, in this chronic infection model, the frequency of cytokine secreting cells was depressed even further in CRC− mice compared to wild-type (Figure 6E), suggesting that the defect in effector cell formation that is established early during initial antigen response in these mice may result in lower T cell numbers throughout the duration of infection.
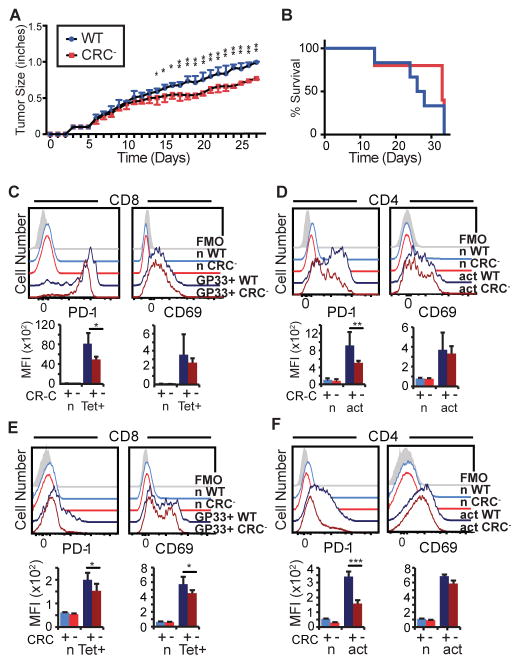
CR-C deletion results in a stronger anti-tumor response
PD-1 has been shown to play an important role in modulating effector functions during anti-tumor immune responses (15, 18, 47). To determine if CR-C-induced PD-1 expression affects cellular function in this inflammatory setting, CRC− mice or WT controls were inoculated with B16.f10-gp33 cells, a melanoma cell line that expresses the LCMV gp33 peptide, making it possible to track tumor-specific CD8 T cells using established LCMV peptide:MHC tetramers. Tumor growth was delayed in CRC− compared to WT mice (Figure 7A). However, CRC− mice ultimately succumbed to the tumor at a comparable rate as WT mice (Figure 7B). Tumor infiltrating lymphocytes (TILs) were isolated and analyzed at day 18. In the CRC− mouse, PD-1 expression was significantly reduced on gp33-specific CD8 TILs (Figure 7C) and highly reduced on CD4 TILs compared to WT mice (Figure 7D). This indicates that CR-C is responsible for inducing maximal PD-1 expression on anti-tumor T cells. T cells in the tumor-draining lymph nodes of WT or CRC− mice were also analyzed, and tumor-specific CD8 T cells identified using MHC tetramer staining for the gp33 epitope. Similar to the TILs, CRC− T cells within the lymph node displayed decreased PD-1 expression in both CD8 (Figure 7E) and CD4 (Figure 7F) populations compared to WT.
Figure 7. Anti-melanoma immune response is improved by CRC deletion.
