Abstract
Salmonella enterica is a gram-negative, facultative intracellular pathogen that causes disease symptoms ranging from gastroenteritis to typhoid fever. A key virulence strategy is the translocation of bacterial effector proteins into the host cell, mediated by the type III secretion systems (TTSSs) encoded in Salmonella pathogenicity island 1 (SPI-1) and SPI-2. In S. enterica serovar Typhimurium LT2, we identified the protein products of STM4157 and STM2137 as novel candidate secreted proteins by comparison to known secreted proteins from enterohemorrhagic Escherichia coli and Citrobacter rodentium. The STM4157 and STM2137 proteins, which we have designated SseK1 and SseK2, respectively, are 61% identical at the amino acid level and differ mainly in their N termini. Western analysis showed that in vitro accumulation and secretion of these proteins in serovar Typhimurium were affected by mutations in the two-component systems SsrA/B and PhoP/Q, which are key mediators of intracellular growth and survival. SPI-2 TTSS-dependent translocation of recombinant SseK1::Cya was evident at 9 h postinfection of epithelial cells, while translocation of SseK2::Cya was not detected until 21 h. Remarkably, the translocation signal for SseK1 was contained within the N-terminal 32 amino acids. Fractionation of infected epithelial cells revealed that following translocation SseK1 localizes to the host cytosol, which is unusual among the currently known Salmonella effectors. Phenotypic analysis of ΔsseK1, ΔsseK2, and ΔsseK1/ΔsseK2 mutants provided evidence for a role that was not critical during systemic infection. In summary, this work demonstrates that SseK1 and SseK2 are novel translocated proteins of serovar Typhimurium.
Salmonella enterica is a gram-negative bacterial species that infects animal and human hosts. The nature and severity of disease is dependent upon the bacterial serovar and the host species (32). For example, S. enterica serovar Typhimurium causes gastroenteritis in calves and humans, whereas it leads to a typhoid-like systemic infection in mice. As a facultative intracellular pathogen, Salmonella possesses virulence determinants that enable it to invade, persist, and replicate within eukaryotic cells by subverting host cell processes. While a significant number of virulence factors are clustered on the virulence plasmid or within large regions (15 to 40 kb) of the chromosome called Salmonella pathogenicity islands (SPI), they are also found scattered throughout the genome in smaller “islets” (reviewed in reference 42). SPIs and pathogenicity islets are characterized by altered DNA base composition and by association with mobile genetic elements, leading to the hypothesis that they have been acquired by horizontal transfer (24, 25).
The two largest pathogenicity islands in the serovar Typhimurium genome, designated SPI-1 and SPI-2, each encode a type III secretion system (TTSS) with structural homology to each other and to the TTSSs of other known pathogens (27, 49; reviewed in references 11 and 30). TTSSs comprise a specialized protein export apparatus that spans the inner and outer bacterial membranes and acts as the secretion machinery for bacterial effectors. A subset of the secreted effectors form a complex called the translocon (11). Within the context of an infection, the TTSS translocates bacterial effector proteins across three membranes and delivers them directly into the target host cell, where they alter or initiate cytoskeletal rearrangements, signal transduction, and vesicular trafficking (reviewed in references 33 and 42).
Importantly, the two TTSSs are differentially expressed and have distinct roles during an infection. The SPI-1-encoded TTSS is expressed during extracellular growth and is required for efficient invasion of host epithelial cells (reviewed in reference 15). The delivery of SPI-1 effectors initiates ruffling of the cell membrane, leading to bacterial uptake into an intracellular compartment termed the Salmonella-containing vacuole (SCV). Subsequently, the SPI-2-encoded TTSS is expressed during the intracellular stage of infection. SPI-2 TTSS mutants are invasion competent but deficient in intracellular survival and replication and, thereby, in systemic progression of the infection (9, 37, 42, 49). A key SPI-2-dependent phenotype observed originally in epithelial-like cell lines (17) and more recently characterized in macrophage-like cell lines (36) is the maturation of the SCV by selective interactions with the endosomal-lysosomal pathway and the formation of tubular membranous extensions called Salmonella-induced filaments (SIF) (22; reviewed in reference 35).
In contrast to the homology between the structural components of the SPI-1 and SPI-2 TTSSs (26), the effectors translocated by each apparatus are not conserved; thus, it is reasonable to assume that a specific complement of translocated effectors is responsible for invasion compared to intracellular growth. Not unexpectedly, the expression of translocated effectors is coordinately regulated with the TTSS apparatus for which they are a substrate, irrespective of their proximity within the chromosome. For example, the translocated effector PipB is encoded in SPI-5 yet is coexpressed with and translocated by the SPI-2 TTSS (34). In general, effectors that are coexpressed with the SPI-2 TTSS, such as SopD2 (5), are not translocated by the SPI-1 TTSS. Notably, there appear to be functionally nonredundant homologues for many of the effectors, as illustrated by the family of Salmonella translocated effectors that shares N-terminal homologous regions (6, 45). However, there is no conserved amino acid sequence motif common to all effectors. Instead, substrate recognition by the TTSS apparatus appears to require two domains in the effector, one that is located within the N-terminal first ∼20 amino acids and a second that is located within the first ∼140 amino acids. The latter is proposed to serve as a chaperone binding site that confers secretion pathway specificity (8, 40).
The functional elucidation of individual SPI-2 effectors, both those encoded in SPI-2 and those scattered throughout the chromosome, has generally focused on their association or colocalization with cytoskeletal components or with membranous structures such as the SCV and SIF (reviewed in references 35 and 62). This is highlighted by research delineating the roles of SifA (4, 59), SseJ (52), SseF (37), and SseG (53), as well as the localization of PipB, PipB2 (36), and SopD2 (5). The function of SpiC/SsaB remains controversial: it may be a structural component of the TTSS, since it is important for the secretion of SseB and the translocation of other SPI-2 effectors (14, 63). On the other hand, other researchers have demonstrated translocation of SpiC/SsaB into the host cytosol (56, 61) and have identified interactions with the host proteins Hook3 (56) and TassC (38), suggesting that this effector plays a role in the selective trafficking of the SCV. A key prerequisite to the translocation of all known SPI-2 effectors is secretion of the translocon components SseB, SseC, and SseD, which are found predominantly on the outer surface of the bacterium and are presumed to contact the host membrane; translocon mutants are defective in translocation but not secretion of the SPI-2 effectors (48).
There is significant interest in more fully elucidating the effector repertoire and molecular basis of the host-pathogen interaction during infection. In this report we identify the proteins encoded by STM4157 and STM2137 in serovar Typhimurium LT2 as novel secreted and translocated proteins, and we propose that they be designated SseK1 and SseK2, respectively. We demonstrate that translocation of SseK1 is dependent upon a functional SPI-2 TTSS and is mediated by the N-terminal 32 amino acid residues. Translocation of SseK2 was detected at low levels and later time points compared to SseK1. Once translocated, SseK1 localized to the host cytosol; this is in contrast to the membrane localization of the majority of currently identified SPI-2 translocated effectors. Phenotypic analysis of the ΔsseK1, ΔsseK2, and ΔsseK1/ΔsseK2 mutants suggested that the role of these putative TTSS effectors is unrelated to the formation of the SCV and SIF, and their function was not apparent in a susceptible murine model.
MATERIALS AND METHODS
Bacterial strains and in vitro growth conditions.
Bacterial strains are listed in Table 1. Bacterial cultures were routinely maintained in Luria-Bertani (LB) liquid medium or on LB agar plates supplemented with the appropriate antibiotic(s) at the following concentrations: chloramphenicol, 30 μg ml−1; kanamycin, 50 μg ml−1; carbenicillin, 50 μg ml−1; streptomycin, 100 μg ml−1.
TABLE 1.
