1. Introduction
The mammalian brain is composed of a very large number of cells belonging to many different cell types, a complexity that poses serious challenges (Kandel et al. 2000). Our current understanding of the development of neural circuits underlying the computation of different visual stimuli remains highly incomplete across species. Insects provide an attractive model system since their nervous system is relatively simple, yet the animals manifest very sophisticated visual behaviors (Collett 2008, Zeil 2012) .
How is neuronal diversity achieved during development of the visual system? What are the genes and pathways defining large numbers of different neuronal cell types? How are these cell types connected to form functional circuits in the optic lobes? Thanks to the development of new molecular genetic tools (del Valle Rodriguez et al. 2012), significant progress has been made towards understanding the development of the Drosophila visual circuitry.
2. The Drosophila visual system
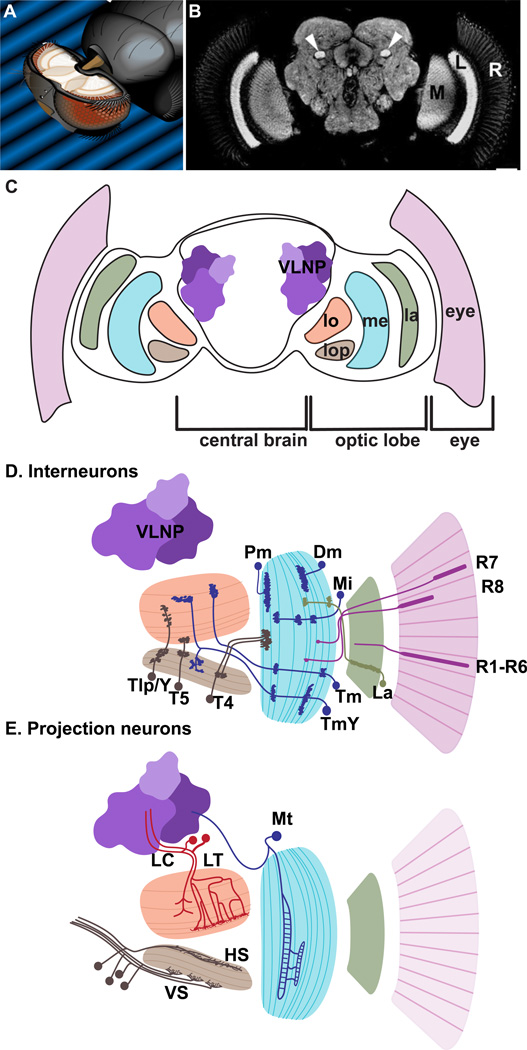
The adult Drosophila visual system contains ~150,000 neurons and glia cells (Chiang et al. 2011). Visual information is detected by the retina, while visual processing occurs in the optic lobes that comprise more than 60% of the brain’s neurons. The optic lobes are the major centers where neuronal computations extract important features from the visual world, such as shape, motion, color, e-vector orientation of polarized light, which are then transmitted to the central brain (CB) (Fig. 1 A-C) (Fischbach and Dittrich 1989, Meinertzhagen and Hanson 1993, Meinertzhagen et al. 2009, Homberg et al. 2011, Otsuna et al. 2014, Silies et al. 2014, Borst and Helmstaedter 2015).
Figure 1. The visual system of Drosophila.
(A) The fly brain in the head; adapted from (Spalthoff et al. 2012). (B) Neuropils of the optic lobes and central brain; from (Krzeptowski et al. 2014). (C) Cross-section of the fly brain indicating the different part of the visual system: eye, lamina (la), medulla (me), lobula (lo), lobula plate (lop) and ventro-lateral neuropils (VNLP). (D) Interneurons of the optic lobe: outer and inner photoreceptors (pink), Lamina neurons (La, green), Medulla interneurons (dark blue): Distal (Dm) and Proximal medulla (Pm); Medulla intrinsic (Mi); unicolumnar Transmedullary (Tm) or multicolumnar TmY; Lobula plate interneurons (T4, T5, Tlp and Y neurons, dark brown). (E) Projections neurons: Medulla tangential (Mt, dark blue); Lobula columnar (LC, red) and tangential/tree-like (LT, red); Lobula plate tangential cells: HS and VS (dark brown). (D, E) adapted from (Erclik et al. 2009)
2.1 The compound eye
The adult Drosophila compound eye is made of ~800 independent unit eyes called ommatidia, corresponding to 800 pixels in the animal’s visual field. Each ommatidium is composed of eight photoreceptor neurons that project into the optic lobe. The organization of the eye will not be further described here as it has been reviewed extensively in the recent past (Paulk et al. 2011, Kumar 2012, Lamb 2013).
2.2 Optic lobe
Fly neurons are organized into approximately 50 areas composed mainly of neuronal processes, called neuropils, with their cell bodies localized at the periphery. Four of these neuropils form the optic lobe: lamina, medulla, and the lobula complex which is further subdivided into the lobula and the lobula plate neuropils (Morante and Desplan 2004). Two major types of neurons can be identified within the optic lobes: “Interneurons” whose cell bodies and projections remain within the optic lobe, and “projection” neurons, which connect the optic lobe to the CB (Fig. 1 D, E) (Hofbauer and Campos-Ortega 1990)
2.2.1 Lamina
Photoreceptors from each ommatidium involved in motion vision (“outer photoreceptors”) first innervate the lamina neuropil, which manifests a columnar organization in which each pixel of the visual field corresponds to one cartridge (Meinertzhagen and Sorra 2001). The lamina is mostly composed of interneurons, whose projections do not leave the optic lobe with their cell bodies located in the lamina cortex region. Lamina neurons can be divided in two populations: Five types of monopolar neurons that contact a single cartridge and project retinotopically into the medulla, and amacrine cells that contact several cartridges within the lamina (Fischbach and Dittrich 1989, Hofbauer and Campos-Ortega 1990, Tuthill et al. 2013).
2.2.2 Medulla
The medulla neuropil receives direct innervation from color (“Inner”) photoreceptors, as well as from lamina monopolar neurons. The medulla neuropil is stratified in ten layers (M1-M10) with the region between layers M1 and M6 referred to as ‘distal medulla’ that receives these external inputs (Fischbach and Dittrich 1989, Morante and Desplan 2008, Takemura et al. 2008). The ‘proximal medulla’ (layers M7 to M10) receives information from the distal medulla and further computes visual information. The medulla neuropil is organized in repetitive columnar units, oriented perpendicular to the ten layers that are reminiscent of lamina cartridges (Fischbach and Dittrich 1989). Retinotopic connections between medulla columns, photoreceptors and lamina cartridges ensure an accurate representation of the visual world (Fischbach and Dittrich 1989, Meinertzhagen and Sorra 2001, Morante and Desplan 2008, Zhu 2013, Chin et al. 2014).
The serpentine layer, which separates the distal and proximal medulla, consists of incoming and outgoing axons of projection neurons, which all connect to more than one medulla column. Different types of projection neurons can be identified based on the location of their cell bodies, their dendritic morphology and their axonal projection patterns (Fischbach and Dittrich 1989, Strausfeld 1989), but the functional contribution of most of these neurons remains unclear.
The medulla contains about 40.000 interneurons, representing the largest neuronal subpopulation in the optic lobe (Fischbach and Dittrich 1989). Their cell bodies are located either within the medulla cortex between the lamina and the medulla neuropils, or within the medulla rim, located below the medulla neuropil near the lobula complex (Fischbach and Dittrich 1989, Bausenwein et al. 1992). Medulla interneurons have been extensively characterized and categorized in over 80 cell types, first by Ramon Y. Cajal (Ramon y Cajal and Sanchez y Sanchez 1915) followed by (Fischbach and Dittrich 1989) and more recently by (Morante and Desplan 2008, Raghu et al. 2009, Varija Raghu et al. 2011, Raghu et al. 2013). They can be subdivided in subcategories based on their projections patterns (Fig. 1.D): Interneurons that project over a large visual field, across many columns, are called tangential (Dm and Pm on Fig. 1D). Columnar neurons projections are mainly parallel to the medulla columns (Mi, Tm and TmY on Fig. 1D). They can be unicolumnar and project within a single medulla column, suggesting that they process information from one visual point in space, or multicolumnar with projections spanning several medulla columns. Only columnar neurons project outside the medulla into the lobula and lobula plate, which represent the main output of the medulla neuropil (Fischbach and Dittrich 1989, Morante and Desplan 2008).
