Abstract
We have used nuclear transplantation to test whether the reprogramming activity of oocytes can reestablish developmental pluripotency of malignant cancer cells. We show here that the nuclei of leukemia, lymphoma, and breast cancer cells could support normal preimplantation development to the blastocyst stage but failed to produce embryonic stem (ES) cells. However, a blastocyst cloned from a RAS-inducible melanoma nucleus gave rise to ES cells with the potential to differentiate into multiple cell types in vivo including melanocytes, lymphocytes, and fibroblasts. Chimeras produced from these ES cells developed cancer with higher penetrance, shorter latency, and an expanded tumor spectrum when compared with the donor mouse model. These results demonstrate that the secondary changes of a melanoma nucleus are compatible with a broad developmental potential but predispose mice to melanomas and other malignant tumors on reactivation of RAS. Our findings serve as a paradigm for studying the tumorigenic effect of a given cancer genome in the context of a whole animal.
Keywords: Cancer, epigenetics, nuclear transfer, reprogramming, pluripotency, embryonic stem cells
Accumulating evidence shows that tumor formation is accompanied by both epigenetic and genetic alterations of the genome (Hahn and Weinberg 2002; Jones and Baylin 2002; Felsher 2003; Egger et al. 2004). Unlike genetic changes, epigenetic changes do not alter the primary DNA sequence and are therefore reversible. Examples of epigenetic modifications are the methylation of DNA and histones, the acetylation/deacetylation of histones, and the packing of chromatin into euchromatic and heterochromatic regions (Li 2002). Epigenetic modifications play an important role during normal development by regulating gene expression through stable activation or silencing of differentiation-associated genes. Similarly, epigenetic changes can promote cell proliferation, inhibit apoptosis, and induce angiogenesis during tumorigenesis by activating oncogenes and silencing tumor suppressor genes (Felsher 2003). For example, the p16 and VHL tumor suppressor genes are frequently silenced in human cancer by methylation of their promoter regions (Jones and Baylin 2002). Moreover, the treatment of tumor cells with methylation- and histone-modifying drugs can inhibit malignancy and this inhibition correlates with the reactivation of important tumor suppressor loci (Jones and Baylin 2002; Egger et al. 2004).
Epigenetic and genetic changes often act in concert during neoplasia. For example, potent oncogenes such as MYC, FOS, and PML-RAR can directly or indirectly interact with proteins that regulate epigenetic modifications such as DNA methyltransferases, histone methyltransferases, and histone acetylases/deacetylases to modulate gene expression (Bakin and Curran 1999; Di Croce et al. 2002; Jones and Baylin 2002; Ogawa et al. 2002; Felsher 2003; Frank et al. 2003). In agreement, animal models deficient for individual components of the epigenetic machinery are more prone to genome instability and cancer. For instance, mice that harbor a hypomorphic allele of the Dnmt1 methyltransferase gene consistently succumb to thymomas, possibly through the induction of chromosomal abnormalities (Gaudet et al. 2003). Likewise, the lack of the histone methyltransferase Suv39h has been associated with an increased tumor risk and genomic instability (Peters et al. 2001). These observations strongly suggest that epigenetic and genetic alterations together contribute to the neoplastic state.
Tumors develop in the context of a particular developmental state, and thus, epigenetics may also influence tumorigenesis through its effects on differentiation. Consistent with this notion, differentiation-inducing drugs can cause regression of some tumors such as all-trans retinoic acid in acute promyelocytic leukemia (Sanz et al. 1998). Moreover, many bona fide oncogenes are tumorigenic only in specific cell lineages, suggesting the requirement for a tissue-specific epigenetic environment that is permissive for an oncogene's tumorigenic potential (Felsher 2003). For example, in a MYC-inducible osteosarcoma mouse model, it has been demonstrated that expression of the MYC oncogene causes tumors in immature osteoblasts, but induces apoptosis in differentiated osteocytes (Jain et al. 2002), an observation that further supports the idea that the differentiation state and thus epigenetic conformation of a tumor cell may determine whether a cell manifests a malignant phenotype or not.
Nuclear transplantation (NT) can reprogram a terminally differentiated cell into a pluripotent embryonic cell that can direct development of an organism (Wilmut et al. 1997; Wakayama et al. 1998). This is accomplished by resetting the epigenetic modifications associated with differentiation to a state equivalent to that of a zygote (Hochedlinger and Jaenisch 2002b), while genetic changes remain unaltered (Hochedlinger and Jaenisch 2002a). Thus, NT provides a tool to selectively reprogram the epigenetic state of a cellular genome without altering its genetic constitution in order to globally analyze the impact of epigenetics on tumorigenesis. Historic experiments in frogs have demonstrated that kidney carcinoma nuclei can be reprogrammed to support early development to the tadpole stage (McKinnell et al. 1969). A similar result was recently obtained in mice where nuclei from a medulloblastoma cell line were able to direct early development, albeit with low efficiency, resulting in arrested embryos (Li et al. 2003). However, these experiments did not unequivocally demonstrate that the clones were derived from cancer cells as opposed to contaminating nontransformed cells (Carlson et al. 1994). Moreover, the experimental setup did not allow the distinction between abnormalities caused by the nuclear transfer procedure versus abnormalities caused by the donor nucleus.
Here, we have taken an alternate approach to investigate whether the reprogramming activity of the oocyte can reverse the cancer phenotype of a tumor genome and establish developmental pluripotency (Fig. 1). Following nuclear transfer of different tumor cells, clones were allowed to develop to blastocysts and then explanted in tissue culture to derive embryonic stem (ES) cells. The resulting ES cells (hereafter denoted as NT ES cells) were then analyzed to confirm the tumor cell origin and tested in multiple assays for their developmental and tumorigenic potential. This modified cloning procedure (1) circumvents abnormalities associated with nuclear transfer (Hochedlinger and Jaenisch 2003) and (2) permits a detailed analysis of the developmental (Hochedlinger and Jaenisch 2002a; Rideout et al. 2002; Eggan et al. 2004) and tumorigenic potential of the reprogrammed nucleus.
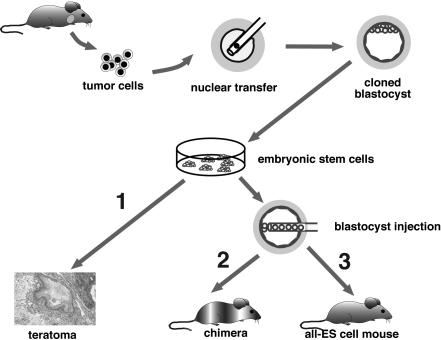
Figure 1.
Two-step cloning procedure to produce mice from cancer cells. Different tumor cells were used as donors for nuclear transfer into enucleated oocytes. Resultant blastocysts were explanted in culture to produce ES cell lines. The tumorigenic and differentiation potential of these ES cells was assayed in vitro by inducing teratomas in SCID mice (1), and in vivo by injecting cells into diploid (2) or tetraploid (3) blastocysts to generate chimeras and entirely ES-cell-derived mice, respectively.
