Abstract
Nitric oxide (NO) is an essential regulator of Drosophila development and physiology. We describe a novel mode of regulation of NO synthase (NOS) function that uses endogenously produced truncated protein isoforms of Drosophila NOS (DNOS). These isoforms inhibit NOS enzymatic activity in vitro and in vivo, reflecting their ability to form complexes with the full-length DNOS protein (DNOS1). Truncated isoforms suppress the antiproliferative action of DNOS1 in the eye imaginal disc by impacting the retinoblastoma-dependent pathway, yielding hyperproliferative phenotypes in pupae and adult flies. Our results indicate that endogenous products of the dNOS locus act as dominant negative regulators of NOS activity during Drosophila development.
Keywords: Nitric oxide, Drosophila, development, signaling, imaginal disc
Nitric oxide (NO) is produced in animal cells by NO synthases (NOSs) in response to a wide range of diverse stimuli. NO acts in an autocrine and paracrine fashion on a large variety of cellular targets to initiate rapid and transient, as well as slow and long-lasting physiological changes (for review, see Ignarro 2000). In mammals, these changes include blood-vessel relaxation, immune response, cell cycle control, and neurotransmission. In Drosophila, NO has been implicated in visual-system development, immunity, behavior, response to hypoxia, osmoregulation, and regulation of cell cycle progression during development (Dow et al. 1994; Kuzin et al. 1996, 2000; Gibbs and Truman 1998; Wingrove and O'Farrell 1999; Nappi et al. 2000; DiGregorio et al. 2001; Teodoro and O'Farrell 2003).
NO is a signaling molecule with diverse targets and with limited spatial and temporal range. The potential of NOS enzymes to trigger NO signaling cascades is restrained by an elaborate network of control mechanisms: transcriptional, posttranscriptional, and translational regulation; posttranslational modifications, such as phosphorylation and palmitoylation; dependence on calcium; subcellular localization; and interactions with other proteins (Papapetropoulos et al. 1999; Wang et al. 1999a; Ignarro 2000; Nathan 2003). These modes of regulation are not unique to the NOS-signaling system; however, the complexity of the regulatory network that controls production of NO makes this system an attractive model for understanding how generation of a signal is regulated in a physiological or developmental context.
A potential mechanism for controlling NO action may be related to the unusual complexity of the genomic organization of the NOS genes. For instance, more than 10 alternative promoters, in addition to multiple alternative splice sites and several polyadenylation sites are used by mammalian neuronal NOS genes (Wang et al. 1999a). The choice of promoters, splice sites, and polyadenylation sites alters the coding capacity of NOS transcripts as well as their translational efficiency (Wang et al. 1999a,b). Likewise, the Drosophila NOS gene (dNOS) encodes a family of at least 10 different transcripts that may code for as many as seven different proteins (Regulski and Tully 1995; Stasiv et al. 2001). Of these mRNAs, which are generated through the use of alternative transcription initiation and splice sites, only one isoform, dNOS1, encodes an enzymatically active protein, DNOS1. The majority of dNOS alternative transcripts encode truncated proteins that lack the crucial C-terminal reductase domain (with binding sites for FMN, FAD, and NADPH) of the full-length DNOS1, but retain the N-terminal oxygenase domain (with binding sites for heme, L-arginine, and tetrahydro-L-biopterine).
All NO synthases are catalytically active as homodimers; their oxygenase domains contain the active center that oxidizes L-arginine to L-citrulline and NO, whereas their reductase domains ensure the flow of electrons required for the catalysis (Stuehr 1999; Alderton et al. 2001). In NOS homodimers, the flow of electrons is directed from the reductase domain of one polypeptide of the dimer to the oxygenase domain of the other member of the dimer (Siddhanta et al. 1998; Sagami et al. 2001). These structural features of NOS suggest a potential regulatory mechanism that could use short NOS isoforms as inhibitors of the activity of the full-length protein. Given the structural similarities between various isoforms of NOS across species, such mechanism could be relevant both for Drosophila and for mammalian NOSs; a number of reports describe alternative transcripts that encode truncated NOS-like proteins (Wang et al. 1999a). However, an experimental model to test this hypothetical mechanism in vivo has not yet been established; thus, the potential biological significance of this notion has not yet been explored.
To understand how an inactive subunit of a multimeric protein may have a dominant negative effect on an important signaling cascade in vivo, we focused on DNOS4, a product of one of the more abundant alternative transcripts of the Drosophila NOS gene. We show that DNOS4 is endogenously expressed in wild-type Drosophila. We demonstrate that ectopic expression of DNOS4 in the imaginal discs of Drosophila larvae suppresses the antiproliferative activity of DNOS1, resulting in hyperproliferative phenotypes in adult flies. DNOS4 is able to form heterodimers with DNOS1 in vitro and in vivo and inhibit production of NO. Together, our results indicate that DNOS4 acts as an endogenous dominant negative regulator of NOS activity during Drosophila development, pointing to a novel mechanism for the regulation of NO production.
Results
dNOS4 is an endogenous alternatively spliced transcript of the Drosophila NOS gene
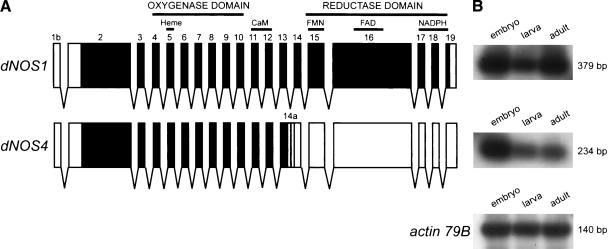
The dNOS locus of Drosophila is subject to complex transcriptional and posttranscriptional regulation (Stasiv et al. 2001). It produces a large variety of mRNA isoforms through the use of multiple promoters and alternative splice sites. Only one of them, dNOS1 (Fig. 1A), codes for the full-length enzymatically active protein. Another abundant alternative transcript of the dNOS gene is the dNOS4 isoform, which retains the entire intron 13 (this 109-nucleotide-long segment is now referred to as exon 14a of dNOS4). The resulting open reading frame is terminated by a stop codon 63 nucleotides into exon 14a (Fig. 1A). It encodes a protein containing 757 amino acids with a predicted molecular mass of 84 kDa (cf. DNOS1 is 1350 amino acids, 150 kDa). DNOS4 contains a unique 21-amino acid-long C-terminal peptide encoded by exon 14a, whereas the preceding 736 amino acids are identical to those of DNOS1. Thus, the DNOS4 protein is a truncated version of DNOS1; it lacks the entire reductase domain, while retaining the oxygenase domain. Semiquantitative RT–PCR analysis indicates that dNOS4 mRNA is expressed in the embryo at levels comparable to those of dNOS1 mRNA; dNOS4 levels are lower in larvae and in adult flies, whereas dNOS1 levels do not change appreciably (Fig. 1B).
Figure 1.
Alternative splicing generates truncated DNOS isoforms. (A) The structure of two major dNOS transcripts, dNOS1 and dNOS4. Regions encoding structural domains and cofactor-binding sites are shown. Exons are numbered and indicated by boxes (black boxes for protein-coding regions, white boxes for 5′- and 3′-UTRs); gray shading highlights last 63 nucleotides (encoded by exon 14a) of the dNOS4 open reading frame. (B) Expression of dNOS1, dNOS4, and actin 79B (taken as a control) transcripts throughout Drosophila development. Total RNA samples were subjected to RT–PCR amplification using transcript-specific primers, followed by Southern blot analysis. Sizes of amplified products are indicated.
Another variant of dNOS mRNA, dNOS7, encodes a protein identical to DNOS4; however, the transcription initiation site for dNOS7 RNA is different from that of dNOS4 (exon 1a vs. exon 1b, respectively). Unlike dNOS4, dNOS7 is exclusively expressed during the larval stage (Stasiv et al. 2001).
