Abstract
Antibody structure prediction has made great strides, but accurately modeling complementarity determining region (CDR) H3 loops remains elusive. Unlike the other five CDR loops, CDR H3 does not adopt canonical conformations and must be modeled de novo. During antibody modeling assessment II (AMA-II) we found that biasing simulations toward kinked conformations enables generating low-RMSD models(1), and since then, we have presented new geometric parameters defining the kink conformation(2). Here, we use these parameters to develop a new biasing constraint. When applied to a benchmark set of high-quality CDR H3 loops, the average minimum RMSD sampled is 0.93 Å, compared to 1.34 Å without the constraint. We then test the performance of the constrained de novo method for homology modeling and rigid-body docking, and present the results for (1) the AMA-II targets; (2) the 2009 RosettaAntibody benchmark set; and (3) the high-quality set.
Introduction
The adaptive immune system in vertebrates is capable of raising antibodies against a countless number of antigens. Additionally, engineered antibodies form various sources are used as therapeutic molecules(3, 4) and biosensors(5–7). In order to optimize specific modes of interactions, rational engineering techniques must be developed. Rational engineering of antibodies requires accurate structural models, but crystallization is not always practical or even possible. Additionally, expressing a large library of mutants in order to assess the energetic implications of specific mutations is time consuming, resource intensive and, in some cases, technically challenging. Computational methods, namely antibody homology modeling, are poised to enable the realization of rational design.
In addition to biotechnology applications, advances in next-generation sequencing techniques(8–10) have made it possible to routinely elucidate the sequences of 104–105 antibodies per individual. Such a vast and complete snapshot of an individual’s antibody repertoire is ripe for extracting an unprecedented amount of immunological information(11). However, sequence analysis and structural analysis will need to be used in concert to produce a fully formed picture. The sheer number of sequences necessitate the use of computational structure prediction methods.
Our goal is to develop methods to improve antibody structure prediction. Generating accurate structural models is critical for producing inputs for additional structure-based simulations such as docking(12), and for computational prediction of binding affinities and dissociation free energies(13, 14). The modeling tool we use, develop and study in this paper is RosettaAntibody. RosettaAntibody’s approach to modeling(15) is to break the structure into eight distinct structural components: the heavy- and light-chain frameworks; CDR loops L1–3; and CDR loops H1–3.
Because the non-H3 CDR loops adopt canonical conformations(16, 17), accurate backbone conformations for them can usually be found in known structures. RosettaAntibody exploits this by selecting templates from curated structural databases using BLAST(18) bit-score for CDRs L1–3, H1 and H2 and the framework regions. Each structural component is defined such that they have overlapping residues that can then be superposed to create a grafted model. An initial VL–VH orientation is also selected from databases, and the grafted heavy and light chains are each superposed to the corresponding chain in the orientation template. After this, the CDR H3 loop is modeled de novo while sampling the VL–VH orientation.
We have presented the performance of RosettaAntibody in Antibody Modeling Assessment II (AMA II)(1, 19). With few exceptions, RosettaAntibody selects templates for the framework regions and the non-H3 CDR loops that have backbone atomic coordinates with sub-angstrom optimal root-mean-squared deviations (RMSDs) from the native structure. Similarly, the other participants in the assessment were generally able to construct models with low RMSDs in the framework and non-H3 CDR loops(20–24). The most difficult aspect of antibody homology remains accurately predicting the VL–VH orientation and the CDR H3 conformation.
Because the CDR H3 loop lies at the interface between the heavy and light chains, incorrect VL–VH orientations can frustrate identifying correct CDR H3 conformations. In the time that has elapsed since AMA II was conducted, progress has been made in predicting VL–VH orientation(25) from sequence by training a random forest model on a set of “fingerprint” residues at the VL–VH interface using ABangle’s six degree-of-freedom description of orientation(26). Further improvement has been achieved with an ensemble approach to modeling VL–VH orientation called multiple-template grafting(27). Similarly, effort has been made to develop a CDR H3-specific loop modeling routine(22, 28, 29). Successful predictions require extremely accurate atomic coordinates for the rest of the FV(23, 28), which may make these tools better-suited for refining crystal structures with poor electron density around the CDR H3 loop than for homology modeling. Another method relies on restraining the dihedral angles of the three N-terminal and four C-terminal residues of the H3 loop based on distributions observed in crystal structures(29).
A large majority – over 80% of known structures – of CDR H3 loops have a C-terminal kink(2, 17, 30–34), a structure that resembles a β bulge in that it disrupts the hydrogen bonding pattern along a β strand and causes the backbone to twist. In AMA II we found that producing low-RMSD models required filtering out non-kinked H3 conformations. However, the scores of the kinked structures, which can be thought of as their free energies of folding, are higher (worse) than some of the extended structures that Rosetta produced. Shirai et al. also used a filter to ensure the predicted H3 loops had appropriate base geometries(22). In response to these findings, we developed new geometric parameters that describe the kink, in particular that the C-terminal loop residues form a pseudo dihedral angle, α101 (100X–103 using Chothia numbering), of 39° and a pseudo bond angle, τ101 (100X–102 using Chothia numbering), of 101°(2). Along with this enhanced description of the kink, we hypothesized that the kink serves to drive CDR H3 diversity and is stabilized by tertiary interactions and not by the sequence of the loop itself. This hypothesis is supported by the recent work by Teplyakov et al.(35) in which four heavy chains and four light chains were combined pairwise to create sixteen antibodies. All sixteen antibodies had the same CDR H3 loop, but the structure of the loop varied considerably.
De novo loop modeling has endured as a challenging problem in part because of the large number of degrees of freedom that need to be sampled, the closure requirement, and the challenges associated with accurately ranking different structures. Additionally, side-chain interactions often play key roles in stabilizing observed loop conformations, potentially complicating low-resolution searches.
CDR H3 loops, in addition to having a variety of lengths, are anchored on two adjacent β strands that are disrupted by the C-terminal kink. In previous work, we found that the simulation tends to continue the β strands well into the loop region, possibly due to the formation of favorable backbone–backbone hydrogen bonds in the low-resolution stage of the search(1).
Complicating the task even further is the most common source of the reference coordinates: crystal structures. Crystals are crowded environments in which each protein molecule is surrounded by several others; crystal contacts may influence the observed conformation within the asymmetric unit. Without the existence of a crystal structure of the same protein in more than one distinct crystal form, it cannot be determined if crystal contacts perturb the conformation of any region of the protein.
Finally, it is not justified to always assume that loop modeling must search for a single set of coordinates. Proteins in physiological conditions are not completely rigid, and estimating the conformational entropy of a loop requires supplying a model to describe the modes of flexibility accessible to the loop(36). Nevertheless, the possible existence of multiple degenerate-energy conformations cannot be dismissed.
In this paper, we use the parameters defined in our previous work(2) to constrain the kink during the course of a simulation. The constraint is tested by predicting H3 conformations on the crystal framework structure across the set of benchmark antibody structures. To limit the uncertainty in the crystallographic coordinates, we have constructed a set of high-resolution H3 loops (described in Materials and Methods). Because our long-term goal is to predict entire FVs from sequence, we also assess CDR H3 modeling on homology modeled frameworks. Finally, we test the ability to dock an antibody with a modeled H3 loop. The results enable us to address long-standing questions about CDR H3 modeling, including the limitations of sampling and ranking candidate conformations, and whether an improved representation of the kink can enable accurate CDR H3 loop structure prediction. Finally, to show the improvements we have made since the initial development of RosettaAntibody, we report the performance of the updated version for all 52 of the structures included in the 2009 study. To place our work into the context of other antibody structure prediction tools, we model the targets from AMA II.
Materials and Methods
Dataset construction
A set of FVs with accurate CDR H3 coordinates was constructed by querying the backend databases of PyIgClassify(37) for structures with a resolution of 2.5 Å or better, a maximum R-value of 0.2, B-factor ≤ 80.0 Å2 for every atom in the structure, only one copy of the FV in the asymmetric unit, and CDR H3 loop-lengths ranging from 9–20 residues using the Honegger–Plückthun-based definition(38). To ensure the set has diverse chemical environments, no two heavy-chain CDR loops are permitted to be identical in sequence. The structures were further filtered to remove antibodies from species other than humans and mice, and modified residues (namely pyroglutamic acid (PCA), a cyclized form of glutamine or glutamic acid). The resulting set of structures contains 49 FVs and is summarized in Table I.
Table I.
