Abstract
Excessive oxidative stress induces dysregulation of functional networks in the retina, resulting in retinal diseases such as glaucoma, age-related macular degeneration, and diabetic retinopathy. Although various therapies have been developed to reduce oxidative stress in retinal diseases, most have failed to show efficacy in clinical trials. This may be due to oversimplification of target selection for such a complex network as oxidative stress. Recent advances in high-throughput technologies have facilitated the collection of multilevel omics data, which has driven growth in public databases and in the development of bioinformatics tools. Integration of the knowledge gained from omics databases can be used to generate disease-related biological networks and to identify potential therapeutic targets within the networks. Here, we provide an overview of integrative approaches in the drug discovery process and provide simple examples of how the approaches can be exploited to identify oxidative stress-related targets for retinal diseases.
1. Introduction
The retina is exposed to chronic oxidative stress (OS) through several mechanisms, including constant exposure to light and reactive oxygen species generated by visual signal transduction pathways [1]. In the healthy state, all cell types in the retina are able to maintain homeostasis under conditions of OS [2]. However, when the balance between pro- and antioxidative signaling is compromised, excessive OS induces dysregulation of functional networks and deleterious changes that result in various retinal diseases, including glaucoma [3–5], age-related macular degeneration (AMD) [6, 7], diabetic retinopathy [8], and retinitis pigmentosa [9]. Due to a combination of lifestyle changes and extended life expectancy, an increasing number of people are at risk for these retinal diseases, and the resulting economic burden imposed on health care systems is increasing accordingly. Various therapies have been developed for retinal diseases [10] with some success, most notably the prostaglandin analogs for glaucoma [11] and antivascular endothelial growth factor (anti-VEGF) agents for AMD [12] and diabetic retinopathy [13]. However, there has been a chronic lack of innovation in drug discovery for retinal diseases [14], as there has been for other diseases [15]. For example, therapies targeting OS in retinal diseases have failed to show efficacy in clinical trials; examples are an antioxidant supplement mixture [16] and a hydroxylamine with antioxidant properties [17]. Thus, there is a clear need for novel approaches to the drug discovery process [18–25]. Here, we first provide an overview of some emerging integrative approaches to therapeutic target discovery and then provide some examples as they relate to OS in retinal diseases.
2. Integrative Approaches in the Search for Therapeutic Targets for OS in Retinal Diseases
2.1. Overview of the Integrative Approaches
Improving our understanding of disease pathogenesis is an important step in the identification of therapeutic targets [14]. Recent technological advances have enabled us to obtain large amounts of multilevel omics data [20, 21]. For example, DNA microarray and next-generation sequencing technologies have made it relatively easy to obtain genomics data, including profiling of single nucleotide polymorphisms, copy number variations (CNVs), and mutations, which can be used for genome-wide association studies (GWAS) to identify diseases associated with these changes. The same technologies have facilitated the collection of epigenomics data, such as profiles of DNA methylation and DNA-binding sites, and of transcriptomics data, such as messenger and noncoding RNA profiles. In addition, advanced mass spectrometry-based technologies have been indispensable for profiling of protein expression and protein-protein interactions (proteomics), intermediary metabolites (metabolomics), and lipids (lipidomics) [26, 27]. The more recently developed clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9) technology has revolutionized the approach to large-scale loss-of-gene-function experiments and phenotypic analysis (phenomics) at both the in vitro and the in vivo levels [28, 29]. In turn, the need to distill these multilevel omics datasets into biological knowledge has led to equally important advances in data curation supported by public databases and novel bioinformatics tools. Some examples are ENCODE for gene expression regulation [30], STRING for protein-protein interactions [31], gene ontology for gene function [32], KEGG for signaling pathways [33], OMIM for human diseases [34], and DrugBank for chemical structures and drug targets [35]. The biological research literature, in combination with text-mining tools such as Agilent Literature Search [36], represents a rich and ever-expanding source of biological knowledge [37]. Integration of multiomics data with existing biological knowledge is essential for generating accurate disease-related networks and for identifying potential therapeutic targets (Figure 1). For example, a recent meta-analysis of omics data identified a number of approved drugs that could potentially be repurposed for the treatment of rheumatoid arthritis (RA) [36]. In that study, RA-associated genes identified by GWAS were prioritized based on eight criteria, including expression quantitative trait loci, protein-protein interactions, pathway analysis, and text mining. This analysis revealed that targets of approved therapies for RA and other indications were significantly enriched in the prioritized genes [38]. Such network-based frameworks generated by integration of multilevel omics data and biological knowledge can be extended to address numerous problems, including interpretation of GWAS data, identification of disease modules located near the drug target, and discovery of disease-disease relationships [39].
