Abstract
The RecQ protein family is a highly conserved group of DNA helicases that play roles in maintaining genomic stability. In this study, we present biochemical and genetic evidence that Escherichia coli RecQ processes stalled replication forks and participates in SOS signaling. Cells that carry dnaE486, a mutation in the DNA polymerase III α-catalytic subunit, induce an RecA-dependent SOS response and become highly filamented at the semirestrictive temperature (38°C). An recQ mutation suppresses the induction of SOS response and the filamentation in the dnaE486 mutant at 38°C, causing appearance of a high proportion of anucleate cells. In vitro, RecQ binds and unwinds forked DNA substrates with a gap on the leading strand more efficiently than those with a gap on the lagging strand or Holliday junction DNA. RecQ unwinds the template duplex ahead of the fork, and then the lagging strand is unwound. Consequently, this process generates a single-stranded DNA (ssDNA) gap on the lagging strand adjacent to a replication fork. These results suggest that RecQ functions to generate an initiating signal that can recruit RecA for SOS induction and recombination at stalled replication forks, which are required for the cell cycle checkpoint and resumption of DNA replication.
Keywords: RecQ DNA helicase, DNA replication, SOS induction, DNA homologous recombination
Progression of replication forks is often blocked following an encounter with either DNA lesions on template strands or other impediments such as bound proteins, secondary structures, or topological stress. Under these circumstances, the efficient resumption of DNA synthesis is essential for the growth and survival of all organisms. The RecQ protein family is a group of highly conserved helicases that includes Escherichia coli RecQ, Saccharomyces cerevisiae Sgs1, Schizosaccharomyces pombe Rqh1, and five gene products in humans (Bachrati and Hickson 2003). Human RecQ homologs include BLM, WRN, and RECQL4, which are implicated in the human hereditary diseases Bloom's syndrome, Werner's syndrome, and Rothmund-Thomson syndrome, respectively (Ellis et al. 1995; Yu et al. 1996; Kitao et al. 1999). The fact that human disorders associated with mutations in different RecQ helicases have distinct features implies that different human RecQ helicases function in distinct cellular processes. However, several lines of evidence indicate that some functions are conserved between the RecQ helicases of lower and higher organisms (Cobb et al. 2002; Bachrati and Hickson 2003). One conserved function among RecQ homologs involves the processing of replication forks when DNA replication has been arrested prior to completion (Murray et al. 1997; Courcelle and Hanawalt 1999; Doe et al. 2000; Frei and Gasser 2000). RecQ family members share a highly conserved helicase domain, but have little homology outside of this region (Bachrati and Hickson 2003). Several RecQ homologs possess a 3′-to-5′ DNA helicase activity that is active on a variety of substrates, including G4 DNA and three- or four-strand DNA structures including D-loops, forked structures, and Holliday junctions (Harmon and Kowalczykowski 1998; Sun et al. 1998; Karow et al. 2000; Mohaghegh et al. 2001; Wu and Maizels 2001; Huber et al. 2002). It is likely, therefore, that the conserved RecQ helicase domain determines the enzyme's DNA substrate specificity and, in many instances, participates during the rescue of stalled replication forks.
E. coli RecQ belongs to the recF recombination pathway (Nakayama et al. 1985). The recF pathway proteins are required for the repair of gapped DNA and resumption of DNA synthesis following UV-induced DNA damage, and to stabilize the nascent strands at stalled forks (Horii and Clark 1973; Kolodner et al. 1985; Tseng et al. 1994; Courcelle et al. 1997). Recently, it was reported that the RecFOR complex loads RecA onto gapped DNA in vitro (Morimatsu and Kowalczykowski 2003). Although unlike recF mutations the recQ mutation does not render cells sensitive to UV irradiation, recent experiments have shown that RecQ together with RecJ play an important role in the resumption of DNA synthesis following UV-induced DNA damage, by degrading the nascent lagging strand of the replication fork (Courcelle and Hanawalt 1999; Courcelle et al. 2003). RecQ also suppresses illegitimate recombination in cells with DNA damage (Hanada et al. 1997). These results indicate that RecQ plays important roles in the resumption of stalled DNA replication and prevention of chromosomal abnormalities.
In this study, we have analyzed the interaction of E. coli RecQ with synthetic DNA substrates resembling DNA replication forks. We demonstrate that RecQ binds leading-strand gapped fork DNA more efficiently than lagging-strand gapped fork DNA or Holliday junction DNA and is capable of converting the ssDNA gaps on the fork from the leading-strand DNA to the lagging-strand DNA by two-step reactions. We also show that RecQ at stalled replication forks acts to induce RecA-dependent SOS signaling. The results suggest that RecQ functions to generate an initiating signal that can recruit RecA for SOS induction and recombination in response to DNA replication interference, which are required for the cell cycle checkpoint and resumption of DNA replication. We propose here that prokaryotic and eukaryotic RecQ helicases play a role in coordinating a cell cycle checkpoint response with recombination and replication.