Structural information for the CDR H3 benchmark set. All structures have R-values lower than 0.2, resolution better than 2.5 Å and maximum B factors lower than 80.0 Å2. To limit variables are being considered, the CDR H3 loops are restricted to range from 9–20 residues in length and to only be derived from human and mouse antibodies. Antibodies crystallized with an antigen bound have an annotation ending with “–Ag” in the Fragment column.
| PDB Code |
R Value |
Res. (Å) |
Max. B factor (Å2) |
Max. H3 B factor (Å2) |
Species | CDR H3 length |
Light chain isotype |
Fragment | pH |
|---|---|---|---|---|---|---|---|---|---|
| 1x9q* | 0.193 | 1.5 | 32.57 | 21.22 | Human | 9 | Κ | scFV–Ag | 4.6 |
| 2d7t | 0.191 | 1.7 | 49.16 | 47.73 | Human | 9 | Κ | FV | 9.3 |
| 3hc4 | 0.162 | 1.62 | 41.11 | 33.5 | Human | 9 | Κ | Fab | 8 |
| 1mlb | 0.181 | 2.1 | 49.63 | 34.78 | Mouse | 9 | κ | Fab | — |
| 2e27* | 0.198 | 1.7 | 51.38 | 31.14 | Mouse | 9 | κ | FV–Ag | 5.7 |
| 3g5y | 0.199 | 1.59 | 40.08 | 35.9 | Mouse | 9 | κ | Fab–Ag | — |
| 3m8o* | 0.154 | 1.55 | 55.22 | 24.48 | Human | 10 | κ | Fab | 7 |
| 1jpt | 0.182 | 1.85 | 49.59 | 29.78 | Humanized Mouse |
10 | κ | Fab | 4.6 |
| 3e8u | 0.188 | 2.1 | 32.5 | 27.11 | Mouse | 10 | κ | Fab–Ag | — |
| 1mqk | 0.136 | 1.28 | 51.66 | 37.15 | Mouse | 11 | κ | FV | 6 |
| 1nlb | 0.197 | 1.6 | 44.78 | 41.47 | Mouse | 11 | κ | Fab | 9 |
| 2adf | 0.192 | 1.9 | 35.22 | 20.68 | Mouse | 11 | κ | Fab–Ag | 4.6 |
| 2fbj | 0.194 | 1.95 | 60.06 | 41.76 | Mouse | 11 | κ | Fab–Ag | — |
| 2w60 | 0.171 | 1.5 | 54.6 | 36.38 | Mouse | 11 | κ | Fab | 8 |
| 3gnm | 0.189 | 2.1 | 46.42 | 43.99 | Mouse | 11 | κ | Fab | 4 |
| 3hnt | 0.199 | 1.8 | 54.27 | 28.18 | Mouse | 11 | κ | Fab–Ag | 7.1 |
| 3v0w | 0.184 | 1.73 | 60.85 | 60.81 | Mouse | 11 | κ | Fab–Ag | 4.6 |
| 1mfa | 0.166 | 1.7 | 69.57 | 41.64 | Mouse | 11 | λ | FV–Ag | — |
| 3mxw | 0.181 | 1.83 | 72.85 | 31.98 | Mouse | 12 | κ | Fab–Ag | — |
| 2xwt | 0.179 | 1.9 | 44.78 | 29.3 | Human | 12 | λ | Fab–Ag | 5 |
| 1dlf | 0.183 | 1.45 | 42.75 | 24.85 | Mouse | 12 | κ | FV | 5.25 |
| 2ypv | 0.183 | 1.8 | 75.36 | 37.38 | Mouse | 12 | κ | Fab–Ag | 8.5 |
| 3ifl | 0.18 | 1.5 | 34.75 | 22.57 | Mouse | 12 | κ | Fab–Ag | 9 |
| 3liz* | 0.178 | 1.8 | 52.33 | 43.85 | Mouse | 12 | κ | Fab–Ag | 7.2 |
| 3oz9 | 0.192 | 1.6 | 55.29 | 31.28 | Mouse | 12 | κ | Fab | 8.5 |
| 3umt | 0.177 | 1.8 | 56.76 | 39.55 | Mouse | 12 | κ | scFV | 9.5 |
| 4h0h | 0.197 | 2 | 65.19 | 37.96 | Mouse | 12 | κ | scFV | 6.5 |
| 4h20 | 0.197 | 1.9 | 45.61 | 20.26 | Mouse | 12 | κ | Fab | 7.4 |
| 4hpy | 0.171 | 1.5 | 55.43 | 42.96 | Human | 13 | λ | Fab–Ag | 6.5 |
| 2v17 | 0.16 | 1.65 | 37.77 | 23.54 | Mouse | 13 | κ | Fab–Ag | 7.5 |
| 3t65 | 0.194 | 1.45 | 63.85 | 34.13 | Mouse | 13 | κ | Fab–Ag | 8 |
| 1oaq | 0.16 | 1.5 | 44.45 | 43.75 | Mouse | 13 | λ | FV | 5 |
| 2vxv | 0.155 | 1.49 | 37.34 | 29.51 | Human | 14 | κ | Fab | 10.5 |
| 3eo9 | 0.191 | 1.8 | 45.44 | 27.45 | Humanized Mouse | 14 | κ | Fab | 5 |
| 3p0y | 0.182 | 1.8 | 76.35 | 25.77 | Human | 14 | κ | Fab–Ag | 8 |
| 1jfq | 0.196 | 1.9 | 57.19 | 40.56 | Mouse | 14 | κ | Fab | 7.5 |
| 2r8s | 0.196 | 1.95 | 63.07 | 34.21 | Human (library) |
14 | κ | Fab–Ag | 5.9 |
| 3i9g | 0.192 | 1.9 | 53.87 | 29.27 | Humanized Mouse |
14 | κ | Fab–Ag | 6 |
| 3giz | 0.198 | 2.2 | 52.12 | 52.12 | Human | 15 | κ | Fab | 6.3 |
| 3go1 | 0.192 | 1.89 | 37.78 | 31.35 | Human | 16 | λ | Fab–Ag | 6.5 |
| 1fns | 0.172 | 2 | 49.64 | 20.8 | Mouse | 16 | κ | Fab–Ag | — |
| 1seq† | 0.194 | 1.78 | 68.08 | 68.08 | Mouse | 16 | κ | Fab | 7.5 |
| 1gig | 0.195 | 2.3 | 53.69 | 34.01 | Mouse | 16 | λ | Fab | — |
| 3mlr | 0.181 | 1.8 | 42.1 | 37.7 | Human | 17 | λ | Fab–Ag | 5.5 |
| 4nzu | 1.37 | 1.2 | 69.1 | 32.8 | Human | 18 | κ | Fab | 4 |
| 3lmj | 0.194 | 2.2 | 58.27 | 58.27 | Human | 18 | λ | Fab | 7.5 |
| 4f57 | 0.188 | 1.7 | 64.7 | 40.56 | Human | 18 | λ | Fab | 6.5 |
| 2fb4 | 0.189 | 1.9 | 39.92 | 39.2 | Human | 19 | λ | Fab | — |
| 3nps | 0.19 | 1.5 | 49.76 | 31.95 | Human | 19 | λ | Fab–Ag | — |
Extended base geometry
Unclear base geometry
Kink constraint
In de novo loop modeling simulations, it is impossible to exhaustively sample all of the structural degrees of freedom. To increase the likelihood of generating CDR H3 models with near-native structures, we constrain(39) two parameters: (1) τ101, the Cα– Cα– Cα pseudo bond angle for the three C-terminal residues; and (2) α101, the Cα–Cα– Cα– Cα pseudo dihedral angle for the three C-terminal residues in the CDR H3 loop and one adjacent residue in the heavy chain framework.
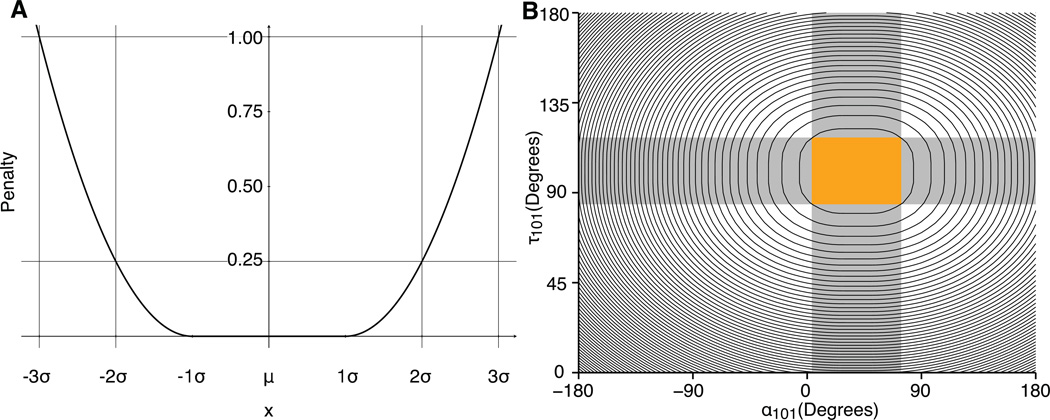
Because an objective of this study is to determine whether or not Rosetta can correctly identify native H3 conformations, it is important not to over-constrain any of the simulations. With this in mind, a FLAT_HARMONIC potential, which has a region wherein no penalty is applied, is a natural choice (Figure 1A). The FLAT_HARMONIC potential is of the form
where µ is the mean, t (tolerance) is the distance from µ with no penalty, and ξ is the scaling factor that controls the penalty that is applied.
