Abstract
Large-scale microparticle arrays (LSMA) are key for material science and bioengineering applications. However, previous approaches suffer from tradeoffs between scalability, precision, specificity, and versatility. Here, we present a porous microwell-based approach to create large-scale microparticle arrays with complex motifs. Microparticles are guided to and pushed into microwells by fluid flow through small open pores at the bottom of the porous well arrays. A scaling theory allows for the rational design of LSMAs to sort and array particles based on their size, shape or modulus. Sequential particle assembly allows for proximal and nested particle arrangements, as well as particle recollection and pattern transfer. We demonstrate the capabilities of the approach by means of three applications: high-throughput single-cell arrays; microenvironment fabrication for neutrophil chemotaxis; and complex, covert tags by the transfer of an upconversion nanocrystal laden LSMA.
Large-scale microparticle arrays (LSMA) have unmatched encoding capacities and rapid decoding capabilities that make them very attractive for a broad range of applications. LSMAs are of utmost interest in biochemical assays1, 2, 3, 4, significantly reducing the amount of time and labor. With high specificity, spatial positioning plays an important role in mimicking biological tissues in vitro to study cellular behavior, such as migration5 and intercellular communication6. The ability to specifically position microparticles enables one to spatially encode chemical or cellular building blocks, resulting in the local control of the microenvironment. For anti-counterfeiting applications, spectrally or graphically coded microparticles are desirable as information carriers because of their high encoding capacity within a small area7, 8, 9. Arrangements of multiple microparticles exponentially increase the potential encoding capacity. However, current approaches to position microscale particles in large arrays cannot simultaneously accomplish the requirements of scalability, precision, specificity, and versatility that will make LSMAs practical. For example, optical tweezers10, 11, 12 provide the high resolution of positioning, but can only be applied to small arrays and small dielectric objects with high object-medium refractive indexes mismatch. Manipulation methods using optoelectronic tweezers13 could accomplish large-scale, high resolution arrangement and sorting, but the technique is unsuitable to generate a heterogeneous pattern containing different types of particles. Magnetic techniques14, 15 have a limitation in versatility because arranged objects should be doped with magnetic materials. Micro-magnetic robotic coding16 overcomes this limitation but becomes unsuitable for large-scale arrangement. Microwells have been used as an assembly template, but are only useful for single particle arrangement and scaling-up efforts resulted in particle arrangements with some degree of randomness1, 3, 4, 17. Recently, geometric docking into conformal microwells enables shape specific positioning, but requires a large particle size and significantly long assembly time5. Therefore, to achieve the full potential of LSMAs, new techniques are required to assemble functional microparticles in precise locations that also have high yields and small error rates.
Here, we develop a porous microwell platform to generate LSMAs with high specificity. Microparticles are steered to the microwells via hydrodynamic forces associated with fluid flow through open pores at the bottom of the microwell. Guided microparticles are inserted into congruent microwells, whereas geometrically mismatched particles are removed in a washing step. Controllable driving forces provide the appropriate magnitude for various microparticles, and have the ideal directionality of a force field (i.e. directed towards the well) to assemble the microparticles. By iterative assembly and washing steps, large-scale particle assembly with high yield is demonstrated within 100 seconds. Shape, size, and modulus sorting is achieved with high specificity. We demonstrate that the technique is also compatible with particle recollection and pattern transfer. As a demonstration, we generate a 2D particle-array-code for anti-counterfeiting applications, which we then transfer to a precise location on a target substrate. We also generate high throughput single cell arrays, and 2D chemokine-releasing particle arrays to study the motility of immune cells in complex-gradient microenvironments.
Design principles for LSMA
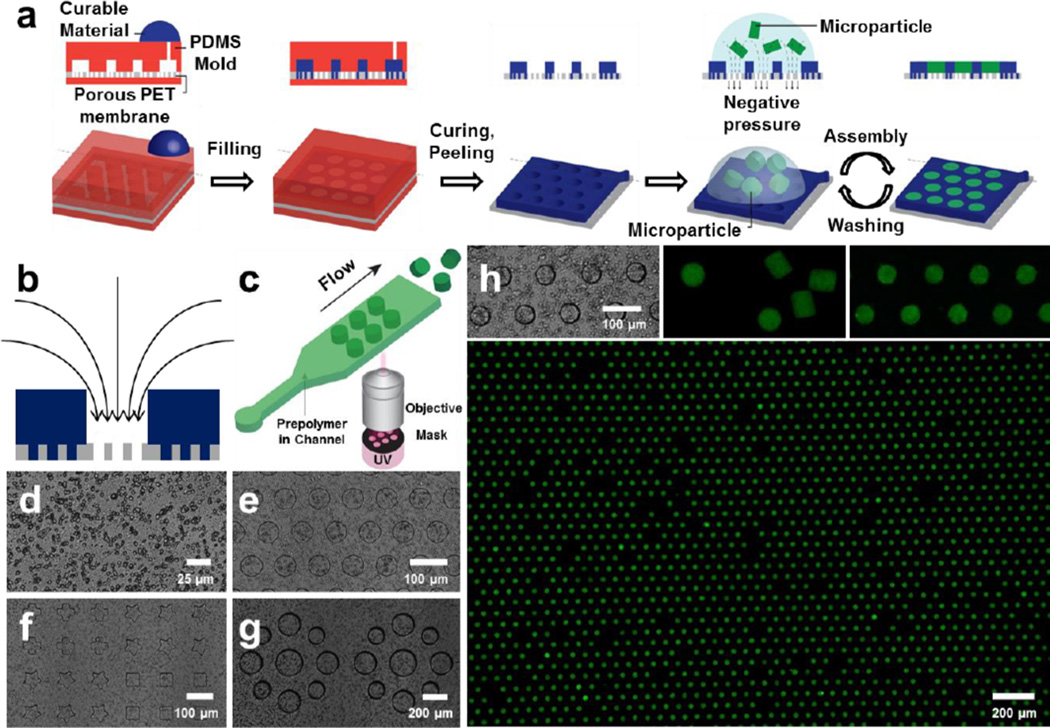
We fabricate porous microwell arrays to generate optimal conditions for particle assembly. We postulate that the ideal force driving for particle assembly should have a direction which points into the assembly template (i.e. microwells). Moreover, the force should have a magnitude which is controllable in order to achieve a high-throughput arrangement without damaging the microparticles during the process. Upon application of a pressure difference across the porous microwells, the resulting driving force in our platform meets these two criteria: flow streamlines point toward open pores inside the microwells, and the magnitude of the driving force is proportional to the applied pressure difference across the well (Fig. 1b). We fabricated the microwell arrays on top of a porous polyethylene terephthalate (PET) membrane (Fig. 1d) and fine-tuned their geometry with the aid of a polydimethylsiloxane (PDMS) mold (Fig. 1a). The mold and the flat PDMS were assembled at the top and bottom of the PET membrane respectively, and a curable material was injected into the mold. Pores that were in contact with the mold were not filled by the curable material. After we cured the microwells, the PDMS mold was peeled away to leave the porous microwell arrays. Both photocurable Norland optical adhesive (NOA) (Fig. 1e, f) and thermal curable PDMS (Fig. 1g) were successfully used to fabricate microwells with distinct shapes, sizes, and arrangements (see Fig. S5, S6 for porous microwells made by a variety of photo/thermal curable materials).