Results
RecQ helicase preferentially binds forked DNA with a gap on the leading strand
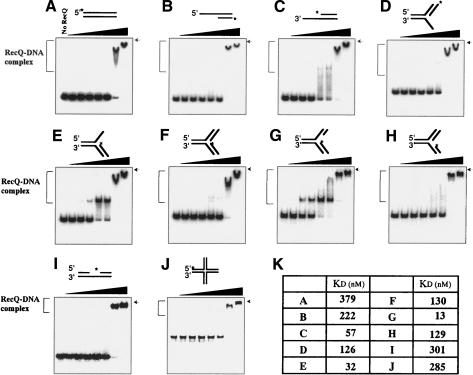
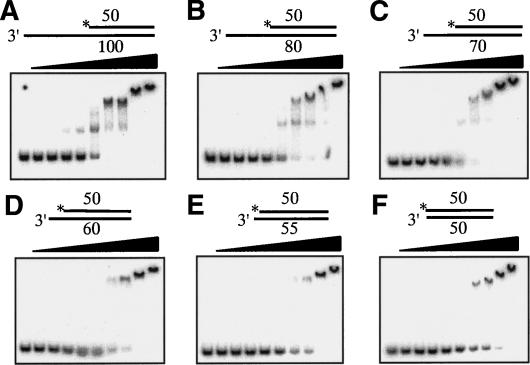
This study examines the interaction between E. coli RecQ helicase and synthetic forked DNA substrates that resemble DNA replication intermediates. DNA binding and DNA helicase activities were measured using 32Plabeled DNA substrates. The substrates included ssDNA and double-stranded DNA (dsDNA) regions of different lengths, or included a Holliday junction structure. DNA substrates were incubated with RecQ at different concentrations, and DNA–RecQ complexes were measured using a gel mobility shift assay. As shown in Figure 1, we observed that two distinct RecQ–DNA complexes were formed with these DNA substrates. Slower migrating complexes were only formed at extremely high concentration and were observed in all DNA substrates used in this study (Figs. 1, 2), indicating the formation of a structure-independent RecQ–DNA complex. These results suggest that RecQ nonspecifically binds DNA at higher protein concentrations and these RecQ–DNA complexes are not physiologically relevant. Faster migrating complexes were observed at lower RecQ concentrations with 3′-overhang, 3′-flap, or leading-strand gapped fork (LeGF) DNA (Fig. 1C,E,G). In complexes involving 3′-overhang DNA, faster migrating species are smeared on the gel, indicating the unstable complexes. Because these complexes were not observed with duplex DNA with 5′-overhang or with blunt ends, junctions between ssDNA and the 5′ terminus of the duplex might be important for the formation of the faster migrating complexes. In addition, they are observed only when the DNA substrate has a 3′-ssDNA region longer than 20 nucleotides (nt) and the binding efficiency increases with increasing the length of the 3′-overhang from 20 to 50 nt (Fig. 2). Thus, RecQ bound 3′-overhang DNA more efficiently than 5′-overhang or blunt-end dsDNA of comparable length, which is consistent with previous studies (Harmon and Kowalczykowski 1998). Moreover, RecQ formed more stable complexes with LeGF and 3′-flap DNA than 3′-overhang DNA, the former mimicking the replication fork structures with ssDNA on the leading-strand DNA (Fig. 1). Especially, the preferential and stable binding to the LeGF DNA substrate (Kd = 13 nM) is not solely due to the presence of ssDNA because it has only an 8-nt ssDNA gap on the leading strand. On the other hand, RecQ bound poorly to lagging-strand gapped fork (LaGF; Kd = 129 nM) and the DNA gap (Kd = 301 nM), both of which have 8-nt DNA gaps also (Fig. 1K). These results demonstrate that RecQ preferentially binds forked DNA with a gap on the leading strand.
Figure 1.
Gel mobility shift analysis of RecQ binding to forked DNA structures. Blunt-end (A), 5′-overhang (B), 3′-overhang (C), 5′-flap (D), 3′-flap (E), forked DNA with no gap (F), LeGF DNA (G), LaGF DNA (H), gapped DNA (I), or HJ DNA (J; 10 nM) was used in binding reactions. RecQ was added at various concentrations (1 nM, 5 nM, 10 nM, 50 nM, 100 nM, 500 nM, and 1000 nM) in the absence of ATP. Protein–DNA complexes were analyzed by 6% nondenaturing PAGE. (*) 32P-labeling. The position of the well on the gel is indicated by arrows. DNA-binding reactions were performed as described in Materials and Methods. (K) Summary of dissociation constants of RecQ with various forms of DNA substrates.
Figure 2.
DNA-binding properties of RecQ depend on ssDNA overhang lengths. Substrates (10 nM) had 3′-overhang lengths of 50 nt (A), 30 nt (B), 20 nt (C), 10 nt (D), 5 nt (E), or 0 nt (F). RecQ concentrations (left to right lanes) were 1 nM, 5 nM, 10 nM, 20 nM, 50 nM, 100 nM, 200 nM, 500 nM, and 1000 nM. (*) 32P-labeling. Experiments are performed as described in Materials and Methods.
Structure-specific unwinding by RecQ helicase
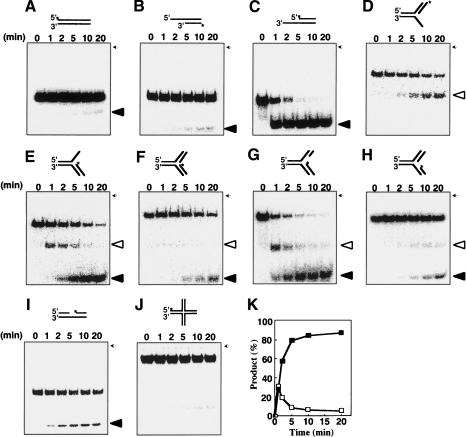
The DNA substrate specificity of RecQ helicase activity was also examined using synthetic forked DNA structures (Fig. 3). At low concentrations of RecQ protein, blunt-ended dsDNA and Holliday junction (HJ) DNA were unwound with very low efficiency (Fig. 3A,J). RecQ unwound DNA substrates with a 3′-overhang significantly more efficiently than DNA substrates with a 5′-overhang (Fig. 3B,C), indicating that the polarity of the RecQ helicase is 3′ to 5′, as previously reported (Umezu et al. 1990). This specificity reflects a preferential affinity of RecQ for 3′-overhang DNA (Fig. 1). Moreover, the kinetics of RecQ helicase activity indicates that it unwinds 3′-flap or LeGF DNA substrates more efficiently than 5′-flap, LaGF, DNA gap, or fork structure without any gap (Fig. 3D–I). When RecQ was added to a reaction containing 3′-flap or LeGF DNA substrate containing 5′-end-labeled nascent lagging strands, a band corresponding to 3′-overhang DNA appeared rapidly (<1 min) and the amount of 3′-overhang DNA substrate decreased as the lagging-strand DNA levels increased (Fig. 3E,G,K). When LeGF DNA substrates containing 5′-end-labeled nascent leading strands were used in this reaction, production of the duplex with 5′-overhang DNA was observed (Supplementary Fig. S1). These results indicate that RecQ efficiently unwinds 3′-flap and LeGF DNA substrates, which is consistent with its binding affinity (Fig. 1) and 3′-to-5′ helicase activity. Moreover, Figure 3E and G demonstrates that RecQ unwinds 3′-flap and LeGF DNA substrates by two distinct steps; RecQ first unwinds the parental strands, translocating 3′ to 5′ ahead of the fork, and subsequently unwinds and releases the nascent lagging strand.
Figure 3.
Time course analysis of RecQ-catalyzed DNA unwinding activity. (A–J) RecQ was incubated at 26°C with blunt-ended (A), 5′-overhang (B), 3′-overhang (C), 5′-flap (D), 3′-flap (E), forked DNA with no gap (F), LeGF DNA (G), LaGF DNA (H), duplex DNA with a gap (I), or HJ DNA (J). Reactions containing 10 nM RecQ, 10 nM DNA, and 1 mM ATP were carried out at 26°C and initiated by the addition of ATP. Aliquots were taken at 1, 2, 5, 10, and 20 min. Deproteinized reaction products except the HJ DNA (9%) were analyzed by 12% nondenaturing PAGE. Open triangles indicate 3′- or 5′-overhang DNA, and closed triangles indicate 32P-labeled ssDNA. (K) The reaction products from G were quantified using a PhosphorImager (Fuji BAS1500) and plotted. Open and filled symbols represent 3′-overhang and 30-mer nascent lagging-strand DNA, respectively. (*) 32P-labeling. The position of the well on the gel is indicated by arrows.