Plasmids and bacterial strains used in this study
| Plasmid or strain | Description | Source |
|---|---|---|
| Plasmids and cloning vectors | ||
| pCR2.1 | Kanr; commercial vector for cloning PCR products | Invitrogen Corp. |
| pACYC184 | Tetr Cmr; low-copy-number cloning vector | New England Biolabs, Inc. |
| pUC18 | Apr; general cloning vector | Fermentas, Inc. |
| pRE112 | Cmr; suicide vector containing sacB1 | 13 |
| pRE112(ΔsseK2) | pRE112 containing in-frame deletion of sseK2 (missing amino acids 7 to 341) | This study |
| pRE112(ΔsseK1) | pRE112 containing in-frame deletion of sseK1 (missing amino acids 7 to 329) | This study |
| pACBC-2HA | pACYC184 containing the PipB ORF and promoter, with C-terminal tandem HA epitope tags | 34 |
| pPipBN180-Cya | pACYC184 expressing the N-terminal 180 amino acids of PipB fused to Cya | 34 |
| pACsseK1::2HA | pACYC184 containing the entire SseK1 ORF and 290 bp of upstream DNA sequence (promoter region) with C-terminal tandem HA tags | This study |
| pACsseK1::cya | pACYC184 containing the entire SseK1 ORF and 290 bp of upstream DNA sequence (promoter region) fused to Cya | This study |
| pACsseK2::2HA | pACYC184 containing the entire SseK2 ORF and 436 bp of upstream DNA sequence (promoter region), with C-terminal tandem HA tags | This study |
| pACsseK2::cya | pACYC184 containing the entire SseK2 ORF and 436 bp of upstream DNA sequence (promoter region) fused to Cya | This study |
| pOG-Ussek1 | pUC18 containing the entire SseK1 ORF and 290 bp of upstream DNA sequence (promoter region) fused to Cya; template for inverse PCR | This study |
| pOG-ATNΔ32 | pACYC184 expressing amino acids 32 to 336 of SseK1 fused to Cya | This study |
| pOG-ATNΔ78 | pACYC184 expressing amino acids 78 to 336 of SseK1 fused to Cya | This study |
| pOG-ATNΔ202 | pACYC184 expressing amino acids 202 to 336 of SseK1 fused to Cya | This study |
| pOG-ATNΔ267 | pACYC184 expressing amino acids 267 to 336 of SseK1 fused to Cya | This study |
| pOG-ATCΔ304 | pACYC184 expressing amino acids 1 to 32 of SseK1 fused to Cya | This study |
| pEGFP-N1(sseK1) | For expression of SseK1 fused to the N terminus of EGFP | This study |
| pEGFP-C1(sseK1) | For expression of SseK1 fused to the C terminus of EGFP | This study |
| E. coli strains | ||
| DH10B | Cloning strain | Invitrogen Corp. |
| SY327λpir | For propagation of pRE112 and derivative plasmids | 46 |
| SM10λpir | Kanr; for mobilizing pRE112(ΔsseK2) and pRE112 (ΔsseK1) into S. Typhimurium SL1344 | 46 |
| Serovar Typhimurium strains | ||
| SL1344 | wt | 28 |
| ΔsseK2 | SL1344, chromosomal deletion of sseK2 (missing amino acids 7 to 341) | This study |
| ΔsseK1 | SL1344, chromosomal deletion of sseK1 (missing amino acids 7 to 329) | This study |
| ΔsseK1/ΔsseK2 | SL1344, chromosomal deletions of both sseK1 and sseK2 | This study |
| ΔsseA | SL1344, chromosomal deletion of sseA | 10 |
| hilA::kan339 | SL1344, defective in regulation of SPI-1 genes | 3 |
| ssrA::mTn5 | SL1344, defective in regulation of SPI-2 genes | 7 |
| phoP::Tn10d-Tc | SL1344, defective in regulation of the PhoP/Q regulon, including SPI-2 genes | 7 |
| ΔssaR | SL1344, defective in structural gene of the SPI-2 type III apparatus | 7 |
| SB103 | SL1344, invA::kan, defective in structural gene of the SPI-1 type III apparatus | 16 |
Growth of Salmonella in LB broth (pH 7.0) was used to approximate SPI-1-inducing conditions in vitro. To induce expression and secretion of SPI-2-regulated proteins, Salmonella was grown in modified magnesium minimal medium (MgM) containing 80 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8), 5 mM KCl, 7.5 mM (NH4)SO4, 0.5 mM K2SO4, 337 μM K2HPO4-KH2PO4 (pH 7.4), 20 mM MgCl2, 38 mM glycerol, and 0.1% Casamino Acids (B. K. Coombes, personal communication). Cultures were grown with aeration for 3 h in LB or 5.5 h in MgM. Equivalent numbers of bacteria were harvested by centrifugation. The culture supernatant was filtered through a 0.2-μm-pore-size HT Tuffryn membrane (Pall Life Sciences, Ann Arbor, Mich.), precipitated by addition of 10% (vol/vol) trichloroacetic acid, and resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye. The corresponding bacterial cell pellets were also resuspended in 1× SDS-PAGE loading dye, and both fractions were subjected to Western analysis as described below. All experiments were performed in triplicate, and representative results are shown.
Molecular biology techniques and construction of plasmids.
Standard molecular biology techniques were used (54), and manufacturer's instructions were followed. As a general cloning strategy, DNA fragments were amplified by PCR (Elongase; Invitrogen Corp., Carlsbad, Calif.) using serovar Typhimurium SL1344 chromosomal DNA as template, ligated into pCR2.1 (Invitrogen Corp.) for sequence confirmation, and then digested with restriction enzymes (New England BioLabs, Inc., Beverly, Mass.) as appropriate for ligation (T4 DNA ligase; Invitrogen Corp.) into a destination vector. Oligonucleotides (Table 2) were purchased from Sigma-Genosys (The Woodlands, Tex.).
TABLE 2.
Oligonucleotides used in this study
| Desig-nation | DNA sequence (5′-3′)a |
|---|---|
| SKC224 | ACGCGTCGACTTTTTCATGGCGGGAAGATTC |
| SKC225 | GAAGATCTCCTCCAAGAACTGGCAGTTAAAC |
| SKC231 | AAGCTTATGATCCCACCATTAAATAATATG |
| SKC232 | GAAGATCTCTGCACATGCCTCGCCCATG |
| SKC233 | ACGCGTCGACTTATTGATTGTTTACGCGGGAC |
| SKC236 | GTCCTAGAATCTCTGCCGCATCGTTCC |
| SKC237 | ACGCGTCGACTCTATTTAATGGTGGGATCAT |
| SKC238 | ACGCGTCGACTCATGGGCGAGGCATGTGCAG |
| SKC239 | GTCGCAAGAAAACTATGCGGAGC |
| SKC240 | GCTCTAGAATGATAGAAGAGGCCCAAAGACC |
| SKC241 | ACGCGTCGACAGCGGCATTAAAACGTGCCAT |
| SKC242 | ACGCGTCGACACTGCCAGTTCTTGGAGGTAATC |
| SKC243 | CGAGCTCTAGACTCTTTAGCAGAAACACGG |
| SKC256 | CTCGAGCTATGATCCCACCATTAAATAGTATGTTCCC |
| SKC257 | GAATTCGCTGCACATGCCTCGCCCATGAAC |
| OG-1 | GGGGGGATCCTTATTGATTGTTTACGCGGGAC |
| OG-2 | CATAACAATTAAATGCTCCATACTTAC |
| OG-3 | CCCCAGATCTAAAGCACAAGGGATAATCTTTTCC |
| OG-4 | GGAAAAGATTATCCCTTGTGC |
| OG-5 | TTTCCGGGACAAAAGGATGC |
| OG-7 | TTGGAGGCGGATAAGGTTGGG |
| OG-8 | CCGGCTTTACTTGCTGGATTAG |
| OG-9 | CCCCAGATCTCTGCACATGCCTCGCCCATG |
Restriction enzyme sites used for cloning of PCR products are underlined.
Plasmids used in this study are listed in Table 1. The sseK1 and sseK2 open reading frames (ORFs) plus promoter regions were cloned in frame with DNA sequence encoding tandem hemagglutinin epitopes (2HA). The sseK1 ORF plus 290 bp of upstream sequence was amplified by PCR using primers SKC233 (SalI site) and SKC232 (BglII site). Similarly, sseK2 coding sequence and 436 bp of upstream DNA were amplified by PCR using primers SKC224 (SalI site) and SKC225 (BglII site). The plasmids pACsseK1::2HA and pACsseK2::2HA were obtained by SalI/BglII digestion of the appropriate pCR2.1 clones, followed by ligation into the SalI-BglII site obtained by digesting pACBC-2HA (34) to release all pipB-specific sequence. Subsequently, the plasmids pACsseK1::cya and pACsseK2::cya were derived by replacing the 2HA coding sequence (removed by BglII/HindIII digestion) with the Bordetella pertussis cya coding sequence obtained as a BglII-HindIII fragment from pPipBN180-Cya (34).