Results
Cancer nuclei can support preimplantation development
We first examined whether cancer nuclei could direct preimplantation development to the blastocyst stage. Nuclei from different murine cancer cells were introduced into enucleated oocytes and subsequently activated to induce cleavage of the embryo. We were unable to generate cloned blastocysts from 63 eggs transplanted with the nuclei from a p53-/- lymphoma (Donehower et al. 1992) and have succeeded in establishing only one cloned blastocyst from ∼500 eggs transplanted with the nuclei from a Moloney murine leukemia virus (MoMLV)-induced leukemia (Table 1; Jaenisch et al. 1981; Stewart et al. 1983). On the other hand, we were able to generate cloned blastocysts at a rate of 1%–12% from a PML-RAR transgene-induced leukemia (Zimonjic et al. 2000), a hypomethylated Chip/c lymphoma (Gaudet et al. 2003), a p53-/- breast cancer cell line (Kuperwasser et al. 2000), and a RAS-inducible melanoma cell line (Table 1; Chin et al. 1999). These results indicate that many cancer cell nuclei can respond to developmental cues in the oocyte environment and support organized cleavage divisions, compaction, and cavitation to form blastocysts (Fig. 2a). From a total of 57 explanted blastocysts, we were able to derive two ES cell lines that were cloned from RAS-inducible melanoma nuclei (Table 1). These NT ES cell lines, R545-1 and R545-2, were then used to address more rigorously the impact of epigenetic reprogramming on developmental and tumorigenic potency.
Table 1.
Efficiencies of deriving cloned blastocysts and ES cell lines from various cancer nuclei
| Type of cancer cell | # Surviving eggs | # Blastocysts (% eggs) | # ES Cell lines (% blastocysts) |
|---|---|---|---|
| MoMLV-1 leukemia (cell line) | 159 | 1 (0.6%) | 0 |
| MoMLV-14 leukemia (primary tumor) | 338 | 0 | 0 |
| PML-RARα leukemia (transplanted tumor) | 337 | 4 (1.2%) | 0 |
| Chip/c lymphoma (cell lines) | 535 | 21 (3.9%) | 0 |
| p53-/- lymphoma (primary tumor) | 63 | 0 | 0 |
| p53-/- breast cancer (cell line) | 189 | 23 (12.2%) | 0 |
| RAS+/ink4a/Arf-/- melanoma (cell lines, p10-15) | 583 | 8 (1.4%) | 2 (25%) |
| RAS+/ink4a/Arf-/- fibroblasts (cell line, p5) | 71 | 10 (14.1%) | 2 (20%) |
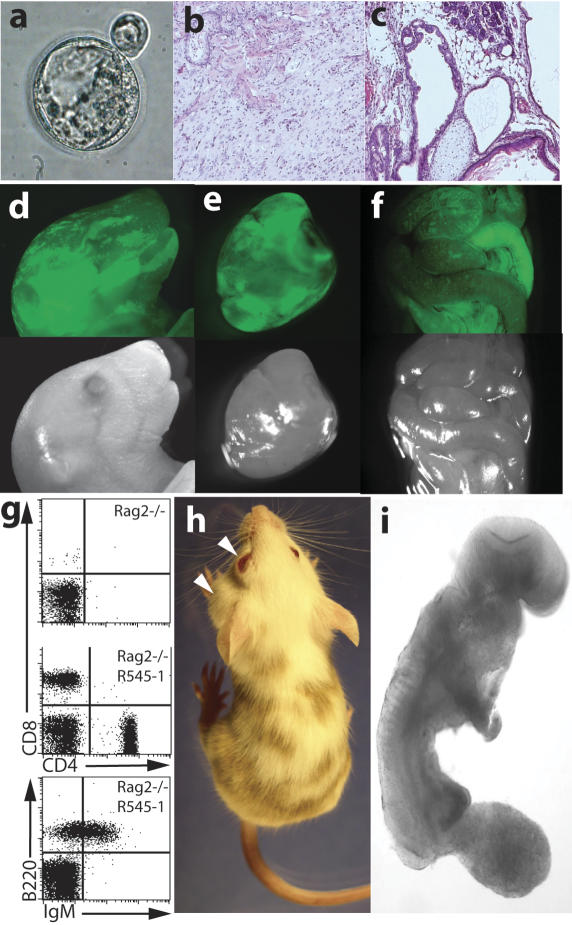
Figure 2.
Analysis of the developmental potential of R545-1 ES cells. (a) A hatching blastocyst derived from a breast cancer cell by nuclear transfer shows a blastocoel cavity, trophectoderm layer, and an inner cell mass. (b,c) H&E staining of teratoma sections produced from R545-1 ES cells shows differentiation into mature neurons, mesenchymal cells, and squamous epithelium (b), and columnar epithelium, chondrocytes, and adipocytes (c). (d–f) Contribution of GFP-labeled R545-1 ES cells to newborn chimeras. Shown on top are the GFP images of the head (d), heart (e), and intestine (f) of one chimera. Below are the same images under phase contrast. (g) FACS analysis of peripheral blood of a Rag2/R545-1 ES cell chimera shows the presence of B cells using antibodies FITC-IgM/PE-B220 and T cells using antibodies FITC-CD4/PE-CD8. (h) Contribution of R545-1 cells to the skin indicates differentiation into melanocytes. Arrows depict spontaneous development of tumors on the eye and neck of chimera #1 (see Table 2). (i) Embryos produced entirely from ES cells by tetraploid complementation develop to E9.5 with obvious tail and limb buds, a closed neural tube, and a beating heart.
In the RAS-inducible melanoma model, the activated RAS transgene is expressed specifically in melanocytes after administration of doxycycline to animals that also carry a null mutation of the ink4a/Arf tumor suppressor locus (genotype: Tyr-rtTA+, Tet-RAS+, ink4a/Arf-/-; Chin et al. 1999). RAS activation results in the formation of melanomas in 25% of doxycycline-treated animals after a latency period of 8–12 wk. Down-regulation of RAS activity following withdrawal of doxycycline causes tumor regression within 2 wk, whereas readministration leads to recurrence of tumors with a shortened latency (Fig. 3a). Because doxycycline-regulated RAS expression is restricted to the melanocytic lineage (Chin et al. 1999), it is reasonable to assume that activated RAS expression is silenced in oocytes and embryos; thus, the status of doxycycline induction prior to nuclear transfer is not expected to influence the outcome of cloning of R545 nuclei. Therefore, R545 donor melanoma cells, resultant NT blastocysts, and NT ES cell lines R545-1 and R545-2, were cultured in the absence of doxycycline. Although both of these NT ES cell lines were derived from the same donor melanoma cell line (Chin et al. 1999), R545-2 was derived from a nontumorigenic subclone of R545.
