Abstract
In recent years, an increase of uropathogenic Escherichia coli (UPEC) strains with Multidrug-resistant (MDR) and Extensively Drug-resistant (XDR) profiles that complicate therapy for urinary tract infections (UTIs) has been observed and has directly impacted costs and extended hospital stays. The aim of this study was to determine MDR- and XDR-UPEC clinical strains, their virulence genes, their phylogenetic groups and to ascertain their relationship with integrons and genetic diversity. From a collection of 500 UPEC strains, 103 were selected with MDR and XDR characteristics. MDR-UPEC strains were mainly associated with phylogenetic groups D (54.87%) and B2 (39.02%) with a high percentage (≥70%) of several fimbrial genes (ecpA, fimH, csgA, and papGII), an iron uptake gene (chuA), and a toxin gene (hlyA). In addition, a moderate frequency (40–70%) of other genes (iutD, tosA, and bcsA) was observed. XDR-UPEC strains were predominantly associated with phylogenetic groups B2 (47.61%) and D (42.85%), which grouped with ≥80 virulence genes, including ecpA, fimH, csgA, papGII, iutD, and chuA. A moderate frequency (40–70%) of the tosA and hlyA genes was observed. The class 1 and 2 integrons that were identified in the MDR- and XDR-UPEC strains were associated with phylogenetic groups D, B2, and A, while the XDR-UPEC strains that were associated with phylogenetic groups B2, D, and A showed an extended-spectrum beta-lactamase (ESBL) phenotype. The modifying enzymes (aadA1, aadB, aacC, ant1, dfrA1, dfrA17, and aadA4) that were identified in the variable region of class 1 and 2 integrons from the MDR strains showed resistance to gentamycin (56.25 and 66.66%, respectively) and trimethoprim-sulfamethoxazole (84.61 and 66.66%, respectively). The MDR- and XDR-UPEC strains were distributed into seven clusters and were closely related to phylogenic groups B2 and D. The diversity analysis by PFGE showed 42.68% of clones of MDR-UPEC and no clonal association in the XDR-UPEC strains. In conclusion, phylogenetic groups including virulence genes are widely associated with two integron classes (1 and 2) in MDR- and XDR-UPEC strains.
Keywords: UPEC, multidrug resistance, virulence genes, phylogenetic groups, PFGE
Introduction
Urinary tract infections (UTIs) are one of the most common bacterial infectious diseases in humans, with ~150 million cases estimated annually worldwide (Flores-Mireles et al., 2015). Uropathogenic Escherichia coli (UPEC) causes 80–90% of community-acquired UTIs and 40–50% of nosocomial-acquired UTIs (Foxman, 2010; Foxman et al., 2012; Toval et al., 2014; Flores-Mireles et al., 2015). UTIs associated with UPEC usually begin as bladder infections (cystitis) but can develop into acute kidney infections (pyelonephritis) and even infections of the bloodstream (urosepsis) (Flores-Mireles et al., 2015). UPEC pathogenesis involves several virulence factors to resist urine flow, to trigger host-bacterial cell signaling pathways, and to establish infection (Siliano et al., 2010; Jadhav et al., 2011; Alteri and Mobley, 2012). FimH (Type 1 fimbriae), EcpA (E. coli Common Pilus), CsgA protein (curli), PapGI, PapGII, and PapGIII variants (P fimbriae) are fimbrial adhesins that participate in UPEC adherence and colonize the bladder epithelium (Mulvey et al., 1998; Lane and Mobley, 2007; Cegelski et al., 2009; Saldaña et al., 2014). Iron uptake proteins (aerobactin, IutD), toxin protein (α-hemolysin, HlyA), type 1 secretion A (TosA), and surface glycan proteins (cellulose and BcsA) participate in UPEC pathogenesis (Gao et al., 2012; Kudinha et al., 2013; Engstrom et al., 2014; Lüthje and Brauner, 2014; Subashchandrabose and Mobley, 2015). UPEC clinical strains are associated with four main phylogenetic groups (A, B1, B2, and D) that are characterized by the existence of genetic markers, such as chuA, yjaA, and the DNA fragment TspE4.C2 (Lee et al., 2016). Phylogenetic groups B2 and D are commonly related to UPEC strains collected from patients with UTIs; however, at a lower frequency, these strains are also related to phylogenetic groups A and B1. In addition, multidrug-resistance (MDR) and virulence factors described in UPEC strains are associated with phylogenetic group B2 (Derakhshandeh et al., 2015; Ferjani et al., 2015). Recently, the MDR-phenotype was found to be related to phylogenetic groups A and D; although, UPEC strains susceptible to most antibiotics are also related to phylogenetic group B2 (Ejrnæs et al., 2011).
The mechanisms that contribute to resistance in UPEC strains are as follows: (1) inactivation of hydrolytic enzymes by β-lactamases; (2) no hydrolytic enzyme inactivation by aminoglycoside acetyl transferase enzymes; (3) permeability alteration through active efflux pumps; (4) inactivation of the target site; and (5) resistance acquired by the horizontal transfer of genetic elements, such as insertion sequences, gene cassettes, integrons, and transposons (Peleg and Hooper, 2010). Integrons generally constitute an integrase gene (intI), an attachment site (attI), and a promoter (Pc) that induces the expression of any suitable integrated gene(s). Additionally, two classes of integrons (classes 1 and 2) have been identified and characterized in MDR-UPEC clinical strains (Stalder et al., 2012; Deng et al., 2015). These integrons carry several genes that encode proteins that participate in antibiotic resistance. These integrons are frequently embedded in mobile DNA elements, such as transposons and conjugative plasmids, to spread horizontally throughout bacterial populations (Stalder et al., 2012; Deng et al., 2015). However, the integrons in MDR-UPEC clinical strains and their relationship with virulence factors and clonality have been poorly studied. In recent years, an increase in UPEC strains with an extensively drug-resistant (XDR) profile complicating UTI therapy has been observed (Dehbanipour et al., 2016; Hadifar et al., 2016). The aim of this study was to identify MDR- and XDR-UPEC clinical strains and their virulence genes and phylogenetic groups and to ascertain their relationship with integrons and their genetic diversity.
Materials and methods
Bacterial strains
A total of 500 UPEC clinical strains were collected from 2010 to 2012 at the Hospital Infantil de México Federico Gómez (HIMFG) from pediatric patients with community- and nosocomial-acquired UTIs. The UPEC strains were selected considering an average bacterial count of ≥100,000 CFU/mL as determined by Kass-Sandford (Kass, 1957). UPEC strains associated with persistent UTIs were isolated from cultures from the same patient taken in different months. UPEC strains were cultured in trypticase soy broth (TSA), MacConkey medium (Mac), Luria-Bertani medium (LB), and Mueller-Hinton Broth (MHB) (Difco-Becton Dickinson, NJ, USA) at 37°C for 24 h according to assay type. Additionally, UPEC strains were stored in cryovials at −70°C with Brain Heart Infusion broth (BHI; Difco-Becton, Dickinson, NJ, USA) containing 25% glycerol (Sigma-Aldrich, St. Louis, MO, USA). Only one isolate from each infectious episode was included in the study, and isolates were considered to come from the same episode if they were collected <30 days apart. Thus, all MDR- and XDR-UPEC strains were isolated from 103 infectious episodes in 71 patients, and 8 patients experienced more than one episode.