Suppression of thermosensitivity of a dnaE486 mutant by recQ mutation
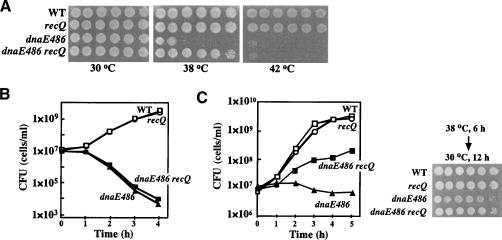
If RecQ is involved in processing stalled DNA replication forks, it is predicted that RecQ will interact genetically with the DNA replication machinery. In E. coli, chromosome replication is performed by DNA polymerase III holoenzyme (Pol III), DNA helicase DnaB, and a DnaG primase. Pol III contains at least 10 different subunits (Kelman and O'Donnell 1995), in which the α-subunit is encoded by the dnaE gene and is responsible for the polymerizing activity of Pol III (Welch and McHenry 1982; Maki et al. 1985). dnaE486 is a mutator allele (S885P; Wechsler and Gross 1971; Konrad 1978; Vandewiele et al. 2002), which causes temperature sensitivity for growth. We studied the effect of recQ mutation on the growth of dnaE486 strains at different temperatures. As shown in Figure 4A and Table 1, wild-type and ΔrecQ cells grew normally at 30°C and 38°C, but dnaE486 cells were severely defective in growth at 38°C. In contrast, we observed that a ΔrecQ::cam mutation suppressed the growth defect of the dnaE486 mutant at 38°C (Fig. 4A; Table 1). ΔrecQ mutation could not suppress the growth defect of the mutant at 42°C (Fig. 4A; Table 1), at which temperature the DNA synthesis activity of Pol III is completely inactivated (Wechsler and Gross 1971). Similar results were also obtained when an recQ61::Tn3 mutation was used (data not shown). This genetic interaction was further confirmed by examining the effect of ΔrecQ mutation on growth rate and cell viability of the dnaE486 mutant at 38°C and 42°C. Cells grown to early logarithmic phase at 30°C were shifted to 38°C or 42°C and plated on LB medium, and then the plates were shifted back to 30°C to allow surviving cells to form colonies (Fig. 4B,C). When incubated at the restrictive temperature (42°C), the plating efficiency of the dnaE486 mutant was rapidly decreased, indicating the loss of viability during prolonged incubation at 42°C (Fig. 4B). The ΔrecQ mutation failed to suppress the lethality of the dnaE486 strain at 42°C. These results suggest that ΔrecQ mutation does not relieve the loss of viability suffered by the DNA Pol III inactivation. In contrast, when incubated at the semirestrictive temperature (38°C), the proportions of dnaE486 cells that could go on to form colonies hardly changed even after culturing for 5 h at 38°C (Fig. 4C), indicating that the dnaE486 cells cease to grow at 38°C, but do not lose viability. Moreover, the absence of RecQ protein evidently suppressed the growth defect of the dnaE486 mutant. These results suggest that RecQ plays a role in stalling the growth of the DNA Pol III mutant cells at 38°C.
Figure 4.
Genetic interaction of RecQ with DNA Pol III. (A) Cells were grown at 30°C in LB medium. Tenfold serial dilutions of cultures (1 × 108 cells/mL) of the indicated mutants were spotted onto LB plates. The plates were incubated at the indicated temperatures. (B) Cells were grown at 30°C to an O.D.600 of 0.2, and the cultures were shifted to 40°C (time 0). Samples were taken at the indicated times, and colony-forming units were counted by plating them on LB plates, which were incubated at 30°C for 24 h. (C) Cells grown at 30°C to an O.D.600 of 0.2 were shifted to 38°C (time 0). (Left) Cells were plated on LB plates following appropriate dilution and incubated for 24 h at 30°C to count colony-forming units. Cells growing at 30°C were spotted as described in A. (Right) The plates were incubated for 6 h at 38°C and then transferred to 30°C.
Table 1.
recQ mutation partially suppresses the temperature sensitivity caused by dnaE486 mutation
| Relevant genotype | 30°Ca | 38°Ca | 42°Ca |
|---|---|---|---|
| wt | 1 | 1.0 ± 0.2 | 1.15 ± 0.25 |
| recQ | 1 | 0.97 ± 0.28 | 1.16 ± 0.2 |
| dnaE486 | 1 | 1.4 × 10-3 ± 0.3 × 10-3 | 3.4 × 10-6 ± 0.2 × 10-6 |
| dnaE486 recQ | 1 | 0.93 ± 0.14 | 3.9 × 10-6 ± 1.1 × 10-6 |
Cells were grown overnight at 30°C, plated on LB plates following appropriate dilutions, and then incubated for 30 h at 30°C, 38°C, or 42°C. Ratios of colony-forming units (CFUs) at 38°C or 42°C relative to CFUs at 30°C are shown
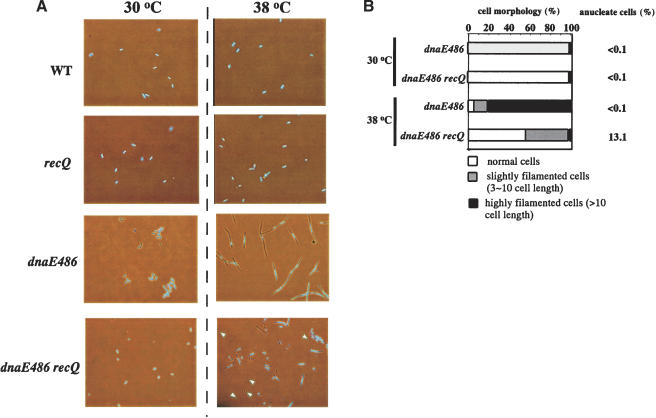
The morphology of the dnaE486 and dnaE486 ΔrecQ cells was analyzed at 6 h after the shift to 38°C (Fig. 5): ∼80% of dnaE486 cells were highly filamented, and the filamentation was much reduced in the dnaE486 ΔrecQ cells. In addition, the ability of ΔrecQ mutation to suppress filamentation in the dnaE486 mutant was associated with appearance of a high proportion of anucleate cells (Fig. 5B). These results are consistent with the interpretation that ΔrecQ mutation allows the replication fork to proceed in the Pol III mutant at 38°C, but that resumption in this situation leads to the loss of genome integrity. Thus, RecQ might interfere with DNA replication fork progression in the Pol III mutant, which may contribute to the faithful recovery of stalled replication forks and thereby the prevention of chromosome loss caused by aberrant DNA replication.