Figure 1. Kink constraint.
Functional form of the constraint used to bias de novo CDR H3 loop modeling simulations toward generating kinked conformations. (A) A plot of the FLAT_HARMONIC potential. (B) A contour plot showing the value of the kink constraint across all values of τ101 and α101. Each contour line represents a 2.0 Rosetta Energy Unit (REU) increase in penalty. The orange box demarcates ± 3.0 σ of the mean of the τ101 and α101 distributions. Throughout the rest of this paper, models falling within this region are classified as “kinked”, models with τ101 and α101 in the gray, shaded regions are classified as “unclear”, and all other models are classified as “extended”.
For the kink parameters α101 and τ101, we designed a penalty schedule with no penalty when the value is within 1.0 σ of the mean and a penalty of 1.0 at 3.0 σ, yielding ξ = 2t. This schedule encourages Rosetta to generate models with kinked H3 loops without forcing the geometry toward the mean values of both parameters. Using the values determined previously(2), the kink constraint for AHo-numbered antibodies is written for input into Rosetta as:
# alpha: pseudo dihedral - last 3 residues in H3 and the following W # mean: 38.85 degrees; SD: 11.75 degrees (in radians) Dihedral CA 136H CA 137H CA 138H CA 139H FLAT_HARMONIC 0.678 0.41 0.205 # tau: pseudo bond angle of the last 3 residues in H3 # mean: 100.9 degrees; SD: 5.57 degrees (in radians) Angle CA 136H CA 137H CA 138H FLAT_HARMONIC 1.761 0.194 0.0972
Figure 1B shows a contour plot of the combined value of the τ101 and α101 constraints, with each line representing an increase in score of 2.0 Rosetta Energy Units (REU). Figure 1B also shows the regions of τ101 and α101 that define kinked (orange; ± 3.0 σ of the mean of both parameters), unclear (gray; ± 3.0 σ of mean of one of the parameters), and extended (white; beyond 3.0 σ of both parameters) conformations. The output score is always derived from the unbiased score function, even when the constraint is employed in the simulation. These definitions are used throughout this study.
De novo loop structure prediction
Next-generation Kinematic Closure (KIC(40)), or NGK, has been developed to further improve the performance of loop modeling in Rosetta. Like KIC, NGK generates diverse loop conformations by drawing φ and ψ torsion angle pairs from Ramachandran distributions for all but three residues in the loop; the torsion angles of the remaining residues are determined analytically by polynomial resultants(41). The approach employed by NGK enhances KIC by using neighbor-dependent Ramachandran maps(42), explicitly sampling ω backbone dihedral angles as well as using a simulated annealing strategy for repulsive and Ramachandran score terms in the all-atom stage. On the same set of loops used to benchmark KIC, NGK generates substantially more near-native models(43), which is why it is used as the starting point in this study. During loop modeling, the backbone coordinates for the non-loop regions of the protein are held fixed.
The flags to run a standard NGK simulation are:
./loopmodel.macosclangrelease
-native input_file.pdb
-s input_file.pdb
-nstruct 500
-loops:loop_file h3.loops
-loops:remodel perturb_kic
-loops:refine refine_kic
-loops:outer_cycles 5
-kic_bump_overlap_factor 0.36
-legacy_kic false
-kic_min_after_repack true
-corrections:score:use_bicubic_interpolation false
-loops:kic_omega_sampling
-loops:kic_rama2b
-allow_omega_move
-loops:ramp_fa_rep
-loops:ramp_rama
-ex1
-ex2
-extrachi_cutoff 0
where h3.loops contains
# FORMAT JSON
{"LoopSet" : [{
"start" : { "resSeq" : 107, "iCode" : " ", "chainID" : "H" },
"stop" : { "resSeq" : 138, "iCode" : " ", "chainID" : "H" },
"extras" : { "extend" : true },
}]
}
We refer readers to the NGK publication(43) for a discussion of the various flags associated with it. NGK simulations with constraints use the above command line with the addition of the following flags to specifcy constraints in both the low-resolution and full atom (“fa”) stages:
-constraints:cst_file kink.constraint
-constraints:cst_weight 1.0
-constraints:cst_fa_file kink.constraint
-constraints:cst_fa_weight 1.0
where the file kink.constraint contains the constraints as shown above.
Scaled scores
In order to compare the results of simulations of different targets, the scores of a set of candidate models are scaled such that a value of 1.0 corresponds to the 95th percentile of scores and a value of 0.0 corresponds to the 5th percentile. After this normalization is completed, the score of the refined native structure is computed on the same scale.
Discrimination score
The discrimination score is used to measure how “funnel-like” a score vs. RMSD plot is, with a lower value being indicative of a more successful simulation. In this study, the reference for RMSD calculations is the set of atomic coordinates of the crystal structure subjected to refinement as described below. As defined by Conway et al.(44), the discrimination score is calculated as:
where r is the RMSD cutoff in Å, Si are the dimensionless scaled scores calculated as described above, and the discrimination score, D, is the sum of the score-differences of the best-scoring models above and below the seven RMSD cutoffs.
Preparation of input structures
The raw crystallographic coordinates of protein structure often do not score favorably within Rosetta, typically because the packed environment of a crystal leads to side-chain conformations that would undesirable in solution and some contacts are classified as steric clashes by Rosetta. To compensate, crystal structures must be relaxed, that is, optimized with respect to the Rosetta scoring function. Relaxation will result in small changes to the atomic coordinates with significant improvements in the score; however, it is important that the backbone coordinates do not vary much, especially in the case of loop modeling. To ensure that only small changes to the backbone are allowed, all coordinates are constrained to their starting positions with a spring potential. The command line used for constrained relax is:
./relax.macosclangrelease
-s input.pdb
-nstruct 500
-relax:constrain_relax_to_start_coords
-relax:coord_constrain_sidechains
-relax:ramp_constraints false
-ex1
-ex2
-use_input_sc
Once the crystallographic coordinates have been optimized, the entire structure can be subjected to fixed-backbone side-chain optimization to further lower the score of the reference structure for loop modeling and to better approximate the side-chain conformations in the free, unbound conformation. The command line used for fixed-backbone side-chain optimization is:
./relax.macosclangrelease
-s lowest_scoring_model_from_previous_simulation.pdb
-nstruct 100
-relax:bb_move false
-ex1
-ex2
-extrachi_cutoff 0
The low-scoring model from this calculation is used as the input structure in the subsequent calculations.
CAPRI criteria
For a model to be considered a high-quality prediction by CAPRI metrics(45), the fraction of native residue–residue contacts recovered (fnat) must be ≥ 0.5 and the Interface RMSD (I_RMSD) or Ligand RMSD (L_RMSD) must be ≤ 1.0 Å. Medium-quality predictions must have fnat ≥ 0.3 and L_RMSD ≤ 5.0 Å or I_RMSD ≤ 2.0 Å, while acceptable predictions have fnat ≥ 0.1 and L_RMSD ≤ 10.0 Å or I_RMSD ≤ 4.0 Å. Models that have fnat ≤ 0.1 or L_RMSD ≥ 10.0 Å and I_RMSD ≥ 4.0 Å are considered incorrect.
Results
High-quality CDR H3 loop benchmark set
Figure 2 shows an example CDR H3 loop with the electron density map for the H3 residues shown in a gray mesh over the residues represented in sticks. At this level of detail, all of the side-chain coordinates are well-defined, and the map even shows a hole in aromatic residues. The level of agreement between the electron density map and the coordinates and the lack of ambiguity in the atomic coordinates suggests that this loop is in a stable conformation in the crystal, making it a prime candidate for loop modeling experiments. We constructed a set of 49 high-quality CDR H3 structures as described in methods. The other loops in the set have similarly well-defined electron density. A limitation of this set is that several structures are in their bound form (annotations ending with “–Ag” in the Fragment column of Table I), and the unbound forms may differ or even be flexible. This danger is mitigated by the fact that usually these loops deviate less than 1.0 Å RMSD between the bound and unbound forms(46).
Figure 2. Electron density of CDR H3 loop of an anti-peptidase S1 antibody.