Antimicrobial susceptibility and phenotypic detection of ESBLs
The minimum inhibitory concentration (MIC) as determined by the MHB microdilution method was used to evaluate the antimicrobial susceptibility of 500 UPEC clinical strains according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2016). MDR strains were defined as having acquired no susceptibility to at least one antibiotic in three or more classes. XDR strains were defined as having non-susceptibility to at least one agent in all but two or fewer antibiotic classes (Magiorakos et al., 2012). The MIC for each antibiotic was compared to the standard values of the CLSI. The antibiotic panel that was used included ampicillin (AM; Sigma-Aldrich, St. Louis, MO, USA), amoxicillin-clavulanate (AMC; Great West Road, Brentford Middlesex, UK), ticarcillin-clavulanate (TIM; Gold Biotechnology, Inc., Ashby Road, St. Louis, MO), piperacillin-tazobactam (TZP; Siemens Medical Solutions USA, Inc., Valley Stream Parkway, Malvern, PA, USA), cephalothin (CF; Eli Lilly and Company, S Harding St, Indianapolis, IN, USA), cefaclor (CEC; Phadia Laboratory Systems, Thermo Scientific, Wyman Street, Waltham, MA, USA), ceftazidime (CAZ; Roselle Rd, Schaumburg, IL, USA), aztreonam (ATM; Bristol-Myers Squibb Corporate, Park Avenue, NY, USA), norfloxacin (NOR), ofloxacin (OFX; MP Biomedicals, Solon, OH, USA), meropenem (MEM), imipenem (IPM; AstraZeneca Pharmaceuticals LP, Wilmington, DE, USA), gentamycin (GM; Schering-Plough Pharmaceuticals, Kenilworth, NJ, USA), ceftriaxone (CRO), trimethoprim-sulfamethoxazole (SXT; Roche, Basel, Switzerland), tetracycline (TE; Heritage Pharmaceuticals Inc., Fieldcrest Avenue, Edison, NJ, USA), and nitrofurantoin (F/M; McKesson Pharmaceutical, One Post Street, San Francisco, CA, USA). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls.
The extended-spectrum beta-lactamases (ESBLs) were phenotypically detected as previously recommended by CLSI using the double-disc synergy test based on the synergistic effect between clavulanic acid (inhibitor of ESBLs) and β-lactam antibiotics (cefotaxime, CRO, CAZ, cefepime, cefpirome, and ATM). Additionally, ESBLs were detected using an individual disk that was tested with/without clavulanic acid (10 μg/mL) and by the Hodge test using Klebsiella pneumoniae ATCC 700603 (ESBL+) and E. coli ATCC 25922 (ESBL-) as control strains (CLSI, 2016).
Phylogenetic groups
DNA was extracted from the MDR- and XDR-UPEC clinical strains cultured in LB using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Woods Hollow Road, Madison, WI, USA) according to the manufacturer's instructions. Multiplex polymerase chain reaction (PCR) assays were used to determine the presence of chuA, yjaA, and one genomic fragment (TspE4.C2) (Clermont et al., 2000) in MDR- and XDR-UPEC clinical strains (Table S1). The PCR products were resolved by electrophoresis on a 1.5% agarose gel (Promega Corporation, Woods Hollow Road, Madison, WI, USA) that was stained with ethidium bromide solution (Sigma-Aldrich, St. Louis, MO, USA) and visualized using a Gel Imaging System (ChemiDoc MP System, Bio-Rad; Mexico). According to the amplicon data, the strains were assigned to phylogenetic groups as follows: group B2 (chuA+/yjaA+), group D (chuA+/yjaA−), group B1 (chuA−/TspE4.C2+), and group A (chuA−/TspE4.C2−) (Clermont et al., 2000).
Identification of virulence genes
Eight virulence-associated genes [three variants (I, II, and III) of papG (PapG), fimH (FimH), bcsA (cellulose), csgA (CsgA), ecpA (EcpA), iutD (aerobactin), hlyA (α-hemolysin), and tosA (type 1 secretion A)] from MDR- and XDR-UPEC clinical strains were identified by multiplex PCR using specific primers (Table S1). CFT073 E. coli was used as a positive control.
Identification of class 1, 2, and 3 integrase genes
Integrons in the MDR- and XDR-UPEC strains were detected by multiplex PCR, which amplified the conserved region of the integrase-encoded genes intI1, intI2, and intI3 using specific primers (Table S1). MDR-UPEC strains 502U1-0412 and 674U-0612 were used as positive controls for class 1 and 2 integrons, respectively.
Sequencing of the amplified integron gene cassettes
The variable regions of class 1 and 2 integrons from the MDR-UPEC clinical strains were amplified by PCR using a high-fidelity Pfu polymerase of Thermo-Fisher Scientific (CA, USA) (Table S1). The amplicons were cleaned and concentrated using the Zymo DNA Clean and Concentrator of Zymo Research (CA, USA) and subjected to next-generation sequencing on a NexSeq500 System (Illumina, CA, USA), which was performed at “Unidad de Secuenciación del Instituto Nacional de Medicina Genómica” (CDMX, Mexico). The sequences were analyzed and assembled using ClustalO, ORF Finder (Open Reading Frame Finder), and BLAST (Basic Local Alignment Search Tool) from the NCBI (National Center of Biotechnology Information) (Sievers et al., 2011; Soleimani et al., 2014).
PFGE analysis in MDR- and XDR-UPEC strains
A phylogenetic analysis of MDR- and XDR-UPEC clinical strains was performed using pulsed-field gel electrophoresis (PFGE) following the specific modifications of the protocols established by the “Laboratorio de Investigación en Bacteriología Intestinal, HIMFG” (Ochoa et al., 2015). Briefly, the samples were digested with 2 U of Xbal (Invitrogen, Life Technologies, Wyman Street, Waltham, MA, USA) at 37°C for 4 h and separated by electrophoresis in a 1.2% ultrapure DNA agarose gel (Bio-Rad, Life Science Research, Hercules, CA, USA) that had been previously dissolved in 0.5X TBE. The samples were run on a CHEF Mapper system (Bio-Rad Life Science Research, Hercules, CA, USA) for 23 h at 200 V (6 V/cm) under two different linear ramped pulse times: 1–10 s and 10–40 s. The gels containing the macrorestriction products were stained for 40 min with 0.5 mg/mL ethidium bromide (Sigma-Aldrich St. Louis, MO, USA) and visualized under UV light using an Imaging System (ChemiDoc MP System®, Bio-Rad, Mexico City, Mexico). A lambda ladder PFGE marker (New England Biolabs, Hertfordshire, England, UK) was used as a molecular weight marker. PFGE patterns were analyzed using NTSYS-pc software (version 2.0, Applied Biostatistics, Inc., NY, USA) with the Sørensen-Dice similarity coefficient and a cluster analysis of the Unweighted Pair Group Method using Arithmetic averages (UPGMA) (Dice, 1945). The clonality of the UPEC strains was evaluated considering the Criteria of Tenover (Tenover et al., 1995).
Statistical analysis
Fisher's exact test and Fisher's PLSD ANOVA were used to identify significant differences in the data at p < 0.05.