Figure 5.
Morphological analysis of wild-type, ΔrecQ, dnaE486, and dnaE486 ΔrecQ mutant cells. (A) Cells were grown to early log phase at 30°C, and then grown for 6 h at 38°C. Cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) and observed under a phase contrast microscope. White arrowheads highlight some anucleate cells. (B) Distribution of cell sizes. Cells grown for 6 h at 38°C were stained with DAPI. The shapes and chromosomal DNA were monitored under a microscope. Proportions of normal cells are indicated by white bars. Slightly (gray bars) and highly (black bars) filamented cells are defined as cells larger than three and 10 cell lengths in size regardless of the number of nucleoids, respectively. At least 300 cells were examined.
The effect of recQ mutation on SOS induction in response to defective DNA replication
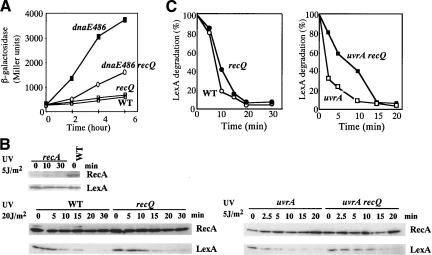
If RecQ helicase generates ssDNA at a stalled replication fork, the ssDNA would recruit RecA and activate an SOS response (Harmon and Kowalczykowski 1998; Humayun 1998), which involves induction of DNA repair genes, inhibition of cell division, and subsequent filamentation. We suggest that the SOS response may be responsible for the cell filamentation observed in the dnaE486 strain at 38°C, and that RecQ plays a role in this process, presumably by increasing the quantity of ssDNA present at stalled replication forks. The RecQ-dependence of the dnaE486-induced SOS response was confirmed by measuring the expression of a UmuC–LacZ fusion protein as a colorimetric indicator of SOS response in dnaE486 and dnaE486 ΔrecQ cells at 38°C. Figure 6A shows that β-galactosidase activity is induced in dnaE486 cells at 6 h after the temperature shift, but it is induced to a significantly lower level in dnaE486 ΔrecQ cells. These results suggest that RecQ functions to generate the signal that activates the SOS response when DNA replication is interfered with, which subsequently blocks cell division and induces repair of DNA damage.
Figure 6.
RecQ is required for proper SOS induction in response to DNA replication interference. (A) Expression of the umuDC::lacZYA fusion gene in the dnaE486 strain grown at the semirestrictive temperature. Overnight cultures of wild-type cells, ΔrecQ, dnaE486, or dnaE486 ΔrecQ mutants were grown at 30°C, diluted into fresh LB medium, and incubated at 38°C. Aliquots were sampled at specific intervals and examined for β-galactosidase activity. Values are averages of the enzyme activities in Miller units of duplicate samples measured after the shift to 38°C. (B) Effect of recQ mutation on UV-induced LexA degradation. Measurements of LexA degradation were performed as described in Materials and Methods. The cells were UV-irradiated (time 0), and aliquots were sampled at intervals. Samples were analyzed by 15% SDS-PAGE, and RecA and LexA proteins were detected by immunoblotting with anti-RecA and anti-LexA antibodies. The reproducibility of the data was confirmed by four independent experiments. (C) The relative amounts of LexA protein shown in B were plotted after they were normalized to the values at time zero. (Open circles) Wild type; (filled circles) recQ61::Tn3; (open squares) uvrA6; (filled squares) uvrA6 recQ61::Tn3.
The role of RecQ in checkpoint signaling was also examined for a UV-induced SOS response. SOS induction was monitored by measuring degradation of LexA, the repressor for the SOS regulon. Figure 6B shows that in wild-type cells, UV-induced LexA degradation was dependent on RecA and was slightly delayed in an recQ61::Tn3 mutant relative to wild-type cells, which was confirmed by quantitative analysis of the amount of LexA protein (Fig. 6C). Because UV-induced DNA lesions are primarily removed from the genome by nucleotide excision repair proteins, SOS signaling was also examined in a uvrA6 mutant, in which DNA replication is frequently blocked and recovery is severely inhibited (Courcelle et al. 1999, 2003). In log-phase cells exposed to 5 J/m2 of UV, LexA degradation was significantly delayed in the uvrA6 recQ61::Tn3 double mutant relative to the uvrA6 single mutant (Fig. 6B,C). These results suggest that RecQ also plays a role in induction of SOS in cells with UV-induced DNA damage.
Discussion
This study characterizes the interaction between E. coli RecQ and forked DNA substrates resembling stalled DNA replication forks and proposes that the action of E. coli RecQ at such structures plays a role in replication restart and in cell cycle checkpoint activation in cells with DNA damage. The evidence for this is as follows: (1) RecQ preferentially binds forked DNA in vitro when the substrate has a gap on the leading strand (Fig. 1). (2) RecQ unwinds the parental strand of such fork moving with a 3′-to-5′ polarity ahead of the fork and subsequently switches back to unwind the lagging strand of a forked substrate in vitro with a 3′-to-5′ polarity releasing the nascent lagging strand (Fig. 3). (3) RecQ is required for a normal SOS signaling in response to DNA replication interference caused by dnaE486 mutation or UV-induced cellular stress. These results suggest that RecQ generates ssDNA gap(s) on the lagging strand of a stalled replication fork; subsequent events (e.g., homologous recombination) promote recovery from DNA damage.
A previous study found that RecQ unwound a diverse set of DNA substrates with equal efficiency, but bound these DNA substrates with different affinities (Harmon and Kowalczykowski 1998). We also found that RecQ bound and unwound all types of DNA substrates used in this study at a high concentration (100 nM), but it did so with different efficiencies at a low concentration (10 nM; Figs. 1, 3; data not shown). It is difficult to compare the previous study with the present study because the assay systems were different; in the previous study, helicase activity was quantified using fluorescent dye displacement, and in this study, the helicase activity was quantified by separating reaction substrate and product by polyacrylamide gel electrophoresis. However, it should be noted that the substrate specificity for RecQ unwinding is largely dependent on RecQ protein concentration. Indeed, it has also been shown that RecQ only partially unwinds dsDNA at low concentration (10 nM), and an increase in the RecQ helicase concentration from 10 nM to 100 nM resulted in a higher rate of DNA unwinding (Harmon and Kowalczykowski 2001). An earlier in vivo study shows that the expression of the recQ gene is regulated by the SOS regulon, and overproduction of RecQ severely inhibits cell growth (Nakayama et al. 1985; Irino et al. 1986) and makes wild-type cells sensitive to UV (data not shown), suggesting that the nonspecific DNA binding and unwinding activity of RecQ observed at higher concentration may interfere with DNA repair and replication. These results support that RecQ has structure specificity for DNA binding and unwinding at physiological concentrations in vivo similar to the one shown in vitro in this study.