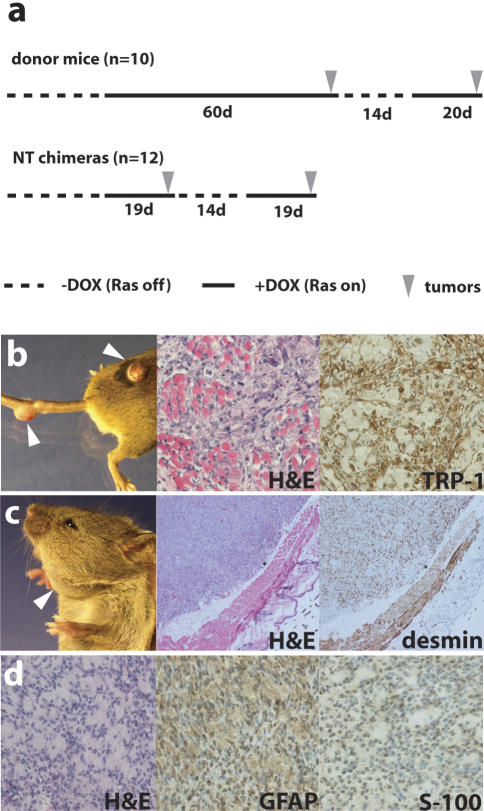
Figure 3.
Cancer phenotype in chimeric mice. (a) Comparison of the average latency period of tumor development in the melanoma donor mice (top) with that in nuclear transfer (NT) chimeras (bottom). Note the similar latency of tumor development in NT chimeras with that in donor mice after readministration of doxycycline (recurrent tumors). (b–d) Representative pictures and immunohistochemistry of tumors that formed in R545-1 NT chimeras. Arrows indicate sites of tumor growth. Melanomas (b), a rhabdomyosarcoma (c), and a malignant peripheral nerve sheath tumor (MPNST; d) were identified by H&E staining and immunohistochemistry with melanocyte-specific TRP-1 or muscle-specific desmin or MPNST-detecting GFAP and S-100 antibodies, respectively.
Teratomas and chimeras generated from melanoma-derived NT ES cells
We first assessed the tumorigenic and differentiation potential of the two NT ES cell lines by their ability to induce tumors in SCID mice. We found that R545-1 but not R545-2 NT ES cells formed typical teratomas with cartilage, adipose, glandular, neuronal, and skin tissue, reflecting a broad developmental potential that was comparable to that of wild-type ES cells (Fig. 2b,c; data not shown). To investigate the developmental potential of R545-1 and R545-2 ES cells in vivo, we produced chimeric embryos by injecting NT ES cells tagged with a constitutively active GFP transgene into blastocysts. Similar to chimeras produced from wild-type ES cells, those derived from the R545-1 NT ES cells showed donor cell contribution to multiple organs including skin, intestine, heart, kidney, lungs, thymus, and liver (Fig. 2d–f). These results demonstrate that in the absence of RAS expression, the R545-1 NT ES cells have the potential to incorporate into most if not all tissues of newborn mice. In contrast, R545-2 ES cells did not generate any chimeras (data not shown). We performed SKY analysis of R545-1 and R545-2 cells to detect potential chromosomal aberrations that might explain the different phenotypes. To our surprise, R545-2 ES cells had a near-tetraploid genotype (data not shown), which is likely the reason for the observed failure to produce teratomas and chimeras. We therefore focused on the R545-1 NT ES cells for the remainder of this study.
To demonstrate functional contribution of R545-1 NT ES cells to adult lineages, we first analyzed the lymphoid compartment in chimeras derived from the injection of R545-1 cells into Rag2-deficient blastocysts. Because Rag2 mutant mice completely lack mature lymphocytes, any B and T cells detected in chimeric animals are derived from the injected ES cells (Chen et al. 1993). Rag2-deficient chimeras derived from the R545-1 NT ES cells had a normal lymphoid compartment as assessed by FACS analysis of peripheral blood using B- and T-cell-specific surface markers such as B220/IgM and CD4/CD8, respectively (Fig. 2g). Similarly, when injected into BALB/C host blastocysts, R545-1 NT ES cells generated chimeras with a 5%–50% coat color chimerism, demonstrating contribution of NT ES cells to the melanocyte lineage (Fig. 2h). Finally, donor-derived fibroblasts isolated from the chimeras showed normal growth characteristics and were propagated in vitro for several passages (data not shown). We were unable to produce germ-line offspring from these chimeras, suggesting that the genetic restrictions of the melanoma donor nucleus interfered with fertility. Moreover, R545-1 NT ES cells were X0 female, whereas the R545 donor mouse was male, indicating loss of the Y chromosome, which has been observed in different normal and NT ES cell lines (Eggan et al. 2002; data not shown). These results show that the oocyte cytoplasm is able to reprogram the epigenetic state of the donor cell nucleus into a pluripotent embryonic state that supports differentiation into multiple somatic cell types including fibroblasts, lymphocytes, and melanocytes.
Autonomous developmental potential of melanoma clones
In a chimera, host cells interact with the transplanted donor ES cells and can potentially complement for noncell-autonomous defects of the transplanted cells. To determine their autonomous developmental potential, we injected R545-1 NT ES cells into tetraploid blastocysts. In this approach, ES cells exclusively give rise to the embryo, whereas tetraploid host cells contribute to the placenta (Wang et al. 1997; Eggan et al. 2001). We detected R545-1 ES cell-derived embryos up to E 9.5 with a beating heart, closed neural tube, and developing limb and tail buds (Fig. 2i). However, embryos at later stages were not recovered. This shows that the secondary changes of the melanoma donor nucleus can support crucial events of early organogenesis but fail to direct full development of a clone, presumably because of irreversible genetic alterations in the melanoma donor genome.
Melanoma formation in chimeras
A total of 12 adult chimeras were generated and observed for tumor development. One animal spontaneously (without doxycycline induction) developed an eye and neck tumor at 3 wk of age that were identified by histological analysis as a melanoma and a rhabdomyosarcoma, respectively (Fig. 2h; Table 2). Cell lines established from the eye and neck tumors produced tumors in the absence of doxycycline when transplanted into SCID mice. RT–PCR analysis on tumor samples using transgene-specific primers confirmed that both tumors constitutively expressed the RAS transgene (data not shown). In contrast, RAS expression was undetectable in fibroblasts isolated from the same animal, demonstrating that the transgene was specifically activated in the tumors.
Table 2.