Results
MDR and XDR profiles in the UPEC strains
According to the resistance profile to 10 classes of antibiotics, 16.40% (82/500) of the UPEC strains were MDR, and 4.20% (21/500) were XDR. Additionally, MDR- and XDR-UPEC strains were isolated from hospitalized patients with complicated UTIs from different wards of HIMFG, such as Nephrology, Urology, PICU (Pediatric Intensive Care Unit), NICU (Neonatal Intensive Care Unit), Oncology, and Pediatrics. From these, 82 MDR-UPEC strains were distributed as follows: 31.70% (26/82) were resistant to three classes of antibiotics, 28.04% (23/82) to four classes, 24.39% (20/82) to five classes, 9.75% (8/82) to six classes, and 6.09% (5/82) to seven classes. Furthermore, in eight patients, 40 MDR-UPEC strains were collected with five isolates from each patient. Moreover, 21 of the XDR-UPEC strains were distributed as follows: 52.38% (11/21) of were resistant to eight classes of antibiotics and 47.61% (10/21) to nine classes (Tables S2, S3). Interestingly, 76.19% (16/21) of XDR-UPEC strains had an ESBL phenotype (p ≤ 0.028) compared to 18.29% (15/82) of MDR-UPEC strains. Class 1 and 2 integrons were identified in 43.90% (36/82) and 2.43% (2/82) of MDR-UPEC strains, respectively. Additionally, 47.61% (10/21) of XDR-UPEC strains presented class 1 integrons, and 9.52% (2/21) presented class 2 integrons. These UPEC strains did not present class 3 integrons.
MDR- and XDR-UPEC strains, phylogenetic groups, and their association with virulence genes
MDR-UPEC strains were distributed into three phylogenetic groups: 54.87% (45/82) to D, 39.02% (32/82) to B2, and 6.09% (5/82) to A; the XDR-UPEC strains were distributed as follows: 47.61% (10/21) to B2, 42.85% (9/21) to D, and 9.52% (2/21) to A. Moreover, phylogenetic group B1 was not identified in MDR- or XDR-UPEC strains. On the other hand, 28.19% (9/32), 11.12% (5/32) and 20% (1/32) of MDR-UPEC strains in groups B2, D, and A, respectively, were associated with the ESBL phenotype. In contrast, 80% (8/10), 66.67% (6/9), and 100% (2/2) of XDR-UPEC strains in groups B2, D, and A, respectively, produced ESBLs.
The tosA gene encoding the type 1 secretion A toxin was distributed at different percentages to phylogenetic groups A, B2, and D of MDR- and XDR-UPEC strains (Table 1). The tosA gene from MDR-UPEC strains associated with phylogenetic group B2 was significantly different (p ≤ 0.0001) from that from XDR-UPEC strains (Table 1). Fimbrial (ecpA, fimH, csgA, and papGII), iron uptake (chuA, iutD) and α-Hemolysin (hlyA) genes from MDR- and XDR-UPEC strains were highly distributed into phylogenetic groups B2 and D (Table 1). Additionally, the papGIII variant gene of the MDR-UPEC strains was only distributed into phylogenetic group B2 and D, while the papGI variant gene was not identified in any UPEC strain (Table 1). The bcsA gene was distributed into phylogenetic groups as follows: 51.10% (23/45) to D, 43.75% (14/32) to B2 and 40% (2/5) to A of MDR-UPEC strains; however, XDR-UPEC strains carrying the bcsA gene were only distributed into phylogenetic group B2 (Table 1).
Table 1.
Phylogenetic groups associated with virulence genes in MDR- and XDR-UPEC clinical strains.
| Variable | MDR-UPEC clinical strains | Total % (n = 82) | XDR-UPEC clinical strains | Total % (n = 21) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Percentage in the phylogenetic groups | Percentage in the phylogenetic groups | |||||||||
| A% (n = 5) | B1% (n = 0) | B2% (n = 32) | D% (n = 45) | A% (n = 2) | B1% (n = 0) | B2% (n = 10) | D% (n = 9) | |||
| SURFACE GLYCANa | ||||||||||
| bcsA | 40 (2) | 0 (0) | 43.75 (14) | 51.10 (23) | 47.56 (39) | 0 (0) | 0 (0) | 20 (2) | 0 (0) | 9.52 (2) |
| FIMBRIAE/ADHESINb | ||||||||||
| ecpA | 100 (5) | 0 (0) | 96.87 (31) | 95.55 (43) | 96.34 (79) | 100 (2) | 0 (0) | 100 (10) | 100 (9) | 100 (21) |
| fimH | 100 (5) | 0 (0) | 100 (32) | 97.76 (44) | 98.78 (81) | 0 (0) | 0 (0) | 90 (9) | 88.89 (8) | 80.95 (17) |
| csgA | 80 (4) | 0 (0) | 96.87 (31) | 97.76 (44) | 96.34 (79) | 100 (2) | 0 (0) | 100 (10) | 100 (9) | 100 (21) |
| papGI | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| papGII | 40 (2) | 0 (0) | 90.62 (29) | 97.76 (44) | 91.46 (75) | 50 (1) | 0 (0) | 100 (10) | 100 (9) | 95.23 (20) |
| papGIII | 0 (0) | 0 (0) | 15.62 (5) | 2.22 (1) | 7.31 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| IRON UPTAKEc | ||||||||||
| iutD | 60 (3) | 0 (0) | 65.62 (21) | 51.10 (23) | 57.31 (47) | 50 (1) | 0 (0) | 100 (10) | 77.76 (7) | 85.71 (18) |
| chuA | 0 (0) | 0 (0) | 100 (32) | 100 (45) | 93.90 (77) | 0 (0) | 0 (0) | 100 (10) | 100 (9) | 90.47 (19) |
| TOXINSd | ||||||||||
| hlyA | 100 (5) | 0 (0) | 75 (24) | 73.32 (33) | 75.60 (62) | 50 (1) | 0 (0) | 40 (4) | 44.43 (4) | 42.85 (9) |
| tosA | 40 (2) | 0 (0) | 62.50 (20) | 15.54 (7) | 35.36 (29) | 50 (1) | 0 (0) | 30 (3) | 55.56 (5) | 42.85 (9) |
Cellulose (bcsA).
E. coli Common Pilus (ecpA), type 1 fimbriae (fimH), curli fimbriae (csgA), P fimbriae, PapG variant I (papGI), PapG variant II (papGII), PapG variant III (papGIII).
Aerobactin (iutD), E. coli haem-utilization-gene (chuA).
α-hemolysin (hlyA), type 1 secretion A (tosA).
Integrons in MDR- and XDR-UPEC strains and their association with phylogenetic groups
MDR-UPEC strains with class 1 integrons were related to phylogenetic groups A, B2, and D [40% (2/5), 43.75% (14/32), and 44.43% (20/45), respectively]; under these same conditions, class 2 integrons were related to phylogenetic groups B2 and D [3.12% (1/32) and 2.22% (1/45), respectively] (Table 2). In the XDR-UPEC strains, class 1 integrons were related to phylogenetic groups B2, D, and A [30% (3/10), 55.56% (5/9), and 100% (2/2), respectively]; in addition, the class 2 integrons were associated with phylogenetic groups B2 and A in 10% (1/10) and 50% (1/2), respectively (Table 2).
Table 2.
