Abstract
Having one and only one centromere per chromosome is essential for proper chromosome segregation during both mitosis and meiosis. Chromosomes containing two centromeres are known as dicentric and often mis-segregate during cell division, resulting in aneuploidy or chromosome breakage. Dicentric chromosome can be stabilized by centromere inactivation, a process which re-establishes monocentric chromosomes. However, little is known about this process in naturally occurring dicentric chromosomes. Using a combination of fluorescence in situ hybridization (FISH) and immunoflourescence combined with FISH (IF-FISH) on metaphase chromosome spreads, we demonstrate that centromere inactivation has evolved on a neo-Y chromosome fusion in the Japan Sea threespine stickleback fish (Gasterosteus nipponicus). We found that the centromere derived from the ancestral Y chromosome has been inactivated. Our data further suggest that there have been genetic changes to this centromere in the two million years since the formation of the neo-Y chromosome, but it remains unclear whether these genetic changes are a cause or consequence of centromere inactivation.
Keywords: Dicentric chromosome fusion, Centromere inactivation, CENP-A, ChIP-seq, Gasterosteus aculeatus, Gasterosteus nipponicus
Introduction
The centromere of a chromosome was first described by Walther Flemming as the primary constriction observed on condensed chromosomes during both mitosis and meiosis (Flemming 1880). This primary constriction is the region of the chromosome where microtubules attach, and it is necessary for the segregation of chromosomes during cell division. Many organisms contain “regional” centromeres, which are found at a single location on each chromosome and comprised of kilobases to megabases of DNA. The sequence of these regional centromeres is often repetitive and AT rich, but the specific sequence varies dramatically among species (Henikoff et al. 2001; Alkan et al. 2011; Melters et al. 2013), and can even vary among centromeres within a species (Nagaki et al. 2004; Piras et al. 2010; Tek et al. 2010; Shang et al. 2010; Gong et al. 2012). Centromeres do share a common epigenetic characteristic, which is the presence of a histone variant called centromere protein A (CENP-A) that replaces histone H3 in centromeric nucleosomes (Palmer et al. 1987; Palmer et al. 1989; Palmer et al. 1991; Sullivan et al. 1994a). The presence of CENP-A is considered the hallmark of an active centromere (Warburton et al. 1997; Heun et al. 2006; Marshall et al. 2008; Allshire and Karpen 2008; Barnhart et al. 2011; Mendiburo et al. 2011; Sekulic and Black 2012; Fukagawa and Earnshaw 2014; McKinley and Cheeseman 2016).
While it is essential that chromosomes have a centromere, in organisms with regional centromeres it is also essential that every chromosome has one and only one active centromere. A normal monocentric chromosome only has a single centromere that can bind microtubules and form a bipolar spindle, allowing normal chromosome segregation. However, a dicentric chromosome has two centromeres that can both bind microtubules, leading to instability during cell division. Barbara McClintock first described this instability and termed it the breakage-fusion-bridge cycle (McClintock 1939; McClintock 1941). If the microtubules from spindle poles on the opposing sides of the cell bind to the two centromeres, the chromosome will be pulled in opposite directions. This leads to merotelic kinetochore attachments where the dicentric chromosome lags in the middle during anaphase as it is being pulled to opposite poles. The physical tension on the chromosome can lead to breakage of the chromosome. When the broken ends are replicated during the next S-phase of the cell cycle, the broken ends can anneal back to each other, re-creating a dicentric. Not only does the breakage-fusion-bridge cycle start again, but the breakage and re-annealing leads to gene loss and gene gain. This is evident in cancer cells where dicentrics are often observed, leading to cells with “increased genetic heterogeneity” (Gisselsson et al. 2000; Gisselsson et al. 2001).
Thus, dicentric chromosomes created by chromosome fusions are predicted to be very unstable. However, chromosome fusion (and fission) events are common during evolution as evidenced by the extensive variation in chromosome number among species. For example, in multicellular eukaryotes, chromosome number ranges from a diploid number of two in the Jack jumper ant (Crosland and Crozier 1986) to 1260 in the Adders tongue fern (van der Burg 2004). In eutherian mammals, the diploid chromosome number ranges from 6 in the Indian muntjac to 102 in the viscacha rat (Gallardo et al. 1999); this diversity results from many independent fusion and fission events (Ferguson-Smith and Trifonov 2007). Even closely related species can vary in chromosome number; for example, the Chinese muntjac has a diploid chromosome number of 46, while the Indian muntjac has a diploid number of 6 in females or 7 in males (Shi et al. 1980; Yang et al. 1995; Wang and Lan 2000). It is speculated that a series of chromosome fusion events between the telocentric chromosomes present in the Chinese muntjac created the much larger Indian muntjac chromosomes (Wang and Lan 2000). Chromosome fusions have also occurred in the primate lineage; human chromosome 2 is a fusion between chimp chromosomes 2a and 2b (Lejeune et al. 1973; IJdo et al. 1991; Avarello et al. 1992). Similar chromosome fusion and fission events are common in many taxa and can account for much of the diversity of karyotype number observed today. Many of these chromosome fusion events must have created dicentric chromosomes and therefore been accompanied by mechanisms to stabilize the chromosome fusions.
To date, however, relatively little is known about the mechanisms involved in the stabilization of dicentric chromosomes during evolution in natural populations. Most of our current understanding of how dicentrics are stabilized comes from studies of artificially-induced or engineered dicentric chromosomes in yeast (Pobiega and Marcand 2010; Sato et al. 2012), fruit flies (Agudo et al. 2000), and human cell lines (Higgins et al. 1999; Higgins et al. 2005; Stimpson et al. 2010; Stimpson et al. 2012), as well as a few examples of dicentric chromosomes in found in plants (Sears and Câmara 1952; Han et al. 2006; Zhang et al. 2010; Gao et al. 2011; Koo et al. 2011; Liu et al. 2015) and human patients (Therman et al. 1989; Maraschio et al. 1990; Fisher et al. 1997; Page and Schaffer 1998; Sullivan and Willard 1998; Lange et al. 2009; Stimpson et al. 2012). In some engineered systems, the dicentric chromosome can simply break in order to re-establish monocentric chromosomes (Pobiega and Marcand 2010; Sato et al. 2012). In other engineered or patient-derived dicentrics, chromosome fusions can be stably inherited because the two centromeres on a fusion chromosome are close enough to each other to act as a single centromere (Koshland et al. 1987; Page and Shaffer 1998; Sullivan and Willard 1998; Higgins et al. 1999; Lange et al. 2009). However, distance between the centromeres does not fully explain the stable maintenance of dicentric chromosomes, and in many cases, there is inactivation of one of the centromeres to re-establish functionally monocentric chromosomes (Stimpson et al. 2012). Centromere inactivation can either occur by genetic or epigenetic inactivation of one centromere. In genetic inactivation, deleterious mutation of the DNA sequence of one centromere prevents it from binding CENP-A. In epigenetic inactivation, the DNA of the inactivated centromere is still present, but CENP-A is not. In either case, the functional consequence of the loss of CENP-A is that the centromere does not recruit kinetochore components and therefore does not attach to mictrotubules during cell division. Although both genetic and epigenetic inactivation have been observed in the systems studied to date (Stimpson et al. 2012), almost nothing is known about the relative contribution of these mechanisms to dicentric stabilization during evolution.