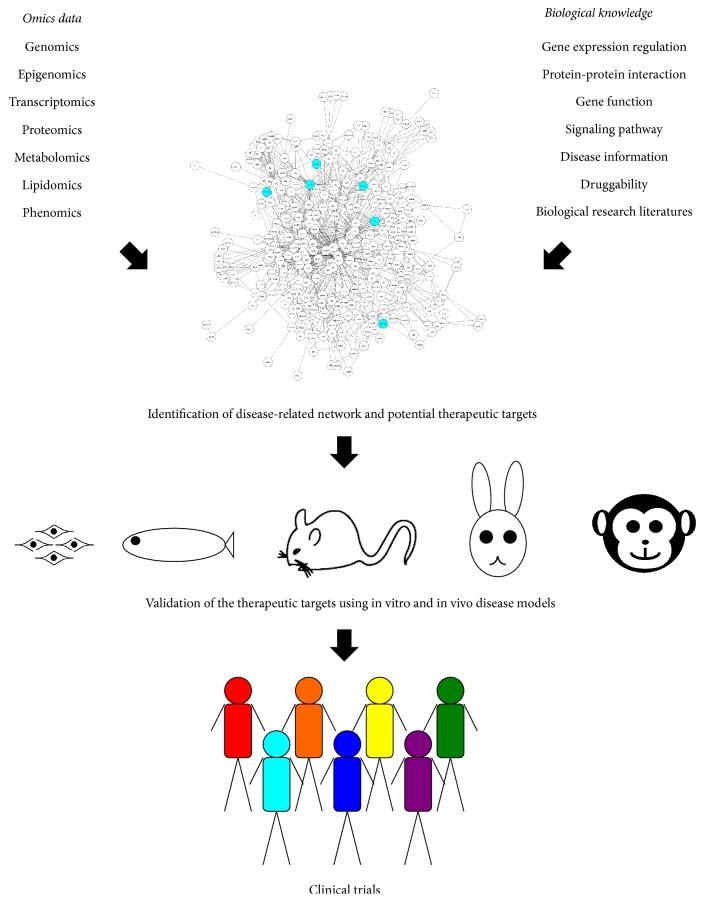
Figure 1.
Integrative approaches to identify therapeutic targets. Integration of multilevel omics data and biological knowledge allows construction of disease-related networks and the discovery of potential therapeutic targets (represented as cyan circles in the network). In vitro and in vivo disease models can be used to validate the therapeutic targets and drugs, and drugs displaying efficacy in preclinical models can then be moved into clinical trials.
Nevertheless, the potential therapeutic targets identified by such integrated approaches will still need to be validated using in vitro and in vivo disease models. Three-dimensional retinal cultures derived from human or mouse embryonic stem cells and induced pluripotent stem cells have been developed and can be used to validate the therapeutic targets for retinal diseases [40–42]. Advances in genome-editing technologies such as transcription activator-like effector nucleases (TALEN) and CRISPR-Cas9 have made it possible to knock out any gene of interest in various species, including teleosts such as zebrafish [43–47]. Indeed, a number of retinal disease models have been developed in zebrafish [48–55], rodents such as mouse and rat, lagomorphs such as rabbit [56–59], and primates such as the common marmoset and cynomolgus monkey [60–62]. Such models have been used successfully to validate the efficacy of therapeutic drugs for retinal diseases, allowing them to be moved forward into clinical trials (Figure 1).
2.2. An Integrative Approach towards Drug Discovery for Glaucoma-Related OS
To illustrate how this integrative approach can be used for OS in retinal diseases, we examined (i) whether glaucoma-associated genes identified by GWAS might be connected through genes related to OS and, if so, (ii) whether the network could identify therapeutic targets to reduce OS in glaucoma. To this end, we used Agilent Literature Search [36], a literature mining tool that can extract biological associations related to a target entity (e.g., gene, mRNA, protein, molecule, chemical, drug, and disease) in a particular context from biomedical literature databases such as OMIM [34] and PubMed [63]. The relationships identified through this analysis (e.g., in our case, glaucoma-associated gene X induces the expression of gene A through interaction with gene B under conditions of OS) can be represented as a network(s) using Cytoscape [64]. If the network of glaucoma-associated gene X and that of glaucoma-associated gene Y share a node(s), these networks can be connected through the node(s), resulting in an integrated network. We used 39 glaucoma-associated genes identified by GWAS [65–67] as the target entities and “oxidative stress” as the designated context in an Agilent Literature Search. The resulting network, shown in Figure 2, contains two glaucoma-associated genes: ATP-binding cassette subfamily A member 1 (ABCA1) and thioredoxin reductase 2 (TXNRD2) (shown in yellow in Figure 2). Gene ontology analysis using DAVID [68] revealed that genes related to “response to oxidative stress” are significantly (p = 2.5 × 10−10) enriched in the network, suggesting that ABCA1 and TXNRD2 are connected through genes related to OS. Superoxide dismutase 2 (SOD2, underlined in red in Figure 2) is another gene related to “response to oxidative stress” and connects to both ABCA1 and TXNRD2.
Figure 2.
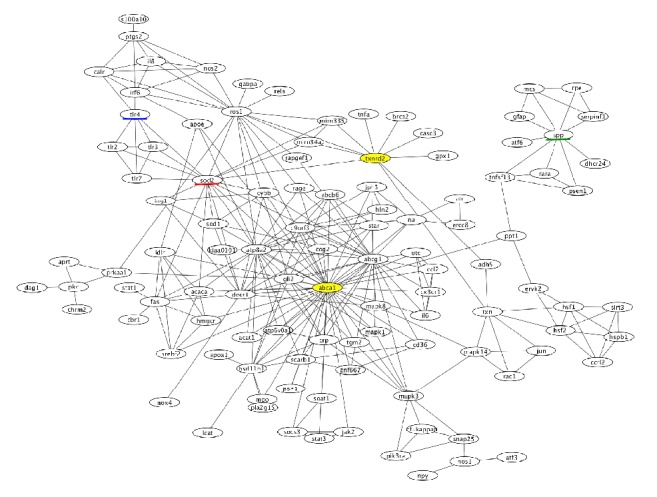
A network related to oxidative stress in glaucoma. An Agilent Literature Search was used as the basis for the network. We used 39 glaucoma-associated genes identified by GWAS as the target molecules and “oxidative stress” as the designated context in the Agilent Literature Search. The glaucoma-associated genes in the network are shown in yellow and other potential therapeutic targets are underlined.