Phylogenetic groups associated with integron classes and the ESBL phenotype in MDR- and XDR-UPEC clinical strains.
| Resistance Mechanisms | MDR-UPEC clinical strains | Total % (n = 82) | XDR-UPEC clinical strains | Total % (n = 21) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Percentage in the phylogenetic groups | Percentage in the phylogenetic groups | |||||||||
| A% (n = 5) | B1% (n = 0) | B2% (n = 32) | D% (n = 45) | A% (n = 2) | B1% (n = 0) | B2% (n = 10) | D% (n = 9) | |||
| intl1 | 40 (2) | 0 (0) | 43.75 (14) | 44.43 (20) | 43.90 (36) | 100 (2) | 0 (0) | 30 (3) | 55.56 (5) | 47.61 (10) |
| intl2 | 0 (0) | 0 (0) | 3.12 (1) | 2.22 (1) | 2.43 (2) | 50 (1) | 0 (0) | 10 (1) | 0 (0) | 9.52 (2) |
| intl3 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| ESBLs | 20 (1) | 0 (0) | 28.12 (9) | 11.12 (5) | 18.29 (15) | 100 (2) | 0 (0) | 80 (8) | 66.67 (6) | 76.19 (16) |
Class 1 integrase (intl1), class 2 integrase (intl2), class 3 integrase (intl3), and extended-spectrum beta-lactamases (ESBLs).
Analysis of class 1 and 2 integron sequences from MDR- and XDR-UPEC strains
A total of 56.52% (26/46) of MDR- and XDR-UPEC strains amplified the variable region of class 1 integrons with a product of ~1800 bp; in addition, 75% (3/4) of the MDR- and XDR-UPEC strains amplified the variable region of class 2 integrons with a product of ~2000 bp (Table 3). The sequencing analysis of ~1800-bp products showed the gene cassettes for aadA1 (aminoglycoside-3′-adenylyltransferase), aadB (aminoglucoside-2′-adenylyl-transferase), aacC (gentamicin-acetyl-transferase), ant1 (streptomycin-3′-adenylyltransferase), dfrA1 (dihydrofolate reductase type 1), and dfrA17 (streptomycin-3′-O-adenylyltransferase); their distribution is shown in Table 3. In contrast, the aadA1, dfrA1, and aadA4 (streptomycin-3′-adenylyl transferase) genes were identified in products of ~2000 bp (Table 3). Finally aadA1, aadB, aacC, ant1, dfrA1, and dfrA17, which encode modifying enzymes within the class 1 and 2 integrons from MDR- and XDR-UPEC strains, were associated with resistance to GM or SXT.
Table 3.
Sequencing of class 1 and 2 integrons and their association with gentamicin and trimethoprim-sulfamethoxazole.
| Clinical strains | Class of Integron | Resistance profile | Phylogenetic groups | Size of gene cassettes (bp) | Identified gene cassettes | Susceptibility to GM μg/mL (R/S) | Susceptibility to SXT μg/mL (R/S) |
|---|---|---|---|---|---|---|---|
| 118U2 | 1 | MDR (4) | D | ~1800 | -dfrA1 (Dihydrofolate reductase type 1) | 8 (R) | >256 (R) |
| 118U4 | 1 | MDR (4) | D | 8 (R) | >256 (R) | ||
| 118U5 | 1 | MDR (4) | D | 8 (R) | >256 (R) | ||
| 11U-0912 | 1 | XDR (8) | B2 | 64 (R) | >256 (R) | ||
| 433U1-0512 | 1 | MDR (4) | D | 0.5 (S) | >256 (R) | ||
| 494U2-0412 | 1 | MDR (5) | D | >256 (R) | 0.5 (S) | ||
| 502U-0412 | 1 | MDR (4) | D | 8 (R) | >256 (R) | ||
| 54U-0612 | 1 | MDR (3) | B2 | 16 (R) | 0.25 (S) | ||
| 647U-0712 | 1 | MDR (5) | D | >256 (R) | >256 (R) | ||
| 909U-0612 | 1 | MDR (5) | D | 2(S) | 0.25 (S) | ||
| 188U-1112 | 1 | MDR (6) | D | ~1800 | -aadA1 (Aminoglycoside-3′-adenylyltransferase) -dfrA1 (Dihydrofolate reductase type 1) |
32 (R) | >256 (R) |
| 440U1 | 1 | MDR (4) | B2 | 2 (S) | >256 (R) | ||
| 440U2 | 1 | MDR (3) | B2 | 2 (S) | >256 (R) | ||
| 440U3 | 1 | MDR (3) | B2 | 2 (S) | >256 (R) | ||
| 440U4 | 1 | MDR (3) | B2 | 2 (S) | >256 (R) | ||
| 440U5 | 1 | MDR (3) | B2 | 2 (S) | >256 (R) | ||
| 945U2-0412 | 1 | XDR (9) | D | >256 (R) | 4 (R) | ||
| 118U3 | 1 | MDR (4) | D | ~1800 | -aadB (Aminoglucoside-2′-adenylyltransferase) -dfrA1 (Dihydrofolate reductase type 1) |
8 (R) | >256 (R) |
| 179U-1012 | 1 | MDR (5) | D | 32 (R) | >256 (R) | ||
| 513U-0912 | 1 | XDR (8) | D | >256 (R) | >256 (R) | ||
| 720U-0712 | 1 | XDR (8) | B2 | 8 (R) | 4 (R) | ||
| 86U-0612 | 1 | MDR (3) | D | >256 (R) | >256 (R) | ||
| 877U-1112 | 1 | XDR (8) | D | >256 (R) | 1 (S) | ||
| 117U1-0512 | 1 | MDR (5) | D | ~1800 | -aacC (Gentamicin-acetyltransferase -dfrA1 (Dihydrofolate reductase type 1) -dfrA17 (Streptomycin-3′-O-adenylyltransferase) -ant1 (Streptomycin-3′-adenylyltransferase) |
0.5 (S) | >256 (R) |
| 118U1 | 1 | MDR (4) | D | 8 (R) | >256 (R) | ||
| 553U-1112 | 1 | MDR (3) | D | 0.5 (S) | >256 (R) | ||
| 11U-0912 | 2 | XDR (8) | B2 | ~2000 | -aadA1 (Aminoglycoside-3′-adenylyltransferase) -dfrA1 (Dihydrofolate reductase type 1) -aadA4 (Streptomycin-3′-adenylyltransferase) |
64 (R) | >256 (R) |
| 188U-1112 | 2 | MDR (6) | D | 32 (R) | >256 (R) | ||
| 674U-0612 | 2 | MDR (4) | B2 | 4 (S) | 2 (S) |
GM, Gentamicin; SXT, Trimethoprim-sulfamethoxazole; S, Sensitive; R, Resistant.