The stickleback family (Gasterosteidae) provides a unique opportunity to study the mechanisms that stabilize natural dicentric chromosomes during evolution because there have been several chromosome fusion events that have occurred within the past 35 million years (Kitano et al. 2009; Ross et al. 2009). Previous work has identified a chromosome fusion involving the ancestral metacentric Y chromosome present in the Pacific Ocean threespine stickleback (Gasterosteus aculeatus). This Y chromosome has fused to the acrocentric chromosome 9 in males of the Japan Sea threespine stickleback (Gasterosteus nipponicus) species within the past two million years, creating an X1X2Y sex chromosome system in which the ancestral X is X1, the unfused chromosome 9 is X2, and the fused chromosome is called the neo-Y (Kitano et al. 2009) (Fig. S1). Because this fusion involved the metacentric Y and an acrocentric chromosome, the neo-Y should have been dicentric upon formation. Preliminary data had suggested that there was no loss of a chromosome arm to bring the two centromeres closer on the Japan Sea neo-Y chromosome, or re-breakage of the fusion chromosome. These data suggested that centromere inactivation has occurred on the Japan Sea neo-Y chromosome.
In this study, we aimed to first determine if centromere inactivation has occurred on the neo-Y, and to then test whether inactivation is due to a genetic or epigenetic mechanism. Both the genetic and epigenetic mechanisms of centromere inactivation ultimately result in only one functional centromere, as defined by the presence of CENP-A. If centromere inactivation has occurred, we expected to see only one region on the neo-Y with CENP-A staining. If no centromere inactivation has occurred, we expected to see two regions of CENP-A staining on the neo-Y. Using CENP-A antibody staining, we determined that the Japan Sea neo-Y has only one active centromere, as defined by presence of CENP-A. In an extension of our previous study (Cech and Peichel 2015), we then performed chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) to determine the centromere sequence in the Japan Sea species. We then used fluorescence in situ hybridization (FISH) to demonstrate that there have likely been genetic changes to the ancestral Y centromere on the Japan Sea neo-Y.
Materials and Methods
Fish use and care
Two species of lab reared threespine stickleback fish were used in this study: Pacific Ocean (Gasterosteus aculeatus) and Japan Sea (Gasterosteus nipponicus). Lab-reared fish were derived from wild caught fish collected in Akkeshi on Hokkaido Island, Japan (Kitano et al. 2007, 2009). Fish were kept in in 3.5% saltwater (3.5 g/l Instant Ocean salt (Spectrum Brands, USA); 0.4 ml/l sodium bicarbonate) in 29-gal aquarium tanks, at 16 °C and 16 hr light / 8 hr dark. Fish were fed once daily with live Artemia nauplii and once daily with frozen Mysis shrimp. All institutional and national guidelines for the care and use of laboratory animals were followed, and all procedures were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee (protocol 1575).
CENP-A immmunoprecipitation followed by high throughput sequencing (ChIP-seq)
Chromatin immunoprecipitation (IP) with the stickleback specific CENP-A antibody (Cech and Peichel 2015) was performed using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads; Cell Signaling Technology, USA). 0.1 g of spleen and liver tissue was taken from three Japan Sea females (one single replicate, and one pooled replicate of two individuals) and four Japan Sea male sticklebacks (two pooled replicates of two individuals each). The chromatin immunoprecipitation was performed at the same time and following the same protocol as for the two Pacific Ocean female samples described in Cech and Peichel (2015). To identify the Japan Sea consensus centromere sequence, analysis was performed using the same methods as in Cech and Peichel (2015). All raw fastq sequence files are available in the NCBI Sequence Read Archive (Pacific Ocean female data: SRP063504; Japan Sea male and female data: SRP081499).
FISH on metaphase spreads and interphase nuclei
Lab reared Pacific Ocean and Japan Sea fish were injected with 10 μl of 1 % colchicine in phosphate-buffed saline (PBS) for 14-16 hours to arrest metaphase cells. Metaphase and interphase cells were obtained following the same protocol described in Ross and Peichel (2008), with the exception that metaphase slides were dried at 50°C, not 37°C. The threespine stickleback centromeric repeat sequence (GacCEN) probe was made as described in Cech and Peichel (2015). Bacterial artificial chromosome (BAC) probes listed in Table 1 were prepared following the protocol in Urton et al. (2011) and labeled with either Alexa-488 or Alexa-568.
Table 1.
List of BACs used in this study
| Chr | BAC | Library | Genomic position (bp) | Cytogenetic position* |
|---|---|---|---|---|
| 9 | 44L12 | CHORI-213 | 497,589 – 729,621 | Distal end of short arm |
| 9 | 35K20 | CHORI-215 | 18,760,858 – 18,914,793 | Distal end of long arm |
| X | 188J19 | CHORI-213 | 2,384,200 – 2,563,816 | Distal end of short arm |
| Y | 188J19 | CHORI-213 | unknown | Distal end of short arm |
| X | 180J08 | CHORI-213 | 3,377,138 – unknown | Proximal end of short arm |
| Y | 180J08 | CHORI-213 | unknown | Proximal end of long arm |
| X | 171H24 | CHORI-213 | 5,465,262 – 5,643,381 | Proximal end of long arm |
| Y | 171H24 | CHORI-213 | unknown | Proximal end of short arm |
| X | 101E08 | CHORI-213 | 11,446,792 – 11,651,387 | Middle of long arm |
| Y | 101E08 | CHORI-213 | unknown | Distal end of long arm |
| Y | 91G03 | CHORI-215 | unknown | Distal end of long arm |
Position is relative to the centromere; proximal is closer to the centromere and distal is further from the centromere. The genomic positions of the BACs on chromosome 9 and the X chromosome were determined by BLASTing the publicly available BAC end sequences (Kingsley and Peichel 2007) to the updated threespine stickleback genome assembly (Glazer et al. 2015). The genomic positions of BACs on the Y chromosome are unknown because a complete genome assembly of the Y chromosome is not available.
Metaphase slides were first washed for 5 min with phosphate-buffered saline Tween-20 (PBST), fixed for 10 min in 4% paraformaldehyde in PBS, then washed again in PBST for 5 min. Subsequent FISH hybridization with only the GacCEN probe or only BAC probes was performed as described in Cech and Peichel (2015). For metaphase slides hybridized with combined GacCEN and BAC probes, 10 μl of the GacCEN probe was first lyophilized at 50°C for four hours. The dried GacCEN probe was then resuspended with either 10 μl of a single BAC probe in hybridization buffer, or 20 μl of equal parts of two BAC probes in hybridization buffer.
