Abstract
Bladder cancer is one of the major cancer types and both environmental factors and genetic background play important roles in its pathology. Kaohsiung is a high industrialized city in Taiwan, and here we focused on this region to evaluate the genetic effects on bladder cancer. Muscarinic acetylcholine receptor M3 (CHRM3) was reported as a key receptor in different cancer types. CHRM3 is located at 1q42-43 which was reported to associate with bladder cancer. Our study attempted to delineate whether genetic variants of CHRM3 contribute to bladder cancer in Chinese Han population in south Taiwan. Five selected SNPs (rs2165870, rs10802789, rs685550, rs7520974, and rs3738435) were genotyped for 30 bladder cancer patients and 60 control individuals and genetic association studies were performed. Five haplotypes (GTTAT, ATTGT, GCTAC, ACTAC, and ACCAC) were found significantly associated with low CHRM3 mRNA level and contributed to increased susceptibility of bladder cancer in Kaohsiung city after rigid 10000 consecutive permutation tests. To our knowledge, this is the first genetic association study that reveals the genetic contribution of CHRM3 gene in bladder cancer etiology.
1. Introduction
Bladder cancer is one of the major cancers worldwide. In 2013, bladder cancer is the eighth and tenth most frequently diagnosed cancer among men and women [1]. About 72571 new bladder cancer cases and 15210 cancer deaths were also reported in 2013 [1]. Although the etiology of cancer has not been fully established, it is mostly accepted as a multifactorial disease involving both environmental and genetic factors [2, 3]. The related risk factors for bladder cancer included interactions between genetic background and environmental factors, for example, tobacco smoking, industrial chemicals (aromatic amine) exposure, chronic infection with Schistosoma species, oxidants, and highly reactive free radicals [3].
Geographically, Kaohsiung is located in south Taiwan at 22.63 degrees north latitude and 120.30 degrees east longitude. The area of Kaohsiung city is 2952 square kilometers containing 1387931 people and is the second most populated city in Taiwan. Kaohsiung is a high industrialized city in Taiwan containing at least 8 industrial areas and 6306 plants in operation (http://erdb.epa.gov.tw/ERDBIndex.aspx; Supplemental Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/4052846). Many types of industries were reported in Kaohsiung city, including semiconductor, optoelectronics, electroplating, steel, shipbuilding, leather, and dyeing. Thousands of industrial wastes were discharged from these plants every day. Therefore, people living in Kaohsiung are under a high exposure level to industrial pollution which has been reported as a key environmental factor for many cancer types, especially cancers in urinary system [4]. As Taiwan Cancer Registry Annual Report 2012 [5], the incidence of bladder cancer in south Taiwan was 9.73 per 100,000 people, which was the highest area in west Taiwan. Notably, most of the new cases of bladder cancer were reported in Kaohsiung city. Therefore, Kaohsiung city is suitable for evaluating the pathological factor of bladder cancer.
Genetic predisposition is an important risk factor related to bladder cancer. Recently, 12 genes were identified to associate with bladder cancer at a genome-wide significant level in European populations from six genome-wide association studies (GWAS) [6–11]. Among these 12 genes, 12 independent single nucleotide polymorphisms (SNPs) have been evaluated and six genes (seven SNPs) showed association with bladder cancer in Chinese populations, including CLPTM1L (rs401681) [12], TP63 (rs710521) [13], TACC3 (rs798766) [14, 15], TERT (rs2736100 and rs2736098) [12, 15], PSCA (rs2294008) [15, 16], and MYC (rs9642880) [15, 17]. However, whether other genes are also playing pathological roles in bladder cancer in the Chinese population remains unknown.
Prior studies revealed that G protein-coupled receptors (GPCRs) pathways regulated kidney and bladder cancer cell lines migration and invasion [18]. GPCRs have enormous potential in biomedical research and drug development resulting in their features and functions to serve as therapeutic targets [19]. Acetylcholine, the first identified neurotransmitter, has been shown to exert lots of its physiological action of GPCRs and modulate neuronal differentiation during early development. Both muscarinic and nicotinic acetylcholine receptors (AChRs) regulate physiological responses widely, including apoptosis, cellular proliferation, and neuronal differentiation [20]. However, the role of muscarinic signaling in neoplasia has exhibited relatively scant attention. Muscarinic acetylcholine receptors are similar to other G protein coupled receptors and have seven transmembrane helical domains connected by three extracellular and three intracellular loops. Muscarinic AChR (CHRM) family comprising five subtypes, M1R to M5R (gene names, CHRM1-CHRM5), regulates cell function via different postreceptor signaling pathways [21]. Robust evidence supporting activation of these receptors plays a critical role in regulation broad-range cancers [22]. Activation of M3R stimulated MEK/ERK signaling to promote cancer cell proliferation in several cancers including brain, breast, colon, and prostate cancers [22].