Genetic diversity of the MDR- and XDR-UPEC strains
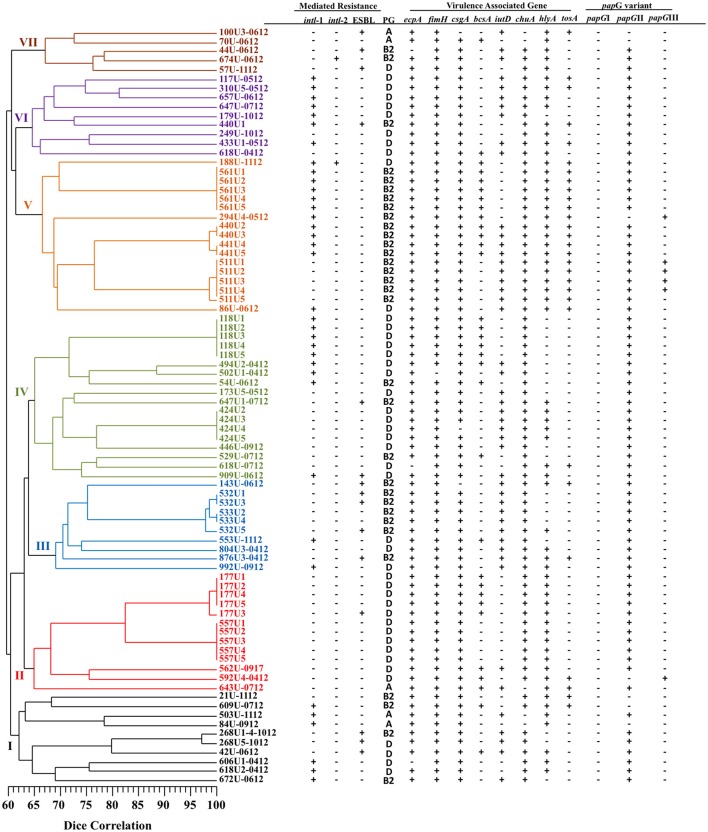
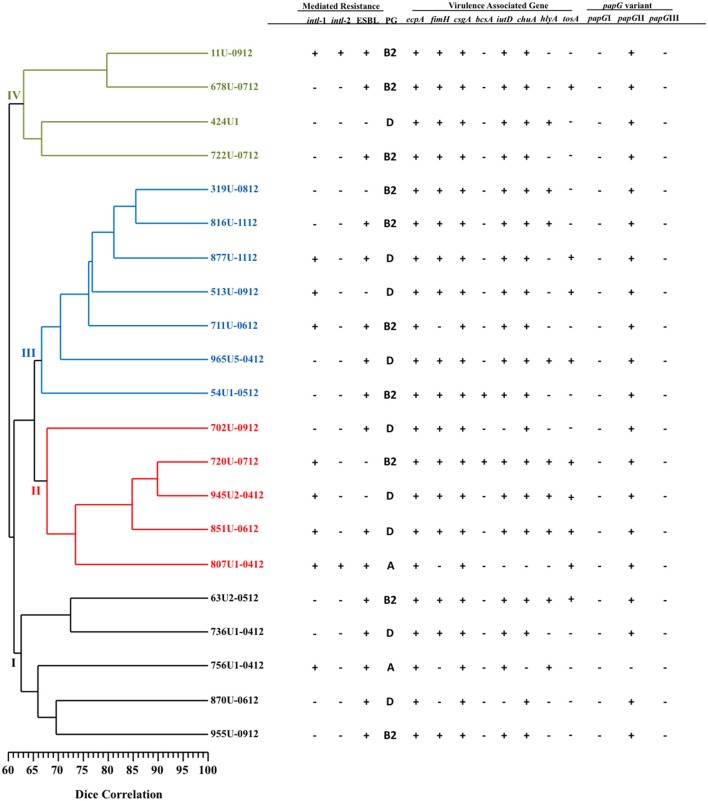
Patterns consisting of 11–21 DNA fragments ranging in size from 48.5 to 436.5 Kb obtained from the PFGE assay of the MDR- (51 pulsotypes) and XDR- (21 pulsotypes) UPEC strains were used to construct two dendrograms with cophenetic correlation coefficients of 0.8124 and 0.8037 (Figures 1, 2). Furthermore, 12.19% (10/82) of the MDR-UPEC strains were distributed into cluster I, 15.85% (13/82) into cluster II, 12.19% (10/82) into cluster III, 21.95% (18/82) into cluster IV, 20.73% (17/82) into cluster V, 10.97% (9/82) into cluster VI, and 5% (5/82) into cluster VII (Figure 1). Of the seven clusters from the dendrogram, four clusters (II, III, IV, and V) grouped MDR-UPEC strains within isolates that were clonally related, collected at different times, and considered persistent; however, in clusters I, VI, and VII, a clonal relationship was not identified. Finally, XDR-UPEC strains were distributed into four clusters: 23.80% (5/21) into cluster I, 23.80% (5/21) into cluster II, 33.33% (7/21) into cluster III, and 19.04% (4/21) into cluster IV (Figure 2).
Figure 1.
PFGE analysis of 82 MDR-UPEC strains associated with virulence genes and their mechanisms of resistance. A diversity analysis was performed using the Sørensen-Dice similarity coefficient in association with the UPGMA algorithm. Additionally, the dendrogram was evaluated using the cophenetic correlation coefficient obtained by the Mantel test, which indicated the dispersion of the data and showed a value of r = 0.8124. The seven clusters identified by PFGE are shown in different colors.
Figure 2.
PFGE analysis of 21 XDR-UPEC strains associated with virulence genes and their mechanisms of resistance. The diversity analysis was performed using the Sørensen-Dice similarity coefficient in association with the UPGMA algorithm. Additionally, the dendrogram was evaluated using the cophenetic correlation coefficient obtained by the Mantel test, which indicated the dispersion of the data and showed a value of r = 0.8037. The four clusters identified by PFGE are shown in different colors.
Clonality, phylogenetic groups, virulence gene, and integrons in the MDR- and XDR-UPEC strains
The four clusters (II, III, IV, and V) grouping MDR-UPEC strains shared the presence of fimbrial genes (ecpA, fimH, csgA, and papGII) and an iron uptake gene (chuA). In clusters I, VI, and VII, a relationship between phylogenetic groups, integrons and virulence genes was not identified. In cluster II, two clonal groups associated with phylogenetic group D were identified: 557U (1–5 clones) characterized by the absence of the bcsA gene and 177U (1, 2, 4, and 5 clones) by the presence of the bcsA gene. In cluster III, two clonal groups associated with phylogenetic group B2 were identified: 532U (1 and 3 clones) characterized by the presence of the ESBL phenotype and 533U (2 and 4 clones) by the lack of the ESBL phenotype. In cluster IV, two clonal groups associated with phylogenetic group D were identified: 424U (2–5 clones) characterized by the presence of the iutD and hlyA genes and 118U (1–5 clones) by the presence of class 1 integrons and the bcsA gene. In addition, 424U4 lacked the csgA gene compared with 424U (2, 3, and 5 clones). In cluster V, four clonal groups [561U (1–5 clones), 440U (2 and 3 clones), 441U (4 and 5 clones), and 511U (1–5 clones)] associated with phylogenetic group B2 and with the tosA and hlyA genes were identified. Class 1 integrons and the bcsA gene were found in three clonal groups (561U, 440U, and 441U) and the iutD gene in three clonal groups (440U, 441U, and 511U). Interestingly, the papGIII gene was only found in clonal group 511U associated with the tosA and hlyA genes (Figure 1).