A) WT or CRC− mice were injected subcutaneously with 106 B16.F10-gp33 melanoma cells, and tumor growth was measured every day by calipers along the largest diameter. N=2 groups of 5 mice for each genotype. B) Survival of mice injected with melanoma as in (a). Mice were allowed to continue growing tumors until tumor size, tumor infection, tumor ulceration/bleeding, or hindered mobility mandated sacrifice. C–D) Mice from (a) were sacrificed at Day 28 after tumor injection. Tumors were collected, and TILs isolated by ficoll gradient centrifugation. Cells were then stained for flow cytometry. E–F) Tumor-draining lymph nodes (inguinal and mesenteric) were harvested from mice in (A) at 28 days after injection, and stained for flow cytometry. Two groups of 5 or 6 mice per genotype were used for these experiments. Error bars represent mean plus standard deviation. Two-way ANOVA was used to calculate statistical significance in (A). Wilcoxon rank-sum was used to calculate significance in (B). Panels C–F used Student’s T tests to calculate statistical significance. * P< 0.05; ** P<0.01; *** P<0.001
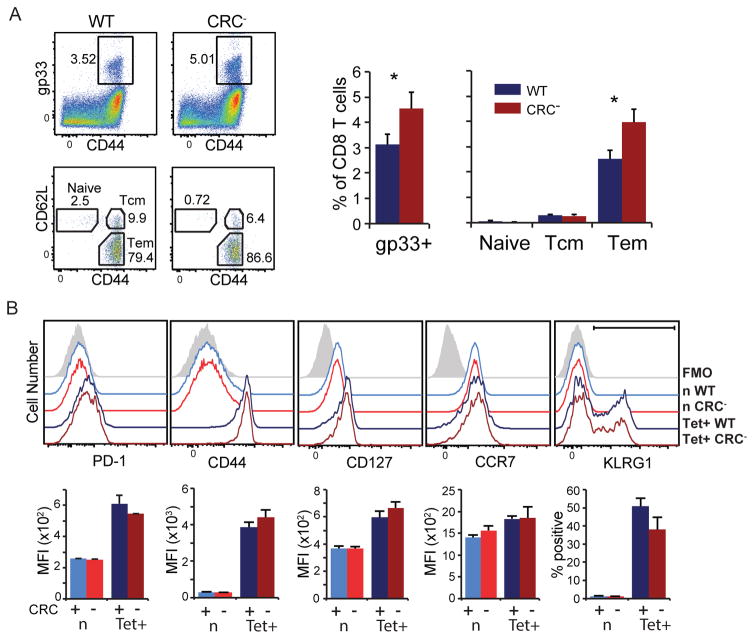
CR-C deletion influences CD8 T cell memory and functionality
PD-1 expression during acute infection was shown to modulate memory formation (33). Because CD8 T cells from CRC− mice expressed less PD-1 and showed fewer functional effector cells after an acute LCMV infection, it is possible that cell differentiation was skewed towards other populations, including memory. To determine if CR-C influences CD8 T cell memory formation and function, mice were infected with LCMV Armstrong and allowed to clear virus. At day 35 after acute infection, approximately 4 weeks after virus was cleared from the serum of these animals, the frequency of LCMV gp33-specific CD8 T cells was assessed by MHC tetramer staining. Compared to the WT, CRC− mice had 50% more LCMV peptide-specific CD8 memory T cells as a proportion of total CD8 T cells, although the total number of CD8 T cells within each genotype was not significantly different (Figure 8A). To characterize the nature of memory cells formed, the frequencies of central memory (Tcm) and effector memory (Tem) T cells were determined as measured by expression of CD44 and CD62L (48). The frequency of Tcm cells (CD44hiCD62Lhi) were slightly decreased in the CRC− mice compared to WT (Figure 8A). Conversely, the frequency of Tem cells (CD44hiCD62Llo) increased from approximately 2.5% of total CD8 T cells in WT mice to 4% in CRC− mice, indicating that the majority of additional memory cells present in these animals were Tem cells (Figure 8A). This shift from Tcm to Tem may indicate a more effective memory response at this time point. To further characterize the nature of the memory response, and to ensure that the antigen-specific cells being analyzed at this time point were in fact memory CD8 T cells and not naïve or effector T cells (49, 50), multiple additional phenotypic markers were analyzed. As expected (25), tetramer positive CD8 T cells from mice 35 days after Armstrong infection expressed only slightly more PD-1 compared to naïve cells (Figure 8B). There was no difference in PD-1 expression between resting memory cells from WT and CRC− animals, indicating that the basal expression of PD-1 on this cell type is not governed by CR-C. Tetramer-specific CD8 T cells from both groups were CD44hi (compared to naïve, Figure 8B), indicating that they were not naïve T cells, and were also CD127hi (compared to FMO and naive) in both mouse strains, indicating that they were not effector T cells despite a predominant expression of CD44. Similarly, memory T cells from both WT and CRC− mice showed a high expression of CCR7 and a large population of KLRG1+ cells, again characterizing them as predominantly Tem cells (Figure 8C).