Coexpression of DNOS1 and DNOS4 inhibits NOS activity in vitro
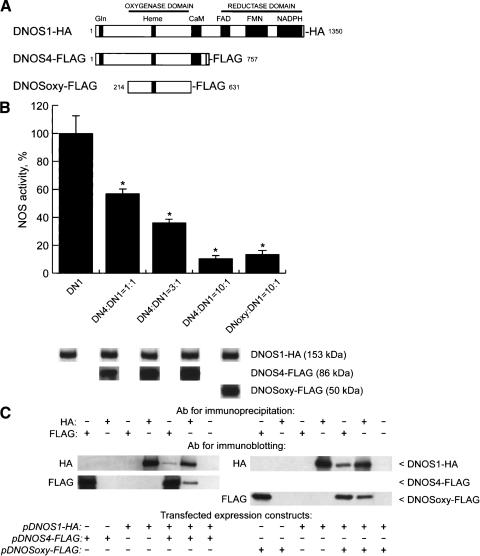
DNOS4 lacks the C-terminal reductase domain that participates in electron transfer during catalysis, while it retains the catalytic N-terminal oxygenase domain, including the critical heme-binding site. DNOS4 also retains a long stretch of glutamine (Gln) residues at the N terminus; such regions have been shown to promote multimerization of proteins (Perutz et al. 1994; Stott et al. 1995; Zoghbi and Orr 2000; note that such Gln-rich region is not present in mammalian NOS proteins). These structural features of DNOS4 predict that (1) DNOS4 itself is incapable of producing NO, (2) it may be capable of forming heterodimers with DNOS1, and (3) heteromers between DNOS1 and DNOS4 will have reduced enzymatic activity. To investigate whether DNOS4 is capable of forming a heteromeric complex with DNOS1 and suppressing NOS activity, and to examine which region of DNOS4 may contribute to its effects on DNOS1, we used expression plasmids for DNOS1 and DNOS4 proteins, each with a short peptide tag fused to its C terminus (Fig. 2A), the influenza virus hemagglutinin (HA) epitope-tagged DNOS1 (pDNOS1-HA), and the synthetic FLAG epitope-tagged DNOS4 (pDNOS4-FLAG; Stasiv et al. 2001). We also generated a plasmid, pDNOSoxy-FLAG, that codes for a shorter version of DNOS4 (residues 214–631) and carries the FLAG epitope at its C terminus (Fig. 2A). It lacks the Gln-rich region at the N terminus, thus representing the “core” oxygenase domain of DNOS, which is highly similar to previously defined oxygenase domains of mammalian NOS proteins (Regulski and Tully 1995; Raman et al. 1998; Fischmann et al. 1999).
Figure 2.
DNOS4 inhibits activity of the full-length DNOS1 enzyme in vitro. (A) DNOS proteins encoded by expression constructs used for transient transfection of 293 cells. Structural domains are indicated. Cofactor-binding sites and the Gln-rich region are shown by black boxes. Gray box indicates C-terminal peptide unique for DNOS4. HA or FLAG indicate epitope tags attached to C termini of expressed DNOS proteins. The number of amino acid residues in each protein (excluding tags) is shown. (B) NOS activity in lysates of transfected 293 cells. The pDNOS1-HA construct (DN1) was either expressed alone or coexpressed with indicated molar excess of a truncated dNOS construct (either pDNOS4-FLAG [DN4] or pDNOSoxy-FLAG [DNoxy]). The total amount of DNA in each transfection was kept constant by addition of carrier DNA. NOS activity was measured by the [3H]arginine–[3H]citrulline conversion assay. Negligible NOS activity in lysates from cells transfected with the expression vector alone was subtracted from NOS activity detected in experimental lysates. The activity data shown are expressed as a percentage of the NOS activity of the full-length enzyme alone (14,330 cpm/min/mg total protein, 100% value). NOS activity in lysates from cells transfected with either pDNOS4-FLAG or pDNOSoxy-FLAG construct alone was not detectable (data not shown). For the detection of expressed DNOS isoforms, an equal amount of total protein (50 μg/lane) from cells transfected with different DNOS encoding constructs was analyzed by SDS-PAGE followed by immunoblotting using either HA antibody of FLAG antibody. The results shown are representative of three independent experiments; all data are mean ± standard error. Statistically significant differences (P < 0.05 according to Student's t-test) between the analyzed samples are marked with an asterisk (*). (C) DNOS4 forms heterodimers with the full-length DNOS1. Shown are the results of coimmunoprecipitation of the full-length DNOS1-HA with truncated DNOS proteins tagged with FLAG epitope. Duplicate samples of protein extracts (200 μg) from transfected 293 cells were immunoprecipitated either with the HA or FLAG antibody (indicated as Ab for immunoprecipitation), followed by SDS-PAGE analysis and immunoblotting using either HA or FLAG antibody (indicated as Ab for immunoblotting). “Transfected expression constructs” indicates dNOS expression constructs used in transfection experiments.
To test the effect of DNOS4 and DNOSoxy proteins on enzymatic activity of the full-length DNOS1, we transiently coexpressed recombinant plasmids in cultured human embryonic kidney cells (293 cells) and measured NOS enzymatic activity in cell-free extracts. NOS activity was undetectable in lysates of untransfected cells and in cells expressing either DNOS4-FLAG or DNOSoxy-FLAG (data not shown), whereas lysates from cells transfected with pDNOS1-HA showed significant levels of NOS activity (Fig. 2B). In contrast, enzymatic activity in cell lysates was decreased when a constant amount of the pDNOS1-HA was cotransfected with increasing amounts of the pDNOS4-FLAG. At a 1:1 molar ratio of plasmids encoding DNOS4 and DNOS1, NOS activity was 57% of the activity seen in lysates from cells transfected by the plasmid encoding full-length DNOS1. At a 3:1 ratio, the activity dropped to 36%, and at 10:1 ratio it was 10% of the control level (Fig. 2B). pDNOSoxy-FLAG, which lacks both the reductase domain and the Gln-rich region, was as effective at suppressing NOS activity (13.5% of control levels when cotransfected with pDNOS1-HA at 10:1 molar ratio) as pDNOS4-HA (which retains the Gln-rich region). Importantly, expression levels of DNOS1 protein were not affected by coexpressed truncated DNOS variants, as determined by immunoblotting using HA-specific antibodies (Fig. 2B). This observation indicates that a decrease of NOS activity was not simply due to a decrease in expression of the DNOS1 protein, but rather was caused by the presence of coexpressed shorter DNOS polypeptides. Furthermore, removal of the Gln-rich region did not alter the inhibitory effect of DNOSoxy on the enzymatic activity of DNOS1, indicating that the Gln-rich stretch does not appreciably contribute to the inhibitory action of DNOS4. Together, these results indicate that truncated enzymatically inactive forms of DNOS can suppress the activity of the full-length DNOS1 protein, thereby acting as dominant negative inhibitors of NO production.
Direct interaction between DNOS1 and DNOS4
To examine the potential of DNOS1 and DNOS4 to form complexes, we performed immunoprecipitation experiments after coexpressing differentially tagged DNOS1 and DNOS4 in 293 cells. Each tagged DNOS protein was immunoprecipitated from extracts of cotransfected cells with antibody specific to its epitope tag (HA for DNOS1 or FLAG for DNOS4). Subsequently, the formation of heteromeric DNOS complexes was examined by immunoblotting using HA-specific antibody for the samples immunoprecipitated with FLAG-specific antibody and vice versa.