Discussion
Uncomplicated UTIs are commonly treated with trimethoprim sulfamethoxazole, ciprofloxacin, and ampicillin. Carbapenems, nitrofutans, and cephalosporins of the 3rd and 4th generations are the most common antibiotics used to treat complicated UTIs (Foxman, 2010). The emergence of MDR- and XDR-UPEC strains has complicated the treatment of UTIs (Dehbanipour et al., 2016). In this study, MDR and XDR-UPEC strains from pediatric patients at HIMFG were analyzed for their phylogeny, integrons, and virulence profile. These strains were mainly associated with phylogenetic groups, B2 and D as described other studies; however, they also have been associated with phylogenetic group A (Ejrnæs et al., 2011; Zhao et al., 2015). These UPEC strains could be considered community strains with predisposing factors that facilitate UTIs in immunocompromised patients, as previously reported (Derakhshandeh et al., 2015; Rodrigues et al., 2015). UPEC strains of Mexican origin are mainly associated with phylogenetic group B2 (López-Banda et al., 2014). Moreover, ESBLs-producing MDR-UPEC strains were mainly associated with phylogenetic groups B2, A, and D, whereas the XDR-UPEC strains were associated with A, B2, and D. The recently reported distribution of UPEC strains associated with these phylogenetic groups agrees with our data (Zhao et al., 2015). Commensal E. coli strains with the ability to cause community-acquired UTIs have been related to phylogenetic group B1, which was not identified in this study (Mosquito et al., 2015). Remarkably, variations in phylogenetic groups are related to geographical area, infection site, and antibiotic resistance (Mokracka et al., 2011). Several adhesins and siderophores from UPEC strains participate in the colonization and persistence of bladder cells (Hannan et al., 2012). The genes that encode adhesins (EcpA, FimH, PapG, and CsgA) and the protein related to iron uptake (siderophores) were widely distributed in MDR- and XDR-UPEC strains associated with two phylogenetic groups (B2 and D) and to a lesser extent with A. Recently, UPEC strains associated with phylogenetic group B2 and D showed a high presence of siderophores, autotransporters protease, and adhesion genes, while a low presence of genes encoding toxins was also observed (López-Banda et al., 2014; Yahiaoui et al., 2015; Zhao et al., 2015). In addition, the PapGII variant gene from MDR- and XDR-UPEC strains associated with phylogenetic groups D and B2 was the most prevalent compared to the papGI and papGIII variant genes. The papGII variant gene from UPEC strains associated with phylogenetic groups A and B2 is the most prevalent gene associated with cystitis in women and pyelonephritis in children and adults (Jantunen et al., 2000; Agarwal et al., 2013). However, four MDR-UPEC strains associated with phylogenetic group B2 and one strain associated with group D were associated with the papGIII variant gene, which correlates with data reported by other authors (Agarwal et al., 2013).
The bcsA gene encoding cellulose is a component of the bacterial extracellular matrix in UPEC strains, and its expression is associated with the csg operon that codes for curli fimbriae (Saldaña et al., 2009). Our data showed the presence of the bcsA and csgA genes in MDR-UPEC strains associated with phylogenetic groups D, B2, and A. A study suggest that the expression of both structures is independent of the strain origin and participates in biofilm formation to protect against different antibiotics (Hung et al., 2013). A deletion in the tosA gene in CFT073 E. coli affected the colonization of the bladder and kidney in a murine model, indicating that TosA adhesin is required for the virulence of UPEC (Vigil et al., 2011a). The distribution of the tosA gene in MDR- and XDR-UPEC strains was associated with three main phylogenetic groups (A, B2, and D), which has been described in E. coli strains isolated from fecal matter, asymptomatic bacteriuria (ABU), cystitis, and pyelonephritis samples (Vigil et al., 2011b). Interestingly, a relationship between the presence of the tosA and hlyA genes was identified in MDR-UPEC strains associated with phylogenetic group B2, while XDR-UPEC strains were related to D and B2. These associations have been described in UPEC strains collected from patients with pyelonephritis (Vigil et al., 2011b). The tosA-positive MDR- and XDR-UPEC strains included in this study showed a wide profile of virulence genes compared with tosA-negative strains and were associated with phylogenetic groups B2 and D. The tosA gene is considered a genetic marker of UPEC strains carrying several virulence genes (Vigil et al., 2011b).
Nosocomial pathogens containing integron classes are related to resistance to different antibiotic groups; however, this association in UPEC strains is poorly studied (Stalder et al., 2012). Class 1 integrons in hospital E. coli strains are the most frequently reported, followed by class 2 integrons (Poey and Laviña, 2014). A total of 24.2% of class 1 integrons in Mexican E. coli strains from different clinical sources were associated with MDR (Acosta-Pérez et al., 2015); similar data were obtained in our study. The highest presence of class 1 integrons was associated with phylogenetic groups D and B2 in MDR- and XDR-UPEC strains; however, several studies described variable results regarding the association of integrons and phylogenetic groups (Gündogdu et al., 2011; Zeighami et al., 2015). Sequencing the variable region of class 1 and 2 integrons from MDR- and XDR-UPEC strains revealed the presence of genes encoding the antibiotic modifying enzymes aadA1, aadB, aacC, ant1, dfrA1, and dfrA17. MDR- and XDR-UPEC strains with resistance to SXT and GM antibiotics were mainly associated with class 1 and 2 integrons, as described by other authors (Solberg et al., 2006; Márquez et al., 2008; El-Najjar et al., 2010; Soleimani et al., 2014; Acosta-Pérez et al., 2015; Yahiaoui et al., 2015). Class 3 integrons are part of the soil/freshwater proteobacteria group, have a poor occurrence rate and were not identified in this study (Deng et al., 2015).
Genetic diversity has been widely used to characterize populations of UPEC strains associated with several diseases of the urinary tract (Luo et al., 2012; Acosta-Pérez et al., 2015; Morales-Espinosa et al., 2016). The recurrence of UTIs at HIMFG was associated with several clones in the MDR-UPEC strains of phylogenetic groups D and B2, which are responsible for a majority of persistent UTIs. In contrast, the primary persistence of recurrent UTIs was associated with phylogenetic group B2, whereas clones associated with phylogenetic group D mainly caused reinfections by UPEC isolates (Luo et al., 2012). The persistence of UPEC strains is related to invasion, bacterial community, and quiescence, including the expression of several virulence genes (Hannan et al., 2012). Four clusters (II, III, IV, and V), two phylogenetic groups (B2 and D), class 1 integrons, and genes encoding fimbrial adhesin and iron uptake are associated with MDR- and XDR-UPEC strains. The bcsA gene and phylogenetic group D, were only related to the 557U clonal group of cluster II and to the 118U group of cluster V. These UPEC strains are associated with catheter-associated bacteremia and are thus likely to function in biofilm formation on the catheter using curli and cellulose (Hung et al., 2013). Clonal group 424U of cluster IV was related to the presence of the iutD and hlyA genes; thus, these genes could be located on the same pathogenicity island, as has been reported for PAI-pheV. Interestingly, cluster V presenting four clonal groups was characterized by the presence of the tosA gene and virulence genes, including hlyA, bcsA, iutD, and papGIII. These data support the hypothesis that the TosA protein is a potential virulence marker in UPEC strains (Vigil et al., 2011a). Nonetheless, ESBL-producing clones associated with phylogenetic group B2 were only localized into the 532U clonal group of cluster III; likewise, the antibiotic resistance profiles showed that resistance to penicillin, beta-lactams, and cephalosporin was included in clonal group B2 (Mohajeri et al., 2014).
In conclusion, a high frequency of genes encoding virulence factors, a broad resistance profile associated with integrons (classes 1 and 2) and the ESBL phenotype are essential elements related to phylogenetic groups (B2 and D), which are distributed specifically in genetic clusters of MDR- and XDR-UPEC clinical strains. Additionally, these attributes confer to bacteria adaptive advantages to colonize, persist and facilitate UTIs caused by UPEC.