Probucol, an approved drug for hyperlipidemia, has been reported to inhibit ABCA1 activity [69] and increase SOD2 activity [70]. Both SOD2 and TXNRD2 are mitochondrial antioxidant stress enzymes [71], and mitochondrial dysfunction has been causally related to glaucoma [3]. Because probucol can ameliorate mitochondrial dysfunction [71], this observation raises the possibility that probucol could be used therapeutically as an OS suppressor for glaucoma. In fact, probucol protects against glutamate-induced cytotoxicity in a neuronal cell line [72] and, intriguingly, glutamate toxicity is one of the main pathogenic mechanisms of normal-tension glaucoma [73–76].
Other genes in the network could also be potential therapeutic targets for glaucoma-associated OS. For example, amyloid β (1–42), which is a proteolytic processing product of amyloid precursor protein (APP, underlined in green in Figure 2), is increased in the optic nerve head of monkeys and humans with glaucoma [77, 78]. Modulation of amyloid β aggregation can reduce apoptosis of retinal ganglion cells in a rat model of glaucoma [79]. OS and amyloid β aggregation exhibit reciprocal stimulation and induce neurodegeneration in both the brain and the retina [80]. In humans, an antibody against amyloid β reduces plaque formation and attenuates clinical decline in Alzheimer's disease [81]. Collectively, these findings suggest that amyloid β is a potential therapeutic target for glaucoma-associated OS.
Toll-like receptor 4 (TLR4, underlined in blue in Figure 2) is another potential target. Oxidized lipoprotein activates macrophages/microglia through TLR4 and promotes inflammation [82, 83]. Expressions of TLR4 and angiotensin II type 1 receptor (AGTR1) are increased in a mouse model of glaucoma, and an AGTR1 antagonist suppresses neurodegeneration in the mouse retina by inhibiting the TLR4-apoptosis signal-regulating kinase 1 pathway [84]. Modulators of TLR4 signaling have already been developed [85, 86], and the studies described here suggest that such modulators could be used to target OS in glaucoma.
2.3. An Integrative Approach towards Drug Discovery for AMD-Related OS
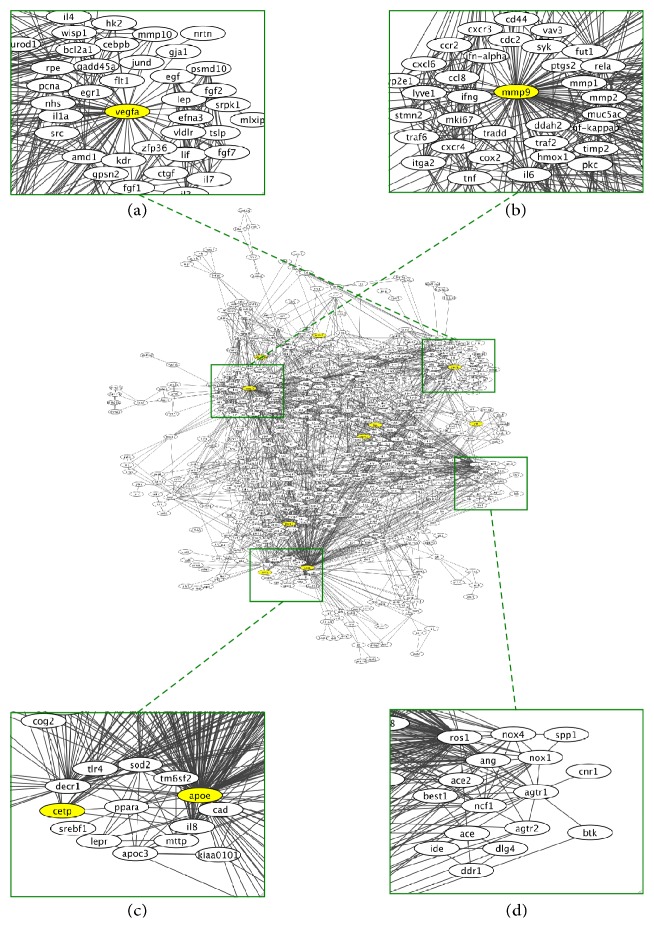
We applied the same approach to identify potential therapeutic targets for AMD-related OS. We used 25 AMD-associated genes identified by GWAS [87, 88] as the target molecules and “oxidative stress” as the designated context in an Agilent Literature Search. The resulting network (shown in Figure 3) contains ten AMD-associated genes (shown in yellow), including vascular endothelial growth factor A (VEGFA, Figure 3(a)) and matrix metallopeptidase 9 (MMP9, Figure 3(b)). Gene ontology analysis using DAVID [68] revealed that genes related to “response to oxidative stress” are significantly (p = 5.2 × 10−16) enriched in the network, suggesting that these ten AMD-associated genes are connected through genes related to OS.