Enzymatic and biological functions of E. coli RecQ helicase at a stalled replication fork
This study shows that ΔrecQ mutation suppresses the temperature-sensitive growth defect of dnaE486 cells at 38°C (Fig. 4). However, the dnaE486 ΔrecQ double mutant accumulates anucleate cells and has a lower proportion of viable cells than wild-type or dnaE486 cells (Fig. 4; data not shown). These results suggest that dnaE486 cells that lack RecQ continue to replicate DNA in an unregulated manner at 38°C, so that DNA synthesis by the mutant DNA Pol III causes genomic instability and cell death. In contrast, dnaE486 cells with active RecQ induce SOS response, restrict DNA replication, and become highly filamented. This study also shows that recQ mutation causes a slight delay in UV-induced SOS response, and nucleotide excision repair-deficient uvrA recQ cells have a significant delay in UV-induced SOS response (Fig. 6). Thus, RecQ is required for the efficient induction of SOS in response to replication block. It is possible that the role of RecQ is to generate ssDNA as part of the SOS signaling process. This would activate RecA, promote cleavage of LexA, induce DNA repair, and inhibit cell division (Humayun 1998). This possibility is consistent with the results that the RecF pathway is necessary for the initiation of recombination repair of ssDNA gaps and that RecQ helicase and RecJ exonuclease cooperatively promote degradation of nascent DNA on the lagging strand at blocked replication forks, thereby allowing RecA to bind to exposed ssDNA (Harmon and Kowalczykowski 1998; Courcelle and Hanawalt 1999; Morimatsu and Kowalczykowski 2003). Although we have not examined the effects of recJ mutation, we observed that a ΔrecA mutation also suppressed the temperature sensitivity and filamentation of the dnaE486 mutant at 38°C (T. Hishida, T. Shibata, and H. Shinagawa, unpubl.), consistent with our conclusion that SOS induction is responsible for the growth arrest and filamentation in the dnaE486 strain grown at 38°C.
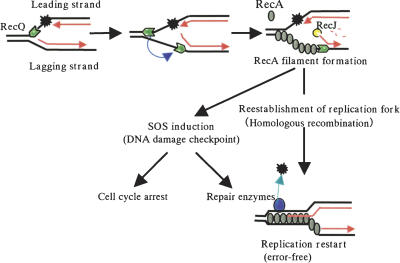
Based on the evidence discussed above, we propose a model for the role of RecQ in replication restart and cell cycle signaling (Fig. 7). This model includes two distinct RecQ-dependent steps. In the first step, RecQ binds an ssDNA gap on the leading strand of a DNA replication fork and unwinds parental dsDNA in front of the fork. In the second step, RecQ switches to the lagging strand and unwinds nascent lagging-strand DNA. On both strands, RecQ moves in the 3′-to-5′ direction. This model predicts that RecQ processes gapped forks into DNA structures that serve to activate RecA and induce the SOS response, which leads to a delay in cell growth, prevention of replication and chromosome segregation, and the expression of specific repair genes. It remains to be determined whether the same RecQ molecule tracks on both strands or the two reactions involves two RecQ molecules. However, this model gives an answer to the questions about how RecQ helicase resolves structures ahead of the fork that impede fork movement (e.g., G4 DNA) and how RecQ degrades nascent DNA on the lagging strand in collaboration with RecJ nuclease. If RecQ acts in vivo in a manner similar to that suggested by our in vitro data, we predict that the unwinding activity of RecQ affects DNA topology and requires the coordinated activity of a DNA topoisomerase because there are no dsDNA ends. It has been shown that the RecQ family of helicases and topoisomerase III (Top III) functionally interacts in many organisms (Gangloff et al. 1994; Harmon et al. 1999; Bennett et al. 2000; Wu et al. 2000). Indeed, recent biochemical evidence has revealed that BLM and Topo III α of human and RecQ helicase and Topo III of E. coli work together to comprise a potent and concerted single-strand DNA passage activity and changes topology of DNA in vitro (Harmon et al. 1999, 2003; Wu and Hickson 2003). Thus, RecQ and Topo III might act in a coordinated manner to process stalled replication forks, resolving topological hinderance and/or decatenating precatenanes that accumulate behind the replication fork during DNA replication. Future studies will be required to investigate the joint action of RecQ and Topo III at stalled forks.
Figure 7.
A model for the roles for RecQ in the rescue of a stalled replication fork. RecQ binds an ssDNA gap on the leading strand and unwinds the template dsDNA ahead of the fork, which might generate positive supercoiling and contribute to the elimination of structures that impede fork movement (e.g., G4 DNA structure). Subsequently, RecQ switches to the lagging strand and creates an ssDNA region by unwinding the lagging strand, which may be an initiating signal for RecA-dependent SOS induction and recombination repair. SOS induction in response to DNA replication interference leads to a delay in cell growth, prevention of replication and chromosome segregation, and the induction of specific repair genes. See text for further details. Red arrowheads indicate the 3′-OH groups of the leading and lagging strands.
Evidence shown here indicates that RecQ is required for the recovery of stalled replication forks. It should be noted that deficiency in RecQ does not cause reduced viability or UV sensitivity (data not shown). These results suggest that not all stalled replication forks are rescued by RecQ, and that stalled DNA replication forks can be rescued by several mechanisms. If RecQ is capable of converting the ssDNA gaps on the fork from the leading-strand DNA to lagging-strand DNA, RecQ may affect the polarity of RecA-catalyzed strand exchange. Indeed, RecA can promote regression of forks with a leading-strand gap but not with a lagging-strand gap in vitro (Robu et al. 2001). Deficiency in RecQ may therefore lead to accumulation of leading-strand gaps, which are substrates for RecA-dependent fork regression. In this case, RecQ may unwind a stalled replication fork to prevent it from becoming a Holliday junction by the regression of a stalled fork with a leading-strand gap, rather than promoting resolution of the junction by reverse branch migration.
In this study, we aimed at determining the role of RecQ at stalled replication forks in vivo and in vitro, and found that RecQ preferentially binds and unwinds the LeGF structure. However, RecQ helicase action may not be solely confined to the LeGF DNA because RecQ also unwinds 3′-overhang DNA (Fig. 3). RecQ is a multifunctional enzyme that acts as an initiator of homologous recombination and a suppressor of illegitimate recombination (Ryder et al. 1994; Hanada et al. 1997; Harmon and Kowalczykowski 1998). Therefore, the above model represents the simplest explanation for the role of RecQ in the rescue of stalled replication forks. RecQ may be involved in other processes like resolution of entangled chromosomes or aberrant recombination events.