Figure 1. Porous microwells for microparticle arrays.
a, Porous microwell fabrication and microparticle assembly. b, Direction of negative pressure induced driving force of porous microwell arrays. c, Synthesis of microparticles by stop flow lithography. d, Porous PET membrane (pore diameter = 3 µm). e–g, Geometry fine-tuned microwell arrays with photo-curable NOA (e, f) and thermal-curable PDMS (g). h, Large-scale (3800), high throughput microparticle assembly: Porous NOA microwell arrays (top-left), PEGDA microparticles (top-middle), assembled PEGDA microparticle arrays in close (top-right) and wide view (bottom).
Microparticles to be assembled were synthesized by stop flow lithography (SFL) with a throughput of 105 particles/h1, 2, 7, 8 (Fig. 1c), and stored in aqueous solution. The particle synthesis throughput can be increased two orders of magnitude by using contact flow lithography18, which is an advanced form of SFL. To show the widespread potential of our platform, we synthesized and assembled various microparticles from two very different sets of prepolymer solutions. Soft, biocompatible microparticles, which are widely used in bioassays and microenvironment fabrication, were synthesized from a poly(ethylene glycol) diacrylate (PEGDA) prepolymer solution. More rigid and thermally stable microparticles for anti-counterfeiting applications were made from a mixture of polyurethane acrylate (PUA) and acrylic acid (AA).
Microparticle assembly in microwells
To generate LSMA, a particle suspension was deposited on top of the microwells, and a negative pressure was applied to the bottom of the array. Microparticles were guided to microwells via fluid flow through the open pores and assembled into shape-matched microwells. Within 10 iterations of assembly and washing, where each cycle takes approximately 10 seconds, large-scale (3800) PEGDA microparticle arrays were successfully generated (Fig. 1h, see Fig. S7–S10 for the largest particle array (15000) and the smallest assembled particles (15 µm)). The applied pressure deformed the soft PEGDA microparticles to squeeze them into slightly smaller but geometrically conformal microwells. Once pushed inside a microwell, the compressed microparticle will push upon the side wall of the microwell which in turn helps to keep a particle in the well during successive wash and assembly steps. This ability to keep particles retained within a well during successive assembly steps, and the ideal driving force directed into the microwells ensure high assembly yield. In Figure 1h, 93.7 (±0.8) % of the microwells were filled with particles, and 87.3 (± 0.7) % of the initially deposited particles were assembled into microwells.
Physics of the LSMA assembly
Ad hoc choice of microwell dimensions, particle attributes and operating conditions would limit the utility of our approach. For example, it is possible to increase the magnitude of the driving force to increase the assembly speed, but eventually particles will be damaged and result in non-specific particle assembly – a particle with a square cross-section could be jammed into a non-conformal cylindrical well. Understanding the physics behind the assembly process allows us to tune the driving force and engineer characteristics of the microwells and microparticles to achieve highly specific assembly of complex motifs. Here, we introduce the key dimensionless parameters governing operation of the devices.
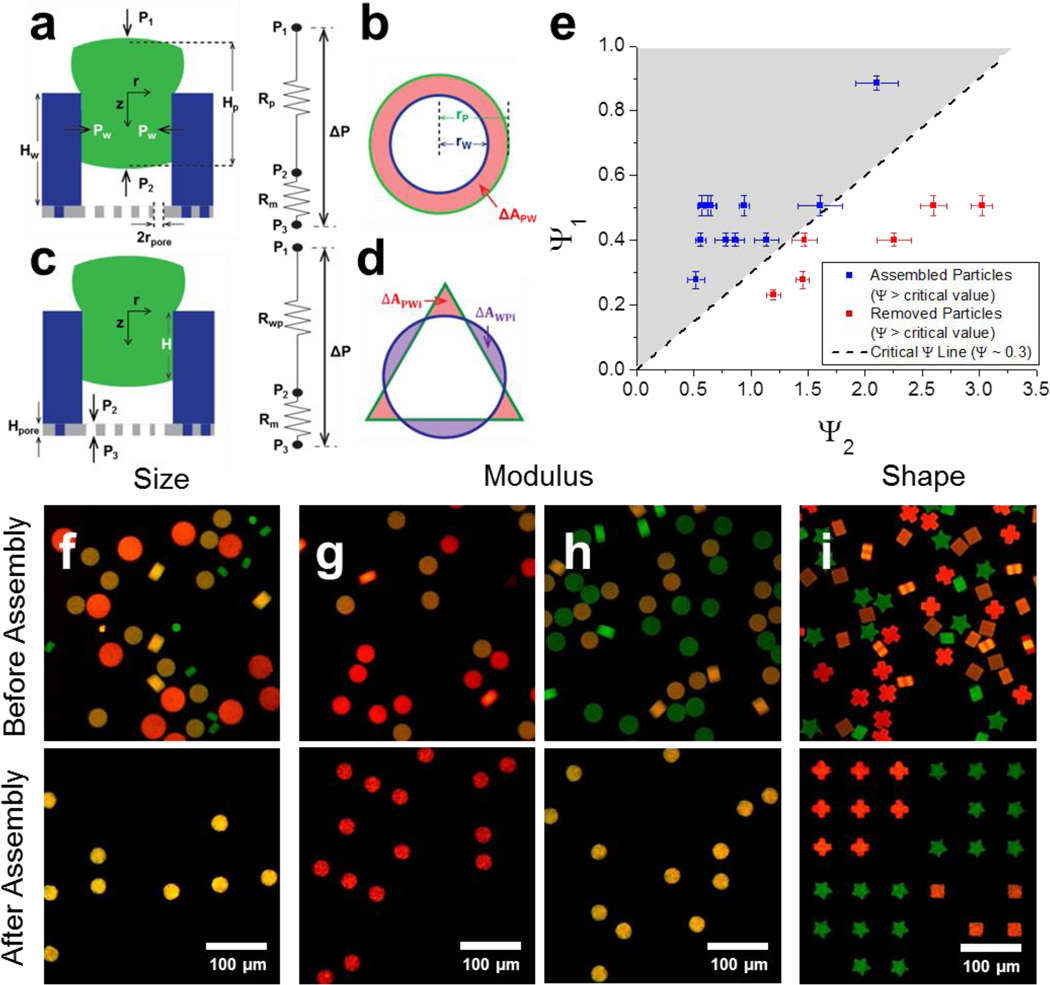
Consider a soft PEGDA particle which conformally blocks the entrance of a microwell. The force originated from the applied pressure difference (Δ) is balanced with the wall friction (Fig. 2a). By using the theory behind the interference fit19 and capillary micromechanics20, 21, scaling analysis is conducted to define the dimensionless number Ψ governing particle assembly which is the ratio of the applied pressure difference to the wall friction (see the supplementary information for the derivation).