We constructed four N-terminal truncations and one C-terminal truncation of the SseK1 protein fused to Cya. The entire sseK1 ORF plus 290 bp of upstream sequence were amplified by PCR using OG-1 (BamHI site) and OG-9 (HindIII site) and cloned into the HincII site of pUC18 (Fermentas, Inc., Hanover, Md.). The resultant plasmid, pOG-UsseK1, served as template for inverse PCRs with the following primer pairs: OG-2 plus OG-4 were used to construct the NΔ32 truncation; OG-2 plus OG-5 were used for the NΔ78 truncation; OG-2 plus OG-7 were used for the NΔ202 truncation; and OG-2 plus OG-8 were used for the NΔ267 truncation. The products of inverse PCR were self-ligated and digested with BamHI-BglII to release the sseK1-specific sequence. To obtain the CΔ304 truncation, we cloned the PCR product obtained with OG-1 plus OG-3 into the HincII site of pUC18, and the sseK1-specific sequence was released as a BamHI-BglII fragment. The plasmids pOG-ATNΔ32, pOG-ATNΔ78, pOG-ATNΔ202, pOG-ATNΔ267, and pOG-ATCΔ304 were constructed from the appropriate BamHI-BglII fragment fused to the BglII-HindIII cya gene, described above, in the BamHI/HindIII sites of pACYC184. All constructs were verified by sequencing.
For transfection experiments, sseK1 was cloned into pEGFP-N1 and pEGFP-C1 (Clontech, BD BioSciences), to obtain SseK1 fused to enhanced green fluorescent protein (EGFP). To generate pEGFP-C1(sseK1), in which SseK1 is fused to the C terminus of EGFP, the sseK1 coding sequence was amplified by PCR using primers SKC256 (XhoI site) and SKC232 and subsequently ligated as a XhoI-BglII fragment into XhoI/BamHI-digested pEGFP-C1. Similarly, to obtain pEGFP-N1(sseK1), in which SseK1 was fused to the N terminus of EGFP, the sseK1 coding sequence was amplified using SKC231 (HindIII site) and SKC257 (EcoRI site) and ligated as a HindIII-EcoRI fragment into the corresponding restriction sites in pEGFP-N1.
Generation of serovar Typhimurium SL1344 mutants.
We engineered unmarked nonpolar internal deletions of the coding sequences for sseK1 and sseK2 by allelic exchange as previously described (13). The cloning strategy was to perform a forced directional ligation of the target suicide plasmid pRE112 (13) with two chromosomal DNA fragments that each comprised at least 1 kb of DNA flanking the deleted ORF, which was replaced by a SalI site. Fragments of 1.4 and 1.33 kb of DNA flanking sseK1 were obtained by PCR using primer pairs SKC236 plus SKC237 (SalI site) and SKC238 (SalI site) plus SKC239, respectively. Similarly, 1.11 and 1.07 kb of DNA flanking sseK2 were obtained by PCR using primer pairs SKC240 plus SKC241 (SalI site) and SKC242 (SalI site) plus SKC243, respectively. The individual PCR products were released from pCR2.1 by restriction digestion with XbaI/SalI or SalI/SacI as appropriate (using XbaI and SacI sites in pCR2.1), while the suicide plasmid pRE112 was digested with XbaI/SacI. The ligation products were electroporated into Escherichia coli SY327 λpir to obtain pRE112(ΔsseK1) and pRE112(ΔsseK2) and confirmed by sequencing. These plasmids were electroporated into E. coli SM10 λpir to generate the conjugative strains for mating with wild-type serovar Typhimurium SL1344, which yielded the mutant strains ΔsseK1 and ΔsseK2 by allelic exchange. The double mutant SL1344 ΔsseK1/ΔsseK2 was obtained by mating between E. coli SM10 λpir (pRE112ΔsseK2) and serovar Typhimurium SL1344 ΔsseK1. The chromosomal deletion(s) was confirmed in all three mutants by PCR and sequencing.
SDS-PAGE, Western analysis, and antibodies.
Samples were resuspended in 1× SDS-PAGE loading buffer and separated on denaturing SDS-PAGE gels containing 10 to 15% (vol/vol) acrylamide (Bio-Rad Laboratories, Hercules, Calif.) prepared according to standard procedures. Proteins were transferred onto Immobilon-P transfer membrane (Millipore Corp., Billerica, Mass.). The membranes were blocked in Tris-buffered saline containing 0.1% (vol/vol) Tween 20 with the addition of 5% (wt/vol) skimmed milk powder for at least 30 min at room temperature (RT). Subsequently, membranes were incubated for at least 1.5 h at RT with the appropriate primary antibodies: mouse monoclonal anti-HA (α-HA; 1/1,000; Covance Laboratories Inc., Madison, Wis.); rabbit polyclonal α-SigD (1/1,000; kind gift from S. Marcus); mouse α-DnaK (1/2,000; Stressgen Biotechnologies Corp., Victoria, British Columbia, Canada); rabbit α-calnexin (1/1,000; Stressgen Biotechnologies Corp.); mouse α-beta-tubulin (1/1,000; Sigma-Aldrich, St. Louis, Mo.). Secondary antibodies were goat α-mouse or goat α-rabbit immunoglobulin G conjugated to horseradish peroxidase (1/2,500; Sigma-Aldrich), which was detected with ECL reagent (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) and captured on Kodak BioMax film (Eastman-Kodak, Rochester, N.Y.). Images were scanned using an Epson Expression 1680 scanner and Adobe Photoshop version 7.0 (San Jose, Calif.).
Tissue culture conditions and bacterial infection.
The human epithelial cell line HeLa (CCL2; passage numbers 10 to 20; American Type Culture Collection) was cultured in Dulbecco's modified Eagle medium (HyClone, Logan, Utah) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco, Invitrogen Corp.) at 37°C in a humidified atmosphere with 5% CO2.
HeLa cells were seeded at 5 × 104 cells ml−1 in 24-well tissue culture plates or, for the fractionation experiments, at 105 cells ml−1 in 100-mm-diameter cell culture dishes and grown for 18 to 24 h prior to bacterial infection. Infection was performed essentially as described elsewhere (58) by using invasive Salmonella that had been subcultured 1/33 from an overnight culture and grown for 3 h without antibiotics. Where appropriate, cells were harvested at the desired time points postinfection (p.i.) by addition of lysis buffer (0.1% [wt/vol] SDS, 1% [vol/vol] Triton X-100 in phosphate-buffered saline [PBS]), and appropriate dilutions were plated on selective medium for bacterial enumeration (CFU counts).
Translocation assay: determination of intracellular cAMP levels.
Serovar Typhimurium SL1344 strains expressing the full-length or truncated SseK1::Cya or SseK2::Cya proteins were obtained by electroporation of the plasmids described above. HeLa cells were seeded in 24-well plates and infected in triplicate. The cyclic AMP (cAMP) enzyme immunoassay (EIA; Amersham Pharmacia Biotech) was performed according to the manufacturer's instructions for determination of intracellular cAMP with the nonacetylation EIA procedure, with the following modification: at 9 or 21 h p.i., cells were washed three times with PBS and lysed in 250 μl of the manufacturer's buffer 1B supplemented with 0.1 M HCl at RT for 10 min with gentle agitation. Samples were neutralized by addition of a 1/10 volume of 1 M NaOH and assayed in duplicate. Protein concentration was assayed using the Bio-Rad protein assay reagent or the Sigma-Aldrich bicinchoninic acid system, with bovine serum albumin as the standard. The entire experiment (infection and EIA) was performed in triplicate. The t test (two-tailed, two-sample unequal variance) was used to determine significant differences between sample means.
Subcellular fractionation of infected HeLa cells.