When HA-tagged DNOS1 is expressed in 293 cells, it can be immunoprecipitated with HA-specific but not with FLAG-specific antibody (Fig. 2C). Conversely, FLAG-tagged DNOS4 can be immunoprecipitated using FLAG-specific, but not HA-specific antibodies. However, if DNOS1-HA and DNOS4-FLAG are coexpressed, each of them can be immunoprecipitated by antibody to either tag (Fig. 2C). Furthermore, when DNOSoxy-FLAG was coexpressed with DNOS1-HA, complexes between these two proteins were detected using the same combination of immunoprecipitation and immunoblotting (Fig. 2C). This suggests that even a part of the oxygenase domain lacking the Gln-rich region, but retaining the heme-binding region, is sufficient to form heteromers with the full-length DNOS1 protein. Finally, when lysates of cells transfected separately with either pDNOS1-HA or pDNOS4-FLAG were combined in vitro, the two proteins did not coimmunoprecipitate with each other (data not shown), indicating that heteromeric DNOS complexes are not formed after cell lysis.
Together, these data point to a possible mechanism for the observed dominant negative effect of DNOS4 on DNOS1 activity, in which formation of DNOS1–DNOS4 heterodimers inhibits NOS activity by reducing formation of enzymatically active homodimers of DNOS1.
Ectopically expressed DNOS4 forms complexes with DNOS1 and inhibits NOS activity in transgenic flies
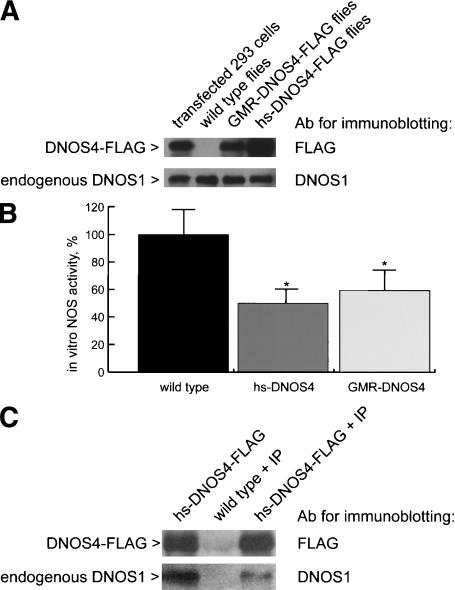
To determine whether DNOS4 can act as a dominant negative regulator of NOS activity in vivo, we generated transgenic flies that express FLAG-tagged DNOS4 under the control either of the heat-shock inducible promoter of the hsp70 gene (hs-DNOS4-FLAG flies), or of the GMR promoter, which is active in all cells of the eye imaginal disc within, and posterior to the morphogenetic furrow (GMR-DNOS4-FLAG flies). We confirmed that DNOS4-FLAG was expressed in transgenic flies; anti-FLAG antibodies detect a protein with an expected mass of 86 kDa in extracts from heads of GMR-DNOS4-FLAG and from hs-DNOS4-FLAG flies, but not in extracts from wild-type flies (Fig. 3A). The mobility of this protein was identical to that of a protein produced in 293 cells after transfection with pDNOS4-FLAG. Importantly, the expression levels of endogenously produced DNOS1 were not affected by ectopic expression of DNOS4 in transgenic flies (Fig. 3A). Furthermore, immunochemical detection with FLAG-specific antibodies showed that DNOS4-FLAG was expressed in the expected pattern within, and posterior to the morphogenetic furrow in the eye imaginal disc of third-instar GMR-DNOS4-FLAG larvae (data not shown).
Figure 3.
Ectopically expressed DNOS4 inhibits NOS activity in transgenic flies and forms complexes with endogenous DNOS1 in vivo. (A) Overexpression of DNOS4-FLAG in transgenic flies (hs-DNOS4-FLAG or GMR-DNOS4-FLAG) does not affect expression of the endogenous DNOS1 protein as determined by immunoblotting using antibody against DNOS1 and antibody against FLAG epitope (Ab for immunoblotting). (B) NOS activity in head extracts of hs-DNOS4-FLAG (hs-DNOS4) and GMR-DNOS4-FLAG (GMR-DNOS4) flies is decreased as compared with that of wild-type flies. NOS activity was measured by the [3H]arginine–[3H]citrulline conversion assay. The activity data are expressed as a percentage of the NOS activity of the full-length enzyme alone (34,271 cpm/min/mg total protein, 100% value). The results shown are representative of three independent experiments; all data are mean ± standard error. Statistical significance (P < 0.05 according to Student's t-test) is indicated with an asterisk (*). (C) In vivo complex formation between the endogenous DNOS1 and ectopically expressed DNOS4-FLAG. Protein complexes were immunoprecipitated (marked as +IP) from head extracts of transgenic (hs-DNOS4-FLAG + IP) and wild-type (wild type + IP) flies using antibody against FLAG epitope, and subsequently analyzed by immunoblotting with either DNOS1-specific or FLAG-specific antibody (Ab for immunoblotting). A control sample (hs-DNOS4-FLAG) was not subjected to immunoprecipitation prior to the immunoblotting analysis.
We next examined NOS activity in extracts from heads of adult flies and found that in hs-DNOS4-FLAG flies and in GMR-DNOS4-FLAG flies activity was 52% and 60%, respectively, of that in control wild-type flies (Fig. 3B). This indicates that endogenous NOS activity is inhibited by ectopically expressed DNOS4, paralleling observations with cultured cells.
To determine whether the observed decrease in NOS activity in transgenic flies is accompanied by in vivo formation of DNOS1–DNOS4 heterodimers, we performed coimmunoprecipitation experiments. Using FLAG-specific antibody, proteins were precipitated from head extracts of adult wild-type or hs-DNOS4-FLAG transgenic flies, and then were analyzed by immunoblotting using anti-DNOS1 or anti-FLAG antibodies. Importantly, full-length DNOS1 protein can be immunoprecipitated with anti-FLAG antibody from extracts of hs-DNOS4-FLAG transgenic flies, but not of wild-type flies (Fig. 3C). This indicates that ectopically expressed DNOS4 can form heterodimeric complexes with endogenous DNOS1 in vivo and suggests that formation of such complexes may explain the dominant negative effect of DNOS4 on NOS activity.
DNOS4 is an endogenous protein product of the Drosophila NOS gene
We have demonstrated (Fig. 1B) that the dNOS4 transcript is expressed in developing and adult Drosophila. To draw conclusions about the role of this isoform in modulating NOS activity in the fly, it is critical to determine whether dNOS4 mRNA is translated and expressed as an endogenous protein. According to the deduced sequence, DNOS4 protein has a unique 21-amino acid-long peptide at its C terminus. We used this peptide to raise a DNOS4-specific polyclonal antibody that was further purified by affinity chromatography on the immobilized recombinant antigen.
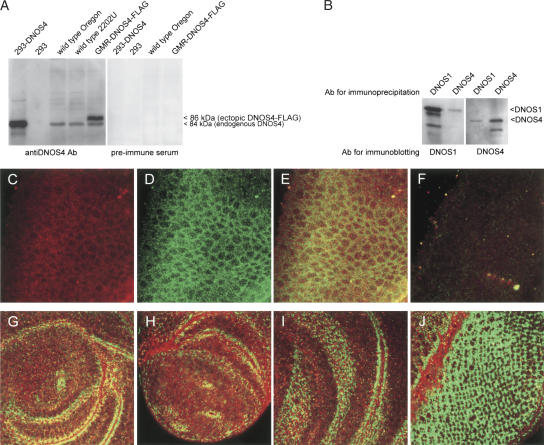
We used this DNOS4-specific antibody to examine expression of DNOS4 protein in wild-type and transgenic flies, and in 293 cells transfected with pDNOS4 (expression plasmid encoding DNOS4 without an added FLAG tag). The antibody recognized a protein with a mobility of 84 kDa in extracts prepared either from heads of wild-type or GMR-DNOS4-FLAG flies, or from 293 cells transfected with the pDNOS4 plasmid, but not in extracts from untransfected 293 cells (Fig. 4A). The electrophoretic mobility of this protein corresponds to the calculated molecular mass for DNOS4. The same antibody was able also to detect the DNOS4-FLAG protein in transgenic flies. This ectopically expressed DNOS4 protein migrates more slowly (∼86 kDa) than the wild-type protein (84 kDa), apparently due to the presence of two copies of the highly charged FLAG epitope at its C terminus. These experiments demonstrate that the DNOS4 protein is expressed in Drosophila as an endogenous product of the dNOS locus.