| (1) |
where ΔP is the applied pressure difference, Ep is the compressive modulus of particle, rp is the radius of particle, rw is the radius of microwell, μ is the friction coefficient between microwell and particle (~ 10−3 for hydrogel particles22), Hp is height of microparticle, and ΔApw (=Σi ΔApw) is the area of particle cross section which sticks out from the cross section of the microwell (Fig. 2a–d). We can recognize that Ψ is the ratio of two dimensionless numbers: (i) the combination of the applied pressure difference and materials’ properties (Ψ1), and (ii) a purely geometric group (Ψ2).
| (2) |
| (3) |
Figure 2. Scaling analysis and characteristic specific positioning.
a–d, Schematics for scaling analysis: Shape matched (a,b) and shape mismatched cases (c,d) at side (a,c) and top views (b,d). Schematics of hydrodynamic resistance of platform are shown next to the side view (a, c). e, Phase diagram, Ψ1 as a function of Ψ2, for particle assembly in shape matched cases. PEGDA particles fit into NOA microwells only when Ψ is larger than critical value (= 0.3). f–i, Characteristic specific positioning of particles based on size (f), modulus (g, h), and shape (i). f, Diameter of green, yellow, and red particles are 30, 60, and 81 µm respectively. Only yellow particles fit into 54 µm microwells. g, h, Modulus of green, yellow, and red particles are 240, 140, and 63 kPa. Diameters of particles are 63 µm (g) and 59 µm (h). Particles with a smaller modulus than critical value in each case can fit into 54 µm microwells. i, Shape of green, yellow, and red particles are star, square, and cross respectively while all particles have same modulus and size. Particles fit into only congruent microwells. Specificities of positioning are 96.6 (± 1.8) % (f), 97.5 (± 2.2) % (g), 95.8 (±5.3) % (h), and 98.8 (±0.8) (i). Error bars represent standard deviation (n = 3, 4).
Microparticles are assembled into microwells only when the dimensionless parameter Ψ is larger than a critical value, i.e. when applied pressure overcomes wall friction. This criterion is confirmed by plotting Ψ1 as a function of Ψ2, and the critical value of Ψ is found as 0.3 (Fig. 2e). This scaling analysis allows us to specifically position particles based on their characteristics. Size specific positioning was successfully demonstrated with high specificity (96.6 % for Fig. 2f). Specificity is defined as the number of correctly assembled particles per the total number of assembled particles. Particles which are slightly larger than microwells were selectively assembled into microwells, whereas too small particles were inserted but then removed in the washing step. As rp increases, Ψ2 increases because ΔApw increases more rapidly than rp, resulting in Ψ smaller than the critical value; particles with too large rp cannot fit into microwells. Likewise, particles of the same size but different modulus can be sorted with high specificity (97.5 % for Fig. 2g, and 95.8 % for Fig. 2h) due to the dependence of Ψ on Ep. Soft particles were successfully sorted from hard particles and positioned in wells (Fig. 2g); as Ep increases, Ψ1 decreases, resulting in Ψ smaller than critical value. As rp decreases, more rigid particles were able to fit into microwell (Fig. 2h, see Fig. S16 for the relationship between rp and Ep).
Even though particles have the same dimension and modulus, they can be specifically positioned depending on another characteristic, shape (Fig. 2i). If the shape of a microparticle is not matched with the microwell shape, the assembly driving force acting on the particles becomes significantly smaller than the shape matched case- because the hydrodynamic resistance of membrane (Rm) is much larger than that of the uncovered area (i.e. gaps) (Rwp) between the microparticle and microwell (Fig. 2c), pressure difference across the particle becomes much smaller than pressure difference across the microwell. A dimensionless number for the shape mismatched case is (see the supplementary information for the derivation)
| (4) |
| (5) |
where Dpore is the number of pores inside a microwell, Apore is the cross section area of pore, Hpore is the height of pore, and ΔAwpi is the cross section area of a microwell not covered by a microparticle (Fig. 2c, d). The ratio of pressure difference across the particle to ΔP is defined as a dimensionless number C, which is also the ratio of particle assembly driving force between shape matched and mismatched cases. Because C is much smaller than 1, high specificity of shape sorting was achieved in Fig. 2i (98.8 %, the phase diagram for Fig. 2i is shown in Fig. S17).
In a bioassay, the sensitivity for target molecule (e.g. microRNA, DNA, protein) detection depends on the porosity and surface area of the particle probe region23, 24. Thus, shape sorted particles with the same gel properties will have the same sensitivities and are useful in multiplexed bioassays.
Encoding capacity of the LSMA is the product of each physical characteristic specifying positioning, size and stiffness, and does not depend on the chemical functionality of particles. To our knowledge, this is the best result to place microparticles at a precise location depending on the largest number of physical characteristics (= 3) with the highest specificity and resulting in the highest encoding capacity.