Infection of HeLa cells was performed as described above, using two 100-mm cell culture plates for each Salmonella strain tested. At 15 to 16 h p.i., the cells were chilled on ice, washed three times with ice-cold PBS, and scraped into 7 ml of PBS containing protease inhibitors (Complete EDTA-free protease inhibitor cocktail tablets; Roche Diagnostics). Following centrifugation for 5 min at 250 × g and 4°C, the supernatant was aspirated and the pellet was washed once and resuspended in HB buffer (250 mM sucrose, 3 mM imidazole, 0.5 mM EDTA; pH 8) also containing protease inhibitors. Mechanical disruption by passage through a 22-gauge needle and fractionation by ultracentrifugation were performed as previously described (21), with an additional wash of the ultracentrifugation pellet to remove cytoplasmic contaminants from the membrane fraction. Fractions were diluted and plated to determine bacterial CFU and then resuspended in 1× SDS-PAGE loading dye for Western analysis. The experiment was performed in triplicate, and representative results are shown.
Immunofluorescence.
HeLa cells were seeded onto 12-mm-diameter glass coverslips 16 to 20 h prior to bacterial infection. For ectopic expression of SseK1-EGFP fusion proteins, HeLa cells were seeded onto 12-mm-diameter coverslips for 6 h prior to transfection with purified plasmid DNA of either pEGFP-C1(sseK1) or pEGFP-N1(sseK1) using FuGENE6 transfection reagent (Roche Diagnostics). At 24 to 40 h posttransfection, samples were washed three times with PBS and fixed in PBS containing 2.5% (vol/vol) paraformaldehyde for 10 min at 37°C. Fixed cells were washed with PBS and then permeabilized and blocked in PBS containing 0.1% Triton X-100 and 10% normal goat serum for 1 h. The coverslips were mounted in Mowiol (Sigma-Aldrich) and viewed on a Zeiss Axioskop2 MOT TV microscope (63× objective).
Bacterial infection of mice.
Female BALB/c mice (6 to 8 weeks old) were purchased from Harlan Laboratories (Indianapolis, Ind.) and housed at the University of British Columbia Animal Facility in sterilized filter-topped cages. Experiments were carried out under specific-pathogen-free conditions according to the standard animal care guidelines and protocols of the UBC Animal Care Committee and Canadian Council on Use of Laboratory Animals. Bacterial cultures were grown overnight in LB broth and diluted in PBS. Groups of five or six mice were infected by intraperitoneal injection with approximately 5 × 104 CFU in 0.3 ml of PBS. The mice were monitored daily, and any that showed extreme distress or became moribund were euthanized.
RESULTS
STM4157 (SseK1) and STM2137 (SseK2) are homologous to secreted proteins from A/E pathogens.
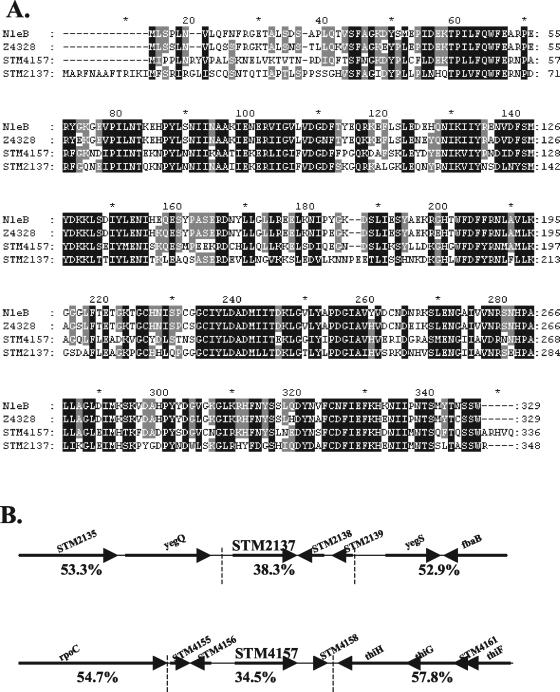
Recently, investigators in our laboratory identified a number of novel Citrobacter rodentium type III secreted proteins with homology to predicted proteins in other attaching and effacing (A/E) pathogens, including enterohemorrhagic E. coli (EHEC) (12). These proteins were of interest for comparison to more-distantly-related pathogens that also possess a TTSS, such as Salmonella. Interestingly, we noted amino acid sequence homology between C. rodentium NleB, EHEC protein Z4328 (NP_289553), and the serovar Typhimurium LT2 proteins STM2137 (NP_461081) and STM4157 (NP_463026). Amino acid sequence identity was 61% between STM2137 and STM4157 and greater than 55% between all four proteins. Amino acid sequence alignment (Fig. 1A) revealed a number of interesting characteristics. Firstly, the sequences differed most in the N termini (approximately the first 24 to 43 amino acids). Specifically, STM2137 had an extension of 13 residues compared to the other proteins. Secondly, there was remarkable conservation throughout the remainder of the protein sequences, with a number of regions of highly conserved amino acids that could represent conserved structural or functional domains. Thirdly, there were no conserved signal peptides or known functional motifs, nor homology to known translocation motifs or domains from other TTSS effectors like SopD2 and other STE proteins [note the absence of the WEK(I/M)XXFF amino acid motif] (6, 45). Standard analyses using PSORT-B (19) predicted a cytoplasmic localization and acidic pI for both STM2137 (39.566 kDa; pI = 6.67) and STM4157 (38.836 kDa; pI = 5.13). The secondary structures as predicted by PSIPRED (31) contained a preponderance of α-helices in the C-terminal regions.
FIG. 1.
STM4157 (SseK1) and STM2137 (SseK2) are homologous to secreted proteins from A/E pathogens and are encoded in pathogenicity islets in serovar Typhimurium. (A) Amino acid sequence alignment of NleB, Z4328, STM2137, and STM4157 was performed by using ClustalW (http://www.ebi.ac.uk/clustalw) with default parameters for all settings and formatted using GeneDoc (http://www.psc.edu/biomed/genedoc). The amino acid sequence of NleB was predicted from the unfinished genome sequence of C. rodentium (www.sanger.ac.uk/projects/microbes). The EHEC homologue is Z4328. The amino acid sequences of STM2137 and STM4157 were predicted from the published genome sequence of serovar Typhimurium LT2 (43). (B) The STM2137 and STM4157 genes are present within low-G+C pathogenicity islets in the serovar Typhimurium LT2 chromosome. The upper and lower regions are not contiguous with each other. Arrows indicate the direction of transcription (5′ to 3′) of known and predicted ORFs. Percent G+C is indicated below each region delimited by the vertical dashed lines.
Diagrammatic representation of the chromosomal regions comprising the STM4157 and STM2137 genes and flanking predicted ORFs in serovar Typhimurium LT2 is shown in Fig. 1B. The STM2137 and STM4157 genes are located outside of the five major pathogenicity islands of serovar Typhimurium LT2 and have altered DNA base composition compared to the whole genome. Based on the spacing and orientation relative to flanking putative ORFs, STM4157 and STM2137 are both predicted to be expressed from their own promoters, presumably located immediately upstream of the coding sequence for each. Given these genomic features and the homology to known secreted proteins, STM4157 and STM2137 were considered excellent candidates for novel translocated effectors of serovar Typhimurium, and we propose that they be referred to as Salmonella secreted effector K1 (SseK1) and SseK2, respectively, according to the conventional nomenclature of Hensel and colleagues (26).
Bacterial expression and secretion of the SseK1 and SseK2 proteins: involvement of SsrA/B and PhoP/Q.
Many effectors are coregulated with the TTSS apparatus by which they are translocated; therefore, we investigated whether the SseK1 and SseK2 proteins were present under SPI-1- and SPI-2-inducing conditions. To facilitate this analysis, we generated the plasmids pACsseK1::2HA and pACsseK2::2HA, in which endogenous promoters drive expression of recombinant proteins containing C-terminal 2HA epitopes. Western blotting against the HA epitope was used to assess SseK1::2HA and SseK2::2HA levels in whole bacteria and culture supernatants following growth of serovar Typhimurium SL1344 in medium that preferentially induces expression of the SPI-1 or SPI-2 TTSS. As another indicator of potential interaction with the TTSS, we assessed recombinant protein levels in isogenic mutants of known SPI-1 and SPI-2 regulatory systems. HilA is a key positive regulator of the SPI-1-encoded TTSS operons (2, 41). The two-component regulatory system SsrA/B, encoded within SPI-2, activates transcription of the SPI-2-encoded TTSS operons as well as genes for SPI-2 translocated effectors that are scattered throughout the chromosome (5, 20, 39). The PhoP/Q regulon in Salmonella includes genes for the extracellular magnesium deprivation response that is essential for intramacrophage survival and formation of the SCV (23).