The FV of an anti-peptidase S1 antibody (PDB accession code 3nps(60)) is shown with the VH domain in cyan and the VL domain in magenta. The electron density of the 19-residue CDR H3 loop is indicated with a mesh contour map within 1.6 Å of the coordinates in the PDB file. The crystal structure has an R-value of 0.190 and a resolution of 1.50 Å. The electron density is clearly resolved across the entire CDR H3 loop, indicating both a high-quality crystal and a stable loop conformation among several symmetric copies of the antibody in the crystal. The crystal structure contains the full Fab bound to the antigen, which may further stabilize the loop’s conformation.
Table I lists all of the loops in the set and includes information on the quality and content of the crystal structure, the species from which the antibody was derived, the length of the loop, and the light chain isotype. In the set, 24 of the 49 structures are crystallized in the bound conformation with their antigen, 40 of the 49 structures are Fabs, six are FVs and the remaining three are scFVs. Eighteen of the structures are of human antibodies, and 11 have λ light chains, making this a diverse set of structures.
The definition of the bounds of the CDR H3 loop differ from the Chothia-based definition(47) used within RosettaAntibody(15), and instead are based on the structure-based Honegger–Plückthun definition(38) used by North et al.(17) and in our previous work on the kink geometry(2). Both definitions end on Chothia residue number 102, but the Chothia-based definition begins at residue 95 while the Honegger–Plückthun-based definition begins at residue 93, making the Honegger–Plückthun CDR H3 loops two residues longer than Chothia loops. In this set, the median and mode of the loop lengths are both 12 residues.
Unconstrained de novo modeling of CDR H3 loops
To establish a baseline, we first assessed whether Rosetta methods create native-like H3 kink structures using established methods. In AMA II(1) NGK was used to model CDR H3 loops on a crystallographic framework. In these cases, a filter was employed to favor kinked structures, using the θbase (α101 in this work) definition developed by Shirai et al. and refined by Kuroda et al.(30–32) Without this penalty, very few kinked structures were produced. From these results it remained unclear if the primary limitation for producing accurate CDR H3 models lies in generating low-RMSD conformations (sampling) or in ranking the candidate models effectively (scoring). Since then, we established the geometric parameters α101 and τ101 to describe the C-terminal kink in CDR H3 loops(2). We now use these parameters to probe model sets to determine if Rosetta modeling failures are in sampling, scoring or both.
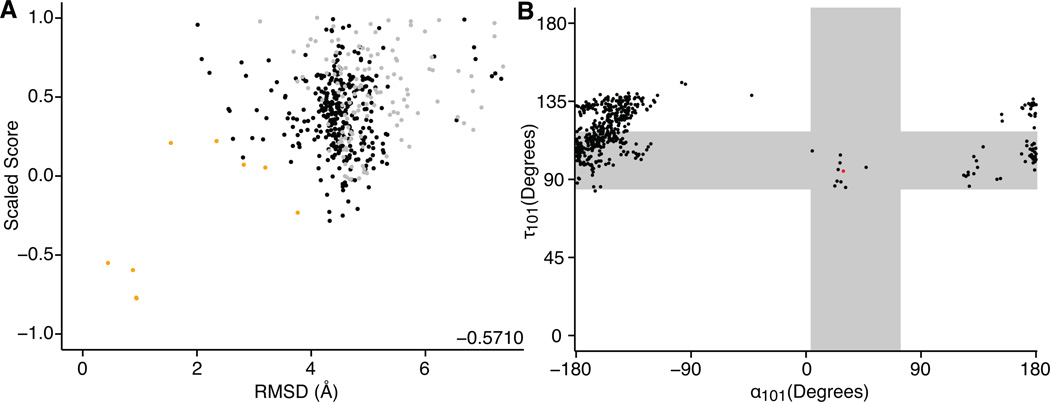
Figure 3 shows the results of a de novo CDR H3 modeling simulation on an anti-citrullinated collagen type II antibody (PDB accession code 2w60(48)). In Figure 3A, a score vs. RMSD plot (funnel plot) shows the models ranked by the scaled score and colored by their base geometry. The kinked models (orange points) make up a small fraction of the structures produced; however, they have lower scores than extended structures at the same RMSD value. The top-ranked models have very low RMSDs, but only three such models were produced. Nonetheless, because the score function successfully separates the near-native and non-native conformations, the discrimination score is −0.5710 (negative discrimination scores indicate success). The scaled score of the refined, native (X-ray) structure is −1.4061, meaning that none of the predicted structures approach near-native scores.
Figure 3. Results of unconstrained de novo NGK.
Results of unconstrained de novo NGK H3 loop prediction on an anti-citrullinated collagen type II antibody (2w60(48)). 2w60 is derived from a mouse and has an 11-residue H3 loop and a κ light chain. (A) Funnel plot showing scaled score (see methods) vs. RMSD of 500 generated models. Orange points correspond to a kinked base geometry, gray points to an unclear base geometry, and black points to an extended base geometry. The discrimination score of −0.5710 is shown in the lower right of the plot area. Very few kinked H3 models are produced, but the top-scoring models have sub-angstrom RMSDs. The refined, native score is −1.4061, meaning that none of the predicted structures have near-native scores. (B) τ101 vs. α101 of 500 generated models. The red point is at the values of the native structure, and the black points correspond to the models. The vast majority of the points have τ101 and α101 values that correspond to extended conformations.
Figure 3B shows the τ101 and α101 values for the models (black) and the crystal structure (red). The gray bars demarcate ± 3.0 σ from the mean of the distribution of each parameter in kinked antibodies as found in our previous work(2). This plot shows a clear preference for NGK to produce H3 loops in the extended conformation, likely because it can form backbone–backbone hydrogen bonds by extended the β strands of the framework in the low-resolution stage of modeling.
Figures S1 and S2 show funnel plots and τ101 vs. α101 plots for the rest of the structures in the dataset. Across the whole set, kinked models represent a small fraction of the models that are produced, but those models tend to have lower RMSDs.
Across the set of antibodies, the native conformation has a significantly better score than the best decoys that are being produced by NGK (Table SI): the average scaled native score for kinked targets is −0.9480 with a standard deviation of 0.5033 (Table II).
Table II.
Summary of de novo loop modeling simulations. The minimum RMSD, scaled native score, average of the ten lowest RMSDs, average RMSD of the 10 top-scoring models and the RMSD of the top-ranked model are shown for unconstrained NGK, constrained NGK and the combined NGK+CCD methods (see Figure S6 for an example). Each value is the average of the values of the 44 kinked targets in the benchmark set. NGK with constraints performs best over the whole set. Only the scaled native score in the combined simulations have superior values, however this affects the ability of the score function to discriminate between near-native and non-native conformations. Per-target values for each type of simulation can be found in Supplemental Information.
| Simulation | Min. RMSD (Å) | Scaled Nat. Score |
Top 10 RMSDs (Å) |
RMSD of Top 10 Scored (Å) |
RMSD of Top 1 Scored (Å) |
|---|---|---|---|---|---|
| Unconstrained | 1.336 | −0.948 | 2.018 | 3.6433 | 3.2174 |
| Constrained | 0.9332 | −0.5264 | 1.2473 | 2.2179 | 2 |
| Combined | 1.3789 | −0.2396 | 1.8398 | 3.4148 | 2.7564 |
The negative discrimination scores indicate that lower scoring conformations are correlated with lower RMSDs, while large, negative scaled native scores indicate that the native conformations score substantially more favorably than the predicted conformations. In this case, the negative discrimination score tells us that if we were to sort our models by score and pick the best-scoring models, we would indeed select the lowest-RMSD models. The large, negative scaled native tells us, however, that the score gap between the refined native and the 5th percentile is, on average, equivalent to the difference between the 5th and 95th percentile of the models. Together, these pieces of information suggest that conformational sampling must be improved or directed.
Constrained de novo modeling of CDR H3 loops
Because the predicted kinked structures have low RMSDs and relaxation of native CDR H3 structures can find substantially lower scores (Figure S3), we tested whether biasing the simulation toward kinked conformations would increase the number of low-scoring and low-RMSD models produced in the course of the simulation. As described in methods, we use the parameters of the kink described in our previous work to develop a kink constraint that can be employed during a simulation (Figure 1). Because the constraint potential is smooth and continuous, the conformation of a structure can be minimized with the constraint enabled.
Figure 4 shows the results of the constrained NGK simulation for anti-citrullinated collagen type II antibody (2w60(48)). The τ101 vs. α101 plot (Figure 4B) shows that the constraint successfully biases the simulation to mostly produce kinked structures. At the same time, many models are not kinked, which indicates that the simulations are not being over-constrained.
Figure 4. Results of constrained de novo NGK.
Results of constrained de novo NGK H3 loop prediction on an anti-citrullinated collagen type II antibody (2w60(48)). (A) Funnel plot showing scaled score vs. RMSD of 500 generated models. Plot colors and annotations are the same as in Figure 3. Many kinked H3 models are produced, and the top-scoring models have sub-angstrom RMSDs. The dashed horizontal line indicates the scaled score of the native structure, and shows that the best-scoring models have a similar score to the native (the scaled score of the refined, native structure is −0.2736). (B) τ101 vs. α101 for 500 generated models. While many of the points have τ101 and α101 values that correspond to kinked conformations, there are still many models that are not kinked. These data suggest that the constraint has an appropriate penalty that can be overcome in cases with favorable interactions.