LSMAs with complex structures
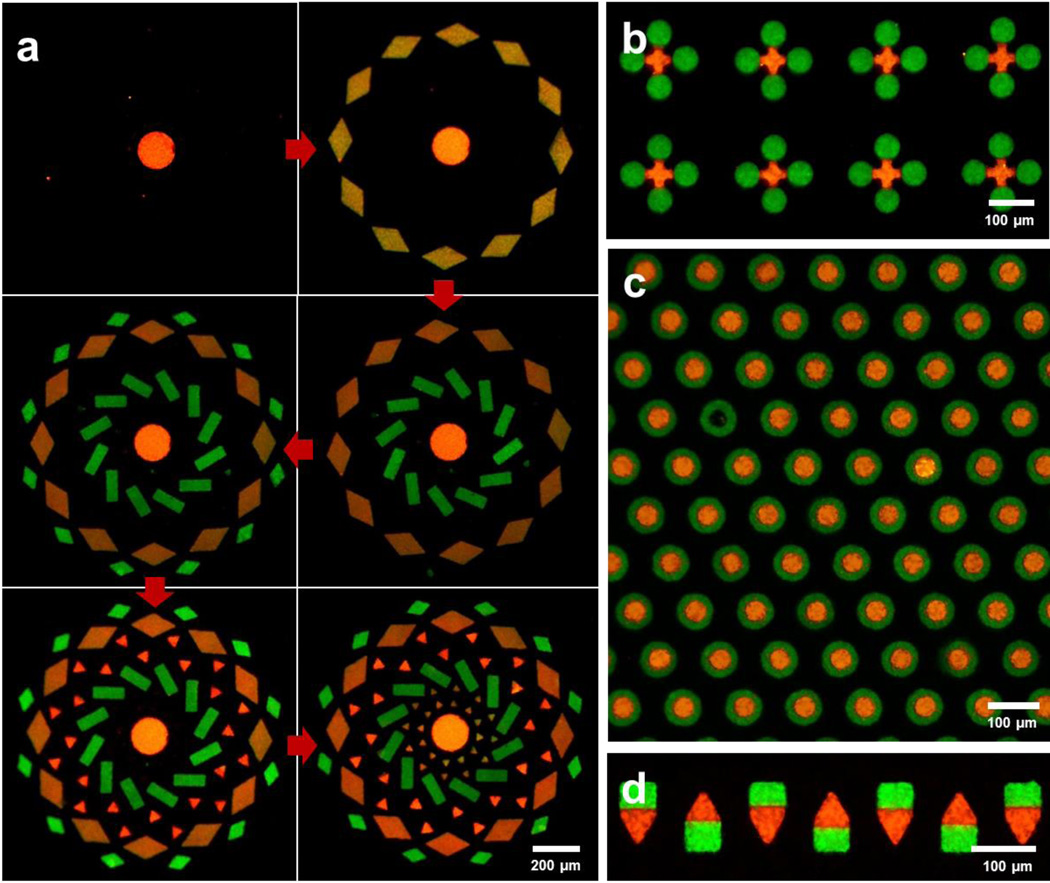
High encoding capacity and persistent positioning of particles in LSMA allowed PEGDA microparticles to be assembled sequentially in order of size to generate the complex motif shown in Figure 3a. With previous approaches, particles did not maintain their positions during iterative assembly5, so it was impossible to assembly microparticles sequentially to generate perfectly filled arrays. Multiple particles can be assembled not only into different microwells, but also at a proximal or nested location inside one microwell. Using sequential assembly, the first set of microparticles was assembled in microwells but did not fully occupy all space in the well, and the second set of microparticles filled the rest of the open space left in a microwell. Particles were located next to each other (Fig. 3b), or inside the hole of another particle (Fig. 3c). These types of arrangement generate the interface of two particles which is critical for the intercellular communication by direct contact6. To our knowledge, this is the first demonstration of non-spherical microparticle assembly resulting in direct particle contact via a microwell approach.
Figure 3. Advanced assembly techniques.
a, Sequential assembly for biocompatible, complex microparticle array fabrication. PEGDA particles are assembled in order of size while pre-assembled particles are remained in microwells. b, c, Sequentially assembly of microparticles at proximal locations. Green particles are assembled before red particle assembly. d, Oriented assembly of Janus microparticles. Particles and microwell arrays were made of PEGDA and NOA 81, respectively.
Thus far, encoding capacity was generated by positioning microparticles at precise locations depending on their characteristics. However, this post-synthesis processing is not the only way to generate codes. Chemically anisotropic microparticles (e.g. Janus or multi-striped) can be synthesized by co-flowing multiple laminar streams in microfluidic channel during the particle synthesis2, 7. In Figure 3d we demonstrate that Janus particles can be arrayed with a specified orientation in the LSMA. Encoding capacity can be exponentially scaled using the combination of the particle’s chemical anisotropy (e.g. chemically distinct stripes) and particle arrangement.
LSMA transfer techniques
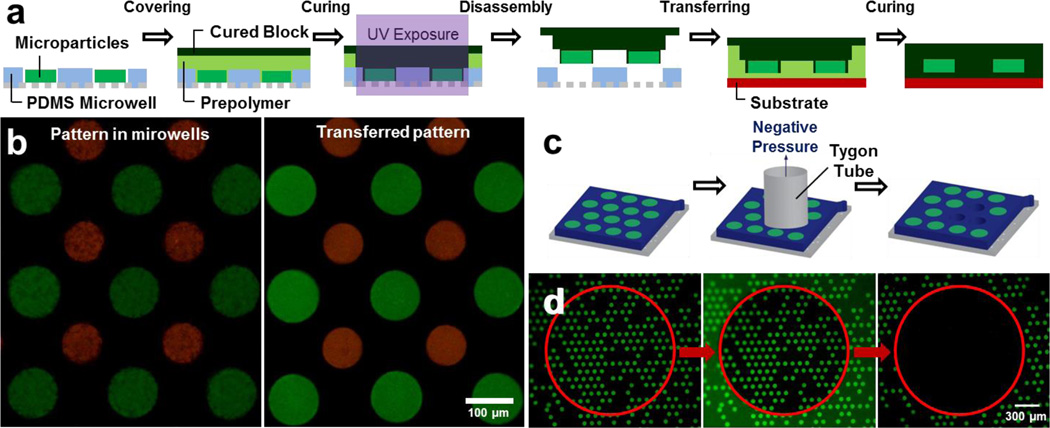
Compared to optoelectronic tweezer13 and microfluidic techniques25, 26, in our approach the arranged particles are not confined in a closed system. Therefore, particle arrays can be readily repositioned via particle recollection or pattern transfer techniques. Assembled microparticles were recollected by shear stress associated with pressure-driven fluid flow through a tygon tube (Fig. 4c). Microparticles which were positioned inside the inner diameter of tygon tube were selectively recollected (Fig. 4d). After recollection, particles restored to their original size with negligible shrinkage (< 2 %).