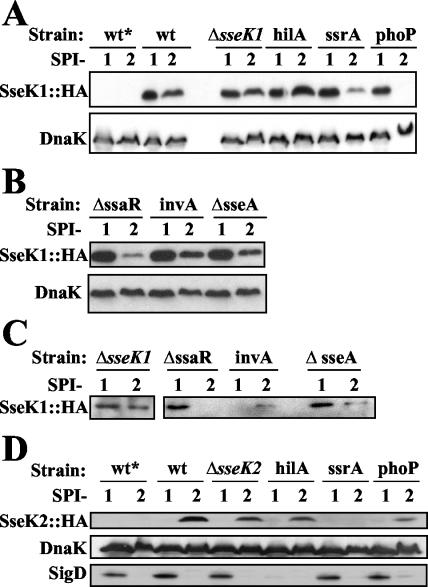
As shown in Fig. 2A, SseK1::2HA was detected as an immunoreactive band corresponding to approximately 40 kDa in the wild-type (wt) bacterial pellet. This confirmed that sseK1 was not a pseudogene and that its promoter was present within the 290 bp immediately upstream of the ORF. In wt Salmonella the SseK1::2HA protein was detected at similar levels under both SPI-1- and SPI-2-inducing conditions. Notably, the presence of SseK1::2HA under SPI-1 conditions was not dependent upon HilA, since the levels were unaffected in the hilA mutant. By comparison, the levels of SseK1::2HA were lower in the ssrA and phoP mutants specifically under SPI-2 conditions, which suggested that protein expression and/or stability was either directly or indirectly dependent upon the SsrA/B and PhoP/Q two-component regulatory systems. Finally, SseK1::2HA levels in the ΔsseK1 mutant were similar to those of the wt strain, making it suitable for use in further studies. These results suggested that the presence of the SseK1 protein may be coordinately regulated with the SPI-2 TTSS.
FIG. 2.
SseK1 and SseK2 protein levels are influenced by mutations affecting the SPI-2 TTSS in serovar Typhimurium SL1344. (A) Detection of SseK1::2HA in bacterial cell pellets obtained from wt and isogenic regulatory mutants. (B) Detection of SseK1::2HA in bacterial cell pellets obtained from TTSS apparatus mutants. (C) Detection of SseK1::2HA in bacterial supernatants (secreted proteins) obtained from TTSS apparatus mutants. (D) Detection of SseK2::2HA in bacterial cell pellets obtained from wt and regulatory mutants. The indicated serovar Typhimurium SL1344 strains lacking (*) plasmid or carrying either pACsseK1::2HA or pACsseK2::2HA were grown under SPI-1- or SPI-2-inducing conditions (indicated by 1 or 2, respectively) as described in Materials and Methods. Samples were analyzed by Western blotting. DnaK was included as an internal control for equal loading in each lane. No DnaK signal was detected in the supernatant (data not shown). SigD was included as a positive control for SPI-1-induced proteins (29).
In addition to the SPI-1 and SPI-2 regulatory mutants described above, we used mutants that specifically abolished assembly of the TTSS apparatus to determine if SseK1 was a substrate for either of the TTSSs. The invA mutant has been shown to be deficient in secretion of PrgI, the main subunit of the SPI-1 TTSS needle (60). The ΔssaR mutant is defective in synthesis of a structural component of the SPI-2-encoded TTSS (7). Since SseA acts as a chaperone for secretion of the SPI-2 translocon components SseB and SseD (51, 64), the ΔsseA mutant is deficient in translocation of SPI-2 effectors (10) but still secretes them into the culture supernatant. The results showed that the SseK1 protein accumulated in all three structural mutants under both SPI-1- and SPI-2-inducing conditions, although it was apparently lower under the latter, particularly in the ΔssaR background (Fig. 2B). Furthermore, analysis of culture supernatants indicated that SseK1::2HA was secreted by the wt strain under both SPI-1- and SPI-2-inducing conditions (Fig. 2C). By comparison, secretion of SseK1 was not detected in the invA mutant specifically under SPI-1-inducing conditions or in the ΔssaR mutant specifically under SPI-2-inducing conditions, despite being present in the corresponding bacterial pellets. Secretion of SseK1 was not abolished in the ΔsseA mutant, although it appeared to decrease under SPI-2-inducing conditions. Since DnaK was not detected in the supernatant, this confirmed that the presence of SseK1 was not due simply to bacterial lysis. Therefore, during in vitro growth SseK1 was a substrate for secretion by both the SPI-1- and the SPI-2-encoded TTSSs.
Similarly, we assessed the accumulation of SseK2::2HA in whole cells and bacterial supernatants under SPI-1- and SPI-2-inducing conditions. As shown in Fig. 2D, SseK2::2HA was detected as an immunoreactive band, corresponding to approximately 38 kDa, in the wt bacterial pellet. Therefore, sseK2 was not a pseudogene, and its promoter was contained within the upstream 436 bp. Furthermore, in contrast to SseK1, SseK2 was detected specifically under SPI-2-inducing conditions, but only after at least 5.5 h of in vitro growth. SseK2 levels in the hilA mutant were comparable to levels in wt cells, which was expected since SseK2 was not present under SPI-1-inducing conditions. In contrast, the SseK2 protein was clearly absent in the ssrA mutant and was present at low levels (often undetectable [data not shown]) in the phoP mutant, suggesting that these regulatory systems directly or indirectly affect the expression and/or accumulation of SseK2. Finally, SseK2::2HA protein levels in the ΔsseK2 mutant were similar to the those in the wt, making it suitable for use in further studies. This pattern of SseK2 protein expression suggested that it was coordinately regulated with the SPI-2 TTSS.
Similar to analysis of SseK1, we tested the hypothesis that SseK2 may be secreted by serovar Typhimurium. The results indicated that SseK2 was present in culture supernatants corresponding to its expression in whole bacteria (data not shown), namely, under SPI-2-inducing conditions in the wt, ΔsseK2, and hilA mutant backgrounds, but not in the ssrA and phoP mutants. Notably, long exposure times were required to enable detection of SseK2, suggesting either that it was secreted at low levels under the growth conditions tested or that, once secreted, SseK2 was not stable in the supernatant.
Altogether, these results indicated that SseK1 and SseK2 are novel secreted proteins and suggested that their secretion may be dependent upon the presence of a functional TTSS apparatus in serovar Typhimurium.
SseK1 and SseK2 are translocated into host cells.
Since we observed TTSS-dependent secretion of SseK1 as well as low levels of secretion of SseK2, it was important to determine whether there was a corresponding translocation of these proteins into host cells during infection. This was examined via the adenylate cyclase (Cya) reporter system (57), which has been widely used to demonstrate TTSS-dependent translocation of a variety of bacterial effectors (34, 55, 56). SseK1 and SseK2 were fused in frame to the Cya domain of the B. pertussis cyclolysin toxin, which catalyzes the formation of cAMP in the presence of the host-supplied cofactor calmodulin. We confirmed expression and enzymatic activity of the recombinant SseK1::Cya and SseK2::Cya proteins in sonicated cellular lysates of serovar Typhimurium (data not shown).