Figure 4.
DNOS4 is endogenously expressed in Drosophila and forms complexes with DNOS1. (A) DNOS4 is endogenously expressed in Drosophila. Head protein extracts from wild-type and transgenic GMR-DNOS4-FLAG flies were analyzed by immunoblotting using DNOS4-specific antibody (anti-DNOS4 Ab) or the preimmune serum. Control lysates from nontransfected 293 cells (293) and from cells transfected with the pDNOS4 expression construct without FLAG tag (293-DNOS4) were probed using the same antibody or the preimmune serum. In addition to the ectopically expressed DNOS4-FLAG protein (86 kDa), which was detected in head extracts of the GMR-DNOS4-FLAG transgenic flies only, DNOS4-specific antibody immunoreacted with 84 kDa protein, which was expressed both in wild-type and in transgenic flies, and had the same mobility as the DNOS4 protein transiently expressed in 293 cells (lane 293-DNOS4). (B) In vivo complex formation between the endogenous DNOS1 and DNOS4 proteins. Protein complexes were immunoprecipitated from wild-type embryo extracts using either DNOS1- or DNOS4-specific antibody (Ab for immunoprecipitation) and cross-analyzed by immunoblotting with either DNOS1- or DNOS4-specific antibody (Ab for immunoblotting). The antibodies used for immunoprecipitation and for immunoblotting are indicated. (C–J) Expression of DNOS1 (green signal) and DNOS4 (red signal) in embryos (C–F), third-instar leg disc (G), pupae (∼3 h after puparium formation) leg disc (H), third-instar wing disc (I), and third-instar eye disc (J). (F) Embryos carrying a homozygous deletion of the dNOS locus were used as a control for the specificity of the antibodies.
To determine whether endogenously produced DNOS1 and DNOS4 proteins form complexes in vivo, we performed coimmunoprecipitation experiments using DNOS1- and DNOS4-specific antibodies. We demonstrated the existence of such complexes by coimmunoprecipitating DNOS1 from extracts of wild-type embryos with DNOS4-specific antibody, and, conversely, by coimmunoprecipitating DNOS4 from the same extracts using antibody to DNOS1 (Fig. 4B).
We next used DNOS1- and DNOS4-specific antibodies to compare the expression patterns of these two DNOS proteins during development and to examine whether they are coexpressed in the same cells in vivo, as indicated by the coimmunoprecipitation experiments. Immunocytochemical detection showed that both proteins are coexpressed in a large number of cells in the embryo (Fig. 4C–E). As a control, to test the specificity of this detection, we used embryos that are homozygous for the Df(2L)69F deletion, which covers a large part of the dNOS locus, including the regions that code for the epitopes recognized by the anti-DNOS1 and anti-DNOS4 antibodies (M. Regulski, Y. Stasiv, T. Tully, and G. Enikolopov, in prep.). We introduced a chromosome that is marked by expression of GFP to be able to recognize the embryos that carry a homozygous deletion of the dNOS gene. We did not detect any specific signals when the mutant embryos were probed with either DNOS1-specific, or DNOS4-specific antibodies (Fig. 4F). Having established the specificity of the antibodies, we next analyzed imaginal discs of the wild-type larvae [note that homozygotes for the Df(2L)69F deletion die in late embryo] and found that in the leg, wing, and eye imaginal discs there are highly specific patterns of expression of DNOS1 and DNOS4, with defined areas of overlap between the two (Fig. 4G–J).
Together, these experiments demonstrate that DNOS4 protein is produced in vivo, is expressed in a highly specific pattern during development, can be found coexpressed with the full-length DNOS1 protein in cells of the imaginal discs of the larvae and in embryo, and forms endogenous complexes with DNOS1. This strongly supports the notion that DNOS4 can act as a negative regulator of DNOS1 activity in the fly.
Ectopic expression of DNOS4 increases the cell number in the eye
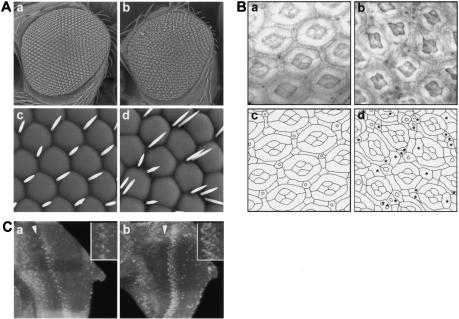
NO acts as an antiproliferative factor during eye development; pharmacological inhibition of NOS activity results in an increase in the number of cells in the ommatidia (Kuzin et al. 1996, 2000). We asked whether DNOS4 can act to suppress the antiproliferative action of NO and affect cell division in the developing eye. Ectopic expression of DNOS4 driven by an eye-specific promoter in GMR-DNOS4-FLAG flies results in a distorted eye phenotype with visible rearrangements of the ommatidia lattice and extra bristles (Fig. 5A). To analyze the changes in more detail, we compared the sections of the retina from wild-type and transgenic pupae. We found that GMR-DNOS4-FLAG flies had extra secondary and tertiary pigment cells as well as extra bristle cells; we did not detect changes in the number of photoreceptor cells. These extra cells distort the regular hexagonal shape of the ommatidia and the general lattice pattern of the retina (Fig. 5B).
Figure 5.
DNOS4 overexpression increases cell number in the adult eye. (A) Scanning electron micrographs of eyes of the wild-type (panels a,c) and transgenic GMR-DNOS4-FLAG (panels b,d) flies. Magnification: panels a,b, 200×; panels c,d, 1000×. GMR-DNOS4-FLAG transgenic flies have a profound eye phenotype with severe rearrangements in ommatidia lattice and appearance of extra bristles. (B) Sections of adult retinas of the wild-type (panel a) and transgenic GMR-DNOS4-FLAG (panel b) flies. (Panels c,d) Schematic drawing representing cell boundaries in panels a and b, respectively. Retina sections show notable changes in the GMR-DNOS4-FLAG flies on an individual ommatidium level, such as extra bristles and primary, secondary, and tertiary pigment cells (marked by asterisks). (C) DNOS4 overexpression in the eye imaginal disc increases number of proliferative cells. BrdU labeling of the eye-antennal imaginal discs from late third instar wild-type (panel a) and transgenic GMR-DNOS4-FLAG (panel b) larvae using fluorescent anti-BrdU antibody. The number of fluorescing nuclei reflects the number and distribution of cells in S phase. Increased number of proliferating cells in the GMR-DNOS4-FLAG vs. wild-type larvae is observed in the areas posterior to the morphogenetic furrow (to the right of the furrow; the furrow is marked by arrowhead); enlarged images of labeled cells near the equator regions of the discs are shown in the insets.