Figure 4. Post-assembly techniques.
a, Schematic of pattern transfer with PDMS microwell arrays. b, Biocompatible PEGDA microparticle arrays before (left) and after (right) the transfer to the glass slide. c, d, Schematic (c) and results (d) of particle recollection. PEGDA microparticles inside red circled area (inner diameter of tygon tube) were selectively recollected from NOA microwells. In the middle image, the inner diameter of the tygon tube is discerned due to the autofluorescence of the tube.
Pattern transfer technique requires oxygen permeable PDMS microwell arrays (Fig. 4a). After particle assembly, prepolymer solution was spread on top of the arrays and covered with a pre-cured polymer block. Ultraviolet light (365 nm) was specifically exposed to the particle arrays which were transferred. Cured prepolymer chemically bonded with the microparticles and pre-cured block, but prepolymer near the PDMS microwells was not cured because oxygen permeability of PDMS generates an oxygen inhibition layer27. Microparticle patterns were transferred with the pre-cured block while their relative positions did not change. Biocompatible PEGDA microparticle arrays were successfully transferred to a glass slide (Fig. 4b).
LSMAs for biological applications
The ability to generate large arrays of cells is important for cell screening applications when the yield and throughput are critical. For example, arranging cells in 2D arrays for the study of cellular processes that progress over time has significant advantages compared to serial approaches, like flow cytometry28. Cells in a 2D array can be analyzed more than once and several cells imaged simultaneously29 for higher yield and to avoid potential differences between the first and last cell analyzed which are typical issues for flow cytometry. Here, we tested the performance of LSMA techniques by trapping mammalian glioma cells (U87) in 25 µm diameter wells, spaced at 25 µm apart. We generated arrays of more than 10,000 cells with high yield. The time required to achieve acceptable yield for each array was approximately 60 seconds (Fig. 5a, Fig. S29, S30), significantly faster than passive cell settling in microwells, which requires between 5 and 40 minutes30, 31. Compared to existing technologies28, 29, 30, 31, our platform provides a way to arrange cells with high throughput and enables massively parallel imaging.
Figure 5. Application of large scale microwell arrays.
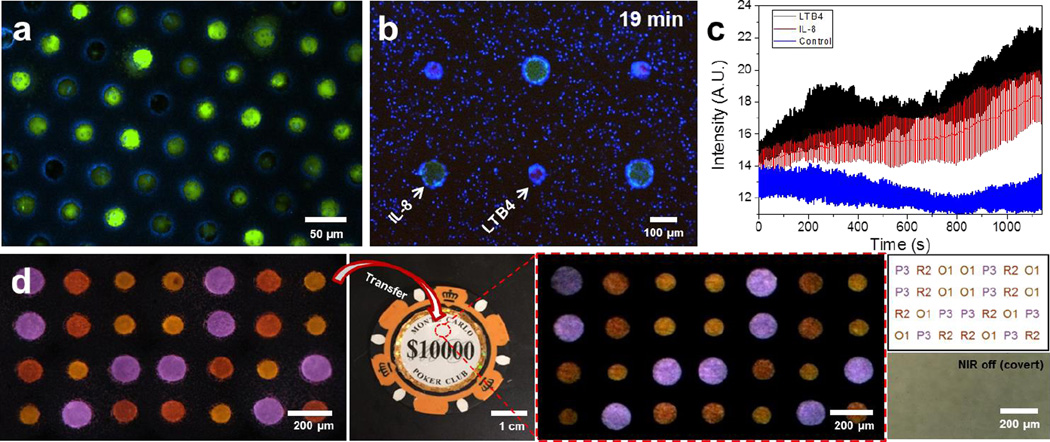
a, High throughput single-cell arrays using glioma cell line (U87) and porous microwells. Cells that are slightly larger than a microwell are squeezed into the microwells. Cells that are smaller than the microwells remain trapped inside the microwells after the washing step. b–c, Larger particles (green), loaded with protein chemoattractant IL-8, and smaller particles (red), loaded with leukotriene chemoattractant LTB4, are arranged in a regular, alternate pattern using LMSA. Human neutrophils (blue) navigate in the heterogeneous microenvironment created by the diffusion of the two chemoattractants from the particles. b, Neutrophils accumulate preferentially around the LTB4 source-particles after 19 minutes. c, Neutrophil accumulation is quantified by the change in average intensity around source-particles over time (blue fluorescence channel). Neutrophil accumulation is slightly faster around LTB4 than IL-8 particles. Control is a region of the array with no particles. Error bars represent standard deviation (n = 3). d, Anti-counterfeiting demonstration. Sequentially assembled 4×7 code was transferred to a target object (poker chip) and decoded into a text form. From the left, images show sequentially assembly UCN-laden microparticles, surface encoded object (poker chip), transferred particle arrays under NIR exposure, decoded results in a text form (top), and bight field image of encoded area (red circled area on poker chip). In decoding results, the alphabet character and number represents color (Table S4) and size of the particles, respectively.
Induced by chemical gradients, leukocytes display typical migratory responses toward infection and inflammation sites32, 33. Multiple cytokines are usually involved, in heterogeneous patterns in tissues34, and distinct patterns of migration are elicited be each cytokine35, resulting in complex leukocyte responses. This complexity is lost in current state of the art in vitro assays that probe leukocyte responses to one cytokine at a time, in uniform gradients, and lack the ability to generate 2D heterogeneous chemical gradients36, 37, 38. Using our platform’s ability to generate complex arrays, we generated a heterogeneous biochemical microenvironment for the study of neutrophil chemotaxis (Fig. 5b–c). Here, we arranged two distinct chemoattractant-laden microparticles loaded with leukotriene B4 (LTB4) and interleukin-8 (IL-8) in alternate patterns with 300 µm spacing. We took advantage of the ability of LSMA techniques to arrange the microparticles quickly before a significant amount of chemoattractant diffuses out of the particles upon wetting. We observed the migration patterns of neutrophils in response to the adjacent LTB4 and IL-8 gradients in the array (Fig. 5b). Neutrophils showed migratory responses with a high directional persistence toward LTB4-laden particles and migratory response with no directionality in the vicinity of IL-8 particles (Fig. S32, S33). The consequence of the two distinct migration patterns was a larger accumulation of neutrophils around the LTB4-laden particles (Fig. 5c). These results are consistent with previous reports of cell migration in competing gradients of LTB4 and IL-839. Moreover, they underline the contributions of distinct migration patterns to steering neutrophil populations precisely towards targets in complex microenvironments. These results also underscore the unique characteristics of high throughput, precision, and large-scale manipulation of individual targets, which clearly surpass the abilities of current techniques employing micropipettes40 or optically trapped beads36.