M3R is the major receptor mediating urinary bladder contraction upon micturition [23]. In normal human urinary bladder, the CHRM3 mRNA expression in bladder areas related to micturition, such as the bladder dome, neck, and trigone [24]. Genetically, CHRM3 gene was mapped to chromosome 1q43. Mutation of CHRM3 showed defects in urinary system, including malformed bladders [25]. Vauhkonen et al. showed that duplication of 1q42-43 was identified in bladder cancer by array CGH analysis [26]. These studies demonstrated that CHRM3 plays a vital role in bladder development and suggested that it may also have pathophysiological role in bladder cancer. However, no solid evidence showed the genetic effect of CHRM3 gene polymorphism on bladder cancer. Thus, we attempted to delineate the genetic association of CHRM3 genotypes and bladder cancer. It is valuable to realize the putative efficiencies in clinical application.
2. Materials and Methods
2.1. Sample Collection
For association analysis, 30 individuals with bladder cancer and 60 noncancer control individuals were recruited in this study (proofed by the institutional review board of Kaohsiung Armed Forces General Hospital). All of them were interviewed and diagnosed by Dr. Chiang-Ting Wang; basic characteristics of both groups were summarized in Table 1. Three milliliter blood samples were collected into EDTA-anticoagulants tubes and their genomic DNA was isolated from blood samples using the DNeasy™ kit (QIAamp DNA Blood Mini kit, Qiagen, Valencia, Germany). Briefly, the blood was digested with 0.5 mg/mL proteinase K in 400 μL cell-lysis solution at 55°C for 24 h until the blood was completely lysed. After adding 200 μL absolute ethanol to the reagents, the mixture was transferred into the DNeasy minicolumn and centrifuged at 8,000 rpm for 1 min. The DNeasy minicolumn was washed with 500 μL washing buffer and centrifuged for 1 min at 8,000 rpm. Finally, the DNA was eluted in a clean 1.5 mL microcentrifuge tube.
Table 1.
Demographic data of individuals collected in this study.
| Patients (n = 31) |
Controls (n = 60) |
pvaluea | |
|---|---|---|---|
| Age (years) | 70.47 ± 12.6 | 53.02 ± 13.66 | 0.001∗ |
| Gender (M/F)b | 25/6 | 48/12 | 0.942 |
| Smoking status (Y/N)c | 7/12 | 17/31 | 0.913 |
| Alcohol drinking (Y/N)c | 4/15 | 11/36 | 0.836 |
| BMI | 25.62 ± 5.08 | 24.95 ± 4.24 | 0.506 |
| Uric acid (mg/day) | 6.72 ± 2.39 | 6.97 ± 2.76 | 0.669 |
Data are presented as mean ± standard deviation.
a Chi-squared tests and student's t-tests were used to perform statistical analysis of differences between the groups. ∗ p < 0.05.
bF, female; M, male.
cY, yes; N, no; information of some individuals is unavailable.
2.2. Tumor Tissue Samples Collection
Thirty bladder cancer tissues of patients and fifty-nine control bladder samples were collected. All samples were frozen in liquid nitrogen immediately after surgical remove and then stored in −70°C.
2.3. Polymorphism Selection and SNPs Genotyping
Five SNPs of CHRM3 gene were selected based on genetic information in HapMap (http://www.hapmap.org/), depending on their genomic distribution and allele frequencies in our population. Allele-specific polymerase chain reaction (AS-PCR) analysis was performed with allele-specific primer sets (Table 2) to genotype alleles of CHRM3 (rs2165870, rs10802789, rs685550, rs7520974, and rs3738435). Briefly, forward primers were designed specifically complementary to each SNP allele and two PCR reactions were conducted with these two specific primer sets in parallel and then detected the allele by using Mycycle™ thermal cycler (Bio-RAD, Hercules, CA, USA). PCR reagents contained 1x PCR Buffer, 1 U of Taq DNA polymerase (TakaRa, Shuzo, Kyoto, Japan), 100 ng of each primer, 200 μmol/L dNTP, and 10 ng of genomic DNA. DNA amplification was through denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 10 min. PCR products were analyzed by 2% agarose gel and also further confirmed via Sanger DNA sequencing.
Table 2.
Primers utilized in this study.
| SNP | Primers, 5′-3′ | Product Size (bp) |
Function |
|---|---|---|---|
| rs2165870 (A/G) |
F: TCCTAGGGCTCTGAAGAATTA | 383 | PCR |
| R: TTCAATTGACATTGCTCAGA | |||
| FA: TGCATCTGTTTACAGCCTTA | 306 | Genotyping | |
| FG: TGCATCTGTTTACAGCCTTG | |||
|
| |||
| rs10802789 (C/T) |
F: CTCGAAGTTAGGCGAGATTT | 397 | PCR |
| R: GTGTGACACGTGTAGGTCAA | |||
| FC: AAAGTGGCATTTCTCTACGC | 213 | Genotyping | |
| FT: AAAGTGGCATTTCTCTACGT | |||
|
| |||
| rs685550 (C/T) |
F: CCACATCAAACGTCGAGAGA | 348 | PCR |
| R: TTGAATCTATGCGCTTGTCG | |||
| FC: ATATAGAGATATTAAACAGC | 241 | Genotyping | |
| FT: ATATAGAGATATTAAACAGT | |||
|
| |||
| rs7520974 (A/G) |
F: TCTCCAAATCAACACTCCTG | 302 | PCR |
| R: CATGTCCTGTAGCACCATTT | |||
| FA: GCTGAAAGAAAGACAAATCA | 175 | Genotyping | |
| FG: GCTGAAAGAAAGACAAATCG | |||
|
| |||
| rs3738435 (C/T) |
F: CAGTGCATGCTACCAATTAT | 361 | PCR |
| R: TGTAGCTGACGAAATGAGTG | |||
| FC: TAAAATAAGAGAATGAACGC | 289 | Genotyping | |
| FT: TAAAATAAGAGAATGAACGT | |||
|
| |||
| CHRM3RT | F: ACCCAGCTCCGAGCAGATGGAC | 339 | RT-qPCR |
| R: CGGCTGACTCTAGCTGGATGGG | |||
| Probe: TGCCTGAGGAGGAGCTGCTGGGGA TGGTGGACTTGGAGAG | |||
SNP: single-nucleotide polymorphism; F: forward; R: reverse.