To determine whether the increase in the number of cells in pupae was related to increased proliferation in the imaginal discs of larvae, we labeled the nuclei of eye-disc cells in S phase of the cell cycle with 5-bromo-2′-deoxyuridine (BrdU). There was a 2.13-fold increase (P < 0.001) in the number of BrdU-positive cells in imaginal discs from GMR-DNOS4-FLAG flies as compared with wild-type flies (Fig. 5C). Extra dividing cells were localized to the region of GMR-driven transgene expression in the morphogenetic furrow and the area posterior to it. The observed increase in the number of dividing cells and the number of cells per ommatidium suggests that introduction of the dominant negative dNOS4 transgene resulted in inhibition of NOS activity and suppression of the antiproliferative effect of NO in the developing eye.
DNOS4 interacts with the retinoblastoma pathway
Because NO interacts with the retinoblastoma (Rb) pathway to control cell division in the developing Drosophila eye (Kuzin et al. 2000), we wanted to determine whether DNOS4 can act to affect signaling via the Rb pathway in the eye.
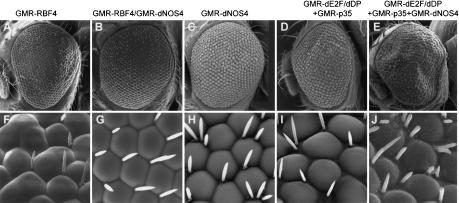
RBF is the Drosophila ortholog of mammalian Rb proteins; like Rb, RBF acts as a negative regulator of cell cycle progression (Du et al. 1996b; Du and Dyson 1999). GMR-RBF flies that carry four copies of the RBF transgene have a profound eye phenotype, due to suppression of cell division by the elevated levels of RBF; some of the pigment cells and bristles are missing and some ommatidia are fused (Fig. 6A,F; see also Du et al. 1996a; Du and Dyson 1999). The RBF phenotype is strongly enhanced by the ectopic overexpression of the dNOS1 transgene (Kuzin et al. 2000). To test the effects of DNOS4 on the action of RBF, we crossed GMR-DNOS4-FLAG flies (Fig. 6C,H) to GMR-RBF flies (Fig. 6A,F) and then backcrossed them to GMR-RBF flies to generate flies bearing one copy of the dNOS4 transgene along with four copies of the RBF transgene. The eyes of the resulting hybrid flies had an almost normal phenotype, without fused ommatidia, and with the usual set of bristles (Fig. 6B,G). Thus, the effect of RBF overexpression was counteracted by DNOS4, indicating that this inhibitor of NOS activity counteracts RBF function in the developing eye.
Figure 6.
Genetic interactions between DNOS4 and components of Drosophila Rb pathway. (A–C,F–H) DNOS4 rescues the RBF eye phenotype. Scanning electron micrographs of adult eyes of indicated genotypes. Magnification: A–C, 200×; F–H, 1000×. GMR-DNOS4-FLAG genotype is indicated as GMR-dNOS4. (D,E,I,J) Manipulation of NOS activity affects dE2F action in the developing eye. Scanning electron micrographs of adult eyes of the indicated genotypes (all transgenes were expressed under control of the GMR promoter). Magnification: D,E, 200×; I,J, 1000×. GMR-DNOS4-FLAG genotype is indicated as GMR-dNOS4.
In mammalian and Drosophila cells, Rb is associated with the E2F transcription-factor complex, whose activity is required for the cells' entry into S phase. Overexpression of E2F overcomes Rb-mediated G1 arrest and induces quiescent cells to enter S phase (Du et al. 1996a; Du and Dyson 1999). Ectopic expression of dE2F/dDP (Drosophila orthologs of mammalian components of the E2F complex) driven by the GMR promoter during development results in generation of extra cells in the adult eye (e.g., multiple extra bristles), particularly when combined with the p35 gene, a baculoviral inhibitor of apoptosis (Hay et al. 1994; Du et al. 1996b; Fig. 6D,I). We crossed GMR-DNOS4-FLAG flies with GMR-dE2F/dDP/GMR-p35 flies and found an even more pronounced eye phenotype, with a larger eye surface, extra ommatidia, and severe rearrangements of the ommatidia lattice (Fig. 6E,J). This indicates that DNOS4 acts to enhance the function of E2F, consistent with the proposed role for DNOS4 as a suppressor of the inhibitory action of NO on cell cycle progression.
Together, these genetic data confirm that development of the Drosophila eye depends on NO or NO-activated signaling pathways interacting with the Rb pathway. They suggest that, by suppressing the antiproliferative action of NO, DNOS4 acts in vivo to suppress the effect of RBF, and to enhance the effect of E2F, on cell cycle regulation.
Discussion
NO is an important signaling molecule for diverse physiological functions in Drosophila. NO regulates cell proliferation in the imaginal discs of developing larvae (Kuzin et al. 1996) and in embryos (Wingrove and O'Farrell 1999), participates in the development of the visual system (Gibbs and Truman 1998; Gibbs 2003), induces vesicle release at the neuromuscular junction of larvae (Wildemann and Bicker 1999), controls epithelial fluid secretion by the Malpigian tubules (Dow et al. 1994; Broderick et al. 2003), triggers the immune response against bacterial pathogens (Nappi et al. 2000), induces arrest of nuclear divisions in early embryo in response to oxygen deprivation (DiGregorio et al. 2001), and mediates hypoxia-dependent exploratory behavioral responses in larvae (Wingrove and O'Farrell 1999) and hypoxia-induced stasis in embryos (Teodoro and O'Farrell 2003). This wide range of processes mediated by NO requires tight regulation of its production in response to different stimuli. To accomplish this, production of NO by NOS is controlled at multiple levels, including transcription and translation, cofactor binding, covalent modifications, and subcellular localization.
We describe here a novel mode of regulation of NOS activity through the synthesis of an endogenous isoform that inhibits DNOS1 in a dominant negative fashion via heterodimerization. This regulator, DNOS4, is encoded by an alternatively spliced version of the dNOS transcript. It is expressed in the imaginal discs of the developing fly in a defined pattern, with areas of conspicuous overlap with cells that express DNOS1. DNOS4 forms heteromeric complexes with the full-length DNOS1 polypeptide and can inhibit NOS activity both in vitro, in cultured cells, and in vivo, in the developing fly. In the eye imaginal disc, where DNOS1 negatively regulates cell cycle progression, ectopic expression of DNOS4 can block the action of DNOS1 and lead to a greater degree of cell proliferation.
Our data further underscore the developmental role of NO as a regulator of cell division in imaginal discs. We show that the action of DNOS4 is reciprocal to RBF and additive or synergistic to dE2F, thus acting to relieve the negative effect of DNOS1 on cell proliferation during eye-disc development. Our results support the hypothesis that the DNOS family of proteins acts to modulate the entry of cells into the S phase of the cell cycle in the eye disc by interacting with the Rb pathway (Kuzin et al. 2000; present study). RBF or E2F proteins themselves are potential direct targets for NO action, for example, via S-nitrosylation of cysteine residues by endogenous NO or NO donors (Stamler et al. 2001); intriguingly, Rb is one of the several proteins identified in an in vivo screen for S-nitrosylated proteins in mammalian cells (Jaffrey et al. 2001). Furthermore, the effects of NO may also be mediated by cGMP signaling, as guanylate cyclase is a crucial direct target of NO in many systems (Ignarro 2000; Russwurm and Koesling 2002) and activated cGMP-phosphodiesterases can serve as an effective counterbalance to the NO signal (Broderick et al. 2003; Friebe and Koesling 2003; Mullershausen et al. 2003).
Structural and kinetic analysis of NOS enzymes provides strong support for the proposed mechanism of NOS inhibition by truncated NOS proteins. Our observations are consistent with the proposed mechanism of NO catalysis, in which electrons flow from the flavins in the reductase domain of one subunit to the heme iron in the oxygenase domain of the other subunit (Siddhanta et al. 1998; Panda et al. 2001, 2002; Sagami et al. 2001). An inhibitory action of truncated products of the NOS gene on NOS activity was demonstrated in transfected cells in culture for the mammalian endothelial and neuronal isoforms as well as for Drosophila NOS (Lee et al. 1995; Phung and Black 1999; Stasiv et al. 2001). We show here how this feature of an endogenous truncated product of the NOS gene can contribute to eye development in Drosophila.