2.2.3 Lobula complex: the lobula
The lobula can be divided into six layers arranged perpendicularly to the columnar structures resulting from the columnar inputs of the medulla neurons (Fischbach and Dittrich 1989). Almost all lobula cells are projection neurons whose cell bodies are located between the CB and the lobula neuropil. Interestingly, despite the cell bodies’ varying distance to the lobula, their projections all merge at the neck of the lobula to form a single fiber tract connecting the lobula to the CB (Fig. 1 E) (Otsuna and Ito 2006). Lobula neurons can be divided into two categories: Columnar neurons (LC) receive visual input from 8–9 ommatidia (Douglass and Strausfeld 2003, Otsuna and Ito 2006, Mu et al. 2012), reminiscent of multicolumnar neurons in the medulla; tangential and treelike neurons (LT) receive input from very large visual fields, similar to medulla tangential neurons (Otsuna and Ito 2006) (Fig. 1 E).
2.2.4 Lobula complex: the lobula plate
Representing the output center of the neural circuits that process motion, the lobula plate neuropil can be divided in four layers containing dendrites that each manifest maximal sensitivity to motion along one of the four cardinal directions (front-to-back, back-to-front, up, and down) (Fischbach and Dittrich 1989, Maisak et al. 2013). Two classes of lobula plate interneurons called T4 and T5 cells can each be further subdivided into four subclasses forming dendrites in only one of the four specific layers (hence termed T4a,b,c,d and T5a,b,c,d). Their role in motion detection has been studied and was reviewed in (Behnia and Desplan 2015, Borst and Helmstaedter 2015). Their cell bodies are all located adjacent to the lobula plate neuropil, underneath medulla rim cell bodies (Fig. 1 C). Two other classes of interneurons have pre-synaptic input in the lobula plate and post-synaptic output in the lobula: the Translobula-plate neurons (Tlp) and Y cells. Both have their cell bodies adjacent to those of T4 and T5 cells. As observed in larger flies, only few intrinsic cells, i.e. which remains within the lobula plate neuropil, can be observed (Fischbach and Dittrich 1989).
The Lobula Plate Tangential Cells (LPTCs) are projection neurons whose characterization has provided great insight into the computation of motion (Hausen 1984, Maisak et al. 2013). In general, LPTCs are sensitive to visual motion in a direction-selective manner (Hausen 1984). Historically, the most extensively studied LTPCs belong to the horizontal (HS) and vertical (VS) system (Fig. 1 E) (Borst and Haag 2002). They receive their inputs from the T4 and T5 neurons and transmit direction-selective visual information to the ventro-lateral neuropils of the CB (Borst and Haag 2002, Silies et al. 2013, Borst 2014, Behnia and Desplan 2015).
2.3 Visual Centers in the CB
The Ventrolateral neuropils (VLNPs, also called optic glomeruli) are located right underneath the optic lobes and can be considered the next step in visual processing after the optic lobes. All fourteen different types of lobula projection neurons project to distinct target regions within the VLNPs (Otsuna and Ito 2006). They each project into the VLNP as one single bundle in a way reminiscent to olfactory glomeruli (Otsuna and Ito 2006, Mu et al. 2012). The availability of highly specific Gal4 lines that label most of the optic lobe cell types (Pfeiffer et al. 2008, Jenett et al. 2012, Li et al. 2014) should allow the determination of the visual function performed by each optic glomerulus (Mu et al. 2012, Aptekar et al. 2015) .
3. Development of the Fly visual system
3.1 The retina
The development of the fly retina is one of the best-understood complex structures. It has been reviewed in many articles (Carthew 2007, Kumar 2012, Treisman 2013) and will not be described further in this review.
3.2 The ventral nerve cord and CB
The ventral nerve cord (VNC) and CB are generated in the embryo from neuroblasts delaminating from the embryonic neuroepithelium. These neuroblasts produce the embryonic nervous system and 10% of the future adult neurons before entering quiescence towards the end of embryonic stages (Fig. 2 A). At late L1/early L2 stages, about 100 neuroblasts start dividing again and produce the remaining 90% of adult neurons. The neuroblasts can be divided in two categories: Type I neuroblasts generate all ventral nerve cord neurons and most of the CB, while 8 Type II neuroblasts generate clones of up to 500 cells giving rise to CB neurons . The formation of neurons from the VNC/CB neuroblasts has been reviewed in (Reichert 2011, Homem and Knoblich 2012, Kang and Reichert 2014).
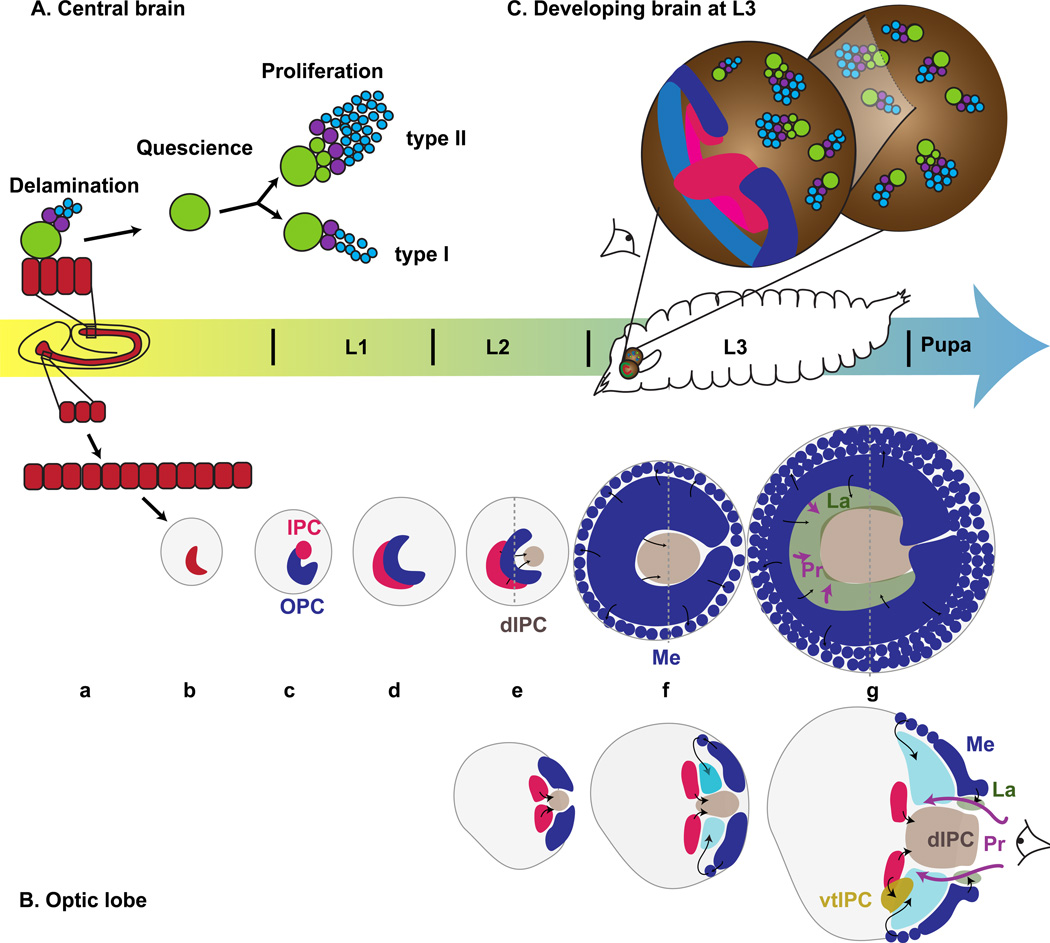
Figure 2. The development of the optic lobe of Drosophila.