2.4. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA (1 μg) was used as templates to generate complementary DNA (cDNA) by using the Moloney Murine Leukemia Virus reverse-transcriptase kit according to the manufacturer's instructions (Life Technologies Inc., Paisley, UK). The expression level of CHRM3 transcript was further quantified by specific primers listed in Table 2. PCR amplifications of CHRM3 transcript were through denaturation at 94°C for 2 min; 28 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 30 sec, extension at 72°C for 1 min, and final extension at 72°C for 10 min. RT-qPCR of CHRM3 and β-actin used TaqMan assays (Applied Biosystems) in a real-time PCR system (StepOne™; Applied Biosystems). The levels of CHRM3 mRNA were measured using the 2−ΔΔCt method and normalized to the expression levels of β-actin.
2.5. Statistical Analysis
Hardy-Weinberg equilibrium (HWE) was tested by online HWE program (http://linkage.rockefeller.edu/ott/linkutil.htm), allele and genotype frequencies examined by Fisher's exact (two-tailed) test, using SPSS version 10.0 software (SPSS for Windows Inc., Chicago, IL). Haplotype construction and the differences in haplotype distributions between patients and controls were analyzed using SNPAlyze (SNPAlyze 4.1; Dynacom Co. Ltd., Kanagawa, Japan), p value < 0.05 constituting statistical significance. Odds ratios with 95% confidence intervals were also computed. Additional rigid 10000 consecutive permutation tests for haplotype association were calculated using SAS 8.0 program (SAS, Cary, NC, USA). Powers were estimated using Power Analysis and Sample Size (PASS) 2005 software (NCSS, Kaysville, UT, USA).
Results of RT-qPCR were analyzed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA), all data were represented as mean ± standard deviation (SD), and a p value < 0.05 was considered as a significant difference.
3. Results and Discussion
Urothelial bladder cancer is a multifactorial disease and its etiology is highly complex. Studies showed that bladder cancer is associated with infection, injury, chronic inflammation, tissue repair, and genetics factors [3, 27]. Current theory suggests that using association analysis will provide acceptable power to detect genetic effects on complex diseases [28]. Many risk-associated genetic variants were linked to contribute a subtle effect to complex diseases, such as cancer, by case-control association studies [28]. Although CHRM3 cholinergic receptor has been reported to promote cancer cell proliferation via stimulating MEK/ERK signaling in several different cancers [22], its role in the bladder cancer have not been estimated yet. Industrial exposure is a very important environmental factor for cancers in urinary system [29]. Kaohsiung city, with high plant density, especially lots of high-polluting industries, such as petroleum, petrochemical, dyeing, steel, and iron industries [8, 30], should be chosen to conduct study on the association between environmental or genetic factors and urinary cancer. In this study, we analyzed association of bladder cancer with CHRM3 polymorphism of Taiwanese population in Kaohsiung city, an industrial city with high exposure level of industrial pollution, to evaluate the CHRM3 genetic effect on etiology of bladder cancer.
Bladder cancer patients and volunteer controls who joined this study were characterized by age, body mass index (BMI), male-to-female ratio, smoking status, alcohol drinking status, and urinalysis. Data were summarized in Table 1. With respect to gender ratio, smoking status, alcohol drinking and uric acid, and the control and bladder cancer groups showed no difference in these analyses. According to the Taiwan Cancer Registry Annual Report 2012, patients aged over 50 years account for over 93% of bladder cancer patients. Although this study does not match the cases and controls for age, the average age (53 yrs) of our control groups is slightly older than 50 years.