The DNOS1 protein carries many regions of strong homology to the mammalian neuronal isoform (Regulski and Tully 1995); this homology includes the part of the oxygenase domain that is involved in dimer formation in mammalian NOSs. However, DNOS1 lacks the N-terminal PDZ domain that is involved in interactions of nNOS with other proteins (e.g., PSD-95 and α-synthrophin). Unlike any other known NOS, the Drosophila enzyme contains an almost uninterrupted stretch of Gln residues at its N terminus (amino acids 24–52). Although Gln-rich motifs may be involved in protein–protein interactions (Perutz et al. 1994; Stott et al. 1995; Zoghbi and Orr 2000), our data suggest that this Gln cluster is not essential for DNOS dimer formation; the DNOSoxy mutant that lacks the Gln-rich stretch, but retains the “core” oxygenase domain, can still effectively complex with the full-length Drosophila NOS and block its activity, whereas the DNOS3 isoform, composed of 200 N-terminal DNOS residues (including the Gln-rich region, but excluding the heme-binding catalytic domain), cannot block the enzymatic activity of DNOS1 (Stasiv et al. 2001).
Is DNOS4 unique in its ability to regulate NOS activity? We had identified five other transcripts that encode inactive truncated DNOS-like proteins (Stasiv et al. 2001), which we showed are capable of suppressing the activity of DNOS1 in vitro. One of these isoforms, dNOS7, encodes a protein identical to DNOS4, but uses a different promoter from dNOS4. The dNOS7 mRNA is exclusively expressed in third-instar larvae (Stasiv et al. 2001). Because we have shown that proliferation of cells in the imaginal discs in the late third instar is controlled by NO (Kuzin et al. 1996), it is possible that DNOS7 attenuates DNOS1 activity at this developmental stage. Other NOS RNA isoforms are differentially expressed during Drosophila development (Stasiv et al. 2001) and the encoded proteins, by analogy to DNOS4, may act to regulate NO production in vivo.
This mode of regulation that we describe for Drosophila also may be involved in regulation of NOS activity in other systems, as similar NOS transcripts have been found in other organisms. Similarly to dNOS4, these alternative NOS transcripts encode truncated polypeptides that lack the reductase domain. For instance, exon insertions and rearrangements that introduce premature stop codons have been reported for NOS genes in mosquito, snail, and humans (Larsson and Phillips 1998; Luckhart and Li 2001; Korneev and O'Shea 2002).
When compared with dNOS1 RNA encoding the full-length product, alternative NOS RNAs encoding truncated proteins might be expected to be in low abundance, perhaps due to nonsense-mediated decay (Wagner and Lykke-Andersen 2002). Furthermore, some of these truncated proteins may be subject to enhanced proteolysis (Kaufman 2002). However, we can clearly identify endogenous dNOS4 and dNOS7 mRNA and the corresponding product, DNOS4 protein, in Drosophila. This suggests that similar truncated proteins may exist in physiologically relevant amounts in other settings. It also suggests that this mode of regulation may be used elsewhere, in other organisms and with other multimeric proteins or in multisubunit signaling complexes. Truncated proteins, in addition to forming inactive heteromers with the functional isoform, may also compete for substrates and cofactors required for catalysis, may interact with intracellular protein partners of the active isoform, and may change the subcellular location of such protein assemblies. Thus, negative control of homodimeric enzymes by truncated isoforms may represent an important regulatory mechanism used in various developmental and physiological processes.
Materials and methods
Fly stocks
To generate transgenic flies expressing DNOS4 in the developing eye, we cloned the protein-coding region of the dNOS4 cDNA fused in-frame at the 3′-end to two copies of the FLAG epitope sequence, into the blunt-ended EcoRI site of the pGMR P-element vector (Hay et al. 1994). Germ-line transformation of 2022U flies was accomplished using standard techniques as described (Kuzin et al. 2000). Three independent transgenic lines P[GMR-DNOS4-FLAG;w+] were generated, one on each chromosome.
To generate transgenic flies carrying the dNOS4 cDNA under the heat-shock promoter, we cloned DNOS4-FLAG open reading frame into the blunt-ended EcoRI site of the linearized pCaSpeR-hs P-element vector. Germ-line transformation was performed as described above. Two independent transgenic lines P[hs-DNOS4FLAG;w+] were generated, one on the second chromosome, and one on the third chromosome.
Stocks used for crosses GMR-RBF4 (second and third chromosome; Du et al. 1996a), GMR-dE2F + GMR-dDP2 (second chromosome; Du et al. 1996b) were gifts from N. Dyson (Massachusetts General Hospital Cancer Center, Charleston, MA) and W. Du (University of Chicago, IL). GMR-p35 (third chromosome; Hay et al. 1994) was a gift from B. Hay (California Institute of Technology, Pasadena, CA).
RNA analysis
A semiquantitative analysis of expression levels of various dNOS alternative splice isoforms using total RNA samples isolated from Drosophila embryos, third-instar larvae, and adult flies was performed by RT–PCR amplification as described previously (Stasiv et al. 2001). For specific detection of the dNOS1 transcript primers from exon 12 (sense, 5′-TGGCAAATCG GAGCAGTATGC-3′) and exon 15 (antisense, 5′-ACCAAAG GTCTCCTCGGTGA-3′) were used. A 234-bp-long fragment of the dNOS4 transcript was amplified using primers from exon 12 (sense, 5′-TGGCAAATCGGAGCAGTATGC-3′) and alternatively spliced exon 14a (antisense, 5′-TTAGAAGGCGGC TGGGCCTAA-3′). As a control, actin 79B RNA was amplified using 5′-CAACAATGTGCTGTCTGGC-3′ (sense) and 5′-CGATCCAGACGGAGTACTTG-3′ (antisense) primers. To eliminate false-positive amplification products, negative (RT minus) controls were amplified by PCR together with the experimental (RT plus) samples. RT–PCR products were size fractionated by agarose gel electrophoresis and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech). After the transfer membranes were hybridized with [γ-32P]ATP oligonucleotide (sense, 5′-TCCTAGGGCACGCATTCAAT-3′) from exon 12 of the dNOS gene or with the actin 79B sense oligonucleotide.
Tissue culture experiments
Expression constructs for the enzymatic activity assays contained the protein-coding regions of dNOS1 or dNOS4 cDNAs, each fused in-frame at the 3′-end to the sequence of either two copies of the HA epitope (pDNOS1-HA construct) or two copies of the FLAG epitope (pDNOS4-FLAG construct) followed by a stop codon, and were described previously (Stasiv et al. 2001). In addition, the synthetic expression construct pDNOSoxy-FLAG, composed of a region encoding the DNOS1 amino acids 214–631, and fused in-frame at the 3′-end to the two copies of FLAG epitope sequence, was generated. The above plasmids were transfected in the 293 cells as described previously (Stasiv et al. 2001). In cotransfection experiments, different molar ratios (10: 1, 3:1, 1:1) between a truncated construct (pDNOS4-FLAG or pDNOSoxy-FLAG) over the full-length DNOS1-HA plasmid (2 μg/transfection) were used. The total amount of DNA used for each transfection was kept constant at 25 μg by the addition of vector DNA. Cells were collected 40 h after transfection, and cell-free protein extracts were prepared as described (Stasiv et al. 2001). Protein concentration in tested lysates was determined using the BCA reagent system (Pierce), and the results were used to normalize the assays. NOS activity in lysates was examined by the [3H]L-arginine–[3H]L-citrulline conversion assay. To compare the expression of the pDNOS1-HA construct across the cotransfection experiments, equal amounts of protein extracts (50 μg/lane) from the transfected cells were analyzed by SDS-PAGE. Coimmunoprecipitation experiments were performed as described (Stasiv et al. 2001). For immunoblotting, either the 12CA5 monoclonal antibody (Field et al. 1988) against HA epitope at a final concentration of 10 μg/mL or the M2 monoclonal antibody against FLAG epitope (Stratagene) at a final concentration of 1 μg/mL were used. Immunoblots were developed using the ECL system (Amersham Pharmacia Biotech). For immunoprecipitation experiments, either the anti-HA tag polyclonal antibody (Upstate Biotechnology) or the anti-FLAG M2-Agarose affinity gel (Sigma) was used.