(A) Central brain: Neuroblasts (green) delaminate from the neuroepithelium (red) during the embryonic stages and generate GMCs (purple) and larval neurons (blue) before they become quiescent. At L1, Type I and Type II neuroblasts are reactivated and exhibit different modes of proliferation to generate adult neurons. (B) Optic lobe: a) Cells in the procephalic regions generate the optic placode neuroepithelium (red), which invaginates (b); c) The optic placode split into inner proliferation center (IPC, pink) and outer proliferation center (OPC, blue) and adopt a crescent shape (d). e) Cells migrate out of the IPC to form the dIPC (light brown). f) The medial edge of the OPC becomes neuroblasts that will produce the medulla (Round blue cells). g) The inner OPC generates Lamina progenitors (La, green) that will produce lamina neurons upon induction by photoreceptors (Pr, pink arrows). The ventral tip of the IPC generates lobula complex neurons (vIPC, dark orange). Lower diagrams are cross sections at the dashed line in upper diagrams. (C) 3D representation of the brain at L3. Central brain neuroblasts and their progeny, OPC (blue) and IPC (pink). The eye symbol shows the view angle of (B). (A and C) adapted from (Homem and Knoblich 2012). (B) adapted from (Nassif et al. 2003)
3.3 The optic lobe
The optic lobe is derived from cells located in the posterior part of the embryonic head (Green et al. 1993, Nassif et al. 2003). During the same time window when neuroblasts of the VNC and CB delaminate, optic lobe precursor cells undergo four rounds of mitosis to form a plate-like structure that is made of densely packed columnar cells called optic placode (Green et al. 1993) (Fig. 2 Ba). After the fourth division, the cells of the developing optic lobe remain mitotically quiescent throughout the rest of embryogenesis (Green et al. 1993).
After all neuroblasts of the CB have delaminated from the head ectoderm, the optic lobe placode becomes attached to the ventro-lateral surface of the brain and invaginates by apical constriction. During this invagination process, the placode adopts a V-like shape, with an anterior and a posterior tip (White and Kankel 1978, Hofbauer and Campos-Ortega 1990, Green et al. 1993) (Fig. 2 Bc).
Upon hatching of the 1st instar larva, a small group of cells at the anterior-dorsal tip of the optic placode detaches, splitting the developing optic lobe in two, creating the Outer Proliferation Center (OPC) and the Inner Proliferation Center (IPC) (Meinertzhagen and Hanson 1993, Younossi-Hartenstein et al. 1996, Nassif et al. 2003) (Fig. 2 Bd).
Toward the end of the second larval instar, the OPC and the IPC both adopt a crescent shape, with the opening of the crescent pointing posteriorly (Fig. 2 Bd). Both OPC and IPC anlagen remain in contact with each other until the end of the second instar, when they become separated by newly generated cells that penetrate into the space between the IPC and OPC (Fig. 2 Be). These cells migrate in from the IPC to form a distant proliferative center called the dIPC. The ventral tip of the IPC also proliferates and generates neurons (Hofbauer and Campos-Ortega 1990, Apitz and Salecker 2015, Neriec et al. Submitted).
From the end of the second instar onwards, cells at the medial edge of the OPC neuroepithelium crescent begin to lose their columnar shape and adherens junctions without delaminating (Fig. 2 Bf). They are converted into neuroblasts in a moving proneural wave (reviewed in (Apitz and Salecker 2014)). These medulla neuroblasts divide asymmetrically to self-renew and generate their progeny composed of neurons and glia (Egger et al. 2007). By 20 hours After Puparium Formation (APF), ~40,000 neurons have been generated, most of which will become medulla cortex neurons.
During the third larval instar, a second proliferation zone develops at the opposite edge of the OPC. This second zone is separated from the future medulla neuroblast by the deep lamina furrow (Fig. 2Bg). By mid-third instar, photoreceptor axons start to project into the developing optic lobe via the optic stalk (Pink arrow on Fig. 2Bg). Lamina neurons are produced and differentiate in response to Hedgehog (Hh) and EGF produced by photoreceptors axons (Selleck et al. 1992, Huang and Kunes 1996, Huang and Kunes 1998, Huang et al. 1998). By 25 hrs APF, it has generated about 6,000 additional cells that become lamina neurons and glia (Apitz and Salecker 2014). The development of the OPC, which generates the lamina and the medulla, has been studied in much detail while the development of the IPC, which generates the lobula and lobula plate, has only recently been the topic of investigations (Apitz and Salecker 2015, Neriec et al. Submitted).
Starting around 25h APF, the developing medulla starts to rotate, being pulled by the lamina (White and Kankel 1978, Langen et al. 2015), and inserting itself right under the lamina neuropil. By 40h APF, the rotation is complete and the optic lobe has adopted its final configuration which the four neuropils: lamina, medulla, lobula and lobula plate (Claire Bertet, Karl Fischbach personal communications).
3.4 Linking development to adult fate
Due to the extreme morphological changes shaping the optic lobe during pupation, the task of linking cell fates between larval and adult neurons remains incomplete. Within the OPC, the cellular mechanisms that lead to the formation of Lamina Precursor Cells, the induction of their differentiation into laminar monopolar neurons by the photoreceptors and their connections have been well studied (Kunes et al. 1993, Huang and Kunes 1996, Huang and Kunes 1998, Dearborn and Kunes 2004). However, how the specification of the five different types of lamina monopolar neurons is achieved remains unknown. The rest of the OPC has been shown to generate medulla cell types as well few lobula and lobula plate neurons (Fig. 3)(Bertet et al. 2014). General mechanisms used to specify different types of neurons have been identified by (Hasegawa et al. 2013, Li et al. 2013b, Sato et al. 2013, Suzuki et al. 2013, Bertet et al. 2014, Erclik et al. Under Review). A recent study has characterized some of the genetic mechanisms involved in the generation of neurons of the lobula plate and medulla rim from the main part of the IPC (Apitz and Salecker 2015, Neriec et al. Submitted). Common mechanisms involved in the generation of neuronal diversity have been identified both in the optic lobe, the ventral nerve cord and the CB. They are similar in many regards to those observed in vertebrates and are detailed in the last section of this review.
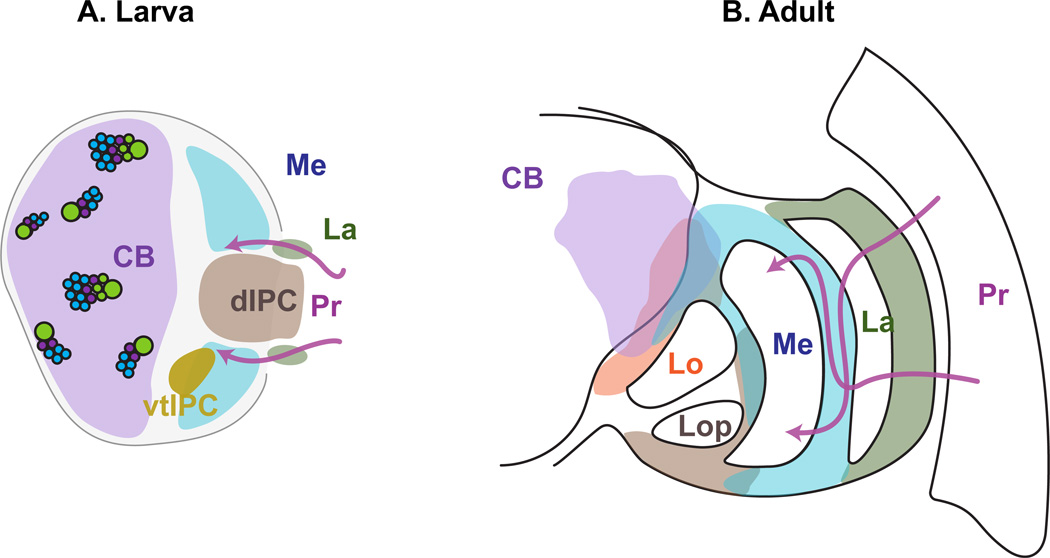
Figure 3. Linking development to adult fate.