LSMAs for anti-counterfeiting applications
To demonstrate the encoding/decoding capacity of LSMA, we generated a 2D arrangement of upconversion nanocrystals (UCNs) laden PUA/AA microparticles for use in anti-counterfeiting. UCNs, synthesized in large quantity41, emit the rationally fine-tuned visible light7, 42 under near-infrared (NIR) exposure via an upconversion process. A covert pattern shows spectrally distinct codes with a single NIR excitation source, resulting in an appealing platform for the security technologies7, 43. A direct ink jet printing method has generated a multicolored, covert pattern, though with a limited precision and resolution43. Soft lithography44 (e.g. microcontact printing) requires a manual alignment of multiple components45, which is not easily achievable. Compared to those conventional patterning methods, our approach is attractive to anti-counterfeiting application because it generates a high resolution, multicomponent pattern by using a prealigned microwell array. For 2D particle arrays, encoding capacity scales as CS × SM, where C is the number of distinctive colors of the UCNs, S is the number of microparticle sizes which is used as a characteristic for specific positioning, and M is the number of microwells. For example, by arranging particles with an individual encoding capacity of 27 (3 different size microparticles and 9 distinct UCN colors) into 36 microwells, we exponentially increased the encoding capacity to 1020 (Fig. S35b) - enough to encode every grain of sand on the Earth (< 1019). Not only a 6 × 6 rectangular pattern, but also other rectangular matrices (Fig. 5d) or even a circular pattern (Fig. S35c) can be used. As particles were located at precise locations and in the same optical plane, particle ensembles can be readily decoded (Fig. 5d, Fig. S36). Also, this approach avoids potential problems such as tilting, overlapping, or touching of particles. The LSMA can be transferred to a target surface (Fig. 5d). Transferred particles were covert without NIR exposure because their refractive index was closely matched with the PUA resin used to transfer and adhere them to the target object. An image of the transferred pattern was taken with an iPhone under NIR exposure and was successfully decoded into a text form within 10 seconds.
Outlook
Our new LSMA platform enables one to position microparticles at precise locations depending on their physical characteristics with high scalability, precision, specificity, and versatility. Assembly in a given microwell is uncoupled from adjacent microwells, leading to a robust, parallelized and scalable process. Our approach is a top-down assembly process wherein the microparticles’ locations are predefined by the microfabrication of the microwell template, ensuring a high precision. Design rules based on a model of the system combined with the ability to fine-tune the properties of the microwells and microparticles, enables the resulting high specificity. The hydrodynamic driving force is not limited by particle composition, allowing our assembly method to be applied to a variety of microparticle chemistries.
We expect that the desirable performance of our approach will expand the utility of microparticles and microparticle arrays in many areas. LSMA allows for the generation of increasingly complex and covert codes than can be transferred to a variety of surfaces to expand the utility of particle-based anti-counterfeiting. LMSA of chemokine loaded particles could help replicate the biochemical heterogeneity of tissues at microscale for a variety of biological studies of immune cell responses, development, or tissue engineering. Complex arrangements of cells have great potential for large-scale studies of cell-cell communication and for the monitoring of cellular response over time29. The fast flow-assisted assembly of LSMA will greatly increase the speed and efficiency of existing microwell-based single cell assays46 or potentially expand them to selectively collect a subpopulation of cells by sorting them based on physical characteristics47. The ability to arrange small objects at proximal locations will pave the way to assemble functional MEMS components, such as microgears48, to tailor acoustic properties of materials49 or to develop new bioassays which take advantage of the proximal location of a single cell and particle within a microwell. We envision that this robust LSMA platform will inspire other particle manipulation techniques and find utility in a variety of material science and bioengineering applications.
Methods
Porous microwell fabrication
Porous microwell arrays are fabricated with a PDMS (Sylgard 184, Dow Corning) mold on top of a porous PET membrane (Millipore, diameter of pore = 3 µm) (Fig. S1, S2). To prepare the mold, PDMS is mixed with curing agents in a 10:1 ratio and cured on a SU-8 (Microchem) master prepared by standard photo-lithography procedures. For PDMS microwell fabrication, the PDMS mold is plasma treated and functionalized with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (Sigma Aldrich) to ensure the detachment of PDMS microwell arrays from the PDMS mold. The PDMS mold and flat PDMS are placed at top and bottom of PET membrane. Curable materials (e.g. NOA 81, PDMS) are placed on top of the inlet of mold. The entire device is placed inside a vacuum desiccator. As vacuum is applied, air trapped in the mold is removed, and curable materials are injected to the mold by the pressure difference. After photo (365 nm light) or thermal (65 °C) curing, the PDMS mold and flat PDMS are removed from the porous microwell arrays. For the anti-counterfeiting demonstration, PDMS microwell arrays were plasma-treated, and functionalized with fluorinated silane to increase the hydrophilicity difference between microwells and microparticles, eliminating any undesired adhesion.
Particle assembly
Microparticle solution dispensed on top of the porous microwell array. Negative pressure is applied at the bottom of porous microwell arrays to generate the driving force for particle assembly. A Kimwipe (Kimberly-Clark) is placed under the NOA microwell arrays to generate the negative pressure due to the capillary wetting (Fig. S2i). For PDMS microwells, vacuum suction is used to generate negative pressure (Fig. S2k). After particle assembly, redundant particles were washed out by water flow using a squeeze bottle (Fig. S2j). Particle assembly and washing steps are repeated to ensure a high yield.
Microparticle synthesis
Microparticles are synthesized in a microfluidic channel via stop flow lithography1, 2, 7, 8. The top part of the PDMS channel (mixed in 10:1 ratio) is cured on SU-8 master overnight at 65 °C, and then placed on the glass slide which is covered by half cured PDMS (mixed in 10:1 ratio, cured for 20 minutes at 65 °C). The assembled device is fully cured overnight at 65 °C. The prepolymer solution is flowed through the channel by compressed air, stopped, and exposed by ultraviolet light (365 nm). Ultraviolet light is patterned by a transparency mask (Fineline) designed in AUTOCAD. 2D extruded-shape microparticles are synthesized by photo-crosslinking. Three steps-flow, stop, exposure- are repeated to synthesize the microparticles in a semi-continuous manner. Flushed microparticles are centrifuged and resuspended several times, and then stored in 1× TET solution (1× tris-EDTA buffer with 0.05% (v/v) Tween-20).