Drosophila protein analysis
Protein extracts from heads of wild-type or transgenic adult flies were prepared as follows. Heads of adult flies were separated on sieves, then homogenized at 4°C in 10 vol of extraction buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5 μM BH4, 1 mM L-arginine, and a standard set of protease inhibitors. Homogenates were cleared by centrifugation for 15 min at 12,000g at 4°C. A similar procedure was used to prepare extracts from embryos. NOS activity in Drosophila cell-free head extracts (prepared as above except for the absence of Triton X-100 in the extraction buffer) was measured using [3H]L-arginine–[3H]L-citrulline conversion assay. For the immunoprecipitation experiments, extracts were incubated with anti-FLAG M2-Agarose affinity gel (Sigma), or with anti-DNOS1- or DNOS4-specific antibodies (below). Immunoprecipitated proteins were separated by SDS-PAGE followed by immunoblotting.
Antibodies
Anti-DNOS1 monoclonal antibody was raised against the C-terminal 180 amino acids of the DNOS protein expressed as a fusion protein in Escherichia coli. Monoclonal antibody was obtained following standard procedures at the CSHL antibody facility. Of 150 candidate hybridoma lines, 10 monoclonal lines were selected; mAb 6/157 line was used for the experiments described in this study.
To generate a DNOS4-specific antibody, a unique DNOS4-specific C-terminal peptide VSTPPRKDHTELINGLGPAAF was synthesized, conjugated to each of two carrier proteins (bovine serum albumin and keyhole limpet hemocyanin), and used to raise DNOS4-specific rabbit polyclonal antibody. The total immunoglobulin G (IgG) fraction was purified from serum of immunized animals using chromatography on proteinA Sepharose 4B (Sigma). The DNOS4-immunoreactive IgG subfraction was subsequently purified by affinity chromatography as follows. Briefly, cDNA encoding the DNOS4 C-terminal peptide was fused in-frame to the 3′-end of the glutathione S-transferase (GST) using the pGEX-2T bacterial expression vector (Amersham Pharmacia Biotech). The resulting 31-kDa GST-DN4 fusion protein was overexpressed in E. coli BL21 strain, purified by affinity chromatography using prepacked GSTrap column (Amersham Pharmacia Biotech), and then immobilized on NHS-activated Sepharose using prepacked HiTrap column (Amersham Pharmacia Biotech). The resulting GST-DN4-Sepharose was used for affinity purification of the DNOS4-specific immunoglobulins. All antibody purification steps with the use of custom-made columns were performed according to a manufacturer's recommendations. This purified polyclonal antibody was used in a dilution 1:100 for the immunoprecipitation and for detection of endogenously and ectopically expressed DNOS4 in Drosophila cell-free extracts.
Immunocytochemistry
Embryos and imaginal discs were immunostained with monoclonal antibody to DNOS1 and polyclonal antibody to DNOS4 (above) using standard procedures and analyzed by confocal microscopy. Anti-DNOS1 and anti-DNOS4 antibodies were detected using Alexa 488- and Alexa 633-conjugated secondary antibodies (Molecular Probes), respectively.
Eye analysis
To identify cells in S phase, BrdU labeling of imaginal discs from the third-instar larvae was performed as described previously (Kuzin et al. 1996). Briefly, imaginal discs were removed, rinsed, and incubated in Schneider's medium supplemented with 50 μg/mL BrdU for 30 min at room temperature. The samples were then fixed in 4% formaldehyde, treated with 1:1 mixture of heptane and formaldehyde, rinsed, depurinated by 1 M HCl, and blocked with 1% sheep serum. Incorporated BrdU was visualized using an anti-BrdU antibody (Beckton-Dickinson) and fluorescein-coupled secondary antibody (Boehringer-Mannheim). Cobalt sulfite staining of retinal sections was performed as described by Wolff and Ready (1991). Scanning electron microscopy was performed at the State University of New York-Stony Brook Imaging Center, essentially as described by Kimmel et al. (1990).
Acknowledgments
We thank Nick Dyson, Wei Du, and Bruce Hay for the GMR-RBF, GMR-dE2FdDP, and GMR-p35 flies; Gregory Rudomen for help with scanning microscopy; and Natasha Peunova and Stephen Hearn for help with confocal microscopy. We are indebted to Alexander Kuzin (NINDS) for his help with generating hs-DNOS4-FLAG transgenic flies. We are grateful to Enikolopov lab members for help and useful discussions and to Julian Banerji for critical reading of the manuscript. This work was funded by the NICHD, NINDS, the Seraph Foundation, the Charles Henry Leach Foundation, and the Donaldson Foundation to G.E. and by MCBRAS to B.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.298004.
References
- Alderton W.K., Cooper, C.E., and Knowles, R.G. 2001. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 357: 593-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick K.E., MacPherson, M.R., Regulski, M., Tully, T., Dow, J.A., and Davies, S.A. 2003. Interactions between epithelial nitric oxide signaling and phosphodiesterase activity in Drosophila. Am. J. Physiol. Cell. Physiol. 285: C1207-C1218. [DOI] [PubMed] [Google Scholar]
- DiGregorio P.J., Ubersax, J.A., and O'Farrell, P.H. 2001. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 276: 1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow J.A., Maddrell, S.H., Davies, S.A., Skaer, N.J., and Kaiser, K. 1994. A novel role for the nitric oxide–cGMP signaling pathway: The control of epithelial function in Drosophila. Am. J. Physiol. 266: R1716-R1719. [DOI] [PubMed] [Google Scholar]
- Du W. and Dyson, N. 1999. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 18: 916-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Vidal, M., Xie, J.E., and Dyson, N. 1996a. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes & Dev. 10: 1206-1218. [DOI] [PubMed] [Google Scholar]
- Du W., Xie, J.E., and Dyson, N. 1996b. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J. 15: 3684-3692. [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa, J., Broek, D., MacDonald, B., Rodgers, L., Wilson, I.A., Lerner, R.A., and Wigler, M. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8: 2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischmann T.O., Hruza, A., Niu, X.D., Fossetta, J.D., Lunn, C.A., Dolphin, E., Prongay, A.J., Reichert, P., Lundell, D.J., Narula, S.K., et al. 1999. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat. Struct. Biol. 6: 233-242. [DOI] [PubMed] [Google Scholar]
- Friebe A. and Koesling, D. 2003. Regulation of nitric oxidesensitive guanylyl cyclase. Circ. Res. 93: 96-105. [DOI] [PubMed] [Google Scholar]
- Gibbs S.M. 2003. Regulation of neuronal proliferation and differentiation by nitric oxide. Mol. Neurobiol. 27: 107-120. [DOI] [PubMed] [Google Scholar]
- Gibbs S.M. and Truman, J.W. 1998. Nitric oxide and cyclic GMP regulate retinal patterning in the optic lobe of Drosophila. Neuron 20: 83-93. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff, T., and Rubin, G.M. 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121-2129. [DOI] [PubMed] [Google Scholar]
- Ignarro L.J. 2000. Nitric oxide: Biology and pathobiology. Academic Press, San Diego, CA.