Tumor developement in chimeric animals
| Chimera | Age when put on dox (days) | Tumor latency (days) | Tumors (sites) | Dox responsiveness | Additional remarks |
|---|---|---|---|---|---|
| #1 | 28a | 0a | Melanoma (eye) | No | Constitutive expression of RAS transgene |
| Rhabdomyosarcoma (neck) | No | Constitutive expression of RAS transgene | |||
| #2 | 22 | 7 | Melanoma (tail, leg) | Yes | |
| #3 | 22 | 9 | Melanoma (anus, tail) | Yes | |
| #4 | 22 | 16 | Melanoma (ear, anus) | Yes | |
| #5 | 22 | 48 | Melanoma (legs) | Yes | |
| #6 | 54 | 12 | Melanoma (skin) | N/A | |
| Rhabdomyosarcoma (abdominal wall) | No | Constitutive expression of RAS transgene | |||
| #7 | 46 | 20 | Melanoma (leg, peritoneum) | Yes | IP tumor compatible with metastatic melanoma, developed during regression period |
| Rhabdomyosarcoma (leg muscle) | No | Developed during regression period | |||
| #8 | 22 | 44 | Melanoma (tail, back, anus) | Yes | |
| Rhabdomyosarcoma (neck) | No | Developed during regression period, constitutive expression of RAS transgene | |||
| #9 | 54 | 12 | Melanoma (head, back) | Yes | |
| #10 | 30 | 15 | Melanoma (tail, anus) | Yes | |
| MPNST (abdominal wall) | No | Tumor developed 3 mo after regression of primary melanoma | |||
| #11 | 30 | 10 | Melanoma (anus) | N/A | |
| #12 | 30 | 10 | Melanoma (leg, thymus) | N/A | Thymic tumor compatible with metastatic melanoma |
| #2-#12 | 18.5 (average) | ||||
| Control #1 | 21 | 98 | Melanoma (anus) | Yes | Re-induction of RAS results in formation of dox-responsive melanoma after 4 wk |
| Control #2 | 21 | 126+ | No tumors | N/A | |
| Control #3 | 21 | 126+ | No tumors | N/A |
Tumors were classified by H&E staining of histological sections and immunohistochemical analysis for melanoma specific markers S-100 and TRP-1, the rhabdomyosarcoma-specific marker desmin, or MPNST markers GFAP and S-100. Responsiveness of tumors to doxycyline was assessed either by regression of primary tumors upon doxycycline withdrawal or by doxycycline dependent growth of established tumor cell lines after transplantation into SCID mice. Expression of the RAS transgene was determined by RT-PCR that specifically detects the transgene but not the endogeneous RAS locus (data not shown). Boxes shaded in dark gray mark tumors of nonmelanocyte origin. (N/A) Not analyzed; (MPNST) Malignant peripheral nerve sheath tumor.
Tumors developed before exposure to doxycycline
The 11 remaining chimeras were fed doxycycline in the drinking water to induce RAS expression in melanocytes and all succumbed to melanomas with an average latency of 19 d after induction (Fig. 3a; Table 2). This 19-d latency is significantly shorter than that observed for de novo melanoma development in the donor mouse model, which is 60 d. However, in the donor animals, melanomas have been shown to recur from minimal residual disease with a similar short latency after doxycycline withdrawal and subsequent readministration, as well as in SCID mice that have been transplanted with established melanoma cell lines (Fig. 3a; Chin et al. 1999). Also, unlike the original donor model, all chimeras developed multiple primary melanoma lesions (Table 2; Fig. 3b). Similar to the donor model, tumors were found on the tail, ear, leg, anus, back, and neck and were identified as melanomas by immunohistochemistry using antibodies against the melanocyte markers S-100 (data not shown) and TRP-1 (Fig. 3b). To assess whether these melanomas were dependent on continuous RAS activity for maintenance as described in the original model, we took four of the mice off doxycycline. In each of the cases, tumors regressed after ∼1–2 wk. Readministration of doxycycline resulted in the rapid reappearance of melanomas within 2–3 wk (Fig. 3a).
High incidence of nonmelanoma tumors in chimeric mice
Surprisingly, we found that 33% of the chimeras developed rhabdomyosarcomas in addition to melanomas, identified by positive immunoreactivity against desmin (Fig. 3c; Table 2). These rhabdomyosarcomas did not regress on doxycycline withdrawal and, similar to the doxycycline-independent melanoma, showed a constitutively expressed RAS transgene in all cases examined (n = 3; data not shown). Furthermore, one chimera that remained tumor-free for 3 mo after regression of the primary melanomas succumbed to a malignant peripheral nerve sheath tumor (MPNST) at the age of 4 mo, as determined by GFAP and S-100-positive staining of sections (Fig. 3d; Table 2). Therefore, the secondary changes of the melanoma nucleus in combination with doxycycline-independent RAS expression can promote tumorigenesis in cellular contexts other than melanocytes, resulting in an expanded tumor spectrum in the chimeric mice.
These biological data show that R545-1 NT ES cell chimeras, derived from a reprogrammed melanoma nucleus, develop cancer with a higher penetrance, reduced latency, and expanded tumor spectrum as compared with the donor mouse model (Fig. 3a; Table 2). This is consistent with the notion that the tumorigenic phenotype of the donor melanoma cells is determined in part by irreversible genetic changes that were unaltered by NT and transferred into NT ES cells and their chimeras.
Chromosomal analysis
To unequivocally determine that R545-1 NT ES cells were derived from a melanoma donor cell rather than a nontransformed cell, we performed two control experiments: (1) High-resolution array-comparative genome hybridization (CGH) to detect genetic changes shared between the melanoma donor cells and the derivative NT ES cell line, and (2) nuclear transfer of tail fibroblasts from a mouse with identical genotype as the donor mouse in order to generate control NT ES cells and chimeras.
Although the CGH profile of the parental R545 melanoma donor cell line was highly heterogeneous (Fig. 4a), the profiles of tumors derived from the injection of R545 cells into SCID mice showed a consistent pattern (Fig. 4a). For example, trisomy 8 with a characteristic 8qter deletion was consistently observed in array-CGH profiles of all SCID tumors derived from R545 donor cells (n = 4; e.g., see Fig. 4a), suggesting that this cytogenetic feature is selected for in the tumorigenic subpopulation of R545 donor cells. This is consistent with previously published results showing that tumor cell lines in culture typically exhibit a highly heterogenous profile of genomic alterations, in contrast to their derivative tumors in explant models (Chang et al. 2003). This likely reflects the increased selective forces in vivo that drive the clonal expansion of specific chromosomal aberrations advantageous for tumor development. We have mapped the 8qter deletion to a 113.36-megabase (MB) and 113.7-MB physical location on chromosome 8 (NCBI Mouse Genome Build 30; Fig. 4c). Deletion in this specific region has not been observed in any other profile of many mouse tumors or cell lines, including over 20 Tyr-rtTA+, Tet-RAS+, ink4a/Arf-/-melanomas (data not shown), arguing against it being a random artifact acquired in vitro.
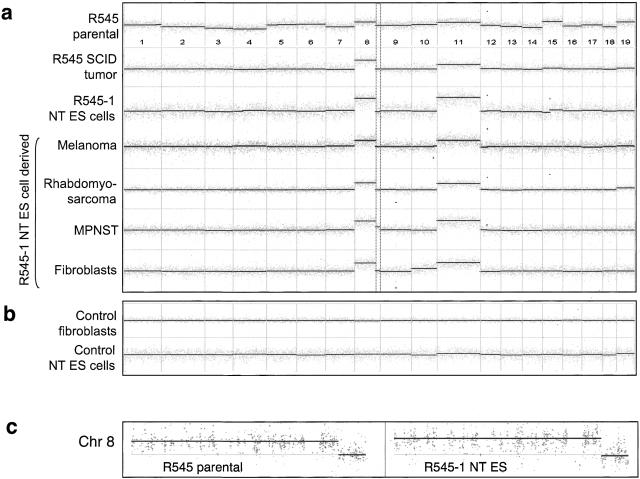
Figure 4.