Figure 8. Memory cell generation in CRC− mice is skewed towards Tem.
WT or CRC− mice were infected with LCMV Armstrong and sacrificed on day 35 after infection. Totally splenocytes were stained for flow cytometry. A) The frequency of gp33-specific cells out of total CD8 T cell population are shown (top). The frequencies of gp33-tetramer+ naïve CD8 T cells (CD62Lhi CD44lo), Tcm (CD62Lhi CD44hi), or Tem (CD62Llo CD44hi) are shown (bottom). The overall frequency of gp33 tetramer+ and subpopulations of these cells as a proportion of total CD8 T cells are shown to the right. B) Flow cytometry histograms of gp33-specific T cells identifying naïve, TSLEC or memory cell markers. Two groups of 5 mice for each genotype were used for these experiments. Each graph represents a single group of 5, shown as mean plus standard deviation. Statistical significance was calculated using Student’s T tests. * P< 0.05; ** P<0.01; *** P<0.001
In addition to measuring the quantity of memory T cells generated following an acute infection, the ability of memory CD8 T cells generated in CRC− mice to respond to secondary exposure to antigen was also assayed using ICCS as a measure of overall functionality. Coinciding with the greater overall frequency of memory cells at day 35, CRC− mice showed a greater proportion of cells secreting IFNγ (Figure 9A). In both WT and CRC− mice the majority of cells responding to gp33 peptide expressed two or more cytokines (Figure 9A), as expected in a memory population. However, in CRC− mice a greater proportion of responding cells were polyfunctional compared to WT (Figure 9A), suggesting an overall increase in the functionality of the memory CD8 T cell population.
Figure 9. Immune memory is more functional in CRC− mice.
A) WT or CRC− mice 35 days after LCMV Armstrong infection were sacrificed and total splenocytes stimulated with no treatment or gp33 peptide in the presence of brefeldin A for 5 hours. IFNγ, TNFα, and IL-2 expressing populations were characterized by ICCS. B) Equivalent numbers of LCMV memory cells from WT (CD45.1 CD90.2) or CRC− (CD45.2 CD90.2) mice were adoptively transferred into WT (CD45.2 CD90.1) hosts, and infected with LMCV Armstrong. Two days after infection, spleens were isolated and stained for flow cytometry. Adoptively transferred WT and CRC− cells were identified from WT host cells (within total splenocytes and analyzed separately within each host. C) 4,000 WT or CRC− gp33-specific memory T cells from day 35 Armstrong-immune mice, or 4000 naïve T cells, were adoptively transferred into WT hosts bearing day 5 B16.F10-gp33 tumors. Subsequent tumor growth was measured daily by calipers along the major and minor diameters of the tumor, and tumor volume was calculated as ½ LxW2. Two groups of 5 mice per genotype were used for melanoma experiments, and 2 groups of 3 mice per genotype for experiments in panels A and B. Each bar represents a mean of a single group of mice. Error bars indicate standard deviation. Student’s T tests were used to calculate significance in (A) and (B). Two-way ANOVA was used to calculate significance in (C). * P< 0.05; ** P<0.01; *** P<0.001
To determine if CR-C played a role in PD-1 expression during a recall response, WT (CD45.1 CD90.2) and CRC− (CD45.2 CD90.2) LCMV immune CD8 T cells were transferred to a naïve WT mouse (CD45.2 CD90.1), which was subsequently challenged with LCMV Armstrong. After two days, the cells were assessed for PD-1 expression. The results showed that PD-1 levels were similar between CRC− and WT cells within the same host (Figure 9B). This suggests CR-C does not play a role in the induction of PD-1 in memory cells.