- Jaffrey S.R., Erdjument-Bromage, H., Ferris, C.D., Tempst, P., and Snyder, S.H. 2001. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3: 193-197. [DOI] [PubMed] [Google Scholar]
- Kaufman R.J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 110: 1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel B.E., Heberlein, U., and Rubin, G.M. 1990. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes & Dev. 4: 712-727. [DOI] [PubMed] [Google Scholar]
- Korneev S. and O'Shea, M. 2002. Evolution of nitric oxide synthase regulatory genes by DNA inversion. Mol. Biol. Evol. 19: 1228-1233. [DOI] [PubMed] [Google Scholar]
- Kuzin B., Roberts, I., Peunova, N., and Enikolopov, G. 1996. Nitric oxide regulates cell proliferation during Drosophila development. Cell 87: 639-649. [DOI] [PubMed] [Google Scholar]
- Kuzin B., Regulski, M., Stasiv, Y., Scheinker, V., Tully, T., and Enikolopov, G. 2000. Nitric oxide interacts with the retinoblastoma pathway to control eye development in Drosophila. Curr. Biol. 10: 459-462. [DOI] [PubMed] [Google Scholar]
- Larsson B. and Phillips, S.C. 1998. Isolation and characterization of a novel, human neuronal nitric oxide synthase cDNA. Biochem. Biophys. Res. Comm. 251: 898-902. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Robinson, L.J., and Michel, T. 1995. Oligomerization of endothelial nitric oxide synthase. Evidence for a dominant negative effect of truncation mutants. J. Biol. Chem. 270: 27403-27406. [DOI] [PubMed] [Google Scholar]
- Luckhart S. and Li, K. 2001. Transcriptional complexity of the Anopheles stephensi nitric oxide synthase gene. Insect Biochem. Mol. Biol. 31: 249-256. [DOI] [PubMed] [Google Scholar]
- Mullershausen F., Friebe, A., Feil, R., Thompson, W.J., Hofmann, F., and Koesling, D. 2003. Direct activation of PDE5 by cGMP: Long-term effects within NO/cGMP signaling. J. Cell. Biol. 160: 719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nappi A.J., Vass, E., Frey, F., and Carton, Y. 2000. Nitric oxide involvement in Drosophila immunity. Nitric Oxide 4: 423-430. [DOI] [PubMed] [Google Scholar]
- Nathan C. 2003. Specificity of a third kind: Reactive oxygen and nitrogen intermediates in cell signaling. J. Clin. Invest. 111: 769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K., Ghosh, S., and Stuehr, D.J. 2001. Calmodulin activates intersubunit electron transfer in the neuronal nitricoxide synthase dimer. J. Biol. Chem. 276: 23349-23356. [DOI] [PubMed] [Google Scholar]
- Panda K., Rosenfeld, R.J., Ghosh, S., Meade, A.L., Getzoff, E.D., and Stuehr, D.J. 2002. Distinct dimer interaction and regulation in nitric-oxide synthase types I, II, and III. J. Biol. Chem. 277: 31020-31030. [DOI] [PubMed] [Google Scholar]
- Papapetropoulos A., Rudic, R.D., and Sessa, W.C. 1999. Molecular control of nitric oxide synthases in the cardiovascular system. Cardiovascular Res. 43: 509-520. [DOI] [PubMed] [Google Scholar]
- Perutz M.F., Johnson, T., Suzuki, M., and Finch, J.T. 1994. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. 91: 5355-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung Y.T. and Black, S.M. 1999. Use of chimeric forms of neuronal nitric-oxide synthase as dominant negative mutants. IUBMB Life 48: 333-338. [DOI] [PubMed] [Google Scholar]
- Raman C.S., Li, H., Martasek, P., Kral, V., Masters, B.S., and Poulos, T.L. 1998. Crystal structure of constitutive endothelial nitric oxide synthase: A paradigm for pterin function involving a novel metal center. Cell 95: 939-950. [DOI] [PubMed] [Google Scholar]
- Regulski M. and Tully, T. 1995. Molecular and biochemical characterization of dNOS: A Drosophila Ca2+/calmodulindependent nitric oxide synthase. Proc. Natl. Acad. Sci. 92: 9072-9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russwurm M. and Koesling, D. 2002. Isoforms of NO-sensitive guanylyl cyclase. Mol. Cell. Biochem. 230: 159-164. [PubMed] [Google Scholar]
- Sagami I., Daff, S., and Shimizu, T. 2001. Intra-subunit and inter-subunit electron transfer in neuronal nitric-oxide synthase: Effect of calmodulin on heterodimer catalysis. J. Biol. Chem. 276: 30036-30042. [DOI] [PubMed] [Google Scholar]
- Siddhanta U., Presta, A., Fan, B., Wolan, D., Rousseau, D.L., and Stuehr, D.J. 1998. Domain swapping in inducible nitricoxide synthase. Electron transfer occurs between flavin and heme groups located on adjacent subunits in the dimer. J. Biol. Chem. 273: 18950-18958. [DOI] [PubMed] [Google Scholar]
- Stamler J.S., Lamas, S., and Fang, F.C. 2001. Nitrosylation. The prototypic redox-based signaling mechanism. Cell 106: 675-683. [DOI] [PubMed] [Google Scholar]
- Stasiv Y., Regulski, M., Kuzin, B., Tully, T., and Enikolopov, G. 2001. The Drosophila nitric-oxide synthase gene (dNOS) encodes a family of proteins that can modulate NOS activity by acting as dominant negative regulators. J. Biol. Chem. 276: 42241-42251. [DOI] [PubMed] [Google Scholar]
- Stott K., Blackburn, J.M., Butler, P.J., and Perutz, M. 1995. Incorporation of glutamine repeats makes protein oligomerize: Implications for neurodegenerative diseases. Proc. Natl. Acad. Sci. 92: 6509-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuehr D.J. 1999. Mammalian nitric oxide synthases. Biochimica et Biophysica Acta 1411: 217-230. [DOI] [PubMed] [Google Scholar]
- Teodoro R.O. and O'Farrell, P.H. 2003. Nitric oxide-induced suspended animation promotes survival during hypoxia. EMBO J. 22: 580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. and Lykke-Andersen, J. 2002. mRNA surveillance: The perfect persist. J. Cell Sci. 115: 3033-3038. [DOI] [PubMed] [Google Scholar]
- Wang Y., Newton, D.C., and Marsden, P.A. 1999a. Neuronal NOS: Gene structure, mRNA diversity, and functional relevance. Crit. Rev. Neurobiol. 13: 21-43. [DOI] [PubMed] [Google Scholar]
- Wang Y., Newton, D.C., Robb, G.B., Kau, C.L., Miller, T.L., Cheung, A.H., Hall, A.V., VanDamme, S., Wilcox, J.N., and Marsden, P.A. 1999b. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc. Natl. Acad. Sci. 96: 12150-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildemann B. and Bicker, G. 1999. Nitric oxide and cyclic GMP induce vesicle release at Drosophila neuromuscular junction. J. Neurobiol. 39: 337-346. [DOI] [PubMed] [Google Scholar]
- Wingrove J.A. and O'Farrell, P.H. 1999. Nitric oxide contributes to behavioral, cellular, and developmental responses to low oxygen in Drosophila. Cell 98: 105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T. and Ready, D.F. 1991. Cell death in normal and rough eye mutants of Drosophila. Development 113: 825-839. [DOI] [PubMed] [Google Scholar]
- Zoghbi H.Y. and Orr, H.T. 2000. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23: 217-247. [DOI] [PubMed] [Google Scholar]