Figure 4A shows a funnel plot for the constrained NGK simulation of 2w60. The fraction of near-native structures has increased dramatically, demonstrating that generating more kinked structures is critical for successful CDR H3 predictions. The dashed horizontal line indicates the scaled score of the native structure, which was below the plotted bounds on the unconstrained plot. That is, the models now have scores near that of the refined, native structure. Therefore, the geometry of the models generated with constraints is more favorable than the geometry of the models generated by the unconstrained method.
Because many more models between 1.0 and 3.0 Å RMSD are generated and those models score more favorably, the discrimination score for the constrained simulation, −0.1721, is worse than the unconstrained case. Figure 4A shows that for many of the models with RMSDs < 2.0 Å, there is a model with RMSD 3.5 Å that scores as well. This result underscores the importance of producing many models even when using constrained NGK.
Figure S4 and S5 show funnel plots and τ101 vs. α101 plots for the rest of the structures in the high-resolution data set. Across the data set, with the exception of five targets (one of which has an unclear base geometry), the scaled native scores appear within the plot bounds, indicating that the models achieve scores close to those of the native structures. Figure S4 shows that the four extended loops in the benchmark (1x9q, 2e27, 3liz, 3m8o) have sets of models that are predominantly kinked. While this is problematic, it appears that, with the exception of 1x9q, these particular targets were also not modeled successfully by unconstrained NGK, which confounds any analysis to determine if the constraint penalty could be overcome when appropriate. Table SII shows numerical results for each target.
The average RMSD of the 10 top-scoring models is lower with constraints in 40 of the 49 targets, and 30 targets have a lower RMSD of the top-scoring model. Without constraints, the average RMSD of the 10 top-scoring models is under 1.0 Å for three targets; this number increases to nine targets when using constraints. When considering only the top-scoring model, seven targets without constraints have RMSDs under 1.0 Å, while thirteen targets with constraints have sub-angstrom RMSDs. Table III shows the lowest RMSD CDR H3 loop produced with and without the constraint for each target in the benchmark set.
Table III.
Comparison of lowest-RMSD model produced from the unconstrained and unconstrained NGK simulations for the high-quality CDR H3 loop benchmark set.
| Target | CDR H3 length |
Min. RMSD unconstrained (Å) |
Min. RMSD constrained (Å) |
|---|---|---|---|
| 1mlb | 9 | 1.0512 | 0.8055 |
| 2d7t | 9 | 0.9517 | 0.8841 |
| 3g5y | 9 | 0.7744 | 0.4713 |
| 3hc4 | 9 | 0.7802 | 0.7545 |
| 1x9q* | 9 | 0.1933 | 2.0763 |
| 2e27* | 9 | 1.3989 | 2.5348 |
| 1jpt | 10 | 0.9972 | 0.7319 |
| 3e8u | 10 | 0.6829 | 0.263 |
| 3m8o* | 10 | 0.8086 | 1.1371 |
| 1mfa | 11 | 1.204 | 0.3159 |
| 1mqk | 11 | 0.8169 | 0.7269 |
| 1nlb | 11 | 0.4001 | 0.3155 |
| 2adf | 11 | 1.4002 | 0.5459 |
| 2fbj | 11 | 0.97 | 0.8883 |
| 2w60 | 11 | 0.445 | 0.3128 |
| 3gnm | 11 | 0.7956 | 0.5754 |
| 3hnt | 11 | 1.2357 | 0.9713 |
| 3v0w | 11 | 0.5647 | 0.4142 |
| 1dlf | 12 | 1.0057 | 0.8847 |
| 2xwt | 12 | 0.3709 | 0.3651 |
| 2ypv | 12 | 1.1427 | 0.5321 |
| 3ifl | 12 | 0.7096 | 0.5588 |
| 3mxw | 12 | 1.1074 | 0.9767 |
| 3oz9 | 12 | 0.573 | 0.5176 |
| 3umt | 12 | 1.2153 | 1.0396 |
| 4h0h | 12 | 1.1919 | 0.6141 |
| 4h20 | 12 | 0.8145 | 0.8209 |
| 3liz* | 12 | 1.3761 | 2.007 |
| 1oaq | 13 | 1.0488 | 0.8637 |
| 2v17 | 13 | 0.7926 | 0.8039 |
| 3t65 | 13 | 2.7751 | 0.9007 |
| 4hpy | 13 | 0.497 | 0.5774 |
| 1jfq | 14 | 1.8429 | 0.563 |
| 2r8s | 14 | 0.6297 | 0.6658 |
| 2vxv | 14 | 1.1711 | 1.1523 |
| 3eo9 | 14 | 3.6195 | 1.2092 |
| 3i9g | 14 | 0.7581 | 0.6414 |
| 3p0y | 14 | 0.4399 | 0.3675 |
| 3giz | 15 | 1.7031 | 0.862 |
| 1fns | 16 | 1.9961 | 1.2479 |
| 1gig | 16 | 2.5965 | 1.9806 |
| 3go1 | 16 | 1.495 | 1.6585 |
| 1seq† | 16 | 3.1272 | 2.8814 |
| 3mlr | 17 | 2.4555 | 2.2999 |
| 3lmj | 18 | 1.8205 | 2.1115 |
| 4f57 | 18 | 1.7381 | 1.3136 |
| 4nzu | 18 | 3.297 | 2.0598 |
| 2fb4 | 19 | 3.0953 | 1.8331 |
| 3nps | 19 | 3.8113 | 2.6624 |
Extended base geometry
Unclear base geometry
Homology modeling with constraints
We tested the constrained method on crystallographic frameworks in order to isolate the loop modeling problem, but our ultimate objective remains predicting antibody structures from sequence. In order to assess the effect of the constraint in the context of homology modeled frameworks, we modified RosettaAntibody to apply constraints to the de novo loop modeling phase and enabled the neighbor-dependent Ramachandran map sampling from NGK. Figure 5 shows cumulative density estimates for CDR H3 loop RMSDs modeled using RosettaAntibody with a kink filter (gray curve) and with the new kink constraint (orange curve) for 2w60. Both methods can generate low-RMSD models of the H3 loop, but with the kink constraint, 1106 of the 2000 models have H3 RMSD < 2.0 Å as opposed to only 796 with the filter.
Figure 5. Modeling CDR H3 on a homology framework.
Modeling CDR H3 on a homology framework for 2w60. A cumulative density estimate of the RMSD of the backbone atoms in the CDR H3 loops of homology models built using the method described in AMA-II(1) (gray) and with the new kink constraint (orange). Dashed vertical lines indicate the fraction of models with RMSD of 1.0 and 2.0 Å or better for each method. The red dashed line shows that 50% of the models produced by the standard method have an RMSD of 3.74 Å or lower, while 50% of the models from the method that exploits the kink constraint have an RMSD of 1.64 Å or lower. Although both methods are successful in producing some low-RMSD models, a significantly larger fraction are produced when using the kink constraint based on τ101 and α101 as opposed to the filter based solely on α101.
The enrichment of low-RMSD models shows that the kink constraint leads to sampling improvements even in cases where RosettaAntibody is already successful. Comparable results can be achieved while generating fewer models, and additional simulation time can be spent performing other stages of modeling, i.e. VL–VH optimization.
With this result, we produced homology models for the entire benchmark set including using the new multi-template VL–VH orientation method(49). Table SIII shows a summary of the H3 accuracy on a homology modeled framework for each target in the set. RosettaAntibody generates sub-angstrom predictions for 26 targets across a variety of loop lengths. The top-scoring model has an H3 RMSD < 1.0 Å for 5 targets. If an ensemble of the top-ten models is considered, as it would be in the case of EnsembleDock(50), 12 targets have sub-angstrom H3 predictions. Notably, all of the loops that were predicted within 1.0 Å are 14 residues or less.
Docking with modeled CDR H3 loops
Successful docking is highly dependent on having accurate models of the bound conformation of each binding partner. To test whether or not the H3 loop conformations predicted using constrained NGK are accurate enough for binding, we focused on 2adf, which is crystallized with its antigen. The CDR H3 loop in 2adf is 11 residues and the constrained NGK simulation has a discrimination score of −0.1758, indicating successful CDR H3 prediction (Table SII). We selected the ten top-scoring models as an ensemble to dock to the bound form of the antigen using EnsembleDock(50). EnsembleDock functions by cycling through a set of distinct backbone conformations after each rigid-body move during the low-resolution stage of docking. Each member of the ensemble is scored, and the best-scoring conformation observed in the low-resolution stage is the starting point for all-atom refinement. EnsembleDock does not resample any of the loop conformations during docking, resulting in all non-H3 RMSDs being 0.0 Å. The kink constraint is not used in this simulation.