Telomere staining
Telomere staining was performed according to manufacturers instructions using the DAKO telomere PNA FISH kit/FITC no. K5325 (Agilent Technologies, USA).
Immunofluorescence-FISH (IF-FISH)
For IF-FISH, 15 Pacific Ocean or Japan Sea embryos at 48 hours post fertilization were dounced in 10 ml 0.56 % potassium chloride using a glass dounce. The remainder of the IF-FISH protocol follows Cech and Peichel (2015).
Microscopy
Images were taken using a Nikon Eclipse 80i microscope (Nikon, Japan) with an automated filter turret (Chroma filters 31000v2 (DAPI), 41001 (FITC), and 41004 (Texas Red); Chroma, USA) using the 100x objective. Following Cech and Peichel (2015), images were pseudo-colored using NIS Elements imaging software (BR 3.00, SP7, Hotfix8, Build 548, Nikon, Japan).
Results
No loss of chromosome arms on the Japan Sea neo-Y fusion chromosome
To determine if the neo-Y fusion was a complete fusion between both chromosomes, or if large portions of each or one of the respective p-arms was lost, we performed two types of FISH experiments. First, we conducted hybridization with BAC probes homologous to the ends of chromosome 9 (44L12) and the Y chromosome (101E8) that are near the fusion site, as well as the end of the Y chromosome (188J19) opposite to the fusion site (Table 1). These probes all hybridize to the neo-Y on Japan Sea male metaphase chromosomes, demonstrating that loss of an entire chromosome arm has not occurred on the neo-Y fusion chromosome (Fig. 1). Hybridization of these same BAC probes on Pacific Ocean male metaphase chromosomes further demonstrates that there have not been gross rearrangements between the Japan Sea and Pacific Ocean sticklebacks on either chromosome 9 or the Y chromosome (Fig. S2).
Fig. 1.
The Japan Sea neo-Y is a fusion between two complete chromosomes. FISH with the ancestral X and Y chromosome BACs 188J19 (purple) and101E08 (green) and the chromosome 9 BAC 44L12 (purple) on a Japan Sea male metaphase spread shows (a) three chromosomes with BAC hybridization each highlighted by square boxes. Higher magnification of the boxed regions in panel (a) shows the neo-Y fusion (b), the X1 chromosome, which is derived from the ancestral X chromosome (c) and the X2 chromosome, which is derived from the ancestral chromosome 9 (d). Scale bar, 5 μm
Second, to test if there was truly no loss of any genetic material, we also used a FISH probe to the 6bp telomere sequence (TTAGGG) found in all vertebrates (Meyne et al. 1989). There is no internal telomere signal on the neo-Y chromosome, indicating that some loss of genetic material encompassing at least the telomeres has occurred on the neo-Y chromosome (Fig. S3). Interestingly, telomere loss can induce chromosome fusions (Gisselsson et al. 2001; Maser and DePinho 2002; Bailey 2006; Pobiega and Marcand 2010; Murnane 2010; Stimpson et al. 2010). However, we do not know whether loss of telomeres is a cause or consequence of the neo-Y fusion. Taken together, these data do suggest that the neo-Y chromosome resulted from a fusion between two nearly complete monocentric chromosomes, creating a dicentric neo-Y chromosome. The neo-Y chromosome is present in all Japan Sea males examined to date, and we have no evidence for dicentric chromosome segregation associated aneuploidy in the Japan Sea males (i.e. no sex ratio bias in the offspring of Japan Sea males; data not shown). Furthermore, there is evidence of only one constriction on the neo-Y in metaphase chromosome spreads (Fig. S3). Thus, we hypothesized that the neo-Y chromosome has been stabilized by centromere inactivation via either genetic or epigenetic inactivation.
Evidence for centromere inactivation on the Japan Sea neo-Y chromosome
Using CENP-A antibody staining in combination with FISH, we determined that only one region of CENP-A staining is found on the neo-Y in multiple independent metaphase spreads from Japan Sea males (Fig. 2). The CENP-A staining was flanked by BAC probes present at the end of the Y near the fusion breakpoint and at the distal end of chromosome 9, indicating that the active centromere is on chromosome 9 (Fig. 2). The CENP-A staining on the neo-Y is in a similar location to the CENP-A staining found on the unfused chromosome 9 (X2), further suggesting that the active centromere on the neo-Y is retained from the ancestral acrocentric chromosome 9 (Fig. 2f).
Fig. 2.
The CENP-A antibody only localizes to one region on the Japan Sea neo-Y chromosome. Panels (a) and (c) show two independent metaphase spreads from Japan Sea embryos stained with the CENPA antibody (green) as well as the X and Y chromosome BAC 101E08 (purple) and the chromosome 9 BAC 35K20 (purple). Panels (b) and (d) are higher magnifications of the boxed regions in panels (a) and (c), highlighting the neo-Y chromosome with the two regions of BAC staining (purple arrowheads) flanking two distinct CENP-A puncta (white arrowhead). The CENP-A staining is located on the chromosome 9-derived part of the neo-Y chromosome. Panel (e) is a higher magnification of the boxed region in (a), highlighting the ancestral X1 chromosome; 101E08 hybridizes to the middle of the long arm of X1 (purple arrowhead), and there is one region of two CENP-A puncta (white arrowhead). Panel (f) is a higher magnification of the boxed region in (c), highlighting the unfused chromosome 9 (X2); 35K20 hybridizes to the end of the long arm of X2 (purple arrowhead), and there is one region of two CENP-A puncta (white arrowhead). Scale bar, 5 μm
To confirm that the ancestral Y chromosome centromere was active before the fusion that created the neo-Y occurred, we performed CENP-A staining in combination with FISH using a BAC probe (91G03; Table 1) that specifically labels the Y chromosome on Pacific Ocean male chromosomes (Fig. S4). We found normal CENP-A staining on the ancestral Y centromere (Fig. S5). This is evidence that the ancestral Y chromosome had an active centromere, and that the Y chromosome centromere was inactivated after the fusion to chromosome 9 in Japan Sea males.
Evidence for genetic inactivation on the Japan Sea neo-Y chromosome
Because we found evidence for only a single active centromere on the Japan Sea neo-Y chromosome, we sought to determine whether there has been genetic inactivation (i.e. deletion or alteration) of the ancestral Y chromosome centromere. We had previously identified the centromere sequence (GacCEN) in the Pacific Ocean threespine stickleback (Cech and Peichel 2015). Although this centromere sequence appears to be present on all autosomes, including chromosome 9 and the X chromosome, we observed very weak hybridization of the centromere sequence to the Y chromosome in the ancestral Pacific Ocean population (Cech and Peichel 2015). It was unclear whether the centromere on the Y chromosome has a completely different sequence, a highly divergent sequence, or a reduced number of repeats. Here, we further explored these possibilities in order to analyze the fate of the ancestral Y chromosome centromere on the Japan Sea neo-Y. We performed additional FISH experiments on Pacific Ocean males with the centromere probe and confirmed that centromere hybridization on the ancestral Y is weak and variable. Over 11 independent FISH experiments, we counted 29 metaphase spreads with positive GacCEN staining, and 21 with negative GacCEN staining (Table 2). This variability is demonstrated by both positive and negative staining on metaphase spreads from the same male (Fig. 3). These data suggest that the Y centromere is likely not a completely different sequence, but instead might represent a divergent repeat sequence and/or a reduced number of the same repeat sequence.