To further characterize the ability of memory T cells formed following acute infection to respond to a secondary challenge, equivalent numbers of CRC− or WT, LCMV gp33-specific memory CD8 T cells were adoptively transferred into host WT mice bearing early stage (5 days post inoculation) B16.f10-gp33 melanoma tumors. The immune function of the memory cells was assayed by their ability to clear or prevent tumor growth. Mice that received WT memory cells showed no decrease in tumor growth compared to mice that received naïve T cells (Figure 9C), indicating that the low number of WT memory cells adoptively transferred was insufficient to respond to and clear the established tumors growing in the host mice. In sharp contrast, mice that received the same number of memory T cells from CRC− donors failed to develop any tumors (Figure 9B), indicating that the memory response from these CRC knockout T cells was significantly stronger on a per cell basis. Collectively, these data indicate that lower initial PD-1 expression during an acute infection, caused here by deletion of the CR-C region, resulted in greater quantity and quality of antigen-specific memory CD8 T cells.
DISCUSSION
CR-C is a complex regulatory element controlling PD-1 expression. Previous work identified and showed that CR-C encoded functional binding sites for NFATc1 (17), NF-κB (22), and FOX01 (23), which were active depending on stimulation conditions and cell type. Although the initial molecular analyses and manipulation of CR-C in in vitro systems showed that its function was likely important for PD-1 expression (17), its role in vivo could only be determined through genetic ablation of the element. Here, CR-C was found to be critical to optimal expression of PD-1 during both acute and chronic LCMV infections in both CD4 and CD8 T cells. Furthermore, the loss of CR-C prior to an acute infection and the corresponding decrease in PD-1 induction resulted in a larger pool of antigen-specific memory CD8 T cells that displayed more enhanced functionality, suggesting that alterations in the timing or full expression of PD-1 during initial T cell activation have a significant influence on T cell differentiation.
In the LCMV Armstrong system, Pdcd1 expression was reduced by ~50% in CRC− mice. This suggests that although CR-C is required for full expression of PD-1 in vivo, distal elements described previously (Figure 1: −3.7, +17.1) (19) or other redundant elements that have not been characterized are critical for its in vivo expression. During chronic infection and in memory CD8 T cells (both resting and during a secondary response), there was little or no difference in PD-1 expression in the CD8 T cells of CRC− animals. This suggests that the distal elements are sufficient to induce PD-1 independently of CR-C in chronic and memory settings. Changes in chromatin architecture or epigenetic modifications of the locus in memory or following chronic exposure could predispose the locus for expression in the absence of CR-C. Unlike CR-C, the −3.7 and +17.1 distal elements bind and respond to STAT3 or STAT4 in addition to NFATc1 and are therefore sensitive to the local or systemic environmental differences attributed to chronic LCMV infection that signal through receptors beyond the TCR (19). Intriguingly, a new element located at −20 kb, identified using a chromatin accessibility assay, is active during chronic LCMV infection (unpublished data) and may over ride the deletion of CR-C. Thus, the redundancy of NFAT responsive elements and the ability to respond to inflammatory cytokines are likely responsible for providing a distinct in vivo control of PD-1 gene expression during chronic versus acute stimulation of CD8 T cells. These diverse modalities of PD-1 regulation may be used to direct the development of T cells into short-lived effector T cells (TSLEC) or exhausted T cells in the context of different immune challenges.
The rapid loss of DNA methylation upstream of CR-B and at CR-C during the initial effector stage of Pdcd1 expression and the regaining of DNA methylation at late effector and memory time points suggested that this epigenetic mechanism was critical to the overall regulation of Pdcd1 (25). The failure of the CR-B upstream sequences to reach their wild-type levels of demethylation during acute CD8 T cell responses in CRC− CD8 T cells suggests that DNA demethylation is mediated by the activity of the CR-C region, with the possibility that NFATc1 itself is responsible for initiating the epigenetic change. The findings also suggest that cytosine methylation of the promoter proximal sequences was not sufficient to completely block induction of Pdcd1 expression. In agreement with this finding is the fact that in macrophages treated with LPS to induce PD-1, DNA methylation at CR-C is not lost, despite the fact that CR-C is used to induce PD-1 (22). In this case, NF-κB was the primary transcription factor binding to CR-C and NFATc1 did not play a role. In the LPS system in macrophages, PD-1 expression was short lived. Together, with previous work (25) these data suggest that prolonged and full expression of PD-1 may require demethylation of the locus.