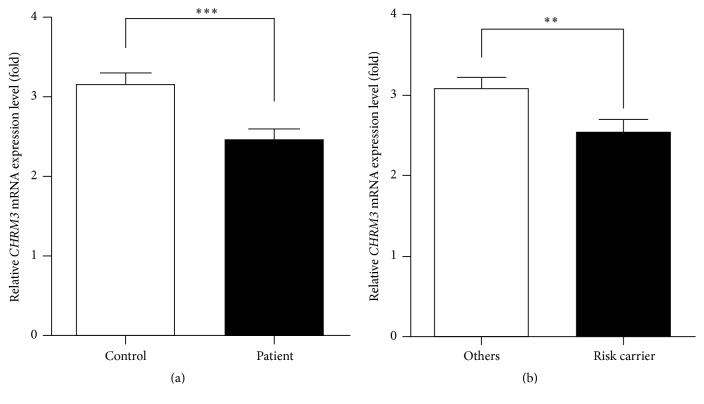
To reveal the expression level of CHRM3 mRNA in bladder tissue of patients and controls, the levels of CHRM3 and β-actin mRNA on 29 patients and 59 controls were measured using real-time qPCR assay. Result showed that the relative level of CHRM3 mRNA was decreased to 0.78-fold (p < 0.0001) in bladder cancer group (Figure 1(a)), implying that CHRM3 expression level might be associated with bladder cancer. To test the genetic association of the CHRM3 polymorphism with bladder cancer, five selected SNPs were genotyped for all our participants. Genotypes of these five SNPs were further confirmed by Sanger DNA sequencing (Supplemental Figure 1). The average genotyping success rate was about 99%. General population-based genotypic and allelic distributions were similar to the data from the Single Nucleotide Polymorphism database (dbSNP) and international HapMap project (https://hapmap.ncbi.nlm.nih.gov/). All SNPs were analyzed to compare allelic and genotypic frequencies between bladder cancer group and control group using the Pearson χ 2 test. There were no significant differences of alleles or genotype distributions in single locus association (Table 3).
Figure 1.
CHRM3 mRNA level is associated with bladder cancer and risk haplotype carrier groups. (a) Bar graph represented the relative CHRM3 mRNA level quantified by real-time qPCR in bladder cancer group and control. CHRM3 mRNA level was decreased to 0.78-fold (p < 0.0001) in bladder cancer group compared to control. The bars represent the mean ± SD. ∗∗∗ p < 0.001. (b) Bar graph represented the relative CHRM3 mRNA level quantified by real-time qPCR in risk carrier and others group. CHRM3 mRNA level was decreased to 0.82-fold (p = 0.0035) compared to others group. The bars represent the mean ± SD. ∗∗ p < 0.01.
Table 3.
Genotype and allele frequencies of each marker analyzed in this study.
| Locus | N | Allelea | χ 2 | p value | Genotypea | χ 2 | p value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1/1 | 1/2 | 2/2 | |||||||
| rs2165870 | P | 30 | 50 | 10 | 0.056 | 0.813 | 22 | 6 | 2 | 0.108 | 0.947 |
| C | 58 | 95 | 21 | 42 | 11 | 5 | |||||
| rs10802789 | P | 30 | 39 | 21 | 0.006 | 0.938 | 12 | 15 | 3 | 0.879 | 0.644 |
| C | 59 | 76 | 42 | 26 | 24 | 9 | |||||
| rs685550 | P | 30 | 42 | 18 | 0.164 | 0.686 | 15 | 12 | 3 | 0.157 | 0.924 |
| C | 59 | 86 | 32 | 32 | 22 | 5 | |||||
| rs7520974 | P | 30 | 31 | 29 | 0.292 | 0.589 | 8 | 15 | 7 | 0.498 | 0.780 |
| C | 59 | 66 | 52 | 20 | 26 | 13 | |||||
| rs3738435 | P | 30 | 39 | 21 | 0.090 | 0.764 | 13 | 13 | 4 | 0.167 | 0.920 |
| C | 59 | 74 | 44 | 23 | 28 | 8 | |||||
a1 indicates the major allele, and 2 indicates the minor allele.
P, patients with bladder cancer; C, control individuals.
Haplotype-based association analysis comprising multiple SNPs can provide additional power for mapping disease genes and also provide insight on factors influencing the dependency among genetic markers and identifying cis-interactions between two or more variants. Therefore, we investigated the genetic associations in haplotype-based analysis for CHRM3 gene with bladder cancer. Haplotype-based genetic association tests were done using the estimated haplotype frequencies. Although there was no single locus association between any CHRM3 polymorphisms with bladder cancer, there was a significantly different distribution of several haplotypes between groups (Table 4). In addition, significant differences on some haplotypes were detected even after rigid 10000 consecutive permutation tests. The distribution of the most common haplotype in control groups, GTTAC, showed a significant protective effect (permutation p = 0.027; OR: 0.35, 95% CI: 0.136–0.903, power = 61% at α = 0.05). And 4 of the demonstrated 16 haplotypes only occurred in control groups (ACTGT, GCCAT, ATTAC, and ACCGC) and also showed protective effect for bladder cancer. On the other hand, haplotype GTTAT (p = 0.029; OR: 3; 95% CI: 1.079–8.344) and ATTGT (p = 0.036; OR: 3.292; 95% CI: 1.027–10.56) are associated with significantly risk effects. However, after rigid permutation analysis, haplotype ATTGT was not significantly associated. There are also three haplotypes only observed in bladder cancer patients (GCTAC, ACTAC, and ACCAC) and showed risk effect for bladder cancer. Significant differences of GCTAC were still detected after rigid consecutive permutation tests (permutation p = 0.044; power = 66% at α = 0.05). Significant differences were also assessed in overall haplotype frequency profiles (total p = 0.0019; global permutation p = 0.0008, power = 99% at α = 0.05).