Conserved functions of RecQ family helicases at stalled replication forks
This study suggests that E. coli RecQ generates ssDNA in cells with a DNA replication defect as part of a cell signaling process that induces SOS response, activates a cell cycle checkpoint, and subsequently permits DNA replication to resume. The eukaryotic homologs of RecQ, including Sgs1, Rqh1, BLM, and WRN, may also perform similar functions. In the yeast S. cerevisiae, sgs1 mutation is associated with a hyperrecombination phenotype and hypersensitivity to HU and MMS in S phase (Gangloff et al. 1994; Mullen et al. 2000). Sgs1 is required for HU-induced activation of Rad53 as a sensor protein, which acts in parallel with Rad24 (Frei and Gasser 2000). In human cells, WRN and RPA colocalize in discrete nuclear foci and interact with p53 when cell growth is arrested (Spillare et al. 1999; Constantinou et al. 2000; Sakamoto et al. 2001). BLM is part of the BASC complex (BRCA-1-associated genome surveillance complex), and BRCA1, BLM helicase, and the RAD50–MRE11–NBS1 complex colocalize to large nuclear foci that contain PCNA when DNA synthesis is inhibited (Wang et al. 2000). Recent study shows that BLM interacts with RAD51 and transports p53 to stalled replication forks (Sengupta et al. 2003). Moreover, it has been reported that ssDNA coated by replication protein A (RPA, a eukaryotic ssDNA-binding protein) is a key structure for checkpoint activation in response to double-strand breaks and replication interference (Zou and Elledge 2003). These findings suggest that E. coli RecQ and eukaryotic RecQ homologs play similar roles in replication-associated homologous recombination and checkpoint activation: ssDNA produced by RecQ helicase activity may be the initiating signal for the checkpoint response in both bacteria and eukaryotes.
The work presented here has uncovered a role of the E. coli RecQ helicase in the integration of the checkpoint response with DNA recombination and DNA replication and provides important clues to understanding the complex interplay between DNA replication, recombination, and checkpoint functions, especially at stalled replication forks.
Materials and methods
Media and growth condition
Standard methods for E. coli genetics and recombinant DNA techniques were as described by Miller (1992) and Sambrook et al. (1989). Unless otherwise stated, cells were grown in Luria broth (LB) or M9 salts-glucose minimal medium (Miller 1992) supplemented with appropriate amino acids and thiamine. Ampicillin (50 μg/mL), tetracycline (10 μg/mL), and kanamycin (30 μg/mL) were used as needed.
Bacterial strains and plasmids
Bacterial strains are listed in Table 2. A ΔrecQ::cat allele was constructed by replacing the SacII–ClaI fragment of the recQ coding region with the chloramphenicol-resistance gene, and it was introduced into an recD strain (D301) as described (Russell et al. 1989). The gene disruption was confirmed by PCR using appropriate primers. The isogenic derivatives of AB1157 strain were constructed by transduction with P1 phage (Miller 1992). pLysS and pSQ211 were used for the overexpression of RecQ (Harmon and Kowalczykowski 1998). pSK1002 was used for SOS induction assays (Shinagawa et al. 1983).
Table 2.
E. coli strains
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| AB1157 | rec+, ruv+, gyr+, dna | Bachmann 1972 |
| E486 | dnaE486 | Wechsler and Gross 1971, NIGa |
| KD1996 | recQ61::Tn3, polA12 | NIG |
| OH1000 | AB1157 but ΔrecQ::Cam | Lab stocks |
| TS1504 | AB1157 but recQ61::Tn3 | AB1157 × P1 KD1996 |
| CAG12204 | Hfr(PO3)btuB3192::Tn10kan | Singer et al. 1989 |
| CAG18436 | zae-502::Tn10 | Singer et al. 1989, NIG |
| HRS2006 | AB1157 but Δ(srl-recA)306::Tn10 | Lab stocks |
| TS1502 | AB1157 but dnaE486 zae-502::Tn10 | AB1157 × P1 TS1501 |
| TS1507 | TS1502 but ΔrecQ::Cam | TS1502 × P1 OH1000 |
| AB1886 | AB1157 but uvrA6 | Brandsma et al. 1988, NIG |
| TS1520 | AB1157 but uvrA6, recQ61::Tn3 | AB1886 × P1 TS1504 |
| TS1501 | E486 but zae-502::Tn10 | E486 × P1 CAG18436 |
NIG: NIG (National Institute of Genetics, Japan) collection
Purification of recombinant RecQ protein
The E. coli BL21 (DE3) strain carrying pLysS and pSQ211 was grown at 37°C to an O.D.600 of ∼0.4 in Luria broth containing ampicillin. Isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 1 mM, and the culture was incubated for 18 h at 18°C. After the cells were harvested, the cells were resuspended in buffer A (20 mM Tris-HCl at pH 8.0, 1 mM EDTA, 100 mM NaCl, 0.1% Triton, 1 mM dithiothreitol [DTT], 10% Glycerol). They were disrupted by sonication and centrifuged at 40,000 × g for 30 min. Polyethyleneimine was added to a final concentration of 0.05%, and the mixture was stirred for 30 min and then centrifuged at 27,000 × g for 15 min. Ammonium sulfate was gradually added to the supernatant to achieve 40% saturation. The mixture was stirred for 30 min and then centrifuged at 27,000 × g for 15 min. The pellet was resuspended in RQ-buffer (20 mM Tris-HCl at pH 8.0, 1 mM EDTA, 1 mM DTT) containing 1 M ammonium sulfate, then centrifuged at 27,000 × g for 15 min, and the pellet was removed. The supernatant was loaded onto HiLoad 16/10 Phenyl Sepharose HP (Amersham Pharmacia), and the proteins were eluted with a 100-mL linear gradient from 1 M to 0 M ammonium sulfate in RQ buffer. The fractions containing RecQ protein were diluted twofold with RQ buffer and loaded onto a Hi-Trap Heparin column (Amersham Pharmacia). The proteins were eluted with a 100-mL linear gradient from 0 M to 1 M NaCl in RQ buffer. RecQ eluted at 300 mM NaCl. The RecQ protein peak fractions were applied to a Superdex 200 column (Amersham Pharmacia). The RecQ protein peak fractions were applied to a Hydroxyapatite column (Bio-Rad), and the proteins were eluted with a 100-mL linear gradient from 20 mM to 600 mM phosphate in phosphate buffer (phosphate at pH 6.8, 1 mM EDTA, 1 mM DTT). The RecQ protein peak fractions were applied to Hi-Trap Heparin to concentrate RecQ protein. The RecQ protein thus purified showed a single band on SDS-PAGE (Supplementary Fig. S2). Protein concentration was determined by the Bradford assay.