CR-C’s in vivo role in regulating PD-1 expression appears to be most critical at early time points following infection. In an acute infection, PD-1 expression by both mRNA and surface protein was abrogated by over 50% in CRC− animals compared to WT. Despite this, CRC− mice and WT mice demonstrated comparable viral clearance in blood by day 8, as well as peak viral titers at day 4. A number of mechanisms may explain why lower PD-1 fails to correlate with improved anti-viral function. First, the decreased level of PD-1 expression during acute infections may be insufficient to inhibit T cell effector functions. Alternatively, in agreement with previous reports (44) and recapitulated here, a loss of PD-1 expression resulted in higher up regulation of other inhibitory receptors, particularly Tigit. This may compensate for the lower PD-1 expression, ultimately yielding a balance in overall cellular activation. As CRC− mice had fewer TSLEC compared to WT, it is possible that early, optimal PD-1 expression may be required to coordinate an effective immune response, as has been previously reported (51), rather than acting as a cellular inhibitor as occurs during chronic stimulation. The decrease in effector CD8 T cells and/or function may be mediated through a number of mechanisms. As previous research has shown, high PD-1 expression on effector T cells slowed migration of T cells through sites containing antigen, and blockade of PD-1 inhibited IFNγ production during a primary response (52). A similar mechanism may be in play here. Alternatively, in promoting a memory cell phenotype, lower PD-1 may drive T cell differentiation away from a TSLEC fate. Differential immunomodulatory signaling through PD-1 may alter the balance between the development of TSLEC and memory precursor cells during the initial immune response, driving cells to differentiate into memory cells rather than effectors (50, 53).
One of the more striking findings here was the ability of CRC− mice to generate a much stronger antigen-specific T cell memory response. This corroborates previous findings that the absence or antibody blockade of PD-1 signaling skews CD8 T cell populations towards a memory phenotype (33, 47, 54, 55). Here, the loss of early PD-1 expression mediated by CR-C deletion resulted in greater number and quality of memory CD8 T cells. Together, these results indicate that the level of PD-1 expression may regulate the nature of the initial immune response; with a decrease approximating 50% PD-1 expression having profound phenotype on T cell memory formation. From a clinical standpoint, this may indicate that a limited blockade in PD-1 signaling during an immune challenge, such as vaccination, may improve memory formation and be a viable protocol for improving vaccination regimens. Furthermore, antigen-specific CRC− memory CD8 T cells, generated here by a primary viral infection, showed a greater ability to combat existing early melanoma tumors upon adoptive transfer into tumor-bearing hosts. Although this experiment utilized an immune response against an exogenous tumor antigen (LCMV-gp33) which may be expressed at higher levels or may be more immunogenic than endogenous tumor epitopes, it nonetheless showed that memory cells lacking CR-C displayed a greater immunological activity. This may suggest future cancer therapies in which PD-1 expression is altered, possibly through CRISPR/Cas9 deletion of CR-C. As CRC− mice showed no increased propensity to autoimmunity and still developed signs of exhaustion during chronic antigen encounter, such an approach may prevent any potential immune-related adverse events (56) associated with current PD-1/PD-L1 checkpoint blockade therapies.