Table 2.
GacCEN staining on Y centromere from Pacific Ocean Y and Japan Sea neo-Y
| Pacific Ocean Y | Japan Sea neo-Y | |
|---|---|---|
| GacCEN positive staining | 29 | 0 |
| GacCEN negative staining | 21 | 26 |
| Number of males | 5 | 6 |
| Number of experiments | 11 | 4 |
The number of metaphase spreads with GacCEN positive and negative staining, the number of males used to generate the metaphase spreads, and the number of independent FISH experiments are indicated.
Fig. 3.
GacCEN staining is weak and variable on the ancestral Pacific Ocean male Y chromosome centromere. FISH with the GacGEN probe (green) and Y chromosome BAC 91G03 (purple) on four different metaphase spreads from the same Pacific Ocean male. Two metaphase spreads show GacCEN staining on the Y chromosome (a-d) while two metaphase spreads lack GacCEN staining on the Y chromosome (e-h). Panels (b, d) show higher magnification of the boxed regions in (a, c) highlighting Y chromosomes with weak GacCEN staining (green arrowhead), and panels (f, h) show higher magnification of the of the boxed regions in (e, g) highlighting Y chromosomes with no GacCEN staining. Scale bar, 5 μm
To determine whether the centromere sequence in the Japan Sea species is the same as in the Pacific Ocean species, we also performed CENP-A ChIP-seq in Japan Sea males and females at the same time we performed the Pacific Ocean CENP-A ChIP-seq (Cech and Peichel 2015). We had previously demonstrated that the Pacific Ocean and Japan Sea CENP-A genes only differ by a single amino acid (Cech and Peichel 2015), which is not in the region of the protein used for the antibody design (Fig. S6). Following previously described methods (Cech and Peichel 2015), we first identified the 500 most abundant sequence clusters in each IP sample and then analyzed the clusters with a fold enrichment of greater than one in the IP relative to the input (Table 3). Using these enriched sequence clusters, we identified a consensus CENP-A associated putative centromere sequence in both Japan Sea males and females. As we found previously, most of the sequence clusters enriched in the IP relative to the input aligned to this consensus sequence (Table 3). The enriched sequence clusters that did not align to the consensus sequence did not align to each other and were among the clusters with the lowest enrichment in each sample (Table 3). Thus, these experiments identified one major CENP-A associated repeat in the Japan Sea males and females; this Japan Sea putative centromere sequence is 98.7% similar to the Pacific Ocean GacCEN and is also 186 bp and 61.2 % AT rich (Fig. S7). Similar to the Pacific Ocean population, the Japan Sea GacCEN probe shows hybridization to the constriction on metaphase spreads (Fig. S8) as well as co-localization with CENP-A (Fig. S9).
Table 3.
Summary of CENP-A ChIP seq data in two Japan Sea male replicates and two Japan Sea female replicates.
| Japan Sea male 1 |
Japan Sea male 2 |
Japan Sea female 1 |
Japan Sea female 2 |
|
|---|---|---|---|---|
| Number of enriched sequence clusters in 500 most abundant | 228 | 230 | 175 | 170 |
| Number of enriched sequence clusters with GacCEN repeat | 220 | 196 | 166 | 97 |
| Average fold enrichment of IP/input | 10.44 | 10.19 | 13.58 | 3.00 |
| Range of fold enrichment across all sequence clusters | 1.00 – 39.29 | 1.00 – 41.62 | 1.03 – 49.36 | 1.00 – 7.24 |
| Range of fold enrichment for clusters without GacCEN | 1.00 – 1.17 | 1.00 – 3.72 | 1.03 – 2.09 | 1.00 – 1.29 |
| Total IP reads | 34,418,733 | 28,294,604 | 23,676,014 | 32,155,558 |
| Total input reads | 32,317,012 | 26,307,950 | 24,794,940 | 38,968,765 |
We performed FISH using the GacCEN probe in combination with BAC probes on Japan Sea metaphase spreads to determine whether it present on the neo-Y. There is strong hybridization of the GacCEN probe to the ancestral chromosome 9 centromere, providing evidence that this centromere has not been deleted on the neo-Y (Fig. 4). Although we also observed strong GacCEN staining on both X chromosomes in Japan Sea females (Fig. S10), there was no hybridization of the GacCEN probe to the ancestral Y centromere on the neo-Y (Fig. 4). In contrast to the variable hybridization of the GacCEN probe to the Y chromosome in Pacific Ocean males (Fig. 3), the lack of GacCEN hybridization to the Y chromosome was consistently observed on 26 different metaphase spreads from 6 independent Japan Sea males over four different experiments (Table 2). Because we do not know the specific sequence of the Y centromere, we cannot determine whether the lack of staining to the Y centromere on the neo-Y is due to deletion of the centromeric DNA on the neo-Y or further divergence of the centromeric DNA on the neo-Y. However, these data do suggest that there has been a genetic alteration to the ancestral Y chromosome centromere on the Japan Sea neo-Y.
Fig. 4.
GacCEN hybridizes to the centromere of chromosome 9 on the Japan Sea neo-Y chromosome. FISH with the GacCEN probe (green), Y chromosome BAC probe 188J19 (purple), and a chromosome 9 BAC probe 44L12 (purple) on a metaphase spread from a Japan Sea male is shown in panel (a). Panel (b) is a magnification of the neo-Y chromosome from the boxed region in panel (a). The GacGEN probe only localizes to the region of the ancestral chromosome 9 centromere on the neo-Y. Scale bar, 5 μm
To further assess whether there has been a deletion of the ancestral Y centromere on the neo-Y, we examined the hybridization pattern of BAC probes flanking the ancestral Y centromere on the neo-Y. Previous work had identified two BAC probes (171H24 and 180J08; Table 1) flanking the centromeric constriction on both the X and Y chromosomes in the Pacific Ocean species (Ross and Peichel 2008). Note that the order of these probes is different between the X and Y chromosomes due to the presence of several inversions on the Y chromosome relative to the X (Ross and Peichel 2008). There is no loss of hybridization with either probe on the Japan Sea neo-Y (Fig. 5), indicating that these large regions flanking the centromere are still present. However, when compared to hybridization on the Japan Sea X, these probes do appear to be closer together on the neo-Y (Fig. 5). Additionally, hybridization with these two probes and the GacCEN probe shows that there is also a slight distance between these two probes on the ancestral Pacific Ocean Y chromosome, along with weak internal centromere staining (Fig. 6b). By contrast, there is no internal GacCEN signal between these two BACs on the Japan Sea neo-Y (Fig. 6e) although there is clear separation between these BACs and internal GacCEN signal on both X chromosomes in Japan Sea females (Fig. S11). These data suggest, but do not definitively prove, that there has been a deletion of the ancestral Y chromosome centromere on the Japan Sea neo-Y. However, the absence of GacCEN hybridzation strongly supports a model in which there has been genetic inactivation of the ancestral Y centromere on the Japan Sea neo-Y. Still, it is important to note that we cannot determine whether genetic changes to this centromere led to its inactivation, or whether genetic changes occurred after an initial phase of centromere inactivation that was mediated by an epigenetic mechanism.