Figure 3.
A network related to oxidative stress in age-related macular degeneration (AMD). An Agilent Literature Search was used as the basis for the network. We used 25 AMD-associated genes identified by GWAS as the target molecules and “oxidative stress” as the designated context in the Agilent Literature Search. The AMD-associated genes in the network are shown in yellow. The subnetworks containing potential therapeutic targets for AMD-related OS are enlarged in (a)–(d).
OS is a major stimulator of VEGFA production and secretion by retinal pigment epithelial cells [89]. Notably, high VEGFA levels also increase oxidative damage, resulting in early degenerative changes in retinal pigment epithelial cells followed by neovascular AMD [90]. Intravitreal injection of anti-VEGF agents can slow the progression of neovascular AMD [12, 89], suggesting that VEGFA is one of the most important therapeutic targets for AMD-related OS.
OS also increases the expression of MMP9 [91] at an early stage of choroidal neovascularization (CNV) [92, 93]. The MMP inhibitors batimastat and marimastat reduce CNV when applied early in the process [93, 94], suggesting a potential therapeutic role for MMP9 inhibitors in AMD.
Other genes in the AMD-OS network may also be therapeutic targets. For example, peroxisome proliferator-activated receptor α (PPARA, Figure 3(c)) is the pharmacological target of fibrates such as fenofibrate, clofibrate, and bezafibrate [35]. PPARA is associated with both antioxidant and anti-inflammatory activities and has previously been suggested as a therapeutic target in AMD [95–97]. In fact, several clinical trials have revealed that fenofibrate can improve diabetic retinopathy [98, 99], which shares some pathophysiological mechanisms with AMD, including OS [100, 101]. These findings suggest that PPARA-activating drugs might have therapeutic utility for AMD.
Several components of the renin-angiotensin system, including angiotensin (ANG), angiotensin I converting enzyme (ACE), ACE2, and angiotensin II receptor type 1 and type 2 (AGTR1 and AGTR2), are shown in the subnetwork (Figure 3(d)). The renin-angiotensin system regulates various biological functions, including OS [102], and is hyperactivated in both AMD and diabetic retinopathy [103, 104]. Several clinical trials have demonstrated some efficacy of renin-angiotensin system inhibitors in slowing the progression of diabetic retinopathy [102, 105]. Thus, the renin-angiotensin system may also be a source of therapeutic targets for AMD-related OS.
3. Conclusion
Here, we provided an overview of some integrative approaches to drug discovery for OS in retinal diseases. We used two simple examples of glaucoma and AMD to illustrate how the approach can identify network hubs as potential therapeutic targets for these retinal diseases. The rapid advances in technology and increasing volume of multilevel omics data continue to create larger and more complex datasets to understand disease-associated biological networks and to build more extensive drug-target networks. Further progress in computational methodology combined with improved in vitro and in vivo disease models will facilitate the prioritization of therapeutic targets in the networks. The integration of multilevel omics data, computational approaches, and validated disease models will thus provide a strong foundation for deciphering the complex mechanisms of OS in retinal diseases and for discovering novel therapies with the greatest potential for efficacy in clinical trials.
Acknowledgments
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (16K08547), the Long-Range Research Initiative of the Japan Chemical Industrial Association (13-PT01-01), and Okasan-Kato Foundation.
Competing Interests
The authors declare that there are no competing interests regarding the publication of this paper.
References
- 1.Nita M., Grzybowski A. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxidative Medicine and Cellular Longevity. 2016;2016:23. doi: 10.1155/2016/3164734.3164734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinazo-Durán M. D., Gallego-Pinazo R., García-Medina J. J., et al. Oxidative stress and its downstream signaling in aging eyes. Clinical Interventions in Aging. 2014;9:637–652. doi: 10.2147/CIA.S52662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chrysostomou V., Rezania F., Trounce I. A., Crowston J. G. Oxidative stress and mitochondrial dysfunction in glaucoma. Current Opinion in Pharmacology. 2013;13(1):12–15. doi: 10.1016/j.coph.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Hong S., Iizuka Y., Lee T., Kim C. Y., Seong G. J. Neuroprotective and neurite outgrowth effects of maltol on retinal ganglion cells under oxidative stress. Molecular Vision. 2014;20:1456–1462. [PMC free article] [PubMed] [Google Scholar]
- 5.Akane M., Shimazawa M., Inokuchi Y., Tsuruma K., Hara H. SUN N8075, a novel radical scavenger, protects against retinal cell death in mice. Neuroscience Letters. 2011;488(1):87–91. doi: 10.1016/j.neulet.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Blasiak J., Petrovski G., Veréb Z., Facskó A., Kaarniranta K. Oxidative stress, hypoxia, and autophagy in the neovascular processes of age-related macular degeneration. BioMed Research International. 2014;2014:7. doi: 10.1155/2014/768026.768026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuruma K., Tanaka Y., Shimazawa M., Mashima Y., Hara H. Unoprostone reduces oxidative stress- and light-induced retinal cell death, and phagocytotic dysfunction, by activating BK channels. Molecular Vision. 2011;17:3556–3565. [PMC free article] [PubMed] [Google Scholar]