Particle recollection
Two tygon tubes are connected to the recollection reservoir. One is connected to a vacuum line, and another one is used as recollection tube. Particle arrays are submerged in 1× TET solution, and the recollection tube is placed on top of targeted area using a xyz stage. The recollection tube is brought into contact with the array, and vacuum is applied to generate the pressure difference. After few seconds, the recollection tube is slightly displaced upward to make a gap between the tube and array. The 1× TET solution is aspirated through this gap and particles are recollected due to the shear stress associated with the fluid flow.
Pattern transfer
PDMS microwell arrays are desired for pattern transfer due to the lubricating layer provided by oxygen inhibition near the PDMS27. Prepolymer solution is dispensed on assembled particle arrays, and pre-cured polymer block is then placed on top. Ultra-violet light (365 nm) exposes to the targeted area from the bottom. In this exposure step, oxygen permeability of PDMS ensures the prepolymer solution does not stick to the PDMS microwell arrays whereas particles and the pre-cured block are covalently connected by cured prepolymer solution. After UV exposure, uncured prepolymer solution is washed out, and pre-cured block is transferred with attached particle arrays. Fluorinated PDMS microwells were plasma-treated to enhance the spreading of the prepolymer solution.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from the National Science Foundation grants CMMI-1120724, a Samsung Scholarship to JJK, and National Institutes of Health (GM092804).This work was supported in part by the MRSEC Program of the National Science Foundation under award number DMR-1419807. Microfabrication was performed at BioMEMS Resource Center (EB002503) and MTL, MIT. The modulus measurement was performed in Prof. Gareth McKinley’s and Prof. Allan S. Myerson’s labs at MIT. We thank Bashar Hamza and Dr. Eugene J. Lim of the BioMEMS Resource Center for fabrication of Si wafer and insightful discussion, and Dr. Lilian C. Hsiao and Dr. H. Burak Eral for the modulus measurement.
Footnotes
Author contributions
J.J.K. designed the research, conducted the majority of the experiments, conducted a scaling analysis, and interpreted data. K.W.B. conceived the project, obtained preliminary results, and interpreted data. J.J.K. and E.R. designed and demonstrated the biological studies. P.S.D. and D.I. designed the research, supervised the study, and interpreted data. J.J.K., P.S.D., and D.I. wrote the manuscript, and all authors commented on the manuscript.
Competing financial interests
Provisional US patent application 61898543 filed 1 November 2013.
References
- 1.Chung SE, Kim J, Oh DY, Song Y, Lee SH, Min S, et al. One-step pipetting and assembly of encoded chemical-laden microparticles for high-throughput multiplexed bioassays. Nat Commun. 2014;5 doi: 10.1038/ncomms4468. [DOI] [PubMed] [Google Scholar]
- 2.Pregibon DC, Toner M, Doyle PS. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 2007;315(5817):1393–1396. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 3.Walt DR. Techview: molecular biology. Bead-based fiber-optic arrays. Science. 2000;287(5452):451–452. doi: 10.1126/science.287.5452.451. [DOI] [PubMed] [Google Scholar]
- 4.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nature biotechnology. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eng G, Lee BW, Parsa H, Chin CD, Schneider J, Linkov G, et al. Assembly of complex cell microenvironments using geometrically docked hydrogel shapes. P Natl Acad Sci USA. 2013;110(12):4551–4556. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci U S A. 2007;104(14):5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Bisso PW, Srinivas RL, Kim JJ, Swiston AJ, Doyle PS. Universal process-inert encoding architecture for polymer microparticles. Nat Mater. 2014;13(5):524–529. doi: 10.1038/nmat3938. [DOI] [PubMed] [Google Scholar]
- 8.Han S, Bae HJ, Kim J, Shin S, Choi SE, Lee SH, et al. Lithographically Encoded Polymer Microtaggant Using High-Capacity and Error-Correctable QR Code for Anti-Counterfeiting of Drugs. Adv Mater. 2012;24(44):5924-+. doi: 10.1002/adma.201201486. [DOI] [PubMed] [Google Scholar]
- 9.Bae HJ, Bae S, Park C, Han S, Kim J, Kim LN, et al. Biomimetic microfingerprints for anti-counterfeiting strategies. Adv Mater. 2015;27(12):2083–2089. doi: 10.1002/adma.201405483. [DOI] [PubMed] [Google Scholar]
- 10.Curtis JE, Koss BA, Grier DG. Dynamic holographic optical tweezers. Opt Commun. 2002;207(1–6):169–175. [Google Scholar]
- 11.Hoogenboom JP, Vossen DLJ, Faivre-Moskalenko C, Dogterom M, van Blaaderen A. Patterning surfaces with colloidal particles using optical tweezers. Appl Phys Lett. 2002;80(25):4828–4830. [Google Scholar]
- 12.Xin HB, Xu R, Li BJ. Optical trapping, driving, and arrangement of particles using a tapered fibre probe. Sci Rep-Uk. 2012;2 doi: 10.1038/srep00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiou PY, Ohta AT, Wu MC. Massively parallel manipulation of single cells and microparticles using optical images. Nature. 2005;436(7049):370–372. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 14.Grzybowski BA, Stone HA, Whitesides GM. Dynamic self-assembly of magnetized, millimetre-sized objects rotating at a liquid-air interface. Nature. 2000;405(6790):1033–1036. doi: 10.1038/35016528. [DOI] [PubMed] [Google Scholar]
- 15.Tasoglu S, Yu CH, Gungordu HI, Guven S, Vural T, Demirci U. Guided and magnetic self-assembly of tunable magnetoceptive gels. Nat Commun. 2014;5 doi: 10.1038/ncomms5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasoglu S, Diller E, Guven S, Sitti M, Demirci U. Untethered micro-robotic coding of three-dimensional material composition. Nat Commun. 2014;5 doi: 10.1038/ncomms4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heo J, Kwon HJ, Jeon H, Kim B, Kim SJ, Lim G. Ultra-high-aspect-orthogonal and tunable three dimensional polymeric nanochannel stack array for BioMEMS applications. Nanoscale. 2014;6(16):9681–9688. doi: 10.1039/c4nr00350k. [DOI] [PubMed] [Google Scholar]