Fig. 5.
Mapping the regions flanking the ancestral Y centromere on the neo-Y fusion. FISH with the X and Y BACs 180J08 (green) and 171H24 (purple) on Japan Sea male interphase nuclei (a) and metaphase spreads (b). Panels (c) and (d) show higher magnifications of the boxed regions in panel (b). Both probes are present on the neo-Y (c) and the ancestral X1 (d), although the probes appear to be closer together on the neo-Y (c) as compared to the ancestral X1 (d). Scale bar, 5 μm
Fig. 6.
GacCEN and centromere flanking regions on the Pacific Ocean X and Y, and Japan Sea neo-Y chromosomes. FISH with the X and Y BACs 180J08 and 171H24 (purple) and GacCEN (green) on a metaphase spread from a Pacific Ocean male is shown in panel (a) and a metaphase spread from a Japan Sea male in (d). Panels (b) and (c) are magnifications of the boxed regions in panel (a) showing two distinct regions of staining (purple arrowheads) flanking weak GacCEN staining (green arrowhead) on the unfused Y (b), and two distinct regions of staining (purple arrowheads) flanking strong GacCEN staining (green arrowhead) on the X (c). Panel (e) is a magnification of the boxed regions in panel (d) showing the two centromere flanking probes (purple arrowheads) are very close together on the neo-Y with no GacCEN staining in between. The strong GacCEN staining (green arrowhead) is from the chromosome 9 centromere on the neo-Y. Scale bar, 5 μm
Discussion
Overall, we have shown that the Japan Sea neo-Y chromosome fusion has been stabilized by centromere inactivation. The ancestral Y centromere has been inactivated as evidenced by loss of CENP-A staining, and the active neo-Y centromere is derived from chromosome 9. While we currently cannot conclusively determine whether the ancestral Y centromere sequence has been deleted or has changed in some other way on the neo-Y, our experiments suggest there have been genetic changes to the Y centromere on the Japan Sea neo-Y. Our evidence for a partial deletion or loss of centromeric DNA is consistent with genetic deletions observed in engineered dicentrics in yeast and humans (Stimpson et al. 2010; Sato et al. 2012) and in human patients (Maraschio et al. 1990; Fisher et al. 1997). However, further experiments are required to identify the specific mechanism of genetic inactivation of the Y centromere on the neo-Y. These experiments are hindered because there is currently not a Y chromosome sequence assembly in threespine stickleback, as is the case in most organisms. However, we are currently working to obtain an assembled sequence of the Pacific Ocean threespine stickleback Y chromosome, which will allow us to test for genetic deletion by identifying the unique sequences at the centromere boundaries. Combining this assembly with CENP-A ChIP-seq experiments in Pacific Ocean males will also allow us to determine whether there are Y-chromosome specific centromeric repeats in the ancestral Pacific Ocean sticklebacks, and whether there have been genetic changes to these repeats on the Japan Sea neo-Y.
It would also be interesting to determine whether additional epigenetic changes have occurred on the inactive centromere. For example, inactive centromeres in wheat show different patterns of histone methylation than those found on active centromeres (Zhang et al. 2010), while the DNA at inactivated centromeres in maize is hypermethylated (Koo et al. 2011). Currently nothing is known about histone modifications or DNA methylation at either active or inactive centromeres in threespine sticklebacks.
Interestingly, the ancestral Y chromosome has independently fused to the acrocentric chromosome 12 in males of the blackspotted stickleback (G. wheatlandi) species within the past 15 million years, creating an independent neo-Y chromosome (Ross et al. 2009). From an evolutionary perspective, it is intriguing that of the seven known species of stickleback, two have undergone fixation of a Y-autosome fusion. It will be particularly interesting to test whether the Y chromosome centromere is also inactivated on the blackspotted neo-Y because previous studies in plants and humans have suggested that there may be differences in centromere size or strength that lead to the preferential inactivation of one centromere on dicentric chromosomes (Sullivan et al. 1994b; Han et al. 2009; Zhang et al. 2010; Liu et al. 2015). Although we do not know if the ancestral Y chromosome centromere in the threespine stickleback is smaller or weaker, we do know that it differs from the centromeres on the rest of the chromosomes (Cech and Peichel 2015), as is also true in humans and mice (Wolfe et al. 1985; Pertile et al. 2009; Miga et al. 2014). Taken together, these data suggest the intriguing possibility Y chromosomes might be better able to tolerate fusions than other chromosomes. Interestingly, Y-autosome fusions are more common that X-autosome fusions in fish and squamate reptiles (Pennell et al. 2015). However, the contribution of centromere strength or centromere inactivation to the establishment and fixation of chromosome fusions during evolution remains to be explored.
Supplementary Material
Fig. S10 Both submetacentric X1 chromosomes in the Japan Sea female show strong GacCEN hybridization. FISH with an X chromosome BAC 188J19 (purple), and GacCEN (green) on a Japan Sea female metaphase spread is in shown in panel (a). Panels (b) and (c) are magnifications of the boxed regions in panel (a), showing the two ancestral X1 chromosomes, with strong GacCEN staining (green arrowhead) consistent with the submetacentric position of the centromere. Scale bar, 5 μm
Fig. S8 The GacCEN probe hybridizes to the centromere on Japan Sea chromosomes. (a) The GacCEN probe hybridizes to a single region on each chromosome in a metaphase spread from a Japan Sea male. Panel (b) shows a magnification of the boxed region in (a), highlighting the hybridization of the GacCEN probe to the primary constriction (white arrowheads) on each chromosome. Scale bar, 5 μm
Fig. S9 The GacCEN probe colocalizes with CENP-A on Japan Sea chromosomes. The GacCEN probe (green) colocalizes with the CENP-A antibody (purple) at distinct puncta in interphase nuclei (a) as well as to a single region on each chromosome in a metaphase spread from a Japan Sea embryo (b). Panel (c) shows a magnification of the metaphase spread shown in (b). Scale bar, 5 μm
Fig. S11 GacCEN and centromere flanking regions on Japan Sea female X1 chromosomes. FISH with the X and Y BACs 180J08 and 171H24 (purple) and GacCEN (green) on a metaphase spread from a Japan Sea female is shown in panel (a). Panels (b) and (c) are magnifications of the boxed regions in (a) showing two distinct regions of BAC hybridization (purple arrowheads) flanking strong GacCEN staining (green arrowhead) on both X1 chromosomes. Scale bar, 5 μm
Fig. S1 Pacific Ocean (ancestral) and Japan Sea (derived) chromosomes used in this study. A fusion between the acrocentric chromosome 9 and the metacentric Y chromosome from the ancestral Pacific Ocean species gave rise to the neo-Y chromosome in the Japan Sea species around two million years ago. The Japan Sea sticklebacks still retain the ancestral submetacentric X chromosome (now termed X1), and the unfused acrocentric chromosome 9 (now termed X2).