Fate of the different neuronal populations found at the L3 larval stage (A) in the adult visual system (B). Pr: Photoreceptors, La: Lamina, Me: Medulla, Lo: Lobula, Lop: Lobula plate, dIPC: distal IPC, vtIPC: Ventral tip of the IPC, CB: Central Brain
The optic lobe projection neurons, including the lobula columnar neurons (LCNs) and the lobula plate tangential cells (LPTCs), as well as the CB neurons of the ventral protocerebrum (VLNPS) are believed to originate mostly from CB neuroblasts. Recent studies have taken advantage of the fact that CB neurons remain close to their birth place and generate reproducible series of neuronal cell fates that represent the clonal units generated from each of the 100 neuroblasts in the CB (Chiang et al. 2011, Ito et al. 2013, Lovick et al. 2013, Wong et al. 2013, Yu et al. 2013) including intermediate progenitors of type II neuroblasts (Wang et al. 2014b).
Thirteen clonal units of neurons projecting into the optic lobe but also into the CB have been described (Ito et al. 2013). These neurons all project into the VLP where they form distinct non-overlapping bulbous masses corresponding to functionally relevant optic glomeruli: Neurons originating from the same neuroblast clone share common projection pattern into specific glomeruli, which is indicative of functional similarities (Otsuna and Ito 2006). This hints towards a direct link between the developmental origin of projection neurons and their adult function. Similarities between the functional connectivity of VLP and olfactory glomeruli have been pointed out (Mu et al. 2012). Extensive work has been undertaken in order to understand the formation of the olfactory glomeruli (For review: (Imai et al. 2010)), and similar future studies on the development of the optic glomeruli will determine if similarities exist between the development of these glomeruli.
4. General Rules of Neuronal Development
The generation of an adult neuron can be described in two main steps. First, neurons are produced by neuronal stem cells. Then, once produced, the pre-specified neurons form axonal and dendritic projections and integrate into developing neural circuits (Fig. 4A). We will review the general mechanisms of neurogenesis observed during the production (4.1) and the pre-specification (4.2) of optic lobe neurons, and compare them with the development of the central brain.
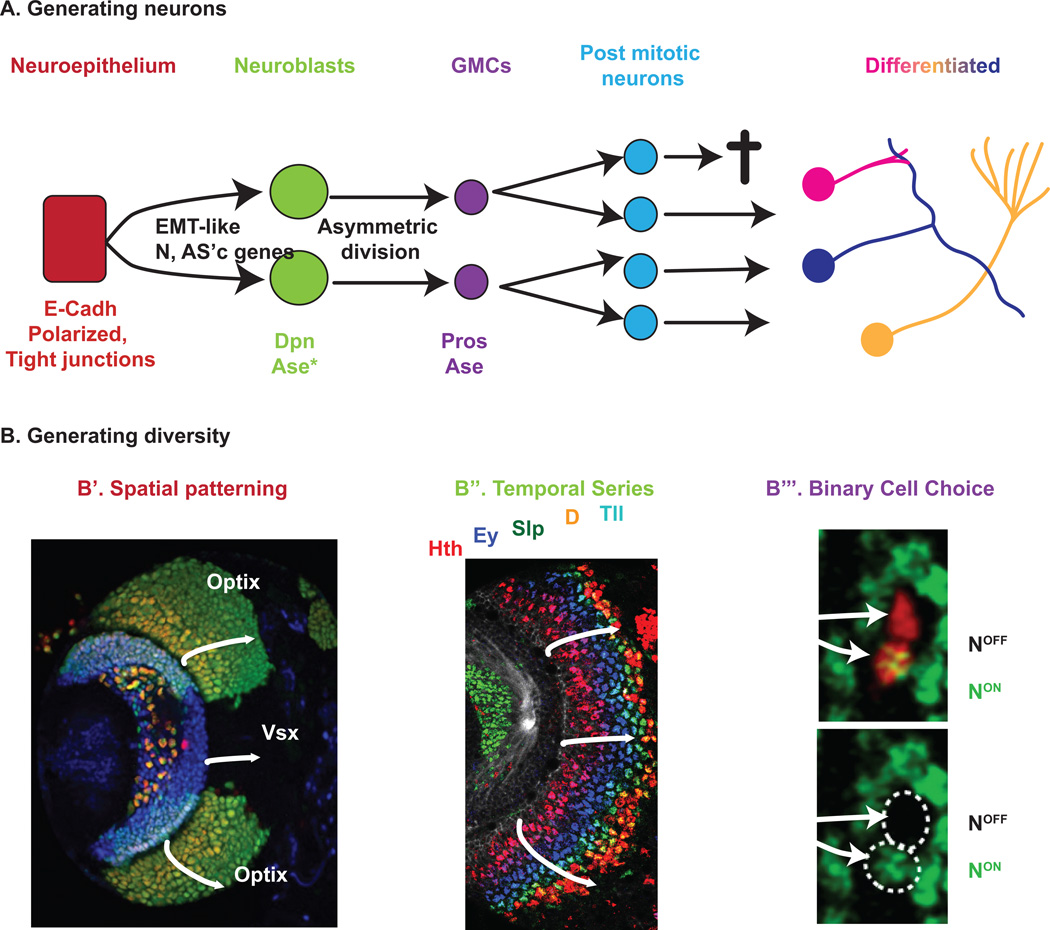
Figure 4. The making of a functional neuronal network.
(A) Common mechanisms for neurogenesis: Polarized neuroepithelial cells (red) undergo EMT-like transition to become Neuroblasts (green) which generate GMCs (purple). GMCs produce post-mitotic neurons (blue) that form adult neuronal circuits. (B) Common mechanisms to generate neuronal diversity: (B’) Spatial patterning of the neuroepithelium illustrated by the expression of Optix and Vsx1 (Gold and Brand 2014). (B”) Temporal patterning of neuroblasts. In the OPC, neuroblasts sequentially express a series of transcription factors, Hth (red), Ey (blue), Slp (green), D (orange) and Tll (not represented) as they age (arrows) (Li et al. 2013). (B”’) Notch-dependent binary cell fate choices during the division of GMCs. Only one of the two daughter cells (red clone, dashed line) receives Notch activity and expresses Apterous (green), while the other does not (Li et al. 2013).
4.1 Production of neurons
4.1.1 Generating neuroblasts from a neuroepithelium
Both the Drosophila VNC/CB and the optic lobes are generated from cells with epithelial-like characteristics; they are organized as monolayers with cell of rectangular shapes connected by adherens junctions. These cells express high levels of DE-Cadherin (E-Cadh) and manifest apico-basal polarity (Doe 1996, Acloque et al. 2009).