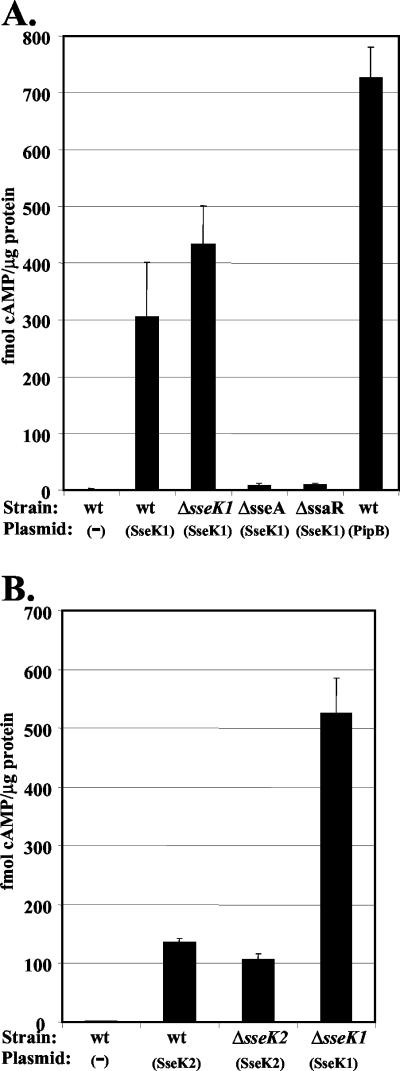
Following infection of HeLa cells with various serovar Typhimurium strains carrying the plasmid pACsseK1::cya, we assayed the intracellular levels of cAMP as a measure of translocation of SseK1::Cya (Fig. 3A). HeLa cells infected with wt Salmonella lacking plasmid (wt control) were used as the negative control, and PipBN180-Cya (PipB) was included as a positive control (34). Analysis of HeLa cell lysates at 1 h p.i. revealed cAMP levels in all samples were similar to the wt control (P > 0.05) (data not shown), indicating that there was no significant translocation of PipB or SseK1 at early time points. In contrast, at 9 h p.i. the wt and ΔsseK1 strains expressing SseK1::Cya yielded higher cAMP levels than the wt control (P < 0.001), which indicated that SseK1 was translocated into HeLa cells. Compared to the wt and ΔsseK1 backgrounds, ΔssaR and ΔsseA yielded significantly lower levels of cAMP (P < 0.001), indicating that translocation of SseK1 is dependent upon a functional TTSS apparatus and assembly of the SPI-2 translocon. This was not attributable to failure of the mutants to replicate within the host cells, since there was no significant difference in bacterial CFU compared to that of the wt at 9 h p.i. (data not shown). We conclude that SseK1 translocation into host cells was mediated via the SPI-2 TTSS.
FIG. 3.
Translocation of SseK1 and SseK2 into host cells. (A) Translocation of SseK1::Cya was assayed at 9 h p.i. of HeLa cells. (B) Translocation of SseK2::Cya was assayed at 21 h p.i. of HeLa cells. As indicated on the x axes, wt serovar Typhimurium SL1344 and derivative strains lacking (−) or carrying the plasmids pACsseK1::cya (sseK1), pPipBN180-Cya (PipB), or pACsseK2::cya (sseK2) were used to infect HeLa cells. The cells were lysed, and intracellular cAMP levels were determined as described in Materials and Methods. Each black bar represents the average of three independent samples analyzed in duplicate, with the standard deviations indicated by the error bars.
Similarly, we assayed translocation of SseK2::Cya. At 9 h p.i. we observed cAMP levels that were not significantly different from those of the negative control (P > 0.05) (data not shown). However, when tested at 21 h p.i. (Fig. 3B), the wt and ΔsseK2 strains expressing SseK2::Cya yielded higher cAMP levels than the wt control (P < 0.05), which indicated that SseK2 was in fact translocated into HeLa cells, albeit at a late stage in infection. Although the levels of translocated SseK2 are low, they are significantly higher than values obtained with a negative control expressing SseA::Cya (10) at 21 h p.i. (P < 0.05) (data not shown). The lower level of translocation of SseK2 relative to that of SseK1 was in agreement with the relative levels of secretion described above. Altogether, these results indicated that both SseK1 and SseK2 were translocated into epithelial cells during infection with serovar Typhimurium.
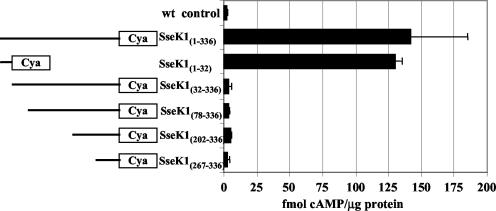
The first 32 amino acids in the N terminus of SseK1 are sufficient to mediate its translocation.
It has been shown that there are at least two domains which target most effectors to the type III apparatus (8); therefore, we decided to investigate which regions are important for translocation of SseK1. Using the Cya assay, we tested a series of four N-terminal truncations and one N-terminal peptide of SseK1, as diagrammed in Fig. 4. Compared to the full-length SseK1, progressive deletions from the N terminus significantly reduced the translocation of Cya fusion proteins into host cells (P < 0.05). Indeed, removal of just the amino-terminal 32 amino acids was sufficient to dramatically reduce translocation to the same level as the wt control (P > 0.05). Remarkably, a construct containing only these first 32 amino acids of SseK1 yielded translocation levels equivalent to those with the full-length protein (P > 0.05). We conclude that N-terminal sequences, specifically within the first 32 amino acid residues, were necessary and sufficient to mediate the translocation of SseK1.
FIG. 4.
The amino-terminal 32 amino acids of SseK1 are necessary and sufficient to mediate its translocation. HeLa cells were infected for 9 h with wt serovar Typhimurium SL1344 lacking plasmid (wt control) or with the ΔsseK1 strain expressing the N- and C-terminally truncated SseK1::Cya recombinant proteins diagrammed on the left. The intracellular cAMP levels, determined using the Cya assay, that correspond to each construct are indicated in the graph on the right. Each black bar represents the average of three independent samples analyzed in duplicate, with standard deviations indicated by error bars. Expression and enzymatic activity of the recombinant proteins were confirmed in sonicated cellular lysates of serovar Typhimurium (data not shown).
Translocated SseK1 localizes to the cytosol of fractionated HeLa cells.
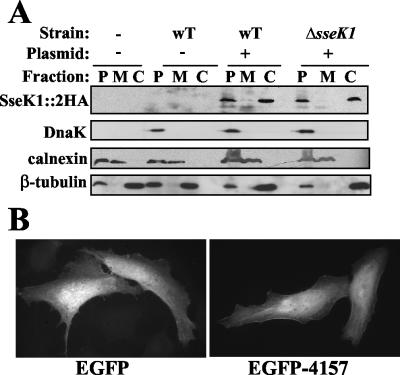
Many of the known SPI-2 effectors have been shown to localize to or associate with host cell membranes (e.g., references 5, 36, and 52), which is ultimately useful as a clue to function. Similarly, we were interested in determining the location of SseK1 once it is translocated into host cells. Therefore, we performed subcellular fractionation (21) following infection of HeLa cells with the wt (pACsseK1::2HA) or ΔsseK1 (pACsseK1::2HA). At 15 h p.i. the cells were fractionated into three fractions: the host membranes (fraction M), the host cytosol components (fraction C), and the pellet (fraction P), which contained whole cells, insoluble host components, and intact bacteria. Western analysis (Fig. 5A) detected SseK1::2HA in the pellet fraction, as expected, and also in the host cytosol fraction, which confirmed the translocation data shown above. Detection of DnaK was included as an indicator of intact bacteria and indicated that the presence of SseK1::2HA in the cytosol was not due to contamination with intact bacterial cells.
FIG. 5.
Translocated SseK1 localizes to the host cytosol. (A) HeLa cells were infected with the indicated serovar Typhimurium SL1344 strains for 15 h prior to mechanical lysis and fractionation by differential centrifugation. Uninfected HeLa cells and HeLa cells infected with wt Salmonella lacking plasmid were included as negative controls. The resultant pellet (P), membrane (M), and cytosol (C) fractions were separated by SDS-PAGE and subjected to Western analysis as described in Materials and Methods. DnaK was included as a marker for intact bacteria, calnexin was included as a marker for host membranes, and β-tubulin was included as a marker for the host cytosol. (B) Ectopic expression of SseK1-EGFP recombinant protein in HeLa cells confirms a cytosolic localization. The left panel shows HeLa cells transfected with the pEGFP-C1 control, while the right panel shows HeLa cells transfected with pEGFP-C1(sseK1) for 40 h.