Synthetic DNA substrates
Oligonucleotides 1 (F3-d47), 2 (49N-2), 3 (d22), 4 (d20), and 5 (d28) were described previously (Komori et al. 2002). The sequences of oligonucleotides 6(49R) and 7(d30-2) are 5′-AGC TATGACCATGATTACGAATTGCTTAATTCGTGCAGGC ATGGTAGCT-3′ and 5′-CTGATACGAATTCGTGCAGGCA TGGTAGCT-3′, respectively. The 5′-overhang, 3′-overhang, 5′-flap, 3′-flap, and blunt end DNAs were made from oligonucleotides 1 + 4, 2 + 3, 1 + 2 + 4, 1 + 2 + 3, and 2 + 6, respectively. Forked DNAs with no gap or with ssDNA on the leading or lagging strand were made from 1 + 2 + 5 + 7, 1 + 2 + 4 + 7, and 1 + 2 + 3 + 5, respectively. The Holliday junction was constructed as described previously (Hishida et al. 1999).
Helicase assay
Reaction mixtures (20 μL) contained RecQ (10 nM), substrate DNA (10 nM), 1 mM ATP, 20 mM Tris-acetate (pH 8.0), 1 mM DTT, and 1 mM Mg-acetate. After incubation for the indicated time at 26°C, reactions were stopped with 5 μL of stop buffer (50 mM Tris-HCl at pH 7.5, 100 mM EDTA, 1% SDS, and 7.5 mg/mL proteinase K) and incubated for 10 min. Reaction products were analyzed by electrophoresis on a 12% polyacrylamide gel (PAGE), except the Holliday junction (9% PAGE), and visualized using a Fuji BAS2000 image analyzer.
DNA-binding assay
Reaction mixtures contained 20 mM Tris-acetate (pH 8.0), 1 mM DTT, 10 nM substrate DNA, and the indicated concentration of RecQ protein. Reactions were incubated at 37°C for 10 min. Products were analyzed on 6% PAGE in TAE buffer and visualized using a Fuji BAS2000 image analyzer. Binding affinity was calculated as the reciprocal of the RecQ concentration at which half the available DNA was bound and half was unbound.
SOS induction
Induction of the SOS response was monitored by measuring synthesis of β-galactosidase (Miller 1992) in strains harboring a umuDC::lacZYA fusion gene on pSK1002 (Shinagawa et al. 1983). Enzyme units are given as a fraction of the O.D.600 of the culture at the time of sampling. The detection of LexA degradation was carried out as described previously (Yasuda et al. 1998). Log-phase cultures were treated with chloramphenicol (100 μg/mL) to inhibit the resynthesis of LexA and incubated for 10 min at 37°C. Then, the cells were UV-irradiated (time 0), and aliquots were sampled at intervals. Samples were analyzed by 15% SDS-PAGE, and RecA and LexA proteins were detected by immunoblotting with anti-RecA and anti-LexA antibodies. The recQ61::Tn3 strain was used in this assay because the ΔrecQ::cam strain is resistant to chloramphenicol.
Western blots
Cells (1 × 108 cells/mL) were lysed in lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% SDS, and bacterial protease inhibitor cocktail (Sigma). After boiling, aliquots of the sample were applied to 15% SDS-PAGE gels. The RecA and LexA proteins were detected with rabbit antibodies raised against RecA and LexA, using the ECL Western blotting analysis system (Amersham Pharmacia).
Morphological analysis
The cells were grown in liquid LB to early logarithmic phase (1 × 107 cells/mL) at 30°C, and grown for 6 h at 38°C.Cellswere harvested, fixed with methanol, and then stained with DAPI (4′,6,-diamino-2-phenylindole). Cell morphology was monitored under a fluorescence microscope (Zeiss Axioplan2) as previously described (Ishioka et al. 1998).
Acknowledgments
We thank B. Michel, K. Umezu, J. Courcelle, J.M. Murray, and R.G. Lloyd for their critical reading of this manuscript. Y.-W.H. was supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists. This work was supported by Grants in Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan to H.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1223804.
Corresponding authors.
References
- Bachmann B.J. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36: 525-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C.Z. and Hickson, I.D. 2003. RecQ helicases: Suppressors of tumorigenesis and premature ageing. Biochem. J. 374 (Pt 3): 577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.J., Noirot-Gros, M.F., and Wang, J.C. 2000. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275: 26898-26905. [DOI] [PubMed] [Google Scholar]
- Brandsma J.A., de Ruijter, M., Brouwer, J., and van de Putte, P. 1988. Identification of the uvrA6 mutation of Escherichia coli. J. Bacteriol. 170: 1012-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek, L., and Gasser, S.M. 2002. RecQ helicases: At the heart of genetic stability. FEBS Lett. 529: 43-48. [DOI] [PubMed] [Google Scholar]
- Constantinou A., Tarsounas, M., Karow, J.K., Brosh, R.M., Bohr, V.A., Hickson, I.D., and West, S.C. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and colocalizes with RPA upon replication arrest. EMBO Rep. 1: 80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J. and Hanawalt, P.C. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262: 543-551. [DOI] [PubMed] [Google Scholar]
- Courcelle J., Carswell-Crumpton, C., and Hanawalt, P.C. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. 94: 3714-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Crowley, D.J., and Hanawalt, P.C. 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol. 181: 916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Donaldson, J.R., Chow, K.H., and Courcelle, C.T. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064-1067. [DOI] [PubMed] [Google Scholar]
- Doe C.L., Dixon, J., Osman, F., and Whitby, M.C. 2000. Partial suppression of the fission yeast rqh1- phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19: 2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N.A., Groden, J., Ye, T.Z., Straughen, J., Lennon, D.J., Ciocci, S., Proytcheva, M., and German, J. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83: 655-666. [DOI] [PubMed] [Google Scholar]
- Frei C. and Gasser, S.M. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes & Dev. 14: 81-96. [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald, J.P., Bendixen, C., Arthur, L., and Rothstein, R. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Ukita, T., Kohno, Y., Saito, K., Kato, J., and Ikeda, H. 1997. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. 94: 3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F.G. and Kowalczykowski, S.C. 1998. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes & Dev. 12: 1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem. 276: 232-243. [DOI] [PubMed] [Google Scholar]
- Harmon F.G., DiGate, R.J., and Kowalczykowski, S.C. 1999. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol. Cell 3: 611-620. [DOI] [PubMed] [Google Scholar]
- Harmon F.G., Brockman, J.P., and Kowalczykowski, S.C. 2003. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J. Biol. Chem. 278: 42668-42678. [DOI] [PubMed] [Google Scholar]
- Hishida T., Iwasaki, H., Yagi, T., and Shinagawa, H. 1999. Role of walker motif A of RuvB protein in promoting branch migration of Holliday junctions. Walker motif a mutations affect ATP binding, ATP hydrolyzing, and DNA binding activities of RuvB. J. Biol. Chem. 274: 25335-25342. [DOI] [PubMed] [Google Scholar]