In the embryonic neuroepithelium, one cell is stochastically singled-out from 6–8 neuroepithelial cells via lateral inhibition mediated by Notch. This cell expresses the highest level of AS-C genes and loses its connections to its neighbors as well as with the apical surface just before delaminating and becoming a neuroblast (For review: (Sanes et al. 2012))
In the OPC, the transition of neuroepithelial cells into neuroblasts occurs in a proneural wave, without the cells undergoing cell movement or delamination: Cells in front of the wave are still neuroepithelial, while cells in the wake of the proneural wave have become neuroblasts. The AS-C protein L(1)sc is expressed in the transition zone between neuroepithelium and neuroblasts and, along with Notch, is necessary for the proneural wave (Egger et al. 2007, Yasugi et al. 2008, Apitz and Salecker 2014). In the IPC, cells undergo EMT and delaminate to later become neuroblasts after migration to the dIPC, most likely also under the control of L(1)sc and Notch (Apitz and Salecker 2015, Neriec et al. Submitted). Further investigations are needed to determine in more details the mechanisms by which L(1)sc and Notch control the timing of neuroepithelium to neuroblast transition in the optic lobe (Egger et al. 2011).
4.1.2 Generating neurons from neuroblasts
New generated neuroblasts undergo asymmetric division which results in one replacement neuroblast and one GMC, which then divides once, generating two post mitotic cells, either neurons or glia. This is the case for 90% of VNC and CB neuroblasts, called type I neuroblasts, and for most neuroblasts of the OPC. The molecular mechanisms involved in such divisions from the neuroblast to the GMCs have been well characterized and reviewed in (Homem and Knoblich 2012, Sousa-Nunes and Somers 2013, Kang and Reichert 2014). Importantly, neuroblasts express Deadpan (Dpn), GMCs express Prospero (Pros) and both express Asense (Ase).
In recent years, examples that differ from such classic models have been characterized. During CB neurogenesis, 8 specialized Type II neuroblasts undergo asymmetric division to produce intermediate neural progenitors instead of GMCs. These intermediate neural progenitors then divide asymmetrically several times to replace themselves and generate one GMC, leading to lineages that produce a large number of neurons (Wallace et al. 2000, Bello et al. 2008, Boone and Doe 2008, Bowman et al. 2008, Bayraktar et al. 2010, Viktorin et al. 2013, Koe et al. 2014). Other neuroblasts (Type 0) of the CB and of the optic lobe generate GMCs that do not divide but instead differentiate directly into one neuron (or glia) (Baumgardt et al. 2014, Bertet et al. 2014). Recently, neuroblasts generated by the IPC have been characterized and display atypical features compared to other neuroblasts. First they migrate between their delamination from the neuroepithelium and their arrival in the dIPC as atypical dividing neuroblasts. Furthermore, these neuroblasts express Ase but not yet Dpn and divide. Finally, they downregulate Ase as they age, thus lacking Ase expression in their GMC progeny (Apitz and Salecker 2015, Neriec et al. Submitted). Further studies remain necessary to characterize how those differences play a role in the formation of neurons.
4.2 Common mechanisms for neuronal specification
Neuronal cell types can be defined based on cell morphology, cell connectivity, marker expression or intrinsic properties such as electrophysiological properties. The specification of neuronal identity occurs in three main steps (Lin and Lee 2012): (a) spatial patterning of the neuroepithelium; (b) temporal patterning of neuroblasts by series of transcription factors; (c) distinct hemi lineages from GMCs (Fig. 4B).
4.2.1 Spatial patterning of the neuroepithelium
The first mechanism to specify different neuronal identities occurs in the neuroepithelium where spatial patterning cues exist early on. A classic example is the fly embryo, where positional cues are provided by dorso-ventral and antero-posterior patterning genes: Gap genes and segment polarity, as well as dorso-ventral and Hox genes establish a molecular coordinate system (Urbach and Technau 2003, Technau 2008). Such Cartesian grid provides a specific identity to any delaminating neuroblast, and this determines the type of neurons that will be produced (Skeath 1999, Skeath and Thor 2003, Technau et al. 2006). In the OPC of the optic lobe, the neuroepithelium crescent is also regionalized (Fig. 4 B’): The tips of the crescent express Wingless (Wg), which are bordered by a region expressing Decapentaplegic (Dpp), and together these two regions express Rx, followed by an Optix region and finally a Vsx-1 expressing region in the center (Gold and Brand 2014, Erclik et al. Under Review) .
Neuroblasts and some of the neurons they produce retain the positional markers of the neuroepithelium in the different regions they are coming from. This can affect their mode of neurogenesis. For instance, young neuroblasts of the Wg region of the OPC do not generate classical Type I like in the rest of the OPC, but instead become Type 0 and generate GMCs that directly differentiate into one neuron (Bertet et al. 2014). More importantly, it ultimately affects the identity of the neurons that are being produced (Bertet et al. 2014, Erclik et al. Under Review). For instance Pm1 and Pm2 neurons are only generated from the Rx regions of the OPC (Erclik et al. Under Review).
4.2.2 Temporal patterning of neuroblasts
The second mechanism by which neuronal diversity is generated temporal patterning of neuroblasts as well as Intermediate Neuronal Progenitors for type II neuroblasts: These cells sequentially express stereotyped temporal series of transcription factors (Reviewed in (Li et al. 2013a)) (Fig. 4 B”). These factors are often transmitted to the post-mitotic neurons, affecting the adult identity of the neuronal progeny (Isshiki et al. 2001, Bayraktar et al. 2010, Li et al. 2013b, Bertet et al. 2014). Despite differences in the transcription factor repertoire used in these temporal series, the general mechanism is very similar: one transcription factor induces expression of the next but also represses expression of the previous; changes in transcription factor expression are correlated with changes in adult neuronal progeny (Li et al. 2013a, Apitz and Salecker 2014). Studies in vertebrates seem to indicate that similar temporal series also play an important role in mammalian neurogenesis, including the mouse retina (Livesey and Cepko 2001, Wang et al. 2014a, Mattar et al. 2015).
4.2.3 Hemi lineages from the GMCs
Finally, a last source of neuronal diversity arises from molecular asymmetry in the final division of GMCs: Indeed, both in the VNC/CB and during optic lobe neuronal development, each GMC generates two post-mitotic cells that receive different levels of the protein Numb, a repressor of Notch signaling. This creates two post-mitotic cells with different Notch pathway activity status: NotchON or NotchOFF (Fig. 4 B”’). These two groups of neuroblast progeny are called hemi-lineages and differ in their adult neuronal identity (Lin et al. 2012, Udolph 2012, Bertet et al. 2014)
In conclusion, spatial patterning in the NE, temporal series of transcription factors in the neuroblasts, and GMC hemi-lineages define three main axes through which neuronal diversity is generated (Lin and Lee 2012). Furthermore, different combinations of transcription factors have also been shown to control cell death or survival (Bertet et al. 2014). For example in the optic lobe, NotchON neurons generated from the Wingless region of the neuroepithelium, undergo apoptosis when they are generated in the time window when the neuroblasts express the transcription factor Eyeless. However, neurons emerging from the same neuroblasts, but during the following Sloppy-paired time window always survive when they are NotchON but die when they are NotchOFF. These cells then adopt specific adult identities (Bertet et al. 2014). In spite of the contribution of spatial and temporal transcription factors, the general rules that dictate the formation of specific adult neuronal morphologies, interneurons vs. projection neurons for example, remain completely unknown.
5. Conclusions and perspectives
Since the first description by Cajal over hundred years ago, the fly visual system has been one of the most important models in developmental neurobiology. Progressing from the eye to the brain, a clearer picture emerges regarding not only the connectome and the processing of visual information in the fly brain, but also the formation of neurons and establishment of such network during development. Among the remaining studies to be done, three main areas emerge of major interests: the integration of the neuronal specification identities at the molecular level, the mechanisms by which neurons integrate within a neuronal circuits and finally the level of evolutionary conservation between insects and human visual systems.