Therefore, it was desirable to visualize the localization of SseK1 within intact host cells. During immunofluorescence analysis, the signal from translocated SseK1::2HA was below the level of detection (data not shown). As an alternative, we engineered chimeric proteins with SseK1 fused to either the C or N terminus of EGFP, for ectopic expression in HeLa cells. At 40 h posttransfection, the SseK1-EGFP chimeras appeared to be localized uniformly throughout the cytoplasm, similar to the EGFP control (Fig. 5B). There was no apparent colocalization with LAMP-1-positive structures, such as endosomes, or with the SCV upon infection with Salmonella (data not shown). Altogether, these results confirmed translocation of SseK1 and indicated that it localized throughout the host cytosol rather than associating with membrane components or other discrete structures within the host cell.
Effects of SseK1 and SseK2 on the virulence of serovar Typhimurium are not evident during infection of tissue culture cells or susceptible mice.
In order to initiate functional analyses of the translocated proteins SseK1 and SseK2, we compared the ΔsseK1, ΔsseK2, and ΔsseK1/ΔsseK2 mutants to the isogenic wt serovar Typhimurium SL1344 strain. Following infection of HeLa cells and RAW 264.7 cells, we performed immunofluorescence to qualitatively assess filamentous bacterial morphology (50) and the typical virulence phenotype of SIF formation (18). There were no apparent differences in bacterial numbers, bacterial filamentation, or formation of SIF at 15 h p.i. (data not shown). These results agreed with the above fractionation analysis, which showed no colocalization of SseK1 with host membranes. Furthermore, we assessed the virulence of the single and double mutants during infection of susceptible (BALB/c) mice. Groups of five or six mice were infected with 5 × 104 bacteria by intraperitoneal injection and monitored daily. All of the mice became ill, requiring euthanization of 100% of mice by day 6. Thus, the loss of sseK1 and/or sseK2 was insufficient to cause detectable attenuation in a susceptible murine host. We conclude that the role of these putative effectors in the virulence of serovar Typhimurium was not apparent under the conditions tested.
DISCUSSION
This report describes the identification and characterization of two new serovar Typhimurium translocated proteins, which are encoded by STM2137 and STM4157 in the annotated genome sequence of serovar Typhimurium LT2 (43). The secretion of these proteins was postulated based on homology to NleB, a newly identified C. rodentium type III secreted protein (12). Since we have demonstrated that these proteins are indeed secreted and/or translocated in a TTSS-dependent manner, we have designated the protein products of STM4157 and STM2137 as SseK1 and SseK2, respectively, according to the conventional nomenclature (26).
The sseK1 and sseK2 genes are located outside of the five serovar Typhimurium pathogenicity islands, in small regions (<5 kb) whose G+C content is significantly lower than the average for the entire genome. The possibility that sseK1 and sseK2 were acquired by horizontal transfer is strengthened by the presence of a third homologous gene, sb26 (corresponding to protein NP_700399), in the ST64B coliform bacteriophage (NC_004313) (47). We propose that the protein encoded by sb26 be referred to as SseK3. In ST64B, the flanking genes are a putative DNA invertase pin protein (sb27) and a probable tail fiber assembly protein (sb25), and their orientation precludes cotranscription with sb26. There is no significant sequence homology between sb25, sb27, and ORFs flanking sseK1 or sseK2. While this study was in progress, the partially completed genome of serovar Typhimurium SL1344 was compared to that of serovar Typhimurium LT2 and revealed that ST64B is present in the former but not the latter strain (D. Goode and N. Brown, personal communication). Interestingly, serovar Typhimurium SL1344, which was used for the work described in this study, is also more virulent than the LT2 strain in the mouse model.
We hypothesize that SseK1, SseK2, and the sb26-encoded protein may comprise a new family of translocated proteins in serovar Typhimurium. In general, many of the known Salmonella effectors are present as multiple, functionally nonredundant proteins with significant amino acid sequence similarities. For example, PipB and PipB2 exhibit different localization profiles following translocation into the host cell, although they are both substrates for the SPI-2 TTSS (36). In contrast, SopD and SopD2 are coregulated with and translocated by different TTSSs (5). Hence, the translocation and/or functions of SseK1, SseK2, and sb26 will not necessarily overlap. Indeed, this is also suggested by the differences in protein accumulation and translocation between SseK1 and SseK2: while both proteins were present under in vitro conditions that induce expression of the SPI-2 TTSS, during an infection the translocation of SseK1 occurred at higher levels and at earlier time points than SseK2.
Analysis of in vitro protein levels revealed that under SPI-2-inducing conditions neither SseK1 nor SseK2 accumulated significantly in mutants lacking the SsrA/B or PhoP/Q two-component regulatory systems. This could result from a direct effect, such as positive regulation of gene expression, or an indirect effect via posttranscriptional mechanisms. Further work will be required to distinguish between these two possibilities. Under SPI-1-inducing conditions in vitro, we observed bacterial expression and secretion of SseK1, suggesting that it may be a substrate for the SPI-1 as well as SPI-2 TTSSs. While this lack of specificity for one or the other TTSS has been observed previously for SspH1 and SlrP, two members of the STE family, the expression of those two proteins is not regulated by SsrA/B or PhoP/Q (45). We hypothesize that during infection of host cells the translocation of SseK1 is specific to the SPI-2 TTSS, since the temporal pattern is characteristic of SPI-2 effectors, namely, it is not detected at 1 h p.i. (which would be expected for SPI-1 effectors) but is apparent at 9 h p.i. and at time points thereafter. Furthermore, translocation of SseK1 was dependent upon the presence of SsaR and SseA, which are critical to the formation of a functional SPI-2 TTSS apparatus and translocon. Specificity of interaction between SseK1 and the SPI-2 TTSS apparatus may therefore be achieved by restricting expression of SseK1 to the intracellular phase during infection. Similarly, we speculate that SseK2 translocation may be dependent upon the SPI-2 TTSS, since it occurs at late time points; however, this could not be tested because of the lack of replication by ΔsseA and ΔssaR mutants at 21 h p.i.
The Cya assay clearly demonstrated that SseK1 and SseK2 were translocated into host cells during infection. Importantly, we discovered that the first 32 amino acids of SseK1 were both necessary and sufficient to mediate its translocation. This is somewhat unusual for Salmonella effectors—in general the translocation domain is much larger and suggests the involvement of chaperones, as demonstrated for a subset of effectors (for example, see reference 40). It is tempting to speculate that the short translocation domain of SseK1 interacts directly with a conserved region of the TTSS apparatus rather than with a specific chaperone, which could explain the in vitro secretion of SseK1 via both the SPI-1 and the SPI-2 TTSSs. By comparison to YopE and YopN of Yersinia, for which the minimal translocation signal occurs within the first 15 amino acids and is affected by mRNA secondary structure in this region (1), it is possible that translocation of SseK1 may be coupled to its translation. Finally, it is remarkable that the translocation signal for SseK1 was contained within the region of the protein where the amino acid sequence is most divergent compared to the other homologues.
Fractionation of infected epithelial cells revealed another somewhat unusual characteristic, namely that the translocated SseK1 protein localizes to the host cytosol. This was also observed during ectopic expression of SseK1-EGFP chimeric proteins within host cells. These results agreed with in silico predictions but were in striking contrast to the membrane localization observed for many of the currently known SPI-2 translocated effectors, with the exception of SpiC/SsaB (38, 56) and effectors such as SspH2 and SseI, which colocalize with cytoskeletal components (44). Since SseK1 apparently remains dispersed throughout the host cytosol, we speculate that it could interact with soluble host factors such as enzymes or even nucleic acids. Further work is required to determine the subcellular localization of translocated SseK2, since SseK2::2HA levels were below the limit of detection during fractionation experiments at 15 and 21 h p.i. (data not shown).