(a) Whole-genome array-CGH profiles of R545 donor melanoma cells (R545 parental, top profile), a SCID tumor derived from R545 melanoma cells (profile #2), R545-1 NT ES cells derived by nuclear transfer from an R545 cell (profile #3), and its chimera derivatives (in order, profiles #4–#7 represent R545-1 melanoma, R545-1 rhabdomyosarcoma, R545-1 MPNST, and R545-1 tail tip fibroblasts, respectively). Dashed box outlines characteristic 8qter deletion. (b) Profiles of nontransformed fibroblasts with an identical genotype to R545 (Tyr-rtTA+, Tet-RAS+, Ink4a/Arf-/-; top profile) and its ES cells derived by nuclear transfer (bottom profile). Gray spots represent median-filtered data (width of three probes); dark lines mark blocks of uniform log2 ratio (and copy number) determined by a segmentation algorithm (see Materials and Methods). (c) Zoom-in view of chromosome 8 profiles for R545-1 parental melanoma (left profile) and R545-1 NT ES cells (right profile). Black points label raw log2 ratios.
Importantly, the profile of the R545-1 NT ES cells shows an identical pattern as the tumors derived from the donor tumor cell line including the characteristic trisomy 8 with the 8qter deletion and trisomy 11 (Fig. 4a,c), providing unequivocal genomic evidence that the R545-1 NT ES cell was cloned from a tumorigenic nucleus of the R545 tumor cell line. Furthermore, these characteristic genomic alterations were present in all R545-1 NT ES cell derivatives, including melanomas, rhabdomyosarcomas, an MPNST, and nontransformed tail-tip fibroblasts derived from chimeras (Fig. 4a). This conserved chromosomal pattern in donor tumor cells and all derivative tumors of NT chimeras suggests that little, if any, additional mutations have occurred in doxycycline-dependent melanomas following reactivation of the RAS oncogene. However, the finding that doxycycline-independent melanomas and all rhabdomyosarcomas showed constitutive expression of the RAS transgene demonstrates that additional genetic changes clearly can occur. Mutations resulting in constitutive RAS expression most likely occurred late in the development of chimeras rather than during nuclear transfer or ES cell derivation because (1) constitutive activation of RAS has been shown to be incompatible with prenatal development (Katsuki et al. 1989) and (2) normal tissue from adult chimeras did not express RAS.
To exclude the possibility that the cancer phenotype was influenced by the NT procedure, we generated two ES cell lines by nuclear transfer from nontransformed tail fibroblasts of mice with an identical genotype (Tyr-rtTA+, Tet-RAS+, ink4a/Arf-/-) as the mouse that gave rise to the R545 tumor (Table 1). Notably, the efficiency of blastocyst formation from fibroblast donors was 10 times higher than that from melanoma donors (Table 1), possibly due to differences in the cell type, the cell cycle status, or the genetic constitution of the respective donor cells. However, the frequency of ES cell derivation from cloned blastocysts was comparable (25% vs. 20%; Table 1). Importantly, no chromosomal abnormalities were found by CGH analysis in either the donor fibroblasts or the derivative NT ES cells (Fig. 4b). As expected, these control NT ES cells when injected into diploid blastocysts were competent to produce coat color chimeras (n = 3), one of which developed a doxycycline-dependent melanoma after a 12-wk latency period (Table 2). This low penetrance (33%) and long latency (12 wk) is similar to that observed in the donor mouse model and contrasts with the high penetrance and short latency of tumor development in the R545-1 NT ES cell-derived chimeras (see earlier). Together, these genetic and biological data confirm that R545-1 NT ES cells are derived from nuclear transfer of a melanoma nucleus and that the secondary changes accrued during melanoma tumorigenesis resulted in the expanded tumor phenotype of NT chimeras.
Discussion
We have used nuclear transfer as a functional assay to determine whether the genome of different cancer cells can be reprogrammed by the oocyte environment into a pluripotent embryonic state. We have shown here that the nuclei of many cancer cells were able to support preimplantation development into normal-appearing blastocysts (Fig. 1a; Table 1) and hence differentiation into the first two cell lineages of the embryo, the epiblast and trophectoderm, without signs of abnormal proliferation. Therefore, the malignant phenotype of these tumor types can be suppressed by the oocyte environment and permit apparently normal early development. Furthermore, ES cells derived from one of the cloned melanoma cells were able to differentiate into most if not all somatic cell lineages in teratomas and chimeras including fibroblasts, lymphocytes, and melanocytes. This occurred despite severe chromosomal changes documented by CGH. Thus, our data suggest that the secondary chromosomal changes associated with malignancy do not necessarily interfere with preimplantation development, ES cell derivation, and a broad nuclear differentiation potential. However, a second NT ES cell line derived from a nontumorigenic subclone of the melanoma cell line was unable to differentiate in the context of teratomas or chimeras, and we correlated this failure to differentiate with the near-tetraploid genotype of the ES cells. This raises the interesting possibility that R545-1 was derived from a rare “cancer stem cell” (Reya et al. 2001; Pardal et al. 2003; Al-Hajj et al. 2004) that carried all of the tumorigenic features of the donor melanoma, whereas R545-2 was derived from a derivative of this cancer stem cell that had undergone additional chromosomal changes that prevented autonomous tumor formation and interfered with differentiation. Alternatively, the subcloning process of the melanoma cell line might have induced these changes and caused the observed phenotype. It remains unclear, however, why melanoma nuclei supported the establishment of ES cells, whereas other cancer nuclei did not. It is possible that different genetic changes present in the other tumor genomes interfered with the reestablishment of pluripotency by nuclear transfer and/or that cell cycle- or cell type-specific differences might have influenced the efficiency of cloning and ES cell derivation, consistent with previous observations (Rideout et al. 2001; Wakayama and Yanagimachi 2001; Hochedlinger and Jaenisch 2002a). Moreover, the use of a doxycycline-inducible system in which the RAS oncogene was regulated temporally in a tissue-specific manner, coupled with the Ink4a/Arf deficiency, which has been shown to facilitate cellular dedifferentiation (Bachoo et al. 2002), may provide a more permissive context for the derivation of pluripotent ES cells from melanoma nuclei compared with the other donor nuclei.
Array-CGH analysis of the donor melanoma cells revealed a heterogeneous population of tumor cells carrying multiple genetic alterations, including trisomy 8 with 8qter deletion and trisomy 11, which were also detected in the NT ES cells as well as in melanomas, rhabdomyosarcomas, an MPNST, and fibroblasts recovered from the chimeras. Although trisomy 8 has been previously observed to arise independently in some ES cells after prolonged in vitro propagation (Liu et al. 1997), trisomy 8 with the characteristic 8qter deletion is a common cytogenetic feature observed in the parental R545 melanoma cell line and all SCID tumors derived from the R545 cell line, suggesting that this chromosomal change was preexisting in the melanoma cells and essential for their tumorigenic phenotype. Consistent with the conclusion that the genetic changes are tumor derived is our observation that neither trisomy 8, the 8qter deletion, nor trisomy 11 were seen in the ES cells derived by nuclear transfer of fibroblast cells from the transgenic donor animals.