- 8.Kowluru R. A., Mishra M. Oxidative stress, mitochondrial damage and diabetic retinopathy. Biochimica et Biophysica Acta—Molecular Basis of Disease. 2015;1852(11):2474–2483. doi: 10.1016/j.bbadis.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Fernández de la Cámara C., Salom D., Sequedo M. D., et al. Altered Antioxidant-Oxidant Status in the Aqueous Humor and Peripheral Blood of Patients with Retinitis Pigmentosa. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0074223.e74223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K., Zhang L., Weinreb R. N. Ophthalmic drug discovery: novel targets and mechanisms for retinal diseases and glaucoma. Nature Reviews Drug Discovery. 2012;11(7):541–559. doi: 10.1038/nrd3745. [DOI] [PubMed] [Google Scholar]
- 11.Donegan R. K., Lieberman R. L. Discovery of molecular therapeutics for glaucoma: challenges, successes, and promising directions. Journal of Medicinal Chemistry. 2016;59(3):788–809. doi: 10.1021/acs.jmedchem.5b00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon S. D., Lindsley K., Vedula S. S., Krzystolik M. G., Hawkins B. S. Anti-vascular endothelial growth factor for neovascular age-related macular degeneration. The Cochrane database of systematic reviews. 2014;8:p. CD005139. doi: 10.1002/14651858.CD005139.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virgili G., Parravano M., Menchini F., Evans J. R. Anti-vascular endothelial growth factor for diabetic macular oedema. The Cochrane Database of Systematic Reviews. 2014;(10) doi: 10.1002/14651858.CD007419.pub4.CD007419 [DOI] [PubMed] [Google Scholar]
- 14.Gower N. J. D., Barry R. J., Edmunds M. R., Titcomb L. C., Denniston A. K. Drug discovery in ophthalmology: past success, present challenges, and future opportunities. BMC Ophthalmology. 2016;16, article 11 doi: 10.1186/s12886-016-0188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay M., Thomas D. W., Craighead J. L., Economides C., Rosenthal J. Clinical development success rates for investigational drugs. Nature Biotechnology. 2014;32(1):40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 16.Bonds D. E., Harrington M., Worrall B. B., et al. Effect of long-chain ω-3 fatty acids and lutein + zeaxanthin supplements on cardiovascular outcomes results of the age-related eye disease study 2 (AREDS2) randomized clinical trial. JAMA Internal Medicine. 2014;174(5):763–771. doi: 10.1001/jamainternmed.2014.328. [DOI] [PubMed] [Google Scholar]
- 17.Wong W. T., Kam W., Cunningham D., et al. Treatment of geographic atrophy by the topical administration of OT-551: results of a phase II clinical trial. Investigative Ophthalmology and Visual Science. 2010;51(12):6131–6139. doi: 10.1167/iovs.10-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plenge R. M., Scolnick E. M., Altshuler D. Validating therapeutic targets through human genetics. Nature Reviews Drug Discovery. 2013;12(8):581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 19.Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discovery Today. 2014;19(2):126–132. doi: 10.1016/j.drudis.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Yang H.-J., Ratnapriya R., Cogliati T., Kim J.-W., Swaroop A. Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Progress in Retinal and Eye Research. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B., Butte A. J. Leveraging big data to transform target selection and drug discovery. Clinical Pharmacology and Therapeutics. 2016;99(3):285–297. doi: 10.1002/cpt.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajorath J. Computer-aided drug discovery. F1000Research. 2015;4 doi: 10.12688/f1000research.6653.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Palczewski K. Systems pharmacology links GPCRs with retinal degenerative disorders. Annual Review of Pharmacology and Toxicology. 2016;56:273–298. doi: 10.1146/annurev-pharmtox-010715-103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal M., Cusick M. E., Barabási A.-L. Interactome networks and human disease. Cell. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segura-Cabrera A., Singh N., Komurov K. An integrated network platform for contextual prioritization of drugs and pathways. Molecular BioSystems. 2015;11(11):2850–2859. doi: 10.1039/c5mb00444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fillet M., Frédérich M. The emergence of metabolomics as a key discipline in the drug discovery process. Drug Discovery Today: Technologies. 2015;13:19–24. doi: 10.1016/j.ddtec.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Dehairs J., Derua R., Rueda-Rincon N., Swinnen J. V. Lipidomics in drug development. Drug Discovery Today: Technologies. 2015;13:33–38. doi: 10.1016/j.ddtec.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Shi J., Wang E., Milazzo J. P., Wang Z., Kinney J. B., Vakoc C. R. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature Biotechnology. 2015;33(6):661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah A. N., Davey C. F., Whitebirch A. C., Miller A. C., Moens C. B. Rapid reverse genetic screening using CRISPR in zebrafish. Nature Methods. 2015;12(6):535–540. doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstein M. B., Kundaje A., Hariharan M., et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43(1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium G. O. Gene ontology consortium: going forward. Nucleic Acids Research. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amberger J. S., Bocchini C. A., Schiettecatte F., Scott A. F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Research. 