5.1 Integration at the molecular level
Albeit cellular mechanisms remain to be elucidated, such as the detailed correlation of larval cell identities to adult neuronal types, developmental studies of the fly visual system already have provided a blueprint for reaching an understanding of this complex process at a molecular level. What are the molecular mechanisms that control the integration of the three axis of neuronal specification (spatial patterning of the neuroepithelium, window of the temporal series of transcription factors and Notch status) for the expression of specific terminal differentiation genes? Recently, the modules that control the expression of a terminal differentiation gene such as the neurotransmitter V-GLUT have been addressed in C. elegans (Serrano-Saiz et al. 2013). What are the modules involved in Drosophila? Future genomic and genetic studies will bring great insights in the mechanisms that control the expression of late terminal differentiation genes in large and complex neuronal circuits such as Drosophila.
Furthermore the production of neurons from induced pluripotent stem cells offer one of the most promising therapeutic approaches for the treatment of neurodegenerative disorders (Liu and Zhang 2011, Hallett et al. 2015). Adult human cells can already be induced into neuronal stem cells and generate neurons in vitro that have been successful transplanted in living animals. However, the similarities and differences between in vitro generated neurons and the in vivo neurons they would replace are a major concern. To address this question, Drosophila offers a valuable model to understand the molecular mechanisms involved in the formation of a neuronal identity and the role of extrinsic vs. intrinsic cues. For example, experiments have shown that type I neuroblasts from the central brain still undergo temporal series when cultivated in vitro (Grosskortenhaus et al. 2005). Similar experiments still remain to be done for other type of neuroblasts, type II and from the optic lobe.
5.2 Circuits and retinotopy
While some aspects of neuronal connections have been identified in the formation of the visual system (Lee et al. 2001, Lee et al. 2003, Sanes and Zipursky 2010, Timofeev et al. 2012), the establishment of connectivity remains largely unknown. A major concept in visual system circuit formation is retinotopy, i.e. the topographic mapping of visual inputs from the retina to optic lobe neurons. Retinotopy starts from the ~800 ommatidia in the eye, each pointing to one point in space and projecting into an equivalent number of cartridges in the lamina, which are therefore representing the same points in space. Similarly, lamina neurons project into ~800 columns of the medulla, in an orderly fashion. Such conservation in the representation of visual information is crucial for any visual system to be able to extract information such as motion and is established very early during development. Due to the direction in which the morphogenetic furrow progresses, lamina neurons acquire their posterior-to-anterior identity based on their time of differentiation. Retinotopy along the dorso-ventral axis is also preserved, when rows of photoreceptors generated at a given time point contact developing lamina neurons, thereby forming cartridges. Therefore, time represents an essential component in the generation of retinotopy, with the induction of the lamina by the photoreceptors representing the critical aspect.
However, how retinotopy is established in the medulla and in the higher processing centers during development remains to be further investigated since photoreceptors do not contribute to generating medulla or lobula complex neurons.
5.3 Evolution
The studies described here have generated broad concepts that start to apply to vertebrate neurogenesis. Whether there is a common ancestry of insect and vertebrate nervous systems is a major question in evolutionary biology (Moroz 2009). Today, the consensus converges towards the concept of a common neuronal origin between flies and vertebrates (Strausfeld 2009, Strausfeld and Hirth 2013b, Strausfeld and Hirth 2013a, Wolff and Strausfeld 2015), yet little is understood about how much is shared between visual processing systems in terms of development.
To address which features of the fly and vertebrate visual system may have been present in their bilaterian ancestor; comparisons must be made at the anatomical and functional levels (Erclik et al. 2009, Sanes and Zipursky 2010). However, while similarities between two adult visual systems might very well be due to evolutionary homology, they could also be the result of convergent evolution due to physical constraints on the neuronal circuits processing visual information (Strausfeld and Hirth 2013b, Strausfeld and Hirth 2013a).
Programs involved in the formation of neuronal circuits are under specific developmental constraints and adult systems that arise from similar developmental programs are more likely to share a common origin. The observed similarities in adult neuronal systems, likely mirroring similarities in developmental programs, strongly suggest a common origin. As one of the best understood neuronal system, in flies and in vertebrates, the visual system is well suited to address the question of homology between flies and mammals (Erclik et al. 2009, Sanes and Zipursky 2010).
References
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz H, Salecker I. A challenge of numbers and diversity: neurogenesis in the Drosophila optic lobe. J Neurogenet. 2014;28(3–4):233–249. doi: 10.3109/01677063.2014.922558. [DOI] [PubMed] [Google Scholar]
- Apitz H, Salecker I. A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nat Neurosci. 2015;18(1):46–55. doi: 10.1038/nn.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aptekar JW, Keles MF, Lu PM, Zolotova NM, Frye MA. Neurons forming optic glomeruli compute figure-ground discriminations in Drosophila. J Neurosci. 2015;35(19):7587–7599. doi: 10.1523/JNEUROSCI.0652-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Salmani BY, Bivik C, MacDonald RB, Gunnar E, Thor S. Global programmed switch in neural daughter cell proliferation mode triggered by a temporal gene cascade. Dev Cell. 2014;30(2):192–208. doi: 10.1016/j.devcel.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Bausenwein B, Dittrich AP, Fischbach KF. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 1992;267(1):17–28. doi: 10.1007/BF00318687. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 2010;5:26. doi: 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Desplan C. Visual circuits in flies: beginning to see the whole picture. Curr Opin Neurobiol. 2015;34:125–132. doi: 10.1016/j.conb.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158(5):1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68(9):1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A. Fly visual course control: behaviour, algorithms and circuits. Nat Rev Neurosci. 2014;15(9):590–599. doi: 10.1038/nrn3799. [DOI] [PubMed] [Google Scholar]
- Borst A, Haag J. Neural networks in the cockpit of the fly. J Comp Physiol A. 2002;188:419–437. doi: 10.1007/s00359-002-0316-8. [DOI] [PubMed] [Google Scholar]
- Borst A, Helmstaedter M. Common circuit design in fly and mammalian motion vision. Nat Neurosci. 2015;18(8):1067–1076. doi: 10.1038/nn.4050. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14(4):535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW. Pattern formation in the Drosophila eye. Curr Opin Genet Dev. 2007;17(4):309–313. doi: 10.1016/j.gde.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, Wu CC, Chen GY, Ching YT, Lee PC, Lin CY, Lin HH, Wu CC, Hsu HW, Huang YA, Chen JY, Chiang HJ, Lu CF, Ni RF, Yeh CY, Hwang JK. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21(1):1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Chin AL, Lin CY, Fu TF, Dickson BJ, Chiang AS. Diversity and wiring variability of visual local neurons in the Drosophila medulla M6 stratum. J Comp Neurol. 2014;522(17):3795–3816. doi: 10.1002/cne.23622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett TS. Insect navigation: visual panoramas and the sky compass. Curr Biol. 2008;18(22):R1058–R1061. doi: 10.1016/j.cub.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Dearborn R, Jr, Kunes S. An axon scaffold induced by retinal axons directs glia to destinations in the Drosophila optic lobe. Development. 2004;131(10):2291–2303. doi: 10.1242/dev.01111. [DOI] [PubMed] [Google Scholar]