The chimeras generated in this study developed melanomas with higher penetrance and shortened latency as compared with the donor model, suggesting that the irreversible chromosomal alterations inherited by the NT ES cells from the donor nucleus contributed to the ultimate tumorigenic potential of the melanocytes. In other words, reacquisition of the epigenetic state of the donor melanoma cell in melanocytes of the chimeras in combination with RAS expression was sufficient to initiate tumor growth against the backdrop of the inherited genetic changes. This notion was supported by the observation that multiple melanomas formed simultaneously at distinct sites in R545-1 NT chimeras with a latency comparable to that required for the development of recurrent tumors or the emergence of explant tumors derived from established melanoma cells (Fig. 3a). Together, these data argue against, although they do not formally exclude, the possibility that doxycycline-dependent melanoma formation in the NT chimeras required the acquisition of additional genetic changes during development of the chimeric animals. In contrast, dox-independent melanomas and rhabdomyosarcomas showed constitutive activation of the RAS transgene, which is likely the result of mutations within the RAS transgene or the rtTA transactivator, consistent with observations in the donor mouse model (Chin et al. 1999). The finding that the dox-independent tumors had an identical CGH pattern to the dox-dependent melanomas and normal tissue from chimeras suggests that the activation of RAS, either by doxycycline induction or mutations targeting the tet-regulatory module, in combination with the melanoma-specific alterations, were sufficient to drive tumor growth in melanocytes and nonmelanocytic tissues.
Our results serve as a paradigm for studying in vivo the consequences of tumor-specific genetic alterations in different tissues. For example, the occurrence of rhabdomyosarcomas in a third of the chimeras suggests that overlapping pathways operate during melanoma and rhabdomyosarcoma evolution, consistent with previous observations (Sharp et al. 2002). Interestingly, rhabdomyosarcoma incidence in humans was found to frequently correlate with a loss of chromosome 16q (Visser et al. 1997; Bridge et al. 2002), the syntenic region of 8q (Fig. 4b,c) in mouse. This genomic region has also been described as a common fragile site, and breakage at this point has been postulated to contribute to tumorigenesis through the loss of a potential tumor suppressor gene (Ludes-Meyers et al. 2003). Similar to rhabdomyosarcoma formation, the development of an MPNST in one chimera suggests that the secondary changes of the melanoma nucleus may be important in the genesis of this type of tumor. Together, these findings demonstrate the general use of NT as a functional assay for characterizing commonalities among different types of cancer. In addition to studying cancer genetics and epigenetics, the nuclear transfer approach should be useful for the analysis of complex genetic disorders such as diabetes in order to characterize and manipulate the multiple alleles affected in these diseases. At present, no other method has the power of amplifying the genome of a single cell with complex genetic alterations into a population of pluripotent ES cells.
This work focused on a subset of murine tumor models with specific genetic alterations and might therefore not be representative of other tumor systems. For example, Ink4a/Arf mutations account for only a proportion of familial and sporadic melanomas (Chin 2003), and other pathways such as the HGF/SF and the PTEN signaling cascades have also been shown to be involved in melanoma formation in mouse and humans. Moreover, because human tumor formation is believed to require more changes, genetic and epigenetic, than in mouse (Hahn and Weinberg 2002), it is possible that the reprogramming of human cancer nuclei by nuclear transfer will give different results from mouse. Barring ethical and legal limitations, the cloning of human cancer nuclei should now be technically feasible (Hwang et al. 2004).
Previous results have demonstrated that the genome of terminally differentiated cells such as B and T cells or mature postmitotic neurons can be reprogrammed to totipotency following NT (Hochedlinger and Jaenisch 2002a; Eggan et al. 2004). The data described in this work argue that the malignant phenotype of at least some cancer cells can be reversed to a pluripotent state despite the presence of irreversible genetic alterations and allow apparently normal differentiation. It is now important to define the epigenetic factors that influence the malignant phenotype to help establish therapeutic strategies for cancer patients.
Materials and methods
Nuclear transfer and ES cell derivation
Nuclear transfer was performed as described previously (Wakayama et al. 1998; Hochedlinger and Jaenisch 2002a; Rideout et al. 2002). Briefly, oocytes were collected from the oviducts of superovulated B6D2F1 female mice (Taconic) and stored in KSOM embryo culture media (Specialty media). Enucleation and nuclear transfer were performed using a piezodriven micromanipulator (Primetech) on a Nikon microscope with inverted optics. Reconstructed embryos were activated for 6 h in calcium-free MCZB medium in the presence of 10 mM Sr2+ and 5 μg/mL of cytochalasin B before replacement by KSOM. Developing blastocysts were treated with Acid Tyrode's (Sigma) to remove the zona pellucida and placed on irradiated murine embryonic fibroblasts in ES cell media supplemented with 1000 U/mL LIF and 50 μM PD98059 MEK1 inhibitor (Cell Signaling). Proliferating outgrowths were dissociated in trypsin and replated on fibroblasts until stable cell lines grew out. Homologous recombination in ES cells was performed as described (Rideout et al. 2002) using a ROSA26-EGFP construct to obtain constitutive transgene expression.
Cell culture and tumor induction
R545 melanoma cells were cultured in RPMI media in the absence of doxycycline. When transplanted into SCID mice, R545 cells formed tumors in a doxycycline-dependent manner within 2–3 wk. Tumor cell lines from NT chimeras were established from small pieces of primary tissue in RPMI media in the absence of doxycycline. R545-1 NT ES cell-derived fibroblasts were produced from either embryonic day 14.5 (E14.5) embryos or from tail biopsies of adult chimeras and selected by the addition of neomycin (Gibco) to the media. For teratoma induction, 5 × 106 ES cells grown in ES cell medium (see earlier) on irradiated murine embryonic fibroblasts were trypsinized into a single cell suspension and preplated for 30 min on a nongelatinized dish to remove fibroblasts. Cells were injected subcutaneously into the flanks of SCID mice. Three weeks later, teratomas were isolated and fixed in formalin for histological analysis. For tumor formation from somatic cells lines, 5 × 106 cells were transplanted subcutaneously into SCID mice that were fed doxycycline (Sigma) in the drinking water at a concentration of 2 mg/mL supplemented with 10 mg/mL of sucrose. Visible tumors were isolated and fixed in formalin.
Chimera production
For diploid blastocyst injections, blastocysts were either flushed from the uteri of day 3.5 pregnant Balb/c mice, or fertilized zygotes were isolated from the oviducts of day 0.5 pregnant B6D2F1 or 129B6F1 Rag2-/- females and allowed to develop to the blastocyst stage in culture. For tetraploid blastocyst injections, two-cell embryos derived from B6D2F1 females were first electro-fused in 0.3 M mannitol/0.3% BSA using the LF-101 cell fusion instrument (Protech International). Between five and 15 ES cells were injected per blastocyst. These were transferred into day 2.5 pseudo-pregnant B6D2F1 recipient females. C-sections were performed 19 d later and pups were fostered with lactating Swiss mice.
FACS analysis
Peripheral blood cells were treated with ACK lysing buffer (0.15 mM NH4Cl, 10 mM KHCO3, and 0.1 mM sodium EDTA at pH 7.2) before FACS analysis to remove red blood cells. We stained 1 × 106 cells with PE-B220 and FITC-IgM antibodies to detect B cells and FITC-CD4 and PE-CD8 antibodies to detect T cells. All antibodies were purchased from Pharmingen. FACS analyses were performed on a Becton Dickinson cell sorter.