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law V., Knox C., Djoumbou Y., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Research. 2014;42(1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vailaya A., Bluvas P., Kincaid R., Kuchinsky A., Creech M., Adler A. An architecture for biological information extraction and representation. Bioinformatics. 2005;21(4):430–438. doi: 10.1093/bioinformatics/bti187. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay S., Palakal M., Maddu K. Multi-way association extraction and visualization from biological text documents using hyper-graphs: applications to genetic association studies for diseases. Artificial Intelligence in Medicine. 2010;49(3):145–154. doi: 10.1016/j.artmed.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Okada Y., Wu D., Trynka G., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menche J., Sharma A., Kitsak M. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347(6224):p. 1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eiraku M., Takata N., Ishibashi H., et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472(7341):51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 41.Nakano T., Ando S., Takata N., et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Reichman S., Terray A., Slembrouck A., et al. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(23):8518–8523. doi: 10.1073/pnas.1324212111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura Y., Okabe S., Sasagawa S., et al. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Frontiers in Pharmacology. 2015;6, article 257 doi: 10.3389/fphar.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasagawa S., Nishimura Y., Sawada H., et al. Comparative transcriptome analysis identifies CCDC80 as a novel gene associated with pulmonary arterial hypertension. Frontiers in Pharmacology. 2016;7:p. 142. doi: 10.3389/fphar.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasagawa S., Nishimura Y., Okabe S., et al. Downregulation of GSTK1 is a common mechanism underlying hypertrophic cardiomyopathy. Frontiers in Pharmacology. 2016;7, article 162 doi: 10.3389/fphar.2016.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura Y., Murakami S., Ashikawa Y., et al. Zebrafish as a systems toxicology model for developmental neurotoxicity testing. Congenital Anomalies. 2015;55(1):1–16. doi: 10.1111/cga.12079. [DOI] [PubMed] [Google Scholar]
- 47.Nishimura Y., Inoue A., Sasagawa S., et al. Using zebrafish in systems toxicology for developmental toxicity testing. Congenital Anomalies. 2016;56(1):18–27. doi: 10.1111/cga.12142. [DOI] [PubMed] [Google Scholar]
- 48.Nakao T., Tsujikawa M., Notomi S., Ikeda Y., Nishida K. The role of mislocalized phototransduction in photoreceptor cell death of retinitis pigmentosa. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0032472.e32472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuruma K., Nishimura Y., Kishi S., Shimazawa M., Tanaka T., Hara H. SEMA4A mutations lead to susceptibility to light irradiation, oxidative stress, and ER stress in retinal pigment epithelial cells. Investigative Ophthalmology and Visual Science. 2012;53(10):6729–6737. doi: 10.1167/iovs.11-9378. [DOI] [PubMed] [Google Scholar]
- 50.Chhetri J., Jacobson G., Gueven N. Zebrafish—on the move towards ophthalmological research. Eye (London) 2014;28(4):367–380. doi: 10.1038/eye.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson R., Tracey-White D., Webster A., Moosajee M. The zebrafish eye—a paradigm for investigating human ocular genetics. Eye. 2016 doi: 10.1038/eye.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasagawa S., Nishimura Y., Kon T., et al. DNA damage reesponse Is involved in the developmental toxicity of mebendazole in zebrafish retina. Frontiers in Pharmacology. 2016;7:p. 57. doi: 10.3389/fphar.2016.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawase R., Nishimura Y., Ashikawa Y., et al. EP300 Protects from Light-Induced Retinopathy in Zebrafish. Frontiers in Pharmacology. 2016;7 doi: 10.3389/fphar.2016.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito Y., Tsuruma K., Shimazawa M., Nishimura Y., Tanaka T., Hara H. Establishment of a drug evaluation model against light-induced retinal degeneration using adult pigmented zebrafish. Journal of Pharmacological Sciences. 2016;131(3):215–218. doi: 10.1016/j.jphs.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Matsubara H., Tanaka T., Nishimura Y., Matsui Y., Yamamoto T., Kondo M. Circadian rhythm of electroretinograms in living zebrafish larvae. Investigative Ophthalmology & Visual Science. 2013;54(15, article 3419) [Google Scholar]
- 56.Jones B. W., Kondo M., Terasaki H., et al. Retinal remodeling in the Tg P347L rabbit, a large-eye model of retinal degeneration. Journal of Comparative Neurology. 2011;519(14):2713–2733. doi: 10.1002/cne.22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirota R., Kondo M., Ueno S., Sakai T., Koyasu T., Terasaki H. Photoreceptor and post-photoreceptoral contributions to photopic ERG a-wave in rhodopsin P347L transgenic rabbits. Investigative Ophthalmology and Visual Science. 2012;53(3):1467–1472. doi: 10.1167/iovs.11-9006. [DOI] [PubMed] [Google Scholar]
- 58.Hasegawa T., Muraoka Y., Ikeda H. O., et al. Neuoroprotective efficacies by KUS121, a VCP modulator, on animal models of retinal degeneration. Scientific Reports. 2016;6, article 31184 doi: 10.1038/srep31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakagami Y., Hatano E., Inoue T., Yoshida K., Kondo M., Terasaki H. Cytoprotective effects of a novel Nrf2 activator, RS9, in Rhodopsin Pro347Leu rabbits. Current Eye Research. 2015;41(8):1–4. doi: 10.3109/02713683.2015.1078362. [DOI] [PubMed] [Google Scholar]