Table 4.
Haplotype frequency and test statistics between bladder cancer patients and controls.
| Haplotypea | Numbers (frequency) | p value | OR (95% CI) | Permutation test |
||
|---|---|---|---|---|---|---|
| Overall | Controls | Patients | pvaluec [W, power] | |||
| G-T-T-G-T | 34 | 25 | 9 | 0.251 | 0.614 (0.266–1.418) | 0.317 |
| G-T-T-A-C | 33 | 27 | 6 | 0.025∗ | 0.350 (0.136–0.903) | 0.027∗[0.17, 61%] |
| G-C-C-G-T | 22 | 15 | 7 | 0.747 | 0.854 (0.328–2.225) | 0.814 |
| G-T-T-A-T | 17 | 7 | 10 | 0.029∗ | 3.000 (1.079–8.344) | 0.036∗[0.17, 61%] |
| G-C-C-A-C | 12 | 5 | 7 | 0.077 | 2.826 (0.857–9.327) | 0.112 |
| A-C-T-G-T | 8 | 8 | 0 | 0.034∗ | —b | 0.056 [0.16, 56%] |
| A-T-T-G-T | 13 | 5 | 8 | 0.036∗ | 3.292 (1.027–10.560) | 0.065 [0.14, 45%] |
| G-T-C-G-T | 7 | 4 | 3 | 0.651 | 1.421 (0.307–6.569) | 0.696 |
| G-C-C-A-T | 3 | 3 | 0 | 0.201 | —b | 0.549 |
| G-T-T-G-C | 6 | 3 | 3 | 0.429 | 1.912 (0.374–9.780) | 0.430 |
| G-C-T-G-T | 6 | 4 | 2 | 0.935 | 0.931 (0.166–5.237) | 1.000 |
| A-T-T-A-C | 4 | 4 | 0 | 0.139 | —b | 0.308 |
| G-C-T-A-C | 3 | 0 | 3 | 0.017∗ | —b | 0.044∗[0.18, 66%] |
| A-C-C-G-C | 2 | 2 | 0 | 0.298 | —b | 0.542 |
| A-C-T-A-C | 1 | 0 | 1 | 0.171 | —b | 0.347 |
| A-C-C-A-C | 1 | 0 | 1 | 0.171 | —b | 0.350 |
|
| ||||||
| Total p value: 1.946E − 3 | ||||||
| Global p value of permutation testd: 0.0008∗∗∗ [0.46, 99%] | ||||||
∗ p < 0.05 and ∗∗∗ p < 0.0001.
aHaplotypes were constructed with CHRM3 SNPs: (from left to right) rs2165870, rs10802789, rs685550, rs7520974, and rs3738435.
bNo OR was calculated for this haplotype.
c10000 permutations were replicated to evaluate permutation p value.
dThe global p value was calculated using a permutation test (n = 10,000).
Interestingly, we found that although the selected CHRM3 gene polymorphisms were not significantly associated with bladder cancer on single locus level, the interaction-considered haplotype-based analysis showed that the CHRM3 gene was significantly associated with bladder cancer. To reveal if these risk haplotype indeed has biological effect, we measure the mRNA expression level on risk haplotype carriers and noncarriers. SNPs located at regulatory regions were considered as functional variants for gene expression and contribute to the disease risk [31, 32]. All of the selected SNPs are located on 5′ introns and may play a role in regulating CHRM3 gene expression [4]. Therefore, we hypothesized that the CHRM3 gene expression level on risk haplotype carriers may differ compared to other individuals. To reveal our hypothesis, we performed cross-analysis of the CHRM3 mRNA qPCR data with risk haplotype analysis. Based on the haplotype data, all of the involved individuals were regrouped into risk haplotype carriers (GTTAT, ATTGT, GCTAC, ACTAC, and ACCAC; odds ratio > 1, 95% CI > 1, or only observed in bladder cancer patients) (n = 28) and others (n = 62), and then their CHRM3 mRNA expression level was compared. Surprisingly, the relative amount of CHRM3 mRNA was significantly reduced to 0.82-fold in the risk haplotype carriers group compared to others (p = 0.0035; Figure 1(b)). These data revealed that at least some of the selected SNPs have biological function and affect the CHRM3 gene expression. To our knowledge, this is the first association study to reveal the contribution of the genetic effect of CHRM3 gene in bladder cancer, and to expose that these risk haplotypes not only are genetically associated but indeed correlate with altered CHRM3 gene expression level, at least among the studied Chinese Han population in Kaohsiung city.
Cholinergic receptors are generally classified into muscarinic or nicotinic categories depending on the binding affinity of two naturally occurring substances, muscarine and nicotine. G proteins regulate extensively biological processes by modulating the activity of adenylyl cyclase, phosphatidylinositol lipid turnover, and ion channels [33]. The G protein- and acetylcholine- (ACh-) binding site are located at intracellular loop and sixth transmembrane domain, respectively [22]. The CHRM family regulates cell function via different postreceptor signaling pathways [21]. The M1R, M3R, and M5R exhibit selectively G proteins of Gq/11 family, which activate phospholipase C-β to initiate the phosphatidylinositol trisphosphate signaling cascade. Activation of these 3 receptors causes phospholipid turnover and alters cell calcium concentration. The M2R and M4R preferentially couple to Gi/o type G proteins that inhibit adenylyl cyclase activity [34]. Activating these two receptors results in suppression of adenylyl cyclase and attenuated levels of cAMP.