Histology
Normal and tumor tissue samples were fixed in 10% buffered formalin for 24 h and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin and serial sections were used for immunohistochemical analysis. Immunostaining was performed using an avidin-biotin immunoperoxidase assay. Primary antibodies were anti-TRP-1 (1:1000 dilution; Santa-Cruz), anti-protein S-100 (1:1500 dilution; Dako), anti-desmin (1:1000; Dako), and anti-GFAP (1:500; Dako). Sections were incubated with primary antibodies for 24 h and subsequently with biotinylated secondary antibodies (Vector Laboratories) for 30 min, followed by incubation with avidin-coupled peroxidase (Vector Laboratories) for 30 min. Diaminobenzidine was used as a chromogen and hematoxylin as the counterstain. For negative controls, primary antibodies were omitted.
Array-CGH profiling on long-oligomer microarrays
Array-CGH profiles were generated and analyzed as described (Brennan et al. 2004). Briefly, genomic DNA was fragmented with DpnII digest and random-prime labeled after purification according to modified protocols (Pollack et al. 1999). Two micrograms of digested DNA was used per labeling reaction, and each sample was dye-swap labeled for two hybridizations against reference. Normal genomic DNA from nonlittermate animals with identical genotype (Tyr-rtTA+, Tet-RAS+, Ink4a/Arf-/-) was used as reference. Labeled DNAs were hybridized onto mouse long-oligonucleotide microarrays (Agilent Technologies) for 18–20 h at 65°C. For detailed labeling and hybridization protocol, see http://genomic.dfci.harvard.edu.
Following hybridization, arrays were washed and scanned on an Agilent scanner and scanned images were analyzed for spot-finding and flagging as well as reporting of Cy3 and Cy5 foreground and background signals for each spot using standard Agilent software designed for the scanner. Custom analytical tools were used to calculate fluorescence ratio calculation (in log2) and oligomer-to-chromosome location mapping (Brennan et al. 2004). The resultant raw array-CGH profiles were analyzed further using a published segmentation algorithm developed by Olshen and Venkatraman (2004) that uses permutation to determine the significance of change points in the raw data in order to identify statistically significant transitions in copy number (Lucito et al. 2003). In this study, significant copy number changes are determined on the basis of segmented profiles only.
Acknowledgments
We are indebted to François Gaudet, Amir Eden, Charlotte Kuperwasser, and Tim Ley for sharing tumor cell lines; to Jessie Dausman and Ruth Flannery for assistance with mouse work; to Christopher Leo and Tali Muller for technical assistance with array-CGH profiles; to Bin Feng for array-CGH analysis; to Marcus Bosenberg for suggestions and critical comments on the project; and to Heinz Linhart, Kathrin Plath, and Erwin F. Wagner for critical reading of the manuscript. Array-CGH profiling is performed at the Arthur & Rochelle Belfer Cancer Genomics Center at the Dana-Farber Cancer Institute. K.H. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds, R.B. by a fellowship from the Lance Armstrong Foundation, C.B. by NIH training grant T32 CA09382 and by the LeBow fund for Myeloma Cure, L.C. by NIH grant RO1 CA93947, and R.J. by NIH grants R37 CA 84198-04 and R350 CA 44339-13.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1213504.
Corresponding authors.
References
- Al-Hajj M., Becker, M.W., Wicha, M., Weissman, I., and Clarke, M.F. 2004. Therapeutic implications of cancer stem cells. Curr. Opin. Genet. Dev. 14: 43-47. [DOI] [PubMed] [Google Scholar]
- Bachoo R.M., Maher, E.A., Ligon, K.L., Sharpless, N.E., Chan, S.S., You, M.J., Tang, Y., DeFrances, J., Stover, E., Weissleder, R., et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269-277. [DOI] [PubMed] [Google Scholar]
- Bakin A.V. and Curran, T. 1999. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 283: 387-390. [DOI] [PubMed] [Google Scholar]
- Brennan C., Zhang, Y., Leo, C., Feng, B., Cauwles, C., Aguirre, A.J., Kim, M., Protopopov, A., and Chin, L. 2004. High-resolution global profiling of genomic alterations with long oligonucleotide microarray. Cancer Res. (in press). [DOI] [PubMed]
- Bridge J.A., Liu, J., Qualman, S.J., Suijkerbuijk, R., Wenger, G., Zhang, J., Wan, X., Baker, K.S., Sorensen, P., and Barr, F.G. 2002. Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer 33: 310-321. [DOI] [PubMed] [Google Scholar]
- Carlson D.L., Sauerbier, W., Rollins-Smith, L.A., and McKinnell, R.G. 1994. Fate of herpesvirus DNA in embryos and tadpoles cloned from Lucké renal carcinoma nuclei. J. Comp. Pathol. 111: 197-203. [DOI] [PubMed] [Google Scholar]
- Chang S., Khoo, C.M., Naylor, M.L., Maser, R.S., and DePinho, R.A. 2003. Telomere-based crisis: Functional differences between telomerase activation and ALT in tumor progression. Genes & Dev. 17: 88-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lansford, R., Stewart, V., Young, F., and Alt, F.W. 1993. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. 90: 4528-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L. 2003. The genetics of malignant melanoma: Lessons from mouse and man. Nat. Rev. Cancer 3: 559-570. [DOI] [PubMed] [Google Scholar]
- Chin L., Tam, A., Pomerantz, J., Wong, M., Holash, J., Bardeesy, N., Shen, Q., O'Hagan, R., Pantginis, J., Zhou, H., et al. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400: 468-472. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Raker, V.A., Corsaro, M., Fazi, F., Fanelli, M., Faretta, M., Fuks, F., Lo Coco, F., Kouzarides, T., Nervi, C., et al. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295: 1079-1082. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey, M., Slagle, B.L., McArthur, M.J., Montgomery Jr., C.A., Butel, J.S., and Bradley, A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215-221. [DOI] [PubMed] [Google Scholar]
- Eggan K., Akutsu, H., Loring, J., Jackson-Grusby, L., Klemm, M., Rideout III, W.M., Yanagimachi, R., and Jaenisch, R. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. 98: 6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K., Rode, A., Jentsch, I., Samuel, C., Hennek, T., Tintrup, H., Zevnik, B., Erwin, J., Loring, J., Jackson-Grusby, L., et al. 2002. Male and female mice derived from the same embryonic stem cell clone by tetraploid embryo complementation. Nat. Biotechnol. 20: 455-459. [DOI] [PubMed] [Google Scholar]
- Eggan K., Baldwin, K., Tackett, M., Osborne, J., Gogos, J., Chess, A., Axel, R., and Jaenisch, R. 2004. Mice cloned from olfactory sensory neurons. Nature 428: 44-49. Epub 2004 Feb 15; doi:10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Egger G., Liang, G., Aparicio, A., and Jones, P.A. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457-463. [DOI] [PubMed] [Google Scholar]