The EnsembleDock results are shown in Figure 6. Points are colored to indicate the Critical Assessment of PRedicted Interactions (CAPRI) quality rating(51) of each model, with gray points corresponding to incorrect structures, orange to acceptable quality, red to medium-quality and blue to high-quality models (see methods). The fnat metric that is used to compute the CAPRI quality is not restricted to the H3 loop and thus the values we report also consider the contributions of non-H3 CDR loops to the interface. The 10 top models by interface score encompass one incorrect model, four acceptable models, one medium-quality model and four high-quality models.
Figure 6. Results of docking an antibody with a modeled H3 loop.
Results of docking an antithrombotic antibody to its antigen (2adf(61)) using EnsembleDock(50) with CDR H3 loops modeled with constrained NGK. Funnel plots showing scaled interface score vs. interface RMSD. Interface score is calculated as the score of the unbound partners subtracted from the score of the complex. Interface RMSD is the RMSD of the backbone atoms of the residues within 8.0 Å of a residue on the other docking partner. The points are colored using the CAPRI quality ratings(51), where gray points correspond to incorrect structures, orange points to acceptable (65 models), red points to medium (8 models), and blue points to high-quality models (4 models). The high-quality models are clearly separated from the other models by interface score as evidenced by the discrimination score of −0.9112 (lower right corner).
The ten models used in the ensemble have scores ranging from −594.19 to −586.90, and the average H3 RMSD is 1.48 Å (Table IV). The top-ranked model has a loop RMSD of 1.53 Å, and the eighth structure in the set has an H3 RMSD of 0.75 Å. As shown in Table IV, the 10 top models do not converge on a single member of the ensemble, showing that considering several models simultaneously is a path forward.
Table IV.
Summary of top 10 models produced by EnsembleDock. Interface score is calculated as the score of the unbound partners subtracted from the score of the complex. Interface RMSD is the RMSD of the backbone atoms of the residues within 8.0 Å of a residue on the other docking partner. Ligand RMSD is the backbone atom RMSD of the antigen after superposing the antibody to the native structure. fnat is the fraction of native residue–residue contacts recovered, where contacting residues are defined as residues on opposite binding partners within 5.0 Å of each other. The H3 RMSD column shows the RMSD of the CDR H3 loop of the model that was ultimately selected by EnsembleDock to generate the docked model. Interestingly, a different member of the ensemble was used in each of the high-quality docked models.
| Rank | Interface Score |
Interface RMSD (Å) |
Ligand RMSD (Å) |
fnat | H3 RMSD (Å) |
CAPRI rating |
|---|---|---|---|---|---|---|
| 1 | −8.79 | 0.43 | 4 | 0.91 | 1.48 | High |
| 2 | −8.51 | 0.54 | 1.78 | 0.86 | 0.75 | High |
| 3 | −8.22 | 0.74 | 2.57 | 0.89 | 1.59 | High |
| 4 | −7.67 | 0.43 | 1.6 | 0.91 | 1.53 | High |
| 5 | −5.54 | 2.72 | 9.25 | 0.26 | 1.69 | Acceptable |
| 6 | −5.55 | 3.5 | 9.49 | 0.29 | 0.75 | Acceptable |
| 7 | −5.53 | 2.28 | 7.56 | 0.57 | 1.54 | Medium |
| 8 | −5.27 | 13.75 | 25.24 | 0.11 | 1.54 | Incorrect |
| 9 | −5.22 | 5.27 | 7.33 | 0.2 | 1.69 | Acceptable |
| 10 | −5.18 | 2.76 | 9.38 | 0.34 | 1.53 | Acceptable |
Big data (set)
In order to assess the progress we have made in the development of RosettaAntibody, we benchmarked the performance for two additional sets of structures: (1) the ten non-rabbit AMA II targets; and (2) the 54 targets in the initial RosettaAntibody study. Table SIV shows the summary of the H3 predictions in the context of a homology-modeled framework for the AMA II targets. In AMA II, RosettaAntibody produced sub-angstrom models for two targets(1). The average minimum RMSD of the ten targets is 1.28 Å. The new constrained method successfully produces sub-angstrom predictions for two of the ten targets, however these models are not ranked in a set of ten top-scoring structures. When considering the ten top-scoring models, six of ten targets are predicted within 2.0 Å RMSD. In AMA-II we were able to rank sub-angstrom predictions in the top-scoring models for two targets. The apparent degradation is due to our use of homology filters in this study to prevent RosettaAntibody from selecting the now-available crystal structures as templates. Additionally, the kink geometry can deviate from native values of τ101 and α101 with little change in overall RMSD, but this deficiency would expose itself during downstream simulations.
The 2009 RosettaAntibody benchmark set provides a more comprehensive view of improvements to de novo CDR H3 structure prediction on homology-modeled frameworks. Table SV shows the per-target summary of modeling and includes the lowest-RMSD achieved for the target in the 2009 study(15). There is a sampling improvement in 49 targets, with average (including the targets with larger RMSDs) improved RMSDs of 0.1 Å for very short loops (4–6 residues), 0.6 Å for short loops (7–9 residues), 0.7 Å for medium loops (10–11 residues), 1.3 Å for long loops (12–14 residues), and 1.1 Å for very long loops (17–22 residues). Three of the five targets with degraded performance had RMSDs of 0.2 Å higher (1dqq: length 5, 0.2 Å → 0.4 Å; 1z3g: length 6, 1.8 Å → 2.0 Å; 1bql: length 7, 1.6 Å → 1.8 Å), one increased by 0.4 Å (2h2h: length 12, 1.6 Å → 2.0 Å), and one increased by 0.6 Å (1bj1: length 14, 1.6 Å → 2.2 Å). Interestingly, 24 of the targets have a lower-RMSD model in the ten top-scoring models than was even sampled in 2009. The smaller change in performance for shorter loops is due to the fact that they were predicted to high accuracy in the initial study, and can only improve marginally, while longer loops improve more substantially.
Discussion
Antibody structure prediction has made great strides, but accurately modeling CDR H3 loops remains elusive due to its variable length, sequence and structure across antibodies. We found that state-of-the-art de novo loop prediction methods fail to generate near-native conformations based on both RMSD and the kink geometry parameters, τ101 and α101. In our previous work(2), we hypothesized that the kink creates the observed structural diversity of CDR H3 loops. In this paper we developed a constraint based on these parameters and tested it on a set of high-quality CDR H3 structures. The effectiveness of this constraint for enabling high-resolution H3 predictions suggests that creating models with native-like values for α101 and τ101 drives better results for the whole loop. We found this constraint is also effective in improving CDR H3 predictions on homology modeled frameworks and producing H3 models of sufficient quality to successfully dock to antigen.
Part of this study required constructing a set of high-resolution CDR H3 loops from crystal structures. Not all CDR H3 loops meet the strict quality cutoffs that were used in this study. It is possible that some of the loops that meet these criteria are simply more stable or rigid than some other H3 loops. If that is the case, these loops may be easier modeling targets. Regardless, since the atomic coordinates of these loops are well-defined, structural comparisons between models and the loops have clear meaning.
Although the atomic coordinates may be well-defined, the static conformation found in crystal structures may not tell a complete story, and this limitation may be particularly pronounced in loop regions. However, we do not believe that all loop regions are extremely flexible; indeed, such a view could lead one to conclude that the entire objective of loop structure prediction is poorly defined at best and a misguided exercise in futility at worst. In the cases of antibodies that have been crystallized in both the bound and free forms (predominantly antibodies with peptide antigens), CDR loop conformations do not change substantially (typically < 1.0 Å RMSD; 37% of CDR H3 loops deviate > 1.0 Å) (46) and, thus, can be treated as rigid. Additional evidence of CDR loop rigidity comes from studies that compare flexibility estimates of naïve and mature antibodies and find that one of the effects of somatic hypermutation is a decrease in loop flexibility(52–54). The degree to which these trends hold for antibodies with protein antigens, or vary as a function of the length of the CDR H3 loop remains to be established.
While there are not enough loops at each length to draw conclusions about prediction performance as a function of length, the longest loop where the average RMSD of the 10 top-scoring models is sub-angstrom without constraints is 13 residues, and the longest with constraints is 14. This difference seems small, but the RMSD of the top-scoring model for some longer loops reveals the extent to which the kink constraint improves the performance of de novo loop modeling. For example, for the 19-residue CDR H3 loop in 2fb4, the RMSD of 14.67 Å without constraints is reduced to 3.63 Å (Table III). While this is a substantial improvement, these models are unlikely to successfully dock. Further improvements might arise from using more cycles of NGK for longer loops, generating more models or incorporating knowledge of additional local structures (e.g. β turns, or additional constraints to the terminal residues of the loop(29)) in addition to the kink into the simulation.