Author contributions
Designed and conceived the experiments: SO and JX. Performed the experiments: SO, VL, FL, JR, and GE. Analyzed the data: SO, VL, AC, JA, MS, and JX. Contributed reagents/materials/analytical tools: SO, AC, VC, Dd, BL, IP, SG, RH, and JX. Wrote and reviewed the manuscript: SO, VL, AC, and JX.
Funding
This work was supported by Public Federal Fund grants HIM/2013/007, HIM/2014/014, and HIM/2014/019 from the Hospital Infantil de México Federico Gómez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Posgrado en Ciencias Químico-Biológicas, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional and Virginia Alcázar López for technical assistance.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02042/full#supplementary-material
References
- Acosta-Pérez G., Ibánez-Cervantes G., Bello-López J. M., Hernández J. M., Hernández-Monta-ez Z., Giono-Cerezo S., et al. (2015). Structural diversity of class 1 integrons in multiresistant strains of Escherichia coli isolated from patients in a hospital in Mexico City. Curr. Microbiol. 71, 501–508. 10.1007/s00284-015-0876-9 [DOI] [PubMed] [Google Scholar]
- Agarwal J., Mishra B., Srivastava S., Srivastava R. (2013). Genotypic characteristics and biofilm formation among Escherichia coli isolates from Indian women with acute cystitis. Trans. R. Soc. Trop. Med. Hyg. 107, 183–187. 10.1093/trstmh/trs090 [DOI] [PubMed] [Google Scholar]
- Alteri C. J., Mobley H. L. (2012). Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 15, 3–9. 10.1016/j.mib.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L., Pinkner J. S., Hammer N. D., Cusumano C. K., Hung C. S., Chorell E., et al. (2009). Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5, 913–919. 10.1038/nchembio.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) (2016). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Six Informational, Supplement. M100-S26. Wayne, PA: Megan L. Tertel. [Google Scholar]
- Dehbanipour R., Rastaghi S., Sedighi M., Maleki N., Faghri J. (2016). High prevalence of multidrug-resistance uropathogenic Escherichia coli strains, Isfahan, Iran. J. Nat. Sci. Biol. Med. 7, 22–26. 10.4103/0976-9668.175020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., et al. (2015). Resistance integrons: class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 14:45. 10.1186/s12941-015-0100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshandeh A., Firouzi R., Motamedifar M., Arabshahi S., Novinrooz A., Boroojeni A. M., et al. (2015). Virulence characteristics and antibiotic resistance patterns among various phylogenetic groups of uropathogenic Escherichia coli isolates. Jpn. J. Infect. Dis. 68, 428–431. 10.7883/yoken.JJID.2014.327 [DOI] [PubMed] [Google Scholar]
- Dice L. R. (1945). Measures of the amount of ecologic association between species. Ecology 26, 297–302. 10.2307/1932409 [DOI] [Google Scholar]
- Dillon B., Thomas L., Mohmand G., Zelynski A., Iredell J. (2005). Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods 62, 221–232. 10.1016/j.mimet.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Ejrnæs K., Stegger M., Reisner A., Ferry S., Monsen T., Holm S. E., et al. (2011). Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence 2, 528–537. 10.4161/viru.2.6.18189 [DOI] [PubMed] [Google Scholar]
- El-Najjar N. G., Farah M. J., Hashwa F. A., Tokajian S. T. (2010). Antibiotic resistance patterns and sequencing of class I integron from uropathogenic Escherichia coli in Lebanon. Lett. Appl. Microbiol. 51, 456–461. 10.1111/j.1472-765X.2010.02926.x [DOI] [PubMed] [Google Scholar]
- Engstrom M. D., Alteri C. J., Mobley H. L. (2014). A conserved PapB family member, TosR, regulates expression of the uropathogenic Escherichia coli RTX nonfimbrial adhesin TosA while conserved LuxR family members TosE and TosF suppress motility. Infect. Immun. 82, 3644–3656. 10.1128/IAI.01608-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani S., Saidani M., Amine F. S., Boutiba Ben Boubaker I., Boubaker I. (2015). A comparative study of antimicrobial resistance rates and phylogenetic groups of community-acquired versus hospital-acquired invasive Escherichia coli. Med. Mal. Infect. 45, 133–138. 10.1016/j.medmal.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Flores-Mireles A. L., Walker J. N., Caparon M., Hultgren S. J. (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284. 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman B. (2010). The epidemiology of urinary tract infection. Nat. Rev. Urol. 7, 653–660. 10.1038/nrurol.2010.190 [DOI] [PubMed] [Google Scholar]
- Foxman B., Wu J., Farrer E. C., Goldberg D. E., Younger J. G., Xi C. (2012). Early development of bacterial community diversity in emergently placed urinary catheters. BMC Res. Notes 5:332. 10.1186/1756-0500-5-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Wang X., Xu H., Xu Y., Ling J., Zhang D., et al. (2012). Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiol. 12:143. 10.1186/1471-2180-12-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündogdu A., Long Y. B., Vollmerhausen T. L., Katouli M. (2011). Antimicrobial resistance and distribution of sul genes and integron-associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J. Med. Microbiol. 60, 1633–1642. 10.1099/jmm.0.034140-0 [DOI] [PubMed] [Google Scholar]
- Hadifar S., Moghoofei M., Nematollahi S., Ramazanzadeh R., Sedighi M., Salehi-Abargouei A., et al. (2016). Epidemiology of multi drug resistant uropathogenic Escherichia coli in Iran: a systematic review and meta-analysis. Jpn. J. Infect. Dis. 18:JJID.2015.652. 10.7883/yoken.jjid.2015.652 [DOI] [PubMed] [Google Scholar]
- Hannan T. J., Totsika M., Mansfield K. J., Moore K. H., Schembri M. A., Hultgren S. J. (2012). Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol. Rev. 36, 616–648. 10.1111/j.1574-6976.2012.00339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C., Zhou Y., Pinkner J. S., Dodson K. W., Crowley J. R., Heuser J., et al. (2013). Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 4, e00645–613. 10.1128/mbio.00645-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav S., Hussain A., Devi S., Kumar A., Parveen S., Gandham N., et al. (2011). Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS ONE 6:e18063. 10.1371/journal.pone.0018063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantunen M. E., Siitonen A., Koskimies O., Wikström S., Kärkkäinen U., Salo E., et al. (2000). Predominance of class II pap G allele of Escherichia coli in pyelonephritis in infants with normal urinary tract anatomy. J. Infect. Dis. 181, 1822–1824. 10.1086/315446 [DOI] [PubMed] [Google Scholar]
- Kass E. H. (1957). Bacteriuria and the diagnosis of infections of the urinary tract; with observations on the use of methionine as a urinary antiseptic. AMA Arch. Intern. Med. 100, 709–714. 10.1001/archinte.1957.00260110025004 [DOI] [PubMed] [Google Scholar]
- Kudinha T., Johnson J. R., Andrew S. D., Kong F., Anderson P., Gilbert G. L. (2013). Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr. Infect. Dis. J. 32, 543–548. 10.1097/INF.0b013e31828ba3f1 [DOI] [PubMed] [Google Scholar]
- Lane M. C., Mobley H. L. (2007). Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 72, 19–15. 10.1038/sj.ki.5002230 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Subhadra B., Son Y. J., Kim D. H., Park H. S., Kim J. M., et al. (2016). Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett. Appl. Microbiol. 62, 84–90. 10.1111/lam.12517 [DOI] [PubMed] [Google Scholar]