Acknowledgments
The authors wish to thank Hanspeter Pircher (Universitätsklinikum Freiburg) for supplying the B16-F10.gp33 melanoma cell line. We thank Dr. Rafi Ahmed (Emory University) for providing LCMV viral strains and for insightful discussions of the work. We thank John D. Altman for helpful critique of the work. We also wish to thank Dr. Arlene Sharpe and her group (Harvard Medical School) for help in the design and generation of the CRCfl/fl mice. We also thank the NIH Tetramer Core Facility (contract HHSN272201300006C) for provision of LCMV-specific MHC-I tetramers gp33, gp276, and np396.
Footnotes
This work was supported by NIH grants RO1 AI113021 and PO1 AI080192 to JMB; RO1 AI110113 and RO1 NS071518 to BE. APRB and RM were supported by NIH T32 AI007610.
CONFLICT OF INTEREST STATEMENTS
The authors have no financial conflicts of interest to declare.
ROLE OF AUTHORS
APRB, YT, BGB, RM, JTL participated in the design, execution, and interpretation of the experiments. BE, and JMB participated in the design of experiments, scientific questions, and interpretation of the data. All authors contributed to the writing and/or editing of the manuscript.
References
- 1.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Kloverpris HN, McGregor R, McLaren JE, Ladell K, Stryhn A, Koofhethile C, Brener J, Chen F, Riddell L, Graziano L, Klenerman P, Leslie A, Buus S, Price DA, Goulder P. Programmed death-1 expression on HIV-1-specific CD8+ T cells is shaped by epitope specificity, T-cell receptor clonotype usage and antigen load. Aids. 2014;28:2007–2021. doi: 10.1097/QAD.0000000000000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaikumar Duraiswamy JDM, DavidMasopust David, Ibegbu Chris C, Wu Hong, Freeman Gordon J, RafiAhmed Rafi. PD-1 expression on memory CD8 and CD4 T-cell subsets in healthy humans. The Journal of Immunology. 2007:178. [Google Scholar]
- 4.Breton G, Chomont N, Takata H, Fromentin R, Ahlers J, Filali-Mouhim A, Riou C, Boulassel MR, Routy JP, Yassine-Diab B, Sekaly RP. Programmed death-1 is a marker for abnormal distribution of naive/memory T cell subsets in HIV-1 infection. Journal of immunology. 2013;191:2194–2204. doi: 10.4049/jimmunol.1200646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Severson JJ, Serracino HS, Mateescu V, Raeburn CD, McIntyre RC, Jr, Sams SB, Haugen BR, French JD. PD-1+Tim-3+ CD8+ T Lymphocytes Display Varied Degrees of Functional Exhaustion in Patients with Regionally Metastatic Differentiated Thyroid Cancer. Cancer Immunol Res. 2015;3:620–630. doi: 10.1158/2326-6066.CIR-14-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang ZN, Zhu ML, Chen YH, Fu YJ, Zhang TW, Jiang YJ, Chu ZX, Shang H. Elevation of Tim-3 and PD-1 expression on T cells appears early in HIV infection, and differential Tim-3 and PD-1 expression patterns can be induced by common gamma -chain cytokines. BioMed research international. 2015;2015:916936. doi: 10.1155/2015/916936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauvin JM, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, Kirkwood JM, Chen TH, Maurer M, Korman AJ, Zarour HM. TIGIT and PD-1 impair tumor antigen-specific CD8+ T cells in melanoma patients. The Journal of clinical investigation. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 9.Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 10.Carter Laura L, LAF, Jussif Jason, Fitz Lori, Deng Bija, Wood Clive R, Collins Mary, Honjo Tasuku, Freeman Gordon J, Carreno Beatriz M. PD-1:PD-L inhibitory pathway affects both CD4+ and CD8+ T cells and is overcome by IL-2. Eur J Immunol. 2002;32:634–643. doi: 10.1002/1521-4141(200203)32:3<634::AID-IMMU634>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 12.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. The New England journal of medicine. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blank C, Brown I, Peterson AC, Spiotto M, Iwai Y, Honjo T, Gajewski TF. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 2004;64:1140–1145. doi: 10.1158/0008-5472.can-03-3259. [DOI] [PubMed] [Google Scholar]