A challenge remains in addressing non-kinked CDR H3 loops. In this study, we applied the kink constraint to all CDR H3 loops without considering whether the native loop has a kink. We made this decision for a few reasons. Most importantly, the vast majority of CDR H3 loops are kinked, and we remain predominantly interested in developing a method to predict those conformations reliably. Second, all attempts to classify the base geometry of the H3 loop by sequence alone have not held up as more structures have been solved, leaving us with no way to determine when we should not apply the constraint. While carrying out this work, we developed several alternate hypotheses to account for non-kinked H3 loops, including using the constraint stochastically with a probability in accordance with the observed populations of kinked loops (that is, each model would have an 85% chance of being constrained). Unfortunately, Rosetta was unable to generate low-RMSD conformations of the non-kinked H3 loops in our dataset even in the absence of the constraint. Because of this, there is no clear way to test alternatives to the approach described in this manuscript at this time.
The sampling performed in the low-resolution stage may lose information of some important interactions that are mediated by side chains. Some modeling methods have been developed that operate in all-atom mode throughout the entire simulation. One such method available in Rosetta is step-wise assembly (SWA)(55), which builds the loop one residue at a time. While this method has shown promise, it is extremely computationally expensive and is therefore not yet well-suited to antibody homology modeling tasks that rebuild the H3 loop while simultaneously sampling VL–VH orientation. Nevertheless, an all-atom loop-modeling routine may enable Rosetta to capture critical side-chain interactions.
In fact, an anti-dansyl antibody, which has been crystallized at two different pH-values (pH 5.25, PDB accession code 1dlf; pH 6.75, 2dlf), has a histidine in its CDR H3 loop(56). Nakasako et al. found that the structure of the antibody remained the same except for the CDR H3 loop, which undergoes a pH-dependent conformational change, presumably controlled by the protonation state of the histidine within the loop. Preliminary tests using pH-aware loop prediction and side-chain packing failed to capture the effect of the protonation state(57), again highlighting the need for side-chain accuracy and increased score function detail.
While this study mostly focused on modeling CDR H3 loops on the crystallographic framework, the ultimate test of the utility of a new loop modeling method in the context of antibody modeling is predicting CDR H3 conformations on a homology framework. We tested the new method on a homology modeled framework by comparing the distribution of CDR H3 RMSDs from the standard method, which uses a filter based on α101, and the new constrained method, which evaluates a potential based on both τ101 and α101. The new constrained method produces a substantially larger fraction of low-RMSD models, which should enable the development of new protocols that focus more time on other aspects of antibody modeling, e.g. VL–VH orientation optimization(27).
The new method can generate sub-angstrom loop predictions for 20 of the 40 very short, short, and medium-length loops in the 2009 benchmark set. While progress is being made, long loops still pose a challenge; on a homology framework, the best-sampled model for all 10 of the long loops in the 2009 benchmark set lies within 1.0–2.0 Å RMSD.
Another goal for antibody structure prediction is to generate models of sufficient quality to be used in downstream applications, namely antibody–antigen docking. To assess the quality of the predicted H3 conformations, we used EnsembleDock with a set of the 10 top-scoring models and the bound conformation of the antigen. The simulation correctly predicts the conformation of the complex. This case is idealized in the sense that the CDR H3 loop was modeled on the crystal framework and the bound form of the antigen was used, but the successful simulation marks a necessary milestone in obtaining reliable blind docked complex predictions from sequence. The success of both the homology modeling and docking simulations is encouraging and these simulations should serve as the starting point for future development of improved antibody—antigen docking methods. In summary, a structure-based kink constraint has enabled accurate de novo structure prediction of the most diverse region of antibodies. We found that performance degrades on homology modeled frameworks, which reinforces the importance of the environment of the loop. Coupled with the chain pairings from Teplyakov et al.(35), there is support for the hypothesis we introduced previously: the C-terminal kink is the key to CDR H3’s diversity and is stabilized by the environment of the loop(2).
Supplementary Material
Acknowledgments
The authors would like to Andrew Leaver-Fay for cutting the two-body Ramachandran sampling method memory requirements from 5 GB to 160 MB, Nick Marze for running the new method on the 2009 RosettaAntibody benchmark set, as well as the Rosetta developers for the continued development of Rosetta. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), as well as the Maryland Advanced Research Computing Center (MARCC).
Funding: This work was supported by National Institutes of Health Grant R01 GM078221 to JJG; APL (acknowledged) is supported by National Institutes of Health Grant R01 GM73151; XSEDE is supported by National Science Foundation grant number ACI-1053575; MARCC is supported by the state of Maryland.
Appendix
Determining the parameters for the flat harmonic potential
Because the penalty should begin after 1.0 σ, t can be set to σ. Now we solve for ξ in order to produce the desired penalty schedule. First, we solve for ξ at 3.0 σ as follows:
and then plug in ξ = 2t and evaluate the penalty at 2.0 σ to check the intermediate value
and we find that setting ξ to 2 σ will exactly produce a reasonable penalty schedule with the useful feature of being a factor of four larger at 3.0 σ than at 2.0 σ.
Mixing CCD and NGK
In addition to KIC and NGK, Rosetta also has implementations of loop modeling methods that use Cyclic Coordinate Descent (CCD(58)) to ensure the continuity of the loop. CCD closure calculates the dihedral angles required to minimize the gap between the loop end points for a single position in the loop, and iterates over each residue and until the loop is closed. In contrast to NGK, CCD-based methods can close loops while maintaining the overall conformation of the starting, open loop. Generally, the CCD-based methods use fragments from known structures for coarse-grained sampling and small perturbations of the conformation of each residue for refinement(59). Because of their object-oriented design, any of the loop modeling methods in Rosetta can be mixed-and-matched within a single simulation. To couple the more conservative CCD refinement with the aggressive NGK sampling, a combined NGK–CCD simulation can be run. The combined simulation uses the same command line as the constrained NGK simulation, but with
-loops:refine refine_kic changed to -loops:refine refine_ccd.
./loopmodel.macosclangrelease
-native input_file.pdb
-s input_file.pdb
-nstruct 500
-loops:loop_file h3.loops
-loops:remodel perturb_kic
-loops:refine refine_ccd
-loops:outer_cycles 5
-kic_bump_overlap_factor 0.36
-legacy_kic false
-kic_min_after_repack true
-corrections:score:use_bicubic_interpolation false
-loops:kic_omega_sampling
-loops:kic_rama2b
-allow_omega_move
-loops:ramp_fa_rep
-loops:ramp_rama
-ex1
-ex2
-extrachi_cutoff 0
The biophysical and numerical details of these flags are described in the original Rosetta kinematic closure papers(40, 43).