- 18.Le Goff GC, Lee J, Gupta A, Hill WA, Doyle PS. High-Throughput Contact Flow Lithography. Advanced Science. 2015 doi: 10.1002/advs.201500149. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamrock BJ, Jacobson BO, Schmid SR. Fundamentals of machine elements. Boston: WCB/McGraw-Hill; 1999. [Google Scholar]
- 20.Guo MY, Wyss HM. Micromechanics of Soft Particles. Macromol Mater Eng. 2011;296(3–4):223–229. [Google Scholar]
- 21.Wyss HM, Franke T, Mele E, Weitz DA. Capillary micromechanics: Measuring the elasticity of microscopic soft objects. Soft Matter. 2010;6(18):4550–4555. [Google Scholar]
- 22.Tominaga T, Takedomi N, Biederman H, Furukawa H, Osada Y, Gong JP. Effect of substrate adhesion and hydrophobicity on hydrogel friction. Soft Matter. 2008;4(5):1033–1040. doi: 10.1039/b716465c. [DOI] [PubMed] [Google Scholar]
- 23.Pregibon DC, Doyle PS. Optimization of encoded hydrogel particles for nucleic acid quantification. Anal Chem. 2009;81(12):4873–4881. doi: 10.1021/ac9005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi NW, Kim J, Chapin SC, Duong T, Donohue E, Pandey P, et al. Multiplexed Detection of mRNA Using Porosity-Tuned Hydrogel Microparticles. Anal Chem. 2012;84(21):9370–9378. doi: 10.1021/ac302128u. [DOI] [PubMed] [Google Scholar]
- 25.Chung SE, Park W, Shin S, Lee SA, Kwon S. Guided and fluidic self-assembly of microstructures using railed microfluidic channels. Nat Mater. 2008;7(7):581–587. doi: 10.1038/nmat2208. [DOI] [PubMed] [Google Scholar]
- 26.Chung SE, Jung Y, Kwon S. Three-dimensional fluidic self-assembly by axis translation of two-dimensionally fabricated microcomponents in railed microfluidics. Small. 2011;7(6):796–803. doi: 10.1002/smll.201001806. [DOI] [PubMed] [Google Scholar]
- 27.Dendukuri D, Panda P, Haghgooie R, Kim JM, Hatton TA, Doyle PS. Modeling of Oxygen-Inhibited Free Radical Photopolymerization in a PDMS Microfluidic Device. Macromolecules. 2008;41(22):8547–8556. [Google Scholar]
- 28.Cruz FC, Koya E, Guez-Barber DH, Bossert JM, Lupica CR, Shaham Y, et al. New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nat Rev Neurosci. 2013;14(11):743–754. doi: 10.1038/nrn3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6(11):1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 30.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77(17):5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 31.Hughes AJ, Spelke DP, Xu ZC, Kang CC, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11(7):U749–U794. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillipson M, Kubes P. The neutrophil in vascular inflammation. Nat Med. 2011;17(11):1381–1390. doi: 10.1038/nm.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 34.Sadik CD, Kim ND, Luster AD. Neutrophils cascading their way to inflammation. Trends Immunol. 2011;32(10):452–460. doi: 10.1016/j.it.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng LG, Qin JS, Roediger B, Wang YL, Jain R, Cavanagh LL, et al. Visualizing the Neutrophil Response to Sterile Tissue Injury in Mouse Dermis Reveals a Three-Phase Cascade of Events. J Invest Dermatol. 2011;131(10):2058–2068. doi: 10.1038/jid.2011.179. [DOI] [PubMed] [Google Scholar]
- 36.Kress H, Park JG, Mejean CO, Forster JD, Park J, Walse SS, et al. Cell stimulation with optically manipulated microsources. Nat Methods. 2009;6(12):905–909. doi: 10.1038/nmeth.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boneschansker L, Yan J, Wong E, Briscoe DM, Irimia D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat Commun. 2014;5 doi: 10.1038/ncomms5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin F, Nguyen CMC, Wang SJ, Saadi W, Gross SP, Jeon NL. Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices. Ann Biomed Eng. 2005;33(4):475–482. doi: 10.1007/s10439-005-2503-6. [DOI] [PubMed] [Google Scholar]
- 39.Heit B, Tavener S, Raharjo E, Kubes P. An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients. J Cell Biol. 2002;159(1):91–102. doi: 10.1083/jcb.200202114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, Bourne HR. Spatial control of actin polymerization during neutrophil chemotaxis. Nat Cell Biol. 1999;1(2):75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Han Y, Lim CS, Lu YH, Wang J, Xu J, et al. Simultaneous phase and size control of upconversion nanocrystals through lanthanide doping. Nature. 2010;463(7284):1061–1065. doi: 10.1038/nature08777. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Liu XG. Upconversion multicolor fine-tuning: Visible to near-infrared emission from lanthanide-doped NaYF4 nanoparticles. J Am Chem Soc. 2008;130(17):5642-+. doi: 10.1021/ja800868a. [DOI] [PubMed] [Google Scholar]
- 43.Meruga JM, Baride A, Cross W, Kellar JJ, May PS. Red-green-blue printing using luminescence-upconversion inks. J Mater Chem C. 2014;2(12):2221–2227. [Google Scholar]
- 44.Xia YN, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 45.Rogers JA, Paul KE, Whitesides GM. Quantifying distortions in soft lithography. J Vac Sci Technol B. 1998;16(1):88–97. [Google Scholar]
- 46.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nat Protoc. 2009;4(5):767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee WC, Shi H, Poon ZY, Nyan LM, Kaushik T, Shivashankar GV, et al. Multivariate biophysical markers predictive of mesenchymal stromal cell multipotency. P Natl Acad Sci USA. 2014;111(42):E4409–E4418. doi: 10.1073/pnas.1402306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shepherd RF, Panda P, Bao Z, Sandhage KH, Hatton TA, Lewis JA, et al. Stop-Flow Lithography of Colloidal, Glass, and Silicon Microcomponents. Adv Mater. 2008;20(24):4734-+. [Google Scholar]
- 49.Martinezsala R, Sancho J, Sanchez JV, Gomez V, Llinares J, Meseguer F. Sound-Attenuation by Sculpture. Nature. 1995;378(6554):241–241. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.