- 60.Shimazawa M., Ito Y., Inokuchi Y., et al. An alteration in the lateral geniculate nucleus of experimental glaucoma monkeys: in vivo positron emission tomography imaging of glial activation. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0030526.e30526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimazawa M., Nakamura S., Miwa M., et al. Establishment of the ocular hypertension model using the common marmoset. Experimental Eye Research. 2013;111:1–8. doi: 10.1016/j.exer.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 62.Shimazawa M., Masuda T., Nakamura S., Miwa M., Nakamura K., Hara H. An experimental model for exudative age-related macular degeneration with choroidal neovascularization using the common marmoset. Current Neurovascular Research. 2015;12(2):128–134. doi: 10.2174/1567202612666150311105814. [DOI] [PubMed] [Google Scholar]
- 63.Patrias K. Citing Medicine: The NLM Style Guide for Authors, Editors, and Publishers. 2nd 2007. [Google Scholar]
- 64.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abu-Amero K., Kondkar A. A., Chalam K. V. An updated review on the genetics of primary open angle glaucoma. International Journal of Molecular Sciences. 2015;16(12):28886–28911. doi: 10.3390/ijms161226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bailey J. N. C., Loomis S. J., Kang J. H., et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nature Genetics. 2016;48(2):189–194. doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khor C. C., Do T., Jia H., et al. Genome-wide association study identifies five new susceptibility loci for primary angle closure glaucoma. Nature Genetics. 2016;48(5):556–562. doi: 10.1038/ng.3540. [DOI] [PubMed] [Google Scholar]
- 68.Huang D. W., Sherman B. T., Tan Q., et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research. 2007;35(2):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Favari E., Zanotti I., Zimetti F., Ronda N., Bernini F., Rothblat G. H. Probucol inhibits ABCA1-mediated cellular lipid efflux. Arteriosclerosis, Thrombosis, and Vascular Biology. 2004;24(12):2345–2350. doi: 10.1161/01.ATV.0000148706.15947.8a. [DOI] [PubMed] [Google Scholar]
- 70.Falk M. J., Polyak E., Zhang Z., et al. Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO Molecular Medicine. 2011;3(7):410–427. doi: 10.1002/emmm.201100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai X.-Y., Ma Y., Ding R., Fu B., Shi S., Chen X.-M. miR-335 and miR-34a promote renal senescence by suppressing mitochondrial antioxidative enzymes. Journal of the American Society of Nephrology. 2011;22(7):1252–1261. doi: 10.1681/ASN.2010040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naito M., Umegaki H., Iguchi A. Protective effects of probucol against glutamate-induced cytotoxicity in neuronal cell line PC12. Neuroscience Letters. 1995;186(2-3):211–213. doi: 10.1016/0304-3940(95)11321-M. [DOI] [PubMed] [Google Scholar]
- 73.Harada T., Harada C., Nakamura K., et al. The potential role of glutamate transporters in the pathogenesis of normal tension glaucoma. The Journal of Clinical Investigation. 2007;117(7):1763–1770. doi: 10.1172/jci30178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bai N., Hayashi H., Aida T., et al. Dock3 interaction with a glutamate-receptor NR2D subunit protects neurons from excitotoxicity. Molecular Brain. 2013;6(1, article no. 22) doi: 10.1186/1756-6606-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Namekata K., Kimura A., Kawamura K., et al. Dock3 attenuates neural cell death due to NMDA neurotoxicity and oxidative stress in a mouse model of normal tension glaucoma. Cell Death and Differentiation. 2013;20(9):1250–1256. doi: 10.1038/cdd.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mastropasqua R., Fasanella V., Agnifili L., et al. Advance in the pathogenesis and treatment of normal-tension glaucoma. Progress in Brain Research. 2015;221:213–232. doi: 10.1016/bs.pbr.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 77.Ito Y., Shimazawa M., Tsuruma K., et al. Induction of amyloid-β1-42 in the retina and optic nerve head of chronic ocular hypertensive monkeys. Molecular Vision. 2012;18:2647–2657. [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta V., You Y., Li J., et al. BDNF impairment is associated with age-related changes in the inner retina and exacerbates experimental glaucoma. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease. 2014;1842(9):1567–1578. doi: 10.1016/j.bbadis.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 79.Salt T. E., Nizari S., Cordeiro M. F., Russ H., Danysz W. Effect of the Aβ aggregation modulator MRZ-99030 on retinal damage in an animal model of glaucoma. Neurotoxicity Research. 2014;26(4):440–446. doi: 10.1007/s12640-014-9488-6. [DOI] [PubMed] [Google Scholar]