Fig. S2 The ancestral state of chromosome 9, and the X and Y in Pacific Ocean stickleback fish. FISH with the ancestral X and Y chromosome BACs 188J19 (purple) and101E08 (green) and the chromosome 9 BAC 44L12 (purple) on a Pacific Ocean male metaphase spread (a) shows the two unfused chromosome 9s, the X chromosome, and the unfused Y chromosome each highlighted with a square box. Higher magnification of the boxed regions in panel (a) shows the X chromosome with two regions of BAC hybridization (b), the unfused Y chromosome with two regions of BAC hybridization (c), and the two unfused chromosome 9s (d, e). Scale bar, 5 μm
Fig. S3 Telomere staining in Japan Sea male and female metaphase spreads. Telomere staining is seen on the ends of chromosomes in (a) Japan Sea female metaphase chromosomes and (b) Japan sea male metaphase chromosomes. The neo-Y is the largest chromosome and is highlighted by a box in panel (b). Panel (c) shows a higher magnification view of the neo-Y with no internal telomere signal. The primary centromeric constriction on the neo-Y is indicated by the white arrowhead. Scale bar, 5 μm
Fig. S4 The BAC clone 91G03 is a Y specific BAC. (a) FISH was performed on a Pacific Ocean male metaphase spread with BACs 101E08 (green), and 91G03 (purple). 101E08 hybridizes to the X and Y chromosome, while 91G03 only hybridizes to the Y. Panel (b) is a magnification of the Y chromosome from (a) showing hybridization of the known sex chromosome BAC 101E08 and BAC 91G03 to the very end of the Y chromosome. Panel (c) is a magnification of the X chromosome from (a), with hybridization of BAC 101E08 to the middle of the long arm, and no hybridization of BAC 91G03. Scale bar, 5 μm
Fig. S5 CENP-A antibody staining on the ancestral Pacific Ocean Y chromosome. (a) A metaphase spread from Pacific Ocean embryos was stained with the CENPA antibody (green) and the Y chromosome specific BAC 91G03 (purple). Panel (b) is a magnification of the boxed region in panel (a), highlighting the Y chromosome with 91G03 staining on the end of the long arm, and two distinct CENP-A puncta hybridizing to the centromere of each sister chromatid on the Y chromosome. Scale bar, 5 μm
Fig. S6 Comparison of the CENP-A protein amino acid sequence between the Pacific Ocean (PO) and Japan Sea (JS) sticklebacks. There is only one amino acid difference (red asterisk) between the two proteins, which is not in the amino acid sequence targeted by the CENP-A antibody (red letters).
Fig. S7 Comparison of the CENP-A associated centromeric sequence between the Pacific Ocean (PO) and Japan Sea (JS) stickleback species. The Japan Sea consensus sequence is 98.9% similar to the GacCEN sequence previously identified in the Pacific Ocean species (Cech and Peichel 2015). The red asterisk denotes the only three nucleotide differences between these two consensus sequences. Nucleotide ambiguities: Y = C or T; R = A or G.
Acknowledgements
We thank Kohta Yoshida for his initial observations, and the Peichel Lab, Sue Biggins and Steve Henikoff for helpful discussions on this project and manuscript. We thank the Fred Hutchinson Cancer Research Center Genomics Shared Resource for help with the ChiP-seq experiment, and Ryan Basom for help with data analysis. This research was supported by a National Science Foundation Graduate Research Fellowship (DGE-1256082), the National Institutes of Health Chromosome Metabolism and Cancer Training Grant (T32 CA009657), a National Institutes of Health grant (R01 GM116853), and the Fred Hutchinson Cancer Research Center.
Abbreviations
- BAC
Bacterial artificial chromosome
- CENP-A
Centromere protein A
- ChIP-seq
Chromatin immunoprecipitation sequencing
- Chr
Chromosome
- DAPI
4’,6’-Diamidino-2-phenlyindole
- FISH
Fluorescence in situ hybridization
- GacCEN
Threespine stickleback (Gasterosteus aculeatus) centromeric repeat sequence
- IF-FISH
Immunofluorescence combined with FISH
- IP
Immunoprecipitation
- PBS
Phosphate-buffered saline
- PBST
Phosphate-buffered saline Tween-20
References
- Agudo M, Abad JP, Molina I, et al. A dicentric chromosome of Drosophila melanogaster showing alternate centromere inactivation. Chromosoma. 2000;109:190–196. doi: 10.1007/s004120050427. [DOI] [PubMed] [Google Scholar]
- Alkan C, Cardone MF, Catacchio CR, et al. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genome Res. 2011;21:137–145. doi: 10.1101/gr.111278.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, et al. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Human Genet. 1992;89:247–249. doi: 10.1007/BF00217134. [DOI] [PubMed] [Google Scholar]
- Bailey SM. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006;34:2408–2417. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PHJL, Stellfox ME, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech JN, Peichel CL. Identification of the centromeric repeat in the threespine stickleback fish (Gasterosteus aculeatus) Chromosome Res. 2015;23:767–779. doi: 10.1007/s10577-015-9495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosland MW, Crozier RH. Myrmecia pilosula, an ant with only one pair of chromosomes. Science. 1986;231:1278–1278. doi: 10.1126/science.231.4743.1278. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev Genet. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- Fisher AM, Al-Gazali L, Pramathan T, et al. Centromeric inactivation in a dicentric human Y;21 translocation chromosome. Chromosoma. 1997;106:199–206. doi: 10.1007/s004120050240. [DOI] [PubMed] [Google Scholar]
- Flemming W. Beitrage zur Kenntniss der Zelle und ihrer Lebenserscheinungen Theil II (in German) Arch Mikros Anat. 1880;18:151–259. [Google Scholar]
- Fukagawa T, Earnshaw WC. Neocentromeres. Curr Biol. 2014;24:R946–R947. doi: 10.1016/j.cub.2014.08.032. [DOI] [PubMed] [Google Scholar]
- Gallardo MH, Bickham JW, Honeycutt RL, et al. Discovery of tetraploidy in a mammal. Nature. 1999;401:341. doi: 10.1038/43815. [DOI] [PubMed] [Google Scholar]
- Gao Z, Fu S, Dong Q, et al. Inactivation of a centromere during the formation of a translocation in maize. Chromosome Res. 2011;19:755–761. doi: 10.1007/s10577-011-9240-5. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Pettersson L, Höglund M, et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Jonson T, Petersén A, et al. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AM, Killingbeck EE, Mitros T, et al. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with genotyping-by-sequencing. G3 (Bethesda) 2015;5:1463–1472. doi: 10.1534/g3.115.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Wu Y, Koblizkova A, et al. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell. 2012;24:3559–3574. doi: 10.1105/tpc.112.100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Gao Z, Birchler JA. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell. 2009;21:1929–1939. doi: 10.1105/tpc.109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Schueler MG, Willard HF. Chromosome engineering: generation of mono- and dicentric isochromosomes in a somatic cell hybrid system. Chromosome. 1999;108:256–265. doi: 10.1007/s004120050376. [DOI] [PubMed] [Google Scholar]
- Higgins AW, Gustashaw KM, Willard HF. Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res. 2005;13:745–762. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, et al. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Östlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the Three-Spined Stickleback. CRC press; Boca Raton: 2007. pp. 41–81. [Google Scholar]
- Kitano J, Mori S, Peichel CL. Phenotypic divergence and reproductive isolation between sympatric forms of Japanese threespine sticklebacks. Biol J Linn Soc. 2007;91:671–685. [Google Scholar]
- Kitano J, Ross JA, Mori S, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Han F, Birchler JA, Jiang J. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 2011;21:908–914. doi: 10.1101/gr.116202.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell LH. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987;48:801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SKM, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune J, Dutrillaux B, Rethoré MO, Prieur M. Comparaison de la structure fine des chromatides d’Homo sapiens et de Pan troglodytes. Chromosoma. 1973;43:423–444. doi: 10.1007/BF00406748. [DOI] [PubMed] [Google Scholar]