- Horii Z. and Clark, A.J. 1973. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: Isolation and characterization of mutants. J. Mol. Biol. 80: 327-344. [DOI] [PubMed] [Google Scholar]
- Huber M.D., Lee, D.C., and Maizels, N. 2002. G4 DNA unwinding by BLM and Sgs1p: Substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 30: 3954-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humayun M.Z. 1998. SOS and Mayday: Multiple inducible mutagenic pathways in Escherichia coli. Mol. Microbiol. 30: 905-910. [DOI] [PubMed] [Google Scholar]
- Irino N., Nakayama, K., and Nakayama, H. 1986. The recQ gene of Escherichia coli K12: Primary structure and evidence for SOS regulation. Mol. Gen. Genet. 205: 298-304. [DOI] [PubMed] [Google Scholar]
- Ishioka K., Fukuoh, A., Iwasaki, H., Nakata, A., and Shinagawa, H. 1998. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells 3: 209-220. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Constantinou, A., Li, J.L., West, S.C., and Hickson, I.D. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. 97: 6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z. and O'Donnell, M. 1995. DNA polymerase III holoenzyme: Structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64: 171-200. [DOI] [PubMed] [Google Scholar]
- Kitao S., Shimamoto, A., Goto, M., Miller, R.W., Smithson, W.A., Lindor, N.M., and Furuichi, Y. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22: 82-84. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Fishel, R.A., and Howard, M. 1985. Genetic recombination of bacterial plasmid DNA: Effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J. Bacteriol. 163: 1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K., Fujikane, R., Shinagawa, H., and Ishino, Y. 2002. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet. Syst. 77: 227-241. [DOI] [PubMed] [Google Scholar]
- Konrad E.B. 1978. Isolation of an Escherichia coli K-12 dnaE mutation as a mutator. J. Bacteriol. 133: 1197-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Horiuchi, T., and Kornberg, A. 1985. The polymerase subunit of DNA polymerase III of Escherichia coli. I. Amplification of the dnaE gene product and polymerase activity of the α subunit. J. Biol. Chem. 260: 12982-12986. [PubMed] [Google Scholar]
- Miller J.H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mohaghegh P., Karow, J.K., Brosh Jr., R.M., Bohr, V.A., and Hickson, I.D. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29: 2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K. and Kowalczykowski, S.C. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol. Cell 11: 1337-1347. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman, V., and Brill, S.J. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.M., Lindsay, H.D., Munday, C.A., and Carr, A.M. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17: 6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K., Irino, N., and Nakayama, H. 1985. The recQ gene of Escherichia coli K12: Molecular cloning and isolation of insertion mutants. Mol. Gen. Genet. 200: 266-271. [DOI] [PubMed] [Google Scholar]
- Robu M.E., Inman, R.B., and Cox, M.M. 2001. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl. Acad. Sci. 98: 8211-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.B., Thaler, D.S., and Dahlquist, F.W. 1989. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J. Bacteriol. 171: 2609-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder L., Whitby, M.C., and Lloyd, R.G. 1994. Mutation of recF, recJ, recO, recQ, or recR improves Hfr recombination in resolvase-deficient ruv recG strains of Escherichia coli. J. Bacteriol. 176: 1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Nishikawa, K., Heo, S.J., Goto, M., Furuichi, Y., and Shimamoto, A. 2001. Werner helicase relocates into nuclear foci in response to DNA damaging agents and colocalizes with RPA and Rad51. Genes Cells 6: 421-430. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sengupta S., Linke, S.P., Pedeux, R., Yang, Q., Farnsworth, J., Garfield, S.H., Valerie, K., Shay, J.W., Ellis, N.A., Wasylyk, B., et al. 2003. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 22: 1210-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H., Kato, T., Ise, T., Makino, K., and Nakata, A. 1983. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene 23: 167-174. [DOI] [PubMed] [Google Scholar]
- Singer M., Baker, T.A., Schnitzler, G., Deischel, S.M., Goel, M., Dove, W., Jaacks, K.J., Grossman, A.D., Erickson, J.W., and Gross, C.A. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53: 1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillare E.A., Robles, A.I., Wang, X.W., Shen, J.C., Yu, C.E., Schellenberg, G.D., and Harris, C.C. 1999. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes & Dev. 13: 1355-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Karow, J.K., Hickson, I.D., and Maizels, N. 1998. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273: 27587-27592. [DOI] [PubMed] [Google Scholar]
- Tseng Y.C., Hung, J.L., and Wang, T.C. 1994. Involvement of RecF pathway recombination genes in postreplication repair in UV-irradiated Escherichia coli cells. Mutat. Res. 315: 1-9. [DOI] [PubMed] [Google Scholar]
- Umezu K., Nakayama, K., and Nakayama, H. 1990. Escherichia coli RecQ protein is a DNA helicase. Proc. Natl. Acad. Sci. 87: 5363-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewiele D., Fernandez de Henestrosa, A.R., Timms, A.R., Bridges, B.A., and Woodgate, R. 2002. Sequence analysis and phenotypes of five temperature sensitive mutator alleles of dnaE, encoding modified α-catalytic subunits of Escherichia coli DNA polymerase III holoenzyme. Mutat. Res. 499: 85-95. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S.J., and Qin, J. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 14: 927-939. [PMC free article] [PubMed] [Google Scholar]
- Wechsler J.A. and Gross, J.D. 1971. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol. Gen. Genet. 113: 273-284. [DOI] [PubMed] [Google Scholar]
- Welch M.M. and McHenry, C.S. 1982. Cloning and identification of the product of the dnaE gene of Escherichia coli. J. Bacteriol. 152: 351-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. and Hickson, I.D. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870-874. [DOI] [PubMed] [Google Scholar]
- Wu X. and Maizels, N. 2001. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 29: 1765-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Davies, S.L., North, P.S., Goulaouic, H., Riou, J.F., Turley, H., Gatter, K.C., and Hickson, I.D. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275: 9636-9644. [DOI] [PubMed] [Google Scholar]
- Yasuda T., Morimatsu, K., Horii, T., Nagata, T., and Ohmori, H. 1998. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 17: 3207-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.E., Oshima, J., Fu, Y.H., Wijsman, E.M., Hisama, F., Alisch, R., Matthews, S., Nakura, J., Miki, T., Ouais, S., et al. 1996. Positional cloning of the Werner's syndrome gene. Science 272: 258-262. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542-1548. [DOI] [PubMed] [Google Scholar]