- 80.Gupta V., Gupta V. B., Chitranshi N., et al. One protein, multiple pathologies: multifaceted involvement of amyloid β in neurodegenerative disorders of the brain and retina. Cellular and Molecular Life Sciences. 2016;73(22):4279–4297. doi: 10.1007/s00018-016-2295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sevigny J., Chiao P., Bussière T., et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 2016;537(7618):50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 82.Stewart C. R., Stuart L. M., Wilkinson K., et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nature Immunology. 2010;11(2):155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi S. H., Sviridov D., Miller Y. I. Oxidized cholesteryl esters and inflammation. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbalip.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Semba K., Namekata K., Guo X., Harada C., Harada T., Mitamura Y. Renin-Angiotensin system regulates neurodegeneration in a mouse model of normal tension glaucoma. Cell Death and Disease. 2014;5(7) doi: 10.1038/cddis.2014.296.e1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suzuki Y., Hattori K., Hamanaka J., et al. Pharmacological inhibition of TLR4-NOX4 signal protects against neuronal death in transient focal ischemia. Scientific Reports. 2012;2, article no. 896 doi: 10.1038/srep00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peri F., Calabrese V. Toll-like receptor 4 (TLR4) modulation by synthetic and natural compounds: an update. Journal of Medicinal Chemistry. 2014;57(9):3612–3622. doi: 10.1021/jm401006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fritsche L. G., Fariss R. N., Stambolian D., Abecasis G. R., Curcio C. A., Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annual Review of Genomics and Human Genetics. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fritsche L. G., Igl W., Bailey J. N., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nature Genetics. 2016;48(2):134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ambati J., Fowler B. J. Mechanisms of age-related macular degeneration. Neuron. 2012;75(1):26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marneros A. G. VEGF-A and the NLRP3 inflammasome in age-related macular degeneration. Advances in Experimental Medicine and Biology. 2016;854:79–85. doi: 10.1007/978-3-319-17121-0_12. [DOI] [PubMed] [Google Scholar]
- 91.Hsieh H.-L., Yang C.-M. Role of redox signaling in neuroinflammation and neurodegenerative diseases. BioMed Research International. 2013;2013:18. doi: 10.1155/2013/484613.484613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lambert V., Wielockx B., Munaut C., et al. MMP-2 and MMP-9 synergize in promoting choroidal neovascularization. FASEB Journal. 2003;17(15):2290–2292. doi: 10.1096/fj.03-0113fje. [DOI] [PubMed] [Google Scholar]
- 93.Grisanti S., Tatar O. The role of vascular endothelial growth factor and other endogenous interplayers in age-related macular degeneration. Progress in Retinal and Eye Research. 2008;27(4):372–390. doi: 10.1016/j.preteyeres.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 94.Zhu W.-H., Guo X., Villaschi S., Francesco Nicosia R. Regulation of vascular growth and regression by matrix metalloproteinases in the rat aorta model of angiogenesis. Laboratory Investigation. 2000;80(4):545–555. doi: 10.1038/labinvest.3780060. [DOI] [PubMed] [Google Scholar]
- 95.Del V Cano M., Gehlbach P. L. PPAR-α ligands as potential therapeutic agents for wet age-related macular degeneration. PPAR Research. 2008;2008:5. doi: 10.1155/2008/821592.821592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herzlich A. A., Tuo J., Chan C.-C. Peroxisome proliferator-activated receptor and age-related macular degeneration. PPAR Research. 2008;2008:11. doi: 10.1155/2008/389507.389507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sur A., Kesaraju S., Prentice H., et al. Pharmacological protection of retinal pigmented epithelial cells by sulindac involves PPAR-α . Proceedings of the National Academy of Sciences of the United States of America. 2014;111(47):16754–16759. doi: 10.1073/pnas.1419576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Knickelbein J. E., Abbott A. B., Chew E. Y. Fenofibrate and diabetic retinopathy. Current Diabetes Reports. 2016;16(10) doi: 10.1007/s11892-016-0786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Moraes G., Layton C. J. Therapeutic targeting of diabetic retinal neuropathy as a strategy in preventing diabetic retinopathy. Clinical & Experimental Ophthalmology. 2016 doi: 10.1111/ceo.12795. [DOI] [PubMed] [Google Scholar]
- 100.Zhou X., Wong L. L., Karakoti A. S., Seal S., McGinnis J. F. Nanoceria inhibit the development and promote the regression of pathologic retinal neovascularization in the Vldlr knockout mouse. PLoS ONE. 2011;6(2) doi: 10.1371/journal.pone.0016733.e16733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen X., Rong S. S., Xu Q., et al. Diabetes mellitus and risk of age-related macular degeneration: a systematic review and meta-analysis. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0108196.e108196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilkinson-Berka J. L., Agrotis A., Deliyanti D. The retinal renin-angiotensin system: roles of angiotensin II and aldosterone. Peptides. 2012;36(1):142–150. doi: 10.1016/j.peptides.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 103.Giese M. J., Speth R. C. The ocular renin-angiotensin system: a therapeutic target for the treatment of ocular disease. Pharmacology and Therapeutics. 2014;142(1):11–32. doi: 10.1016/j.pharmthera.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 104.Hernández C., Dal Monte M., Simó R., Casini G. Neuroprotection as a therapeutic target for diabetic retinopathy. Journal of Diabetes Research. 2016;2016:18. doi: 10.1155/2016/9508541.9508541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Phipps J. A., Jobling A. I., Greferath U., Fletcher E. L., Vessey K. A. Alternative pathways in the development of diabetic retinopathy: the renin-angiotensin and kallikrein-kinin systems. Clinical and Experimental Optometry. 2012;95(3):282–289. doi: 10.1111/j.1444-0938.2012.00747.x. [DOI] [PubMed] [Google Scholar]