- Liu Y, Su H, Pang J, et al. Sequential de novo centromere formation and inactivation on a chromosomal fragment in maize. Proc Natl Acad Sci USA. 2015;112:E1263–1271. doi: 10.1073/pnas.1418248112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraschio P, Zuffardi O, Caiulo A, et al. Deletion of specific sequences or modification of centromeric chromatin are responsible for Y chromosome centromere inactivation. Human Genet. 1990;85:491–494. doi: 10.1007/BF00194222. [DOI] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 2016;17:16–29. doi: 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14:R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, et al. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Newton J, Jain M, et al. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP. Telomere loss as a mechanism for chromosome instability in human cancer. Cancer Res. 2010;70:4255–4259. doi: 10.1158/0008-5472.CAN-09-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Page SL, Shaffer LG. Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosome Res. 1998;6:115–122. doi: 10.1023/a:1009286929145. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, et al. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Margolis RL. Biochemical analysis of CENP-A, a centromeric protein with histone-like properties. Prog Clin Biol Res. 1989;318:61–72. [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Trong HL, et al. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Kirkpatrick M, Otto SP, et al. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 2015;11:1005237. doi: 10.1371/journal.pgen.1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertile MD, Graham AN, Choo KHA, Kalitsis P. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 2009;19:2202–2213. doi: 10.1101/gr.092080.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Magnani E, et al. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010;6:e1000845. doi: 10.1371/journal.pgen.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S, Marcand S. Dicentric breakage at telomere fusions. Genes Dev. 2010;24:720–733. doi: 10.1101/gad.571510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Peichel CL. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics. 2008;179:2173–2182. doi: 10.1534/genetics.108.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, et al. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Masuda F, Takayama Y, et al. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol. 2012;22:658–667. doi: 10.1016/j.cub.2012.02.062. [DOI] [PubMed] [Google Scholar]
- Sears ER, Câmara A. A transmissible dicentric chromosome. Genetics. 1952;37:125–135. doi: 10.1093/genetics/37.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Black BE. Molecular underpinnings of centromere identity and maintenance. Trends Biochem Sci. 2012;37:220–229. doi: 10.1016/j.tibs.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Toyoda A, et al. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ye Y, Duan D. Comparative cytogenetic studies on the red muntjac, Chinese muntjac, and their F1 hybrids. Cytogenet Cell Genet. 1980;26:22–27. doi: 10.1159/000131417. [DOI] [PubMed] [Google Scholar]
- Stimpson KM, Song IY, Jauch A, et al. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 2010;6:e1001061. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Matheny JE, Sullivan BA. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res. 2012;20:595–605. doi: 10.1007/s10577-012-9302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994a;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Wolff DJ, Schwartz S. Analysis of centromeric activity in Robertsonian translocations: implications for a functional acrocentric hierarchy. Chromosoma. 1994b;103:459–467. doi: 10.1007/BF00337384. [DOI] [PubMed] [Google Scholar]
- Tek AL, Kashihara K, Murata M, Nagaki K. Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon. Chromosome Res. 2010;18:337–347. doi: 10.1007/s10577-010-9119-x. [DOI] [PubMed] [Google Scholar]
- Therman E, Susman B, Denniston C. The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations. Ann Hum Genet. 1989;53:49–65. doi: 10.1111/j.1469-1809.1989.tb01121.x. [DOI] [PubMed] [Google Scholar]
- Urton JR, McCann SR, Peichel CL. Karyotype differentiation between two stickleback species (Gasterosteidae) Cytogenet Genome Res. 2011;135:150–159. doi: 10.1159/000331232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg WJ. Ophioglossum reticulatum L. Record from PROTA4U. In: Grubben GJH, Denton OA, editors. PROTA (Plant Resources of Tropical Africa / Ressources végétales de l’Afrique tropicale) Wageningen; Netherlands: [Accessed 21 June 2016]. 2004. http://www.prota4u.org/search.asp. [Google Scholar]
- Wang W, Lan H. Rapid and parallel chromosomal number reductions in muntjac deer inferred from mitochondrial DNA phylogeny. Mol Biol Evol. 2000;17:1326–1333. doi: 10.1093/oxfordjournals.molbev.a026416. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Wolfe J, Darling SM, Erickson RP, et al. Isolation and characterization of an alphoid centromeric repeat family from the human Y chromosome. J Mol Biol. 1985;182:477–485. doi: 10.1016/0022-2836(85)90234-7. [DOI] [PubMed] [Google Scholar]
- Yang F, Carter NP, Shi L, Ferguson-Smith MA. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma. 1995;103:642–652. doi: 10.1007/BF00357691. [DOI] [PubMed] [Google Scholar]
- Zhang W, Friebe B, Gill BS, Jiang J. Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma. 2010;119:553–563. doi: 10.1007/s00412-010-0278-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S10 Both submetacentric X1 chromosomes in the Japan Sea female show strong GacCEN hybridization. FISH with an X chromosome BAC 188J19 (purple), and GacCEN (green) on a Japan Sea female metaphase spread is in shown in panel (a). Panels (b) and (c) are magnifications of the boxed regions in panel (a), showing the two ancestral X1 chromosomes, with strong GacCEN staining (green arrowhead) consistent with the submetacentric position of the centromere. Scale bar, 5 μm
Fig. S8 The GacCEN probe hybridizes to the centromere on Japan Sea chromosomes. (a) The GacCEN probe hybridizes to a single region on each chromosome in a metaphase spread from a Japan Sea male. Panel (b) shows a magnification of the boxed region in (a), highlighting the hybridization of the GacCEN probe to the primary constriction (white arrowheads) on each chromosome. Scale bar, 5 μm
Fig. S9 The GacCEN probe colocalizes with CENP-A on Japan Sea chromosomes. The GacCEN probe (green) colocalizes with the CENP-A antibody (purple) at distinct puncta in interphase nuclei (a) as well as to a single region on each chromosome in a metaphase spread from a Japan Sea embryo (b). Panel (c) shows a magnification of the metaphase spread shown in (b). Scale bar, 5 μm
Fig. S11 GacCEN and centromere flanking regions on Japan Sea female X1 chromosomes. FISH with the X and Y BACs 180J08 and 171H24 (purple) and GacCEN (green) on a metaphase spread from a Japan Sea female is shown in panel (a). Panels (b) and (c) are magnifications of the boxed regions in (a) showing two distinct regions of BAC hybridization (purple arrowheads) flanking strong GacCEN staining (green arrowhead) on both X1 chromosomes. Scale bar, 5 μm
Fig. S1 Pacific Ocean (ancestral) and Japan Sea (derived) chromosomes used in this study. A fusion between the acrocentric chromosome 9 and the metacentric Y chromosome from the ancestral Pacific Ocean species gave rise to the neo-Y chromosome in the Japan Sea species around two million years ago. The Japan Sea sticklebacks still retain the ancestral submetacentric X chromosome (now termed X1), and the unfused acrocentric chromosome 9 (now termed X2).