- López-Banda D. A., Carrillo-Casas E. M., Leyva-Leyva M., Orozco-Hoyuela G., Manjarrez-Hernández Á. H., Arroyo-Escalante S., et al. (2014). Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Biomed. Res. Int. 2014:959206. 10.1155/2014/959206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Ma Y., Zhao Q., Wang L., Guo L., Ye L., et al. (2012). Similarity and divergence of phylogenies, antimicrobial susceptibilities, and virulence factor profiles of Escherichia coli isolates causing recurrent urinary tract infections that persist or result from reinfection. J. Clin. Microbiol. 50, 4002–4007. 10.1128/JCM.02086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje P., Brauner A. (2014). Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Microb. Physiol. 65, 337–372. 10.1016/bs.ampbs.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Márquez C., Labbate M., Raymondo C., Fernández J., Gestal A. M., Holley M., et al. (2008). Urinary tract infections in a South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. J. Clin. Microbiol. 46, 3417–3425. 10.1128/jcm.00835-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohajeri P., Darfarin G., Farahani A. (2014). Genotyping of ESBL producing Uropathogenic Escherichia coli in west of Iran. Int. J. Microbiol. 2014:276941. 10.1155/2014/276941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokracka J., Koczura R., Jabłońska L., Kaznowski A. (2011). Phylogenetic groups, virulence genes and quinolone resistance of integrón-bearing Escherichia coli strains isolated from a wastewater treatment plant. Antonie Van Leeuwenhoek 99, 817–824. 10.1007/s10482-011-9555-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Espinosa R., Hernández-Castro R., Delgado G., Mendez J. L., Navarro A., Manjarrez A., et al. (2016). UPEC strain characterization isolated from Mexican patients with recurrent urinary infections. J. Infect. Dev. Ctries 10, 317–328. 10.3855/jidc.6652 [DOI] [PubMed] [Google Scholar]
- Mosquito S., Pons M. J., Riveros M., Ruiz J., Ochoa T. J. (2015). Diarrheagenic Escherichia coli phylogroups are associated with antibiotic resistance and duration of diarrheal episode. Sci. World J. 2015:610403. 10.1155/2015/610403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. A., Lopez-Boado Y. S., Wilson C. L., Roth R., Parks W. C., Heuser J., et al. (1998). Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282, 1494–1497. 10.1126/science.282.5393.1494 [DOI] [PubMed] [Google Scholar]
- Ochoa S. A., Cruz-Córdova A., Rodea G. E., Cazares-Domínguez V., Escalona G., Arellano-Galindo J., et al. (2015). Phenotypic characterization of multidrug-resistant Pseudomonas aeruginosa strains isolated from pediatric patients associated to biofilm formation. Microbiol. Res. 172, 68–78. 10.1016/j.micres.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Hooper D. C. (2010). Hospital-acquired infections due to Gram-negative bacteria. N. Engl. J. Med. 362, 1804–1813. 10.1056/NEJMra0904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poey M. E., Laviña M. (2014). Integrons in uropathogenic Escherichia coli and their relationship with phylogeny and virulence. Microb. Pathog. 77, 73–77. 10.1016/j.micpath.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Rodrigues C., MacHado E., Pires J., Ramos H., Novais, Peixe L. (2015). Increase of widespread A, B1 and D Escherichia coli clones producing a high diversity of CTX-M-types in a Portuguese hospital. Future Microbiol. 10, 1125–1131. 10.2217/fmb.15.38 [DOI] [PubMed] [Google Scholar]
- Saldaña Z., De la Cruz M. A., Carrillo-Casas E. M., Durán L., Zhang Y., Hernández-Castro R., et al. (2014). Production of the Escherichia coli common pilus by uropathogenic E. coli is associated with adherence to HeLa and HTB-4 cells and invasion of mouse bladder urothelium. PLoS ONE 9:e101200. 10.1371/journal.pone.0101200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña Z., Xicohtencatl-Cortes J., Avelino F., Phillips A. D., Kaper J. B., Puente J. L., et al. (2009). Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11, 992–1006. 10.1111/j.1462-2920.2008.01824.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliano P. R., Rocha L. A., Medina-Pestana J. O., Heilberg I. P. (2010). The role of host factors and bacterial virulence genes in the development of pyelonephritis caused by Escherichia coli in renal transplant recipients. Clin. J. Am. Soc. Nephrol. 5, 1290–1297. 10.2215/CJN.06740909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O. D., Ajiboye R. M., Riley L. W. (2006). Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 44, 1347–1351. 10.1128/JCM.44.4.1347-1351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani N., Aganj M., Ali L., Shokoohizadeh L., Sakinc T. (2014). Frequency distribution of genes encoding aminoglycoside modifying enzymes in uropathogenic E. coli isolated from Iranian Hospital. BMC Res. Notes 7:842. 10.1186/1756-0500-7-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder T., Barraud O., Casellas M., Dagot C., Ploy M. C. (2012). Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119. 10.3389/fmicb.2012.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S., Mobley H. L. T. (2015). Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol. Spectr. 3:UTI-0015-2012. 10.1128/microbiolspec.uti-0015-2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiba M. R., Yano T., da Silva Leite D. (2008). Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Rev. Inst. Med. Trop. Sao Paulo 50, 255–260. 10.1590/S0036-46652008000500001 [DOI] [PubMed] [Google Scholar]
- Toval F., Köhler C. D., Vogel U., Wagenlehner F., Mellmann A., Fruth A., et al. (2014). Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 52, 407–418. 10.1128/JCM.02069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil P. D., Alteri C. J., Mobley H. L. (2011a). Identification of in vivo-induced antigens including an RTX family exoprotein required for uropathogenic Escherichia coli virulence. Infect. Immun. 79, 2335–2344. 10.1128/IAI.00110-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil P. D., Stapleton A. E., Johnson J. R., Hooton T. M., Hodges A. P., He Y., et al. (2011b). Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. MBio 2, e00066–11. 10.1128/mBio.00066-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiaoui M., Robin F., Bakour R., Hamidi M., Bonnet R., Messai Y. (2015). Antibiotic resistance, virulence, and genetic background of community-acquired uropathogenic Escherichia coli from Algeria. Microb. Drug Resist. 21, 516–526. 10.1089/mdr.2015.0045 [DOI] [PubMed] [Google Scholar]
- Zeighami H., Haghi F., Masumian N., Hemmati F., Samei A., Naderi G. (2015). Distribution of integrons and gene cassettes among uropathogenic and diarrheagenic Escherichia coli isolates in Iran. Microb. Drug Resist. 21, 435–440. 10.1089/mdr.2014.0147 [DOI] [PubMed] [Google Scholar]
- Zhao R., Shi J., Shen Y., Li Y., Han Q., Zhang X., et al. (2015). Phylogenetic distribution of virulence genes among ESBL-producing uropathogenic Escherichia coli isolated from long-term hospitalized patients. J. Clin. Diagn. Res. 9, DC01–DC04. 10.7860/jcdr/2015/13234.6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.