Muscarinic receptors are well defined and long-established as apparatus in neuronal system [35]. Likewise, muscarinic receptors are expressed in various organ systems including gastrointestinal tract, brain, eye, heart, lung bronchioles, urinary bladder, and uterus [36]. The pathological roles of muscarinic receptors have also been reported in 1991. Muscarinic receptors are conditional oncogenes by activating downstream phosphatidylinositol hydrolysis when they are expressed in cells capable of proliferation [37]. Muscarinic receptor was also linked to brain, breast, colon, lung, prostate cancer, and melanoma [22]. Robust evidence supports the idea that activation of these receptors plays a critical role in regulation broad-range cancers [22]. It has been found that CHRM3 receptors are abundantly expressed on 60% of studied colon cancer cell lines (6/10) [38]. In one small study, five of the eight (62%) showed an up to 8-fold expression level of CHRM3 mRNA compared with the matched normal tissue [39]. Activation of M3R stimulated MEK/ERK signaling to promote cancer cell proliferation in several cancers including brain, breast, colon, and prostate cancers [22]. Postmuscarinic receptor signaling cascade included NF-κB, AKT, and cyclin D1 signaling pathways resulting in augment cell survival in diverse cancers. These pathways raise gene transcription favoring cell proliferation and survival, both hallmarks of neoplasia. Furthermore, CHRM3 knockout mice were shown to attenuate intestinal tumor number and size, revealing an oncogenic role of CHRM3 in cancer progression [40]. These studies suggested a functional role of CHRM3 in cancer and further support the result from our genetic study. Although studies showed CHRM3 mRNA are overexpressed in colon cancer and activation of M3R stimulated MEK/ERK signaling to promote cancer cell proliferation, less study was performed to reveal the role of CHRM3 in bladder cancer. The rodent cancer model showed that increased fecal bile acids promote colon dysplasia through interacting with muscarinic receptors [41]. Therefore, the overreactive signaling of muscarinic receptors may be caused by increasing the amount of the receptor or its ligands [42]. Our data showed a minor reduction of CHRM3 mRNA in risk haplotype carriers group and bladder cancer samples, distinct from finding on colon cancer studies (Figure 1(b)). The reason may result from different pathological mechanism, different clinicopathological features of the studied patients, tissue-specific expression of CHRM3 mRNA, or small sample size. It is not uncommon that single gene has multifaceted role in different cancer types. For example, PRL-3 gene plays a tumor suppressor role in lung cancer, distinct from observation on colon and gastric cancers [43]. Recently, one study revealed that CHRM3 expression level was correlated with some clinicopathological features, such as FIGO stage, vascular invasion, and lymphatic metastasis [44]. Majority of our cancer patients belong to the FIGO stage I with no regional lymph node and distant metastasis that should fit into the low expression group. However, the analyzed cell lines and cancer patients in colon cancer studies are more clinical heterogeneity and may lead to different CHRM3 expression patterns. One of the other possibilities is the cell type-specific regulation of CHRM3 mRNA expression. The cell type of bladder cancer samples in our study is all urothelial carcinoma, also known as transitional cell carcinoma (TCC), and the most common type of colon cancer is adenocarcinoma. Gene regulation machinery and their role on the carcinogenesis could be different in different cell types. And the actual roles of CHRM-3 in bladder cancer etiology are needed to be clarified.
4. Conclusions
Evidence from our genetic and functional studies suggested that some risk haplotypes were significantly associated with low CHRM3 mRNA level and contributed to increased susceptibility of bladder cancer in Kaohsiung city. In conclusion, results from other research groups and ours confirm the importance of muscarinic signaling in bladder cancer, which further suggests that intervention of muscarinic signaling may provide novel therapeutic or prognostic strategies for this disease. Although the association of bladder cancer and CHRM3 is significant, we could not completely exclude a possibility of false-positives because of small sample size or other factors (e.g., confounders with other medical conditions and adjustment of other risk factors). However, we have provided additional power estimation and rigid permutation test to support our conclusion. Moreover, besides the SNPs explored in this study, there may have been other polymorphisms associated with bladder cancer; a systematic association study with a large sample size is considerable in the following study.
Supplementary Material
The Supplementary Material included one supplementary table and one supplementary figure as follows. Supplementary Table1: Summary of plants and populations on different areas in Taiwan, 2012. Supplementary Figure S1: Sequencing results of CHRM3 SNPs.
Acknowledgments
This work was funded by Kaohsiung Armed Forces General Hospital (101-051 to Chiang-Ting Wang), the Ministry of Science and Technology of Taiwan (NSC 102-2320-B-039-053 to Ya-Huey Chen; MOST 103-2320-B-022-001 and MOST 105-2628-B002-001-MY3 to Tsung-Ming Chen), the Joint Research Project of NSYSU/NKMU (#105-P017 to Tsung-Ming Chen), and the Joint Research Project of NCHU/CCH (103S0514 to Yi-Mei Joy Lin). The authors thank Kaohsiung Armed Forces General Hospital for their support and Miss Ya-Fen Shiao (Asia-Pacific Biotech Developing Inc.) for assistance in gene polymorphism analysis. They also appreciate Mr. Charles Wagamon for the critical reading and editing of the manuscript and all people participating in this investigation.