- Felsher D.W. 2003. Cancer revoked: Oncogenes as therapeutic targets. Nat. Rev. Cancer 3: 375-380. [DOI] [PubMed] [Google Scholar]
- Frank S.R., Parisi, T., Taubert, S., Fernandez, P., Fuchs, M., Chan, H.M., Livingston, D.M., and Amati, B. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4: 575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F., Hodgson, J.G., Eden, A., Jackson-Grusby, L., Dausman, J., Gray, J.W., Leonhardt, H., and Jaenisch, R. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489-492. [DOI] [PubMed] [Google Scholar]
- Hahn W.C. and Weinberg, R.A. 2002. Rules for making human tumor cells. N. Engl. J. Med. 347: 1593-1603. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K. and Jaenisch, R. 2002a. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature 415: 1035-1038. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Nuclear transplantation: Lessons from frogs and mice. Curr. Opin. Cell Biol. 14: 741-748. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N. Engl. J. Med. 349: 275-286. [DOI] [PubMed] [Google Scholar]
- Hwang W.S., Ryu, Y.J., Park, J.H., Park, E.S., Lee, E.G., Koo, J.M., Chun, H.Y., Lee, B.C., Kang, S.K., Kim, S.J., et al. 2004. Evidence of a pluripotent human embryonic stem cell line derived from a cloned blastocyst. Science 303: 1669-1674. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Jahner, D., Nobis, P., Simon, I., Lohler, J., Harbers, K., and Grotkopp, D. 1981. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell 24: 519-529. [DOI] [PubMed] [Google Scholar]
- Jain M., Arvanitis, C., Chu, K., Dewey, W., Leonhardt, E., Trinh, M., Sundberg, C.D., Bishop, J.M., and Felsher, D.W. 2002. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297: 102-104. [DOI] [PubMed] [Google Scholar]
- Jones P.A. and Baylin, S.B. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3: 415-428. [DOI] [PubMed] [Google Scholar]
- Katsuki M., Kimura, M., Hata, J., Takahashi, R., Nozawa, S., Yokoyama, M., Izawa, M., Sekiya, T., Nishimura, S., and Nomura, T. 1989. Embryonal tumors from transgenic mouse zygotes carrying human activated c-Ha-ras genes. Mol. Biol. Med. 6: 567-572. [PubMed] [Google Scholar]
- Kuperwasser C., Hurlbut, G.D., Kittrell, F.S., Dickinson, E.S., Laucirica, R., Medina, D., Naber, S.P., and Jerry, D.J. 2000. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am. J. Pathol. 157: 2151-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3: 662-673. [DOI] [PubMed] [Google Scholar]
- Li L., Connelly, M.C., Wetmore, C., Curran, T., and Morgan, J.I. 2003. Mouse embryos cloned from brain tumors. Cancer Res. 63: 2733-2736. [PubMed] [Google Scholar]
- Liu X., Wu, H., Loring, J., Hormuzdi, S., Disteche, C.M., Bornstein, P., and Jaenisch, R. 1997. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev. Dyn. 209: 85-91. [DOI] [PubMed] [Google Scholar]
- Lucito R., Healy, J., Alexander, J., Reiner, A., Esposito, D., Chi, M., Rodgers, L., Brady, A., Sebat, J., Troge, J., et al. 2003. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 13: 2291-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludes-Meyers J.H., Bednarek, A.K., Popescu, N.C., Bedford, M., and Aldaz, C.M. 2003. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet. Genome Res. 100: 101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell R.G., Deggins, B.A., and Labat, D.D. 1969. Transplantation of pluripotential nuclei from triploid frog tumors. Science 165: 394-396. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro, K., Gaubatz, S., Livingston, D.M., and Nakatani, Y. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132-1136. [DOI] [PubMed] [Google Scholar]
- Olshen A. and Venkatraman, E. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics (in press). [DOI] [PubMed]
- Pardal R., Clarke, M.F., and Morrison, S.J. 2003. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer 3: 895-902. [DOI] [PubMed] [Google Scholar]
- Peters A.H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323-337. [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Perou, C.M., Alizadeh, A.A., Eisen, M.B., Pergamenschikov, A., Williams, C.F., Jeffrey, S.S., Botstein, D., and Brown, P.O. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23: 41-46. [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison, S.J., Clarke, M.F., and Weissman, I.L. 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105-111. [DOI] [PubMed] [Google Scholar]
- Rideout III W.M., Eggan, K., and Jaenisch, R. 2001. Nuclear cloning and epigenetic reprogramming of the genome. Science 293: 1093-1098. [DOI] [PubMed] [Google Scholar]
- Rideout W.M., Hochedlinger, K., Kyba, M., Daley, G.Q., and Jaenisch, R. 2002. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell 109: 17-27. [DOI] [PubMed] [Google Scholar]
- Sanz M.A., Martin, G., and Diaz-Mediavilla, J. 1998. All-trans-retinoic acid in acute promyelocytic leukemia. N. Engl. J. Med. 338: 393-394. [DOI] [PubMed] [Google Scholar]
- Sharp R., Recio, J.A., Jhappan, C., Otsuka, T., Liu, S., Yu, Y., Liu, W., Anver, M., Navid, F., Helman, L.J., et al. 2002. Synergism between INK4a/ARF inactivation and aberrant HGF/SF signaling in rhabdomyosarcomagenesis. Nat. Med. 8: 1276-1280. [DOI] [PubMed] [Google Scholar]
- Stewart C., Harbers, K., Jahner, D., and Jaenisch, R. 1983. X chromosome-linked transmission and expression of retroviral genomes microinjected into mouse zygotes. Science 221: 760-762. [DOI] [PubMed] [Google Scholar]
- Visser M., Sijmons, C., Bras, J., Arceci, R.J., Godfried, M., Valentijn, L.J., Voute, P.A., and Baas, F. 1997. Allelotype of pediatric rhabdomyosarcoma. Oncogene 15: 1309-1314. [DOI] [PubMed] [Google Scholar]
- Wakayama T. and Yanagimachi, R. 2001. Mouse cloning with nucleus donor cells of different age and type. Mol. Reprod. Dev. 58: 376-383. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Perry, A.C., Zuccotti, M., Johnson, K.R., and Yanagimachi, R. 1998. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394: 369-374. [DOI] [PubMed] [Google Scholar]
- Wang Z.Q., Kiefer, F., Urbanek, P., and Wagner, E.F. 1997. Generation of completely embryonic stem cell-derived mutant mice using tetraploid blastocyst injection. Mech. Dev. 62: 137-145. [DOI] [PubMed] [Google Scholar]
- Wilmut I., Schnieke, A.E., McWhir, J., Kind, A.J., and Campbell, K.H. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810-813. [DOI] [PubMed] [Google Scholar]
- Zimonjic D.B., Pollock, J.L., Westervelt, P., Popescu, N.C., and Ley, T.J. 2000. Acquired, nonrandom chromosomal abnormalities associated with the development of acute promyelocytic leukemia in transgenic mice. Proc. Natl. Acad. Sci. 97: 13306-13311. [DOI] [PMC free article] [PubMed] [Google Scholar]