Fig. S2 The ancestral state of chromosome 9, and the X and Y in Pacific Ocean stickleback fish. FISH with the ancestral X and Y chromosome BACs 188J19 (purple) and101E08 (green) and the chromosome 9 BAC 44L12 (purple) on a Pacific Ocean male metaphase spread (a) shows the two unfused chromosome 9s, the X chromosome, and the unfused Y chromosome each highlighted with a square box. Higher magnification of the boxed regions in panel (a) shows the X chromosome with two regions of BAC hybridization (b), the unfused Y chromosome with two regions of BAC hybridization (c), and the two unfused chromosome 9s (d, e). Scale bar, 5 μm
Fig. S3 Telomere staining in Japan Sea male and female metaphase spreads. Telomere staining is seen on the ends of chromosomes in (a) Japan Sea female metaphase chromosomes and (b) Japan sea male metaphase chromosomes. The neo-Y is the largest chromosome and is highlighted by a box in panel (b). Panel (c) shows a higher magnification view of the neo-Y with no internal telomere signal. The primary centromeric constriction on the neo-Y is indicated by the white arrowhead. Scale bar, 5 μm
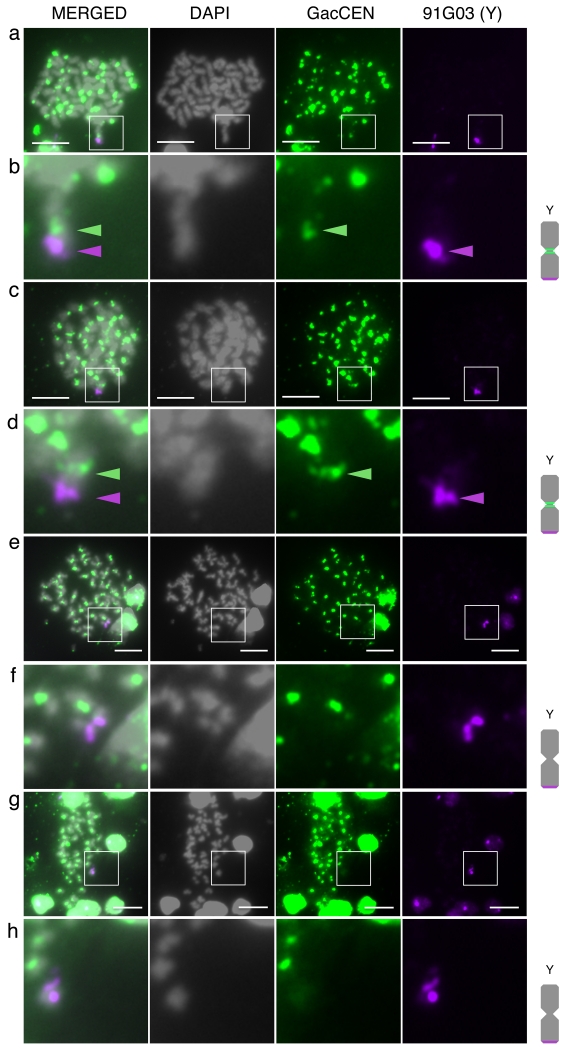
Fig. S4 The BAC clone 91G03 is a Y specific BAC. (a) FISH was performed on a Pacific Ocean male metaphase spread with BACs 101E08 (green), and 91G03 (purple). 101E08 hybridizes to the X and Y chromosome, while 91G03 only hybridizes to the Y. Panel (b) is a magnification of the Y chromosome from (a) showing hybridization of the known sex chromosome BAC 101E08 and BAC 91G03 to the very end of the Y chromosome. Panel (c) is a magnification of the X chromosome from (a), with hybridization of BAC 101E08 to the middle of the long arm, and no hybridization of BAC 91G03. Scale bar, 5 μm
Fig. S5 CENP-A antibody staining on the ancestral Pacific Ocean Y chromosome. (a) A metaphase spread from Pacific Ocean embryos was stained with the CENPA antibody (green) and the Y chromosome specific BAC 91G03 (purple). Panel (b) is a magnification of the boxed region in panel (a), highlighting the Y chromosome with 91G03 staining on the end of the long arm, and two distinct CENP-A puncta hybridizing to the centromere of each sister chromatid on the Y chromosome. Scale bar, 5 μm
Fig. S6 Comparison of the CENP-A protein amino acid sequence between the Pacific Ocean (PO) and Japan Sea (JS) sticklebacks. There is only one amino acid difference (red asterisk) between the two proteins, which is not in the amino acid sequence targeted by the CENP-A antibody (red letters).
Fig. S7 Comparison of the CENP-A associated centromeric sequence between the Pacific Ocean (PO) and Japan Sea (JS) stickleback species. The Japan Sea consensus sequence is 98.9% similar to the GacCEN sequence previously identified in the Pacific Ocean species (Cech and Peichel 2015). The red asterisk denotes the only three nucleotide differences between these two consensus sequences. Nucleotide ambiguities: Y = C or T; R = A or G.