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA: Cancer Journal for Clinicians. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Hirao Y., Kim W.-J., Fujimoto K. Environmental factors promoting bladder cancer. Current Opinion in Urology. 2009;19(5):494–499. doi: 10.1097/MOU.0b013e32832eb4ef. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L., Davison D. D., Adams J., et al. Biomarkers in bladder cancer: translational and clinical implications. Critical Reviews in Oncology/Hematology. 2014;89(1):73–111. doi: 10.1016/j.critrevonc.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Abente G., Aragones N., Ramis R., et al. Municipal distribution of bladder cancer mortality in Spain: possible role of mining and industry. BMC Public Health. 2006;6(1, article 17):1. doi: 10.1186/1471-2458-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taiwan Cancer Registry Annual Report. Taipei City, Taiwan: Health Promotion Administration, Ministry of Health and Welfare; 2012. [Google Scholar]
- 6.Kiemeney L. A., Thorlacius S., Sulem P., et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nature Genetics. 2008;40(11):1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X., Ye Y., Kiemeney L. A., et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nature Genetics. 2009;41(9):991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothman N., Garcia-Closas M., Chatterjee N., et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nature Genetics. 2010;42(11):978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiemeney L. A., Sulem P., Besenbacher S., et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nature Genetics. 2010;42(5):415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafnar T., Vermeulen S. H., Sulem P., et al. European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Human Molecular Genetics. 2011;20(21):4268–4281. doi: 10.1093/hmg/ddr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Closas M., Ye Y., Rothman N., et al. A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Human Molecular Genetics. 2011;20(21):4282–4289. doi: 10.1093/hmg/ddr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gago-Dominguez M., Jiang X., Conti D. V., et al. Genetic variations on chromosomes 5p15 and 15q25 and bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Carcinogenesis. 2011;32(2):197–202. doi: 10.1093/carcin/bgq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern M. C., Van Den Berg D., Yuan J.-M., et al. Sequence variant on 3q28 and urinary bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Cancer Epidemiology Biomarkers and Prevention. 2009;18(11):3057–3061. doi: 10.1158/1055-9965.epi-09-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M., Chu H., Yan F., et al. Chromosome 4p16.3 variant modify bladder cancer risk in a Chinese population. Carcinogenesis. 2011;32(6):872–875. doi: 10.1093/carcin/bgr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Z., Hu Q., Chen Z., et al. Systematic evaluation of bladder cancer risk-associated single-nucleotide polymorphisms in a chinese population. Molecular Carcinogenesis. 2013;52(11):916–921. doi: 10.1002/mc.21932. [DOI] [PubMed] [Google Scholar]
- 16.Wang S., Tang J., Wang M., Yuan L., Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a chinese population. Carcinogenesis. 2010;31(4):621–624. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 17.Wang M., Wang M., Zhang W., et al. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30(6):991–996. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- 18.Schäfer B., Gschwind A., Ullrich A. Multiple G-protein-coupled receptor signals converge on the epidermal growth factor receptor to promote migration and invasion. Oncogene. 2004;23(4):991–999. doi: 10.1038/sj.onc.1207278. [DOI] [PubMed] [Google Scholar]
- 19.Tang X.-L., Wang Y., Li D.-L., Luo J., Liu M.-Y. Orphan G protein-coupled receptors (GPCRs): biological functions and potential drug targets. Acta Pharmacologica Sinica. 2012;33(3):363–371. doi: 10.1038/aps.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resende R. R., Adhikari A. Cholinergic receptor pathways involved in apoptosis, cell proliferation and neuronal differentiation. Cell Communication and Signaling. 2009;7, article 20 doi: 10.1186/1478-811x-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonner T. I., Buckley N. J., Young A. C., Brann M. R. Identification of a family of muscarinic acetylcholine receptor genes. Science. 1987;237(4814):527–532. doi: 10.1126/science.3037705. [DOI] [PubMed] [Google Scholar]
- 22.Shah N., Khurana S., Cheng K., Raufman J.-P. Muscarinic receptors and ligands in cancer. American Journal of Physiology—Cell Physiology. 2009;296(2):C221–C232. doi: 10.1152/ajpcell.00514.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler C. J., Griffiths D., De Groat W. C. The neural control of micturition. Nature Reviews Neuroscience. 2008;9(6):453–466. doi: 10.1038/nrn2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chess-Williams R. Muscarinic receptors of the urinary bladder: detrusor, urothelial and prejunctional. Autonomic and Autacoid Pharmacology. 2002;22(3):133–145. doi: 10.1046/j.1474-8673.2002.00258.x. [DOI] [PubMed] [Google Scholar]