References
- 1.Weitzner BD, Kuroda D, Marze N, Xu J, Gray JJ. Blind prediction performance of RosettaAntibody 3.0: Grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins. 2014;82:1611–1623. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weitzner Brian D, Dunbrack Roland L, Gray Jeffrey J. The Origin of CDR H3 Structural Diversity. Structure. 2015;23:302–311. doi: 10.1016/j.str.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buss NA, Henderson SJ, McFarlane M, Shenton JM, de Haan L. Monoclonal antibody therapeutics: history and future. Curr Opin Pharmacol. 2012;12:615–622. doi: 10.1016/j.coph.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Reichert JM. Antibodies to watch in 2016. MAbs. 2016;8:197–204. doi: 10.1080/19420862.2015.1125583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 6.Van Weemen BK, Schuurs AHWM. Immunoassay using antigen-enzyme conjugates. FEBS Lett. 1971;15:232–236. doi: 10.1016/0014-5793(71)80319-8. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Tack D, Hughes RA, Ellington AD, Gray JJ. Structure-based non-canonical amino acid design to covalently crosslink an antibody-antigen complex. J Struct Biol. 2013;185:215–222. doi: 10.1016/j.jsb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, D\”orner T, Andrews SF, Wilson PC, Hunicke-Smith SP, Willson CG, Ellington AD, Georgiou G, D$\backslash$”orner T. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, Georgiou G. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc Natl Acad Sci U S A. 2016;113:E2636–E2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLOS Comput Biol. 2010;6:e1000644. doi: 10.1371/journal.pcbi.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marillet S, Boudinot P, Cazals F. High-resolution crystal structures leverage protein binding affinity predictions. Proteins. 2016;84:9–20. doi: 10.1002/prot.24946. [DOI] [PubMed] [Google Scholar]
- 14.Vangone A, Bonvin AM. Contacts-based prediction of binding affinity in protein-protein complexes. Elife. 2015;4:e07454. doi: 10.7554/eLife.07454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody FV regions and application to antibody-antigen docking. Proteins. 2009;74:497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 17.North B, Lehmann A, Dunbrack JRL. A new clustering of antibody CDR loop conformations. J Mol Biol. 2011;406:228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Almagro JC, Teplyakov A, Luo J, Sweet RW, Kodangattil S, Hernandez-Guzman F, Gilliland GL. Second Antibody Modeling Assessment (AMA-II) Proteins. 2014;82:1553–1562. doi: 10.1002/prot.24567. [DOI] [PubMed] [Google Scholar]
- 20.Fasnacht M, Butenhof K, Goupil-Lamy A, Hernandez-Guzman F, Huang H, Yan L. Automated antibody structure prediction using Accelrys tools: results and best practices. Proteins. 2014;82:1583–1598. doi: 10.1002/prot.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier JK, Labute P. Assessment of fully automated antibody homology modeling protocols in molecular operating environment. Proteins. 2014;82:1599–1610. doi: 10.1002/prot.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirai H, Ikeda K, Yamashita K, Tsuchiya Y, Sarmiento J, Liang S, Morokata T, Mizuguchi K, Higo J, Standley DM, Nakamura H. High-resolution modeling of antibody structures by a combination of bioinformatics, expert knowledge, and molecular simulations. Proteins. 2014;82:1624–1635. doi: 10.1002/prot.24591. [DOI] [PubMed] [Google Scholar]
- 23.Zhu K, Day T, Warshaviak D, Murrett C, Friesner R, Pearlman D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins. 2014;82:1646–1655. doi: 10.1002/prot.24551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berrondo M, Kaufmann S, Berrondo M. Automated Aufbau of antibody structures from given sequences using Macromoltek’s SmrtMolAntibody. Proteins. 2014;82:1636–1645. doi: 10.1002/prot.24595. [DOI] [PubMed] [Google Scholar]
- 25.Bujotzek A, Dunbar J, Lipsmeier F, Schäfer W, Antes I, Deane CM, Georges G. Prediction of VH–VL domain orientation for antibody variable domain modeling. Proteins. 2015 doi: 10.1002/prot.24756. [DOI] [PubMed] [Google Scholar]
- 26.Dunbar J, Krawczyk K, Leem J, Baker T, Fuchs A, Georges G, Shi J, Deane CM. SAbDab: the structural antibody database. Nucleic Acids Res. 2013;42:D1140–D1146. doi: 10.1093/nar/gkt1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marze NA, Lyskov S, Gray JJ. Improved prediction of antibody VL–VH orientation. Protein Eng Des Sel. 2016 doi: 10.1093/protein/gzw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu K, Day T. Ab initio structure prediction of the antibody hypervariable H3 loop. Proteins. 2013;81:1081–1089. doi: 10.1002/prot.24240. [DOI] [PubMed] [Google Scholar]
- 29.Finn JA, Koehler Leman J, Willis JR, Cisneros A, 3rd, Crowe JE, Jr, Meiler J. Improving Loop Modeling of the Antibody Complementarity-Determining Region 3 Using Knowledge-Based Restraints. PLoS One. 2016;11:e0154811. doi: 10.1371/journal.pone.0154811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirai H, Kidera A, Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996;399:1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- 31.Shirai H, Kidera A, Nakamura H. H3-rules: identification of CDR-H3 structures in antibodies. FEBS Lett. 1999;455:188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda D, Shirai H, Kobori M, Nakamura H. Structural classification of CDR-H3 revisited: a lesson in antibody modeling. Proteins. 2008;73:608–620. doi: 10.1002/prot.22087. [DOI] [PubMed] [Google Scholar]
- 33.Morea V, Tramontano A, Rustici M, Chothia C, Lesk AM. Antibody structure, prediction and redesign. Biophys Chem. 1997;68:9–16. doi: 10.1016/s0301-4622(96)02266-1. [DOI] [PubMed] [Google Scholar]
- 34.Morea V, Tramontano A, Rustici M, Chothia C, Lesk AM. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J Mol Biol. 1998;275:269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- 35.Teplyakov A, Obmolova G, Malia TJ, Luo J, Muzammil S, Sweet R, Almagro JC, Gilliland GL. Structural diversity in a human antibody germline library. MAbs. 2016 doi: 10.1080/19420862.2016.1190060. Advanced access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chirikjian GS. Modeling loop entropy. Methods Enzymol. 2011;487:99–132. doi: 10.1016/B978-0-12-381270-4.00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adolf-Bryfogle J, Xu Q, North B, Lehmann A, Dunbrack RL. PyIgClassify: a database of antibody CDR structural classifications. Nucleic Acids Res. 2014;43:D432–D438. doi: 10.1093/nar/gku1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honegger A, Pluckthun A. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol. 2001;309:657–670. doi: 10.1006/jmbi.2001.4662. [DOI] [PubMed] [Google Scholar]
- 39.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutsias EA, Seok C, Wester MJ, Dill KA. Resultants and loop closure. Int J Quantum Chem. 2006;106:176–189. [Google Scholar]
- 42.Ting D, Wang G, Shapovalov M, Mitra R, Jordan MI, Dunbrack RL, Dunbrack RL., Jr Neighbor-dependent Ramachandran probability distributions of amino acids developed from a hierarchical Dirichlet process model. PLOS Comput Biol. 2010;6:e1000763. doi: 10.1371/journal.pcbi.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein A, Kortemme T. Improvements to robotics-inspired conformational sampling in Rosetta. PLOS ONE. 2013;8:e63090. doi: 10.1371/journal.pone.0063090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janin J. Assessing predictions of protein-protein interaction: the CAPRI experiment. Protein Sci. 2005;14:278–283. doi: 10.1110/ps.041081905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sela-Culang I, Alon S, Ofran Y. A systematic comparison of free and bound antibodies reveals binding-related conformational changes. J Immunol. 2012;189:4890–4899. doi: 10.4049/jimmunol.1201493. [DOI] [PubMed] [Google Scholar]
- 47.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 48.Uysal H, Bockermann R, Nandakumar KS, Sehnert B, Bajtner E, Engström A, Serre G, Burkhardt H, Thunnissen MMGM, Holmdahl R. Structure and pathogenicity of antibodies specific for citrullinated collagen type II in experimental arthritis. J Exp Med. 2009;206:449–462. doi: 10.1084/jem.20081862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marze N, Gray JJ. Improved prediction of antibody VL–VH orientation. Protein Engineering, Design & Selection. 2016 doi: 10.1093/protein/gzw013. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J Mol Biol. 2008;381:1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Méndez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 52.Babor M, Kortemme T. Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins. 2009;75:846–858. doi: 10.1002/prot.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong SE, Sellers BD, Jacobson MP. Effects of somatic mutations on CDR loop flexibility during affinity maturation. Proteins. 2011;79:821–829. doi: 10.1002/prot.22920. [DOI] [PubMed] [Google Scholar]
- 54.Sinha N, Smith-Gill SJ. Molecular dynamics simulation of a high-affinity antibody-protein complex: the binding site is a mosaic of locally flexible and preorganized rigid regions. Cell Biochem Biophys. 2005;43:253–273. doi: 10.1385/CBB:43:2:253. [DOI] [PubMed] [Google Scholar]
- 55.Das R. Atomic-accuracy prediction of protein loop structures through an RNA-inspired ansatz. PLOS ONE. 2013;8:e74830. doi: 10.1371/journal.pone.0074830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakasako M, Takahashi H, Shimba N, Shimada I, Arata Y. The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the F$_\textV$ fragment from an anti-dansyl monoclonal antibody. J Mol Biol. 1999;291:117–134. doi: 10.1006/jmbi.1999.2931. [DOI] [PubMed] [Google Scholar]
- 57.Weitzner BD. Chemical & Biomolecular Engineering. Johns Hopkins University; 2015. Next-generation antibody modeling. [Google Scholar]
- 58.Canutescu AA, Dunbrack RL., Jr Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohl CA, Strauss CE, Chivian D, Baker D. Modeling structurally variable regions in homologous proteins with Rosetta. Proteins. 2004;55:656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 60.Schneider EL, Lee MS, Baharuddin A, Goetz DH, Farady CJ, Ward M, Wang C-I, Craik CS. A reverse binding motif that contributes to specific protease inhibition by antibodies. J Mol Biol. 2012;415:699–715. doi: 10.1016/j.jmb.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Staelens S, Hadders MA, Vauterin S, Platteau Ce, De Maeyer M, Vanhoorelbeke K, Huizinga EG, Deckmyn H. Paratope determination of the antithrombotic antibody 82D6A3 based on the crystal structure of its complex with the von Willebrand factor A3-domain. J Biol Chem. 2006;281:2225–2231. doi: 10.1074/jbc.M508191200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.