- del Valle Rodriguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat Methods. 2012;9(1):47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Spindle Orientation and Asymmetric Localization in Drosophila: Both Inscuteable? Cell. 1996;86(5):695–697. doi: 10.1016/s0092-8674(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Douglass JK, Strausfeld NJ. Anatomical organization of retinotopic motion-sensitive pathways in the optic lobes of flies. Microsc Res Tech. 2003;62(2):132–150. doi: 10.1002/jemt.10367. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly (Austin) 2011;5(3):237–241. doi: 10.4161/fly.5.3.15640. [DOI] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, McInnes RR, Lipshitz HD. Eye evolution at high resolution: the neuron as a unit of homology. Dev Biol. 2009;332(1):70–79. doi: 10.1016/j.ydbio.2009.05.565. [DOI] [PubMed] [Google Scholar]
- Erclik T, Li X, Bertet C, Baumert R, Ng J, Chen Z, Del A, Rodriguez V, Senderowicz L, Negre N, White P, Desplan C. Selective integration of spatial and temporal inputs generates neural diversity and establishes retinotopy in the Drosophila medulla. Nature. (Under Review) [Google Scholar]
- Fischbach KF, Dittrich APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258(3):441–475. [Google Scholar]
- Gold KS, Brand AH. Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev. 2014;9:18. doi: 10.1186/1749-8104-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Hartenstein AY, Hartenstein V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 1993;273(3):583–598. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8(2):193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Hallett Penelope J, Deleidi M, Astradsson A, Smith Gaynor A, Cooper O, Osborn Teresia M, Sundberg M, Moore Michele A, Perez-Torres E, Brownell A-L, Schumacher James M, Spealman Roger D, Isacson O. Successful Function of Autologous iPSC-Derived Dopamine Neurons following Transplantation in a Non-Human Primate Model of Parkinson’s Disease. Cell Stem Cell. 2015;16:269–274. doi: 10.1016/j.stem.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa E, Kaido M, Takayama R, Sato M. Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Dev Biol. 2013;377(1):90–99. doi: 10.1016/j.ydbio.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Hausen K. Photoreception and Vision in Invertebrates. Vol. 74. Springer US: M. A. Ali; 1984. The Lobula-Complex of the Fly: Structure, Function and Significance in Visual Behaviour; pp. 523–559. [Google Scholar]
- Hofbauer A, Campos-Ortega Ja. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux Arch Dev Biol. 1990;198:264–274. doi: 10.1007/BF00377393. [DOI] [PubMed] [Google Scholar]
- Homberg U, Heinze S, Pfeiffer K, Kinoshita M, Jundi Bel. Central neural coding of sky polarization in insects. Philos Trans R Soc Lond B Biol Sci. 2011;366(1565):680–687. doi: 10.1098/rstb.2010.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CCF, Knoblich Ja. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kunes S. Hedgehog, transmitted along retinal axons, triggers neurogenesis in the developing visual centers of the Drosophila brain. Cell. 1996;86(3):411–422. doi: 10.1016/s0092-8674(00)80114-2. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kunes S. Signals transmitted along retinal axons in Drosophila: Hedgehog signal reception and the cell circuitry of lamina cartridge assembly. Development. 1998;125(19):3753–3764. doi: 10.1242/dev.125.19.3753. [DOI] [PubMed] [Google Scholar]
- Huang Z, Shilo BZ, Kunes S. A retinal axon fascicle uses spitz, an EGF receptor ligand, to construct a synaptic cartridge in the brain of Drosophila. Cell. 1998;95(5):693–703. doi: 10.1016/s0092-8674(00)81639-6. [DOI] [PubMed] [Google Scholar]
- Imai T, Sakano H, Vosshall LB. Topographic Mapping — The Olfactory System. CSH Perspect Biol. 2010:1–19. doi: 10.1101/cshperspect.a001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106(4):511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito M, Masuda N, Shinomiya K, Endo K, Ito K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr Biol. 2013;23(8):644–655. doi: 10.1016/j.cub.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SCB, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM. Principles of Neural Science. 4th. McGraw-Hill Medical; 2000. [Google Scholar]
- Kang KH, Reichert H. Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tissue Res. 2014:33–45. doi: 10.1007/s00441-014-1914-9. [DOI] [PubMed] [Google Scholar]
- Koe CT, Li S, Rossi F, Wong JJL, Wang Y, Zhang Z, Chen K, Aw SS, Richardson HE, Robson P, Sung WK, Yu F, Gonzalez C, Wang H, Jing J, Wong L, Wang Y, Zhang Z, Chen K, Aw SS, Richardson HE, Wong JJL, Wang Y, Zhang Z, Chen K, Aw SS, Richardson HE, Robson P, Sung WK, Yu F, Gonzalez C, Wang H. The Brm-HDAC3-Erm repressor complex suppresses dedifferentiation in Drosophila type II neuroblast lineages. eLIFE. 2014;2014:1–19. doi: 10.7554/eLife.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeptowski W, Gorska-Andrzejak J, Kijak E, Gorlich A, Guzik E, Moore G, Pyza EM. External and circadian inputs modulate synaptic protein expression in the visual system of Drosophila melanogaster. Front Physiol. 2014;5:102. doi: 10.3389/fphys.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. Building an ommatidium one cell at a time. Dev Dyn. 2012;241:136–149. doi: 10.1002/dvdy.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S, Wilson C, Steller H. Independent guidance of retinal axons in the developing visual system of Drosophila. J Neurosci. 1993;13(2):752–767. doi: 10.1523/JNEUROSCI.13-02-00752.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TD. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog Retin Eye Res. 2013;36:52–119. doi: 10.1016/j.preteyeres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, Hiesinger PR. The Developmental Rules of Neural Superposition in Drosophila. Cell. 2015;162(1):120–133. doi: 10.1016/j.cell.2015.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30(2):437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Lee RC, Clandinin TR, Lee C-H, Chen P-L, Meinertzhagen IA, Zipursky SL. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci. 2003;6:557–563. doi: 10.1038/nn1063. [DOI] [PubMed] [Google Scholar]
- Li HH, Kroll Jason R, Lennox Sara M, Ogundeyi O, Jeter J, Depasquale G, Truman James W. A GAL4 Driver Resource for Developmental and Behavioral Studies on the Larval CNS of Drosophila. Cell Rep. 2014;8:897–908. doi: 10.1016/j.celrep.2014.06.065. [DOI] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C. Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol. 2013a;105:69–96. doi: 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013b;498(7455):456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biol. 2012;10(11):e1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lee T. Generating neuronal diversity in the Drosophila central nervous system. Dev Dyn. 2012;241(1):57–68. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang S-C. Specification of neuronal and glial subtypes from human pluripotent stem cells. CMLS. 2011;68:3995–4008. doi: 10.1007/s00018-011-0770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2(2):109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Developmental Biology. 2013;384(2):228–257. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisak MS, Haag J, Ammer G, Serbe E, Meier M, Leonhardt A, Schilling T, Bahl A, Rubin GM, Nern A, Dickson BJ, Reiff DF, Hopp E, Borst A. A directional tuning map of Drosophila elementary motion detectors. Nature. 2013;500(7461):212–216. doi: 10.1038/nature12320. [DOI] [PubMed] [Google Scholar]
- Mattar P, Ericson J, Blackshaw S, Cayouette M. A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron. 2015;85(3):497–504. doi: 10.1016/j.neuron.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 1363–1491. [Google Scholar]
- Meinertzhagen IA, Sorra KE. Chapter 3: Synaptic organization in the fly’s optic lamina: few cells, many synapses and divergent microcircuits. Prog Brain Res. 2001;131:53–69. doi: 10.1016/s0079-6123(01)31007-5. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Takemura SY, Lu Z, Huang S, Gao S, Ting CY, Lee CH. From form to function: the ways to know a neuron. J Neurogenet. 2009;23(1–2):68–77. doi: 10.1080/01677060802610604. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. Building a projection map for photoreceptor neurons in the Drosophila optic lobes. Sem in Cell & Dev Biol. 2004;15(1):137–143. doi: 10.1016/j.semcdb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Curr Biol. 2008;18(8):553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL. On the independent origins of complex brains and neurons. Brain Behav Evol. 2009;74(3):177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L, Ito K, Bacon JP, Strausfeld NJ. Optic glomeruli and their inputs in Drosophila share an organizational ground pattern with the antennal lobes. J Neurosci. 2012;32(18):6061–6071. doi: 10.1523/JNEUROSCI.0221-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol. 2003;455(4):417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Neriec N, Dua W, Mehmet O, Texeira Sousa F. Pinto, Li X, Erclik T, Bertet C, Mancini G, Rodriguez A. delValle, Hiesinger R, Desplan C. Cell migration and neuronal diversity during optic lobe neurogenesis in Drosophila. eLIFE. (Submitted) [Google Scholar]