- 16.Bally AP, Austin JW, Boss JM. Genetic and Epigenetic Regulation of PD-1 Expression. Journal of immunology. 2016;196:2431–2437. doi: 10.4049/jimmunol.1502643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol. 2008;181:4832–4839. doi: 10.4049/jimmunol.181.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao G, Deng A, Liu H, Ge G, Liu X. Activator protein 1 suppresses antitumor T-cell function via the induction of programmed death 1. Proc Natl Acad Sci U S A. 2012;109:15419–15424. doi: 10.1073/pnas.1206370109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin JW, Lu P, Majumder P, Ahmed R, Boss JM. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J Immunol. 2014;192:4876–4886. doi: 10.4049/jimmunol.1302750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HY, Lee SW, Seo SK, Choi IW, Choi I. Interferon-sensitive response element (ISRE) is mainly responsible for IFN-alpha-induced upregulation of programmed death-1 (PD-1) in macrophages. Biochimica et biophysica acta. 2008;1779:811–819. doi: 10.1016/j.bbagrm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Terawaki S, Chikuma S, Shibayama S, Hayashi T, Yoshida T, Okazaki T, Honjo T. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. Journal of immunology. 2011;186:2772–2779. doi: 10.4049/jimmunol.1003208. [DOI] [PubMed] [Google Scholar]
- 22.Bally AP, Lu P, Tang Y, Austin JW, Scharer CD, Ahmed R, Boss JM. NF-kappaB Regulates PD-1 Expression in Macrophages. J Immunol. 2015;194:4545–4554. doi: 10.4049/jimmunol.1402550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staron MM, Gray SM, Marshall HD, Parish IA, Chen JH, Perry CJ, Cui G, Li MO, Kaech SM. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity. 2014;41:802–814. doi: 10.1016/j.immuni.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 25.Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, Boss JM, Ahmed R. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youngblood B, Noto A, Porichis F, Akondy RS, Ndhlovu ZM, Austin JW, Bordi R, Procopio FA, Miura T, Allen TM, Sidney J, Sette A, Walker BD, Ahmed R, Boss JM, Sekaly RP, Kaufmann DE. Cutting edge: Prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. J Immunol. 2013;191:540–544. doi: 10.4049/jimmunol.1203161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu P, Youngblood BA, Austin JW, Rasheed Mohammed AU, Butler R, Ahmed R, Boss JM. Blimp-1 represses CD8 T cell expression of PD-1 using a feed-forward transcriptional circuit during acute viral infection. The Journal of experimental medicine. 2014;211:515–527. doi: 10.1084/jem.20130208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura H, Honjo T, Minato N. Facilitation of beta selection and modification of positive selection in the thymus of PD-1-deficient mice. J Exp Med. 2000;191:891–898. doi: 10.1084/jem.191.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blank C, Brown I, Marks R, Nishimura H, Honjo T, Gajewski TF. Absence of programmed death receptor 1 alters thymic development and enhances generation of CD4/CD8 double-negative TCR-transgenic T cells. J Immunol. 2003;171:4574–4581. doi: 10.4049/jimmunol.171.9.4574. [DOI] [PubMed] [Google Scholar]