- 25.Weber S., Thiele H., Mir S., et al. Muscarinic acetylcholine receptor m3 mutation causes urinary bladder disease and a prune-belly-like syndrome. American Journal of Human Genetics. 2011;89(5):668–674. doi: 10.1016/j.ajhg.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vauhkonen H., Böhling T., Eissa S., Shoman S., Knuutila S. Can bladder adenocarcinomas be distinguished from schistosomiasis-associated bladder cancers by using array comparative genomic hybridization analysis? Cancer Genetics and Cytogenetics. 2007;177(2):153–157. doi: 10.1016/j.cancergencyto.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Karin M., Greten F. R. NF-κB: Linking inflammation and immunity to cancer development and progression. Nature Reviews Immunology. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 28.Cardon L. R., Bell J. I. Association study designs for complex diseases. Nature Reviews Genetics. 2001;2(2):91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 29.Clin B., Pairon J.-C. Medical follow-up for workers exposed to bladder carcinogens: the French evidence-based and pragmatic statement. BMC Public Health. 2014;14:p. 1155. doi: 10.1186/1471-2458-14-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y.-M., Lin W.-Y., Chan C.-C. The impact of petrochemical industrialisation on life expectancy and per capita income in Taiwan: an 11-year longitudinal study. BMC Public Health. 2014;14(1, article 247) doi: 10.1186/1471-2458-14-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T.-M., Kuo P.-L., Hsu C.-H., et al. Microsatellite in the 3′ untranslated region of human fibroblast growth factor 9 (FGF9) gene exhibits pleiotropic effect on modulating FGF9 protein expression. Human mutation. 2007;28(1):p. 98. doi: 10.1002/humu.9471. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y.-M. J., Chao S.-C., Chen T.-M., Lai T.-J., Chen J.-S., Sun H. S. Association of functional polymorphisms of the human tryptophan hydroxylase 2 gene with risk for bipolar disorder in Han Chinese. Archives of General Psychiatry. 2007;64(9):1015–1024. doi: 10.1001/archpsyc.64.9.1015. [DOI] [PubMed] [Google Scholar]
- 33.Malbon C. C. G proteins in development. Nature Reviews Molecular Cell Biology. 2005;6(9):689–701. doi: 10.1038/nrm1716. [DOI] [PubMed] [Google Scholar]
- 34.Kruse A. C., Hu J., Pan A. C., et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482(7386):552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valenstein E. S. The War of The Soups and The Sparks : The Discovery of Neurotransmitters and The Dispute Over How Nerves Communicate. New York, NY, USA: Columbia University Press; 2005. [Google Scholar]
- 36.Abrams P., Andersson K.-E., Buccafusco J. J., et al. Muscarinic receptors: their distribution and function in body systems, and the implications for treating overactive bladder. British Journal of Pharmacology. 2006;148(5):565–578. doi: 10.1038/sj.bjp.0706780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutkind J. S., Novotny E. A., Brann M. R., Robbins K. C. Muscarinic acetylcholine receptor subtypes as agonist-dependent oncogenes. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(11):4703–4707. doi: 10.1073/pnas.88.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frucht H., Jensen R. T., Dexter D., Yang W.-L., Xiao Y. Human colon cancer cell proliferation mediated by the M3 muscarinic cholinergic receptor. Clinical Cancer Research. 1999;5(9):2532–2539. [PubMed] [Google Scholar]
- 39.Yang W.-L., Frucht H. Cholinergic receptor up-regulates COX-2 expression and prostaglandin E2 production in colon cancer cells. Carcinogenesis. 2000;21(10):1789–1793. doi: 10.1093/carcin/21.10.1789. [DOI] [PubMed] [Google Scholar]
- 40.Raufman J.-P., Shant J., Xie G., et al. Muscarinic receptor subtype-3 gene ablation and scopolamine butylbromide treatment attenuate small intestinal neoplasia in Apcmin/+ mice. Carcinogenesis. 2011;32(9):1396–1402. doi: 10.1093/carcin/bgr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narisawa T., Magadia N. E., Weisburger J. H., Wynder E. L. Promoting effect of bile acids on colon carcinogenesis after intrarectal instillation of N methyl N' nitro N nitrosoguanidine in rats. Journal of the National Cancer Institute. 1974;53(4):1093–1097. doi: 10.1093/jnci/53.4.1093. [DOI] [PubMed] [Google Scholar]
- 42.von Rosenvinge E. C., Raufman J.-P. Muscarinic receptor signaling in colon cancer. Cancers. 2011;3(1):971–981. doi: 10.3390/cancers3010971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin S. Y., Lee Y. X., Yu S. L., et al. Phosphatase of regenerating liver-3 inhibits invasiveness and proliferation in non-small cell lung cancer by regulating the epithelial-mesenchymal transition. Oncotarget. 2016;7(16):21799–21811. doi: 10.18632/oncotarget.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Li J., Wen S., et al. CHRM3 is a novel prognostic factor of poor prognosis in patients with endometrial carcinoma. American Journal of Translational Research. 2015;7(5):902–911. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary Material included one supplementary table and one supplementary figure as follows. Supplementary Table1: Summary of plants and populations on different areas in Taiwan, 2012. Supplementary Figure S1: Sequencing results of CHRM3 SNPs.