Functional analysis of SseK1 and SseK2 was initiated by assessing serovar Typhimurium SL1344 single and double mutants lacking these proteins. In standard replication assays, the intracellular growth of these mutants was not significantly different from growth of the isogenic wt strain (data not shown). Deletion of sseK1 and/or sseK2 did not have a detectable effect on SCV and SIF formation in epithelial cells, nor on the ability to cause typhoid-like disease and death in a susceptible murine host. It has been previously observed that the loss of a single effector does not necessarily alter bacterial virulence (e.g., references 27 and 34). Possibly, the absence of SseK1 and/or SseK2 may have been complemented by sb26, which will be addressed by the triple mutant. Alternatively, the role of these translocated proteins may not be manifested in a model of systemic infection in susceptible mice. The absence of S. enterica serovar Typhi homologues and the presence of homologues in A/E pathogens, which remain extracellular and attach to the apical surface of epithelial cells in the host intestine, raise the possibility that SseK1 and SseK2 may actually be more important in the development of gastroenteritis than in systemic disease.
Acknowledgments
This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR) and the Howard Hughes Medical Institute (HHMI) to B.B.F. B.B.F. is a CIHR Distinguished Investigator, an HHMI International Research Scholar, and the University of British Columbia Peter Wall Distinguished Professor. O.G. is the recipient of postdoctoral fellowships from the Canadian Association of Gastroenterology/CIHR/Ferring Pharmaceuticals and from the Michael Smith Foundation for Health Research.
We thank Wanyin Deng for indicating the homology to secreted proteins of C. rodentium, Nat Brown for noting the presence of sb26 in serovar Typhimurium strain SL1344, Phil Hardwidge for advice on statistical analyses, and members of the Finlay lab for helpful discussion and critical reading of the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj, V., C. Hwang, and C. A. Lee. 1995. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumell, J. H., S. Kujat-Choy, N. F. Brown, B. A. Vallance, L. A. Knodler, and B. B. Finlay. 2003. SopD2 is a novel type III secreted effector of Salmonella typhimurium that targets late endocytic compartments upon delivery into host cells. Traffic 4:36-48. [DOI] [PubMed] [Google Scholar]
- 6.Brumell, J. H., S. L. Marcus, and B. B. Finlay. 2000. N-terminal conservation of putative type III secreted effectors of Salmonella typhimurium. Mol. Microbiol. 36:773-774. [DOI] [PubMed] [Google Scholar]
- 7.Brumell, J. H., C. M. Rosenberger, G. T. Gotto, S. L. Marcus, and B. B. Finlay. 2001. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 3:75-84. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 10.Coombes, B. K., N. F. Brown, S. Kujat-Choy, B. A. Vallance, and B. B. Finlay. 2003. SseA is required for translocation of Salmonella pathogenicity island-2 effectors into host cells. Microbes Infect. 5:561-570. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 12.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 14.Freeman, J. A., C. Rappl, V. Kuhle, M. Hensel, and S. I. Miller. 2002. SpiC is required for translocation of Salmonella pathogenicity island 2 effectors and secretion of translocon proteins SseB and SseC. J. Bacteriol. 184:4971-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 16.Galan, J. E., and R. Curtiss III. 1991. Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun. 59:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-del Portillo, F., and B. B. Finlay. 1995. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J. Cell Biol. 129:81-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Intracellular replication of Salmonella within epithelial cells is associated with filamentous structures containing lysosomal membrane glycoproteins. Infect. Agents Dis. 2:227-231. [PubMed] [Google Scholar]
- 19.Gardy, J. L., C. Spencer, K. Wang, M. Ester, G. E. Tusnady, I. Simon, S. Hua, K. deFays, C. Lambert, K. Nakai, and F. S. L. Brinkman. 2003. PSORT-B: improving protein subcellular localization prediction for gram-negative bacteria. Nucleic Acids Res. 31:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garmendia, J., C. R. Beuzon, J. Ruiz-Albert, and D. W. Holden. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385-2396. [DOI] [PubMed] [Google Scholar]
- 21.Gauthier, A., M. de Grado, and B. B. Finlay. 2000. Mechanical fractionation reveals structural requirements for enteropathogenic Escherichia coli Tir insertion into host membranes. Infect. Immun. 68:4344-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorvel, J. P., and S. Meresse. 2001. Maturation steps of the Salmonella-containing vacuole. Microbes Infect. 3:1299-1303. [DOI] [PubMed] [Google Scholar]
- 23.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 25.Hansen-Wester, I., B. Stecher, and M. Hensel. 2002. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect. Immun. 70:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hensel, M., J. E. Shea, B. Raupach, D. Monack, S. Falkow, C. Gleeson, T. Kubo, and D. W. Holden. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155-167. [DOI] [PubMed] [Google Scholar]
- 27.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 28.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 29.Hong, K. H., and V. L. Miller. 1998. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J. Bacteriol. 180:1793-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 32.Kingsley, R. A., and A. J. Baumler. 2002. Pathogenicity islands and host adaptation of Salmonella serovars. Curr. Top. Microbiol. Immunol. 264:67-87. [PubMed] [Google Scholar]
- 33.Knodler, L. A., J. Celli, and B. B. Finlay. 2001. Pathogenic trickery: deception of host cell processes. Nat. Rev. Mol. Cell Biol. 2:578-588. [DOI] [PubMed] [Google Scholar]
- 34.Knodler, L. A., J. Celli, W.-D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089-1103. [DOI] [PubMed] [Google Scholar]
- 35.Knodler, L. A., and O. Steele-Mortimer. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587-599. [DOI] [PubMed] [Google Scholar]
- 36.Knodler, L. A., B. A. Vallance, M. Hensel, D. Jackel, B. B. Finlay, and O. Steele-Mortimer. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49:685-704. [DOI] [PubMed] [Google Scholar]
- 37.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 38.Lee, A. H., M. P. Zareei, and S. Daefler. 2002. Identification of a NIPSNAP homologue as host cell target for Salmonella virulence protein SpiC. Cell Microbiol. 4:739-750. [DOI] [PubMed] [Google Scholar]
- 39.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S. H., and J. E. Galan. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483-495. [DOI] [PubMed] [Google Scholar]
- 41.Lostroh, C. P., and C. A. Lee. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of pprgH from Salmonella pathogenicity island 1. J. Bacteriol. 183:4876-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2:145-156. [DOI] [PubMed] [Google Scholar]
- 43.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 44.Miao, E. A., M. Brittnacher, A. Haraga, R. L. Jeng, M. D. Welch, and S. I. Miller. 2003. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48:401-415. [DOI] [PubMed] [Google Scholar]
- 45.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mmolawa, P. T., H. Schmieger, and M. W. Heuzenroeder. 2003. Bacteriophage ST64B, a genetic mosaic of genes from diverse sources isolated from Salmonella enterica serovar Typhimurium DT64. J. Bacteriol. 185:6481-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberger, C. M., and B. B. Finlay. 2002. Macrophages inhibit Salmonella Typhimurium replication through MEK/ERK kinase and phagocyte NADPH oxidase activities. J. Biol. Chem. 277:18753-18762. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Albert, J., R. Mundy, X.-J. Yu, C. R. Beuzon, and D. W. Holden. 2003. SseA is a chaperone for the SseB and SseD translocon components of the Salmonella pathogenicity-island-2-encoded type III secretion system. Microbiology 149:1103-1111. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 53.Salcedo, S. P., and D. W. Holden. 2003. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22:5003-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shotland, Y., H. Kramer, and E. A. Groisman. 2003. The Salmonella SpiC protein targets the mammalian Hook3 protein function to alter cellular trafficking. Mol. Microbiol. 49:1565-1576. [DOI] [PubMed] [Google Scholar]
- 57.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 58.Steele-Mortimer, O., M. St-Louis, M. Olivier, and B. B. Finlay. 2000. Vacuole acidification is not required for survival of Salmonella enterica serovar Typhimurium within cultured macrophages and epithelial cells. Infect. Immun. 68:5401-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 60.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galan. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 63.Yu, X.-J., J. Ruiz-Albert, K. E. Unsworth, S. Garvis, M. Liu, and D. W. Holden. 2002. SpiC is required for secretion of Salmonella pathogenicity island 2 type III secretion system proteins. Cell Microbiol. 4:531-540. [DOI] [PubMed] [Google Scholar]
- 64.Zurawski, D. V., and M. A. Stein. 2003. SseA acts as the chaperone for the SseB component of the Salmonella pathogenicity island 2 translocon. Mol. Microbiol. 47:1341-1351. [DOI] [PubMed] [Google Scholar]