- 31.Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc Natl Acad Sci U S A. 2001;98:13866–13871. doi: 10.1073/pnas.231486598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 33.Allie SR, Zhang W, Fuse S, Usherwood EJ. Programmed death 1 regulates development of central memory CD8 T cells after acute viral infection. Journal of immunology. 2011;186:6280–6286. doi: 10.4049/jimmunol.1003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frebel H, Nindl V, Schuepbach RA, Braunschweiler T, Richter K, Vogel J, Wagner CA, Loffing-Cueni D, Kurrer M, Ludewig B, Oxenius A. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. The Journal of experimental medicine. 2012;209:2485–2499. doi: 10.1084/jem.20121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 36.Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- 38.Kersh AE, Edwards LJ, Evavold BD. Progression of relapsing-remitting demyelinating disease does not require increased TCR affinity or epitope spread. Journal of immunology. 2014;193:4429–4438. doi: 10.4049/jimmunol.1401456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prevost-Blondel A, Zimmermann C, Stemmer C, Kulmburg P, Rosenthal FM, Pircher H. Tumor-infiltrating lymphocytes exhibiting high ex vivo cytolytic activity fail to prevent murine melanoma tumor growth in vivo. Journal of immunology. 1998;161:2187–2194. [PubMed] [Google Scholar]
- 40.Matloubian M, Somasundaram T, Kolhekar SR, Selvakumar R, Ahmed R. Genetic basis of viral persistence: single amino acid change in the viral glycoprotein affects ability of lymphocytic choriomeningitis virus to persist in adult mice. The Journal of experimental medicine. 1990;172:1043–1048. doi: 10.1084/jem.172.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura H, Agata Y, Kawasaki A, Sato M, Imamura S, Minato N, Yagita H, Nakano T, Honjo T. Developmentally regulated expression of the PD-1 protein on the surface of double-negative (CD4−CD8−) thymocytes. Int Immunol. 1996;8:773–780. doi: 10.1093/intimm/8.5.773. [DOI] [PubMed] [Google Scholar]
- 44.Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. The Journal of experimental medicine. 2015;212:1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter L, Fouser LA, Jussif J, Fitz L, Deng B, Wood CR, Collins M, Honjo T, Freeman GJ, Carreno BM. PD-1:PD-L inhibitory pathway affects both CD4(+) and CD8(+) T cells and is overcome by IL-2. European journal of immunology. 2002;32:634–643. doi: 10.1002/1521-4141(200203)32:3<634::AID-IMMU634>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 46.Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- 47.Ribas A, Shin DS, Zaretsky J, Frederiksen J, Cornish A, Avramis E, Seja E, Kivork C, Siebert J, Kaplan-Lefko P, Wang X, Chmielowski B, Glaspy JA, Tumeh PC, Chodon T, Pe’er D, Comin-Anduix B. PD-1 Blockade Expands Intratumoral Memory T Cells. Cancer Immunol Res. 2016;4:194–203. doi: 10.1158/2326-6066.CIR-15-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Faassen H, Saldanha M, Gilbertson D, Dudani R, Krishnan L, Sad S. Reducing the stimulation of CD8+ T cells during infection with intracellular bacteria promotes differentiation primarily into a central (CD62LhighCD44high) subset. Journal of immunology. 2005;174:5341–5350. doi: 10.4049/jimmunol.174.9.5341. [DOI] [PubMed] [Google Scholar]
- 49.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talay O, Shen CH, Chen L, Chen J. B7-H1 (PD-L1) on T cells is required for T-cell-mediated conditioning of dendritic cell maturation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2741–2746. doi: 10.1073/pnas.0813367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honda T, Egen JG, Lammermann T, Kastenmuller W, Torabi-Parizi P, Germain RN. Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity. 2014;40:235–247. doi: 10.1016/j.immuni.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charlton JJ, Chatzidakis I, Tsoukatou D, Boumpas DT, Garinis GA, Mamalaki C. Programmed death-1 shapes memory phenotype CD8 T cell subsets in a cell-intrinsic manner. Journal of immunology. 2013;190:6104–6114. doi: 10.4049/jimmunol.1201617. [DOI] [PubMed] [Google Scholar]
- 56.Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB, Powderly JD, Leming PD, Lipson EJ, Puzanov I, Smith DC, Taube JM, Wigginton JM, Kollia GD, Gupta A, Pardoll DM, Sosman JA, Hodi FS. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]