- Otsuna H, Ito K. Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol. 2006;497(6):928–958. doi: 10.1002/cne.21015. [DOI] [PubMed] [Google Scholar]
- Otsuna H, Shinomiya K, Ito K. Parallel neural pathways in higher visual centers of the Drosophila brain that mediate wavelength-specific behavior. Front in Neur Circ. 2014;8 doi: 10.3389/fncir.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulk A, Millard SS, van Swinderen B. Vision in Drosophila: Seeing the World Through a Model’s Eyes. Ann Rev of Entomol. 2011;58:313–332. doi: 10.1146/annurev-ento-120811-153715. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105(28):9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu SV, Claussen J, Borst A. Neurons with GABAergic phenotype in the visual system of Drosophila. J Comp Neurol. 2013;521(1):252–265. doi: 10.1002/cne.23208. [DOI] [PubMed] [Google Scholar]
- Raghu SV, Joesch M, Sigrist SJ, Borst A, Reiff DF. Synaptic Organization of Lobula Plate Tangential Cells in Drosophila: Dα7 Cholinergic Receptors. J Neurogenet. 2009;23(1–2):200–209. doi: 10.1080/01677060802471684. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S, Sanchez y Sanchez D. Contribucion al conociemento de los centros nerviosos de los insectos. Trabajos del Laboratorio de Investigaciones biológicas de la Universidad de Madrid. 1915 [Google Scholar]
- Reichert H. Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results Probl Cell Differ. 2011;53:529–546. doi: 10.1007/978-3-642-19065-0_21. [DOI] [PubMed] [Google Scholar]
- Sanes DH, Reh TA, Harris WA. In: 1 - Neural induction. Development of the Nervous System. Third. Sanes DH, Reh TA, Harris WA, editors. London: Academic Press; 2012. pp. 1–22. [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66(1):15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suzuki T, Nakai Y. Waves of differentiation in the fly visual system. Dev Biol. 2013;380(1):1–11. doi: 10.1016/j.ydbio.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Selleck SB, Gonzalez C, Glover DM, White K. Regulation of the G1-S transition in postembryonic neuronal precursors by axon ingrowth. Nature. 1992;355(6357):253–255. doi: 10.1038/355253a0. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell. 2013;155(3):659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Gohl D, Fisher Y, Freifeld L, Clark D, Clandinin TR. Modular Use of Peripheral Input Channels Tunes Motion-Detecting Circuitry. Neuron. 2013;79:111–127. doi: 10.1016/j.neuron.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Gohl DM, Clandinin TR. Motion-Detecting Circuits in Flies: Coming into View. Ann Rev of Neuro. 2014;37(1):307–327. doi: 10.1146/annurev-neuro-071013-013931. [DOI] [PubMed] [Google Scholar]
- Skeath JB. At the nexus between pattern formation and cell-type specification: The generation of individual neuroblast fates in the drosophila embryonic central nervous system. BioEssays. 1999;21:922–931. doi: 10.1002/(SICI)1521-1878(199911)21:11<922::AID-BIES4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Somers WGG. Mechanisms of asymmetric progenitor divisions in the Drosophila central nervous system. In: Hime G, Abud H, editors. Transcriptional and Translational Regulation of Stem Cells SE - 6. Vol. 786. Dordrecht, Springer Netherlands: 2013. pp. 79–102. [DOI] [PubMed] [Google Scholar]
- Spalthoff C, Gerdes R, Kurtz R. Neuronal representation of visual motion and orientation in the fly medulla. Front Neural Circuits. 2012;6:72. doi: 10.3389/fncir.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld N. Insect Vision and Olfaction: Common Design Principles of Neuronal Organization. Neurobiology of Sensory Systems. 1989:319–353. [Google Scholar]
- Strausfeld NJ. Brain organization and the origin of insects: an assessment. Proc Biol Sci. 2009;276(1664):1929–1937. doi: 10.1098/rspb.2008.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ, Hirth F. Deep homology of arthropod central complex and vertebrate basal ganglia. Science. 2013a;340(6129):157–161. doi: 10.1126/science.1231828. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ, Hirth F. Homology versus convergence in resolving transphyletic correspondences of brain organization. Brain Behav Evol. 2013b;82:215–219. doi: 10.1159/000356102. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kaido M, Takayama R, Sato M. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol. 2013;380(1):12–24. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol. 2008;509(5):493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau GM. Advances in Experimental Medicine and Biology. Brain development in Drosophila melanogaster. Preface. Adv Exp Med Biol. 2008;628:v–vi. [PubMed] [Google Scholar]
- Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev Dyn. 2006;235:861–869. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- Timofeev K, Joly W, Hadjieconomou D, Salecker I. Localized netrins act as positional cues to control layer-specific targeting of photoreceptor axons in Drosophila. Neuron. 2012;75(1):80–93. doi: 10.1016/j.neuron.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE. Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol. 2013;2(4):545–557. doi: 10.1002/wdev.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill J, Nern A, Holtz S, Rubin G, Reiser MB. Contributions of the 12 Neuron Classes in the Fly Lamina to Motion Vision. Neuron. 2013;79:128–140. doi: 10.1016/j.neuron.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udolph G. Notch signaling and the generation of cell diversity in Drosophila neuroblast lineages. Adv Exp Med Biol. 2012;727:47–60. doi: 10.1007/978-1-4614-0899-4_4. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Early steps in building the insect brain: Neuroblast formation and segmental patterning in the developing brain of different insect species. Arthr Struc Dev. 2003;32:103–123. doi: 10.1016/S1467-8039(03)00042-2. [DOI] [PubMed] [Google Scholar]
- Varija Raghu S, Reiff DF, Borst A. Neurons with cholinergic phenotype in the visual system of Drosophila. J Comp Neurol. 2011;519(1):162–176. doi: 10.1002/cne.22512. [DOI] [PubMed] [Google Scholar]
- Viktorin G, Riebli N, Reichert H. A multipotent transit-amplifying neuroblast lineage in the central brain gives rise to optic lobe glial cells in Drosophila. Dev Biol. 2013;379(2):182–194. doi: 10.1016/j.ydbio.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Wallace K, Liu TH, Vaessin H. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis. 2000;26(1):77–85. doi: 10.1002/(sici)1526-968x(200001)26:1<77::aid-gene10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Wang S, Sengel C, Emerson MM, Cepko CL. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell. 2014a;30(5):513–527. doi: 10.1016/j.devcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-C, Yang JS, Johnston R, Ren Q, Lee Y-J, Luan H, Brody T, Odenwald WF, Lee T. Drosophila intermediate neural progenitors produce lineage-dependent related series of diverse neurons. Development. 2014b;141:253–258. doi: 10.1242/dev.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Kankel DR. Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Dev Biol. 1978;65(2):296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- Wolff GH, Strausfeld NJ. Genealogical correspondence of mushroom bodies across invertebrate phyla. Curr Biol. 2015;25(1):38–44. doi: 10.1016/j.cub.2014.10.049. [DOI] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V. Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Dev Biol. 2013;384(2):258–289. doi: 10.1016/j.ydbio.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135(8):1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370(3):313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Yu H-h, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao J-c, Wu GY-y, Peng H, Myers G, Lee T. Clonal development and organization of the adult Drosophila central brain. Curr biol. 2013;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeil J. Visual homing: an insect perspective. Curr Opin Neurobiol. 2012;22(2):285–293. doi: 10.1016/j.conb.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Zhu Y. The Drosophila visual system: From neural circuits to behavior. Cell Adh & Migr. 2013;7(4):333–344. doi: 10.4161/cam.25521. [DOI] [PMC free article] [PubMed] [Google Scholar]