Abstract
A Mycobacterium tuberculosis strain disrupted in the AraC homologue Rv1931c was isolated. The mutant strain exhibited reduced survival both in macrophages and in a mouse infection model, with survival being restored on complementation with the Rv1931c gene. These results suggest that Rv1931c regulates genes important for virulence of M. tuberculosis.
Transcriptional regulators of the AraC family are widespread among bacteria and regulate genes having diverse functions, ranging from carbon metabolism to stress responses to virulence (4, 7). Six members of this family (Rv1317c [alkA], Rv1395, Rv1931c, Rv3082c [virS], Rv3736, and Rv3833) have been identified in the genome of Mycobacterium tuberculosis by sequence similarity to the defining DNA binding domain. The first of these, Rv1317c (alkA), has additional domains conferring a role in the repair of alkylated DNA, with the regulatory functions residing at the N terminus. A mutant strain of H37Rv containing a defined mutation in Rv1317c, which also affects the downstream gene Rv1316c, did not show any significant differences in bacillary loads in the lungs, liver, or spleen following infection of BALB/c mice (3). The remaining five homologues possess the AraC family motif at their C termini, a typical location for this family of regulatory proteins, and appear to be straightforward transcriptional regulators. Interestingly, a transposon mutation was identified in one of these, Rv1395, from a signature-tagged mutagenesis screen of strain Mt103, which was confirmed to show attenuation in lungs upon infection of BALB/c mice with the individual strain (1). Subsequently, Rv1395 has been shown to regulate the expression of a cytochrome P450-encoding gene transcribed divergently from it (10). Another member of this AraC family, Rv3082c (virS), has been suggested to play a role in M. tuberculosis pathogenesis on the basis of sequence similarities (5) and has also been shown to regulate an operon transcribed divergently from it (15). The AraC homologue Rv1931c is annotated as a probable transcriptional regulator in the TubercuList database (http://genolist.pasteur.fr/TubercuList) and has been predicted to be nonessential by Himar 1-based transposon mutagenesis in strain H37Rv (14). In order to assess whether Rv1931c plays a role in virulence in M. tuberculosis, it was targeted for mutagenesis and the ability of the resulting strain to grow and survive in macrophages and in mice was determined.
A DNA fragment containing the coding sequence of the target gene along with approximately 2 kb of the flanking region was generated by PCR from M. tuberculosis genomic DNA and cloned into Escherichia coli plasmid pUC19. Four hundred ninety-four base pairs of the 777-bp coding sequence of Rv1931c was then deleted, and a unique cloning site was simultaneously introduced by inverse PCR and religation of the PCR product. A kanamycin resistance gene from pUC4K was then inserted into this site. Finally, the disrupted fragment containing the flanking regions plus the kan gene was cloned into the streptomycin counterselection vector ptrpA-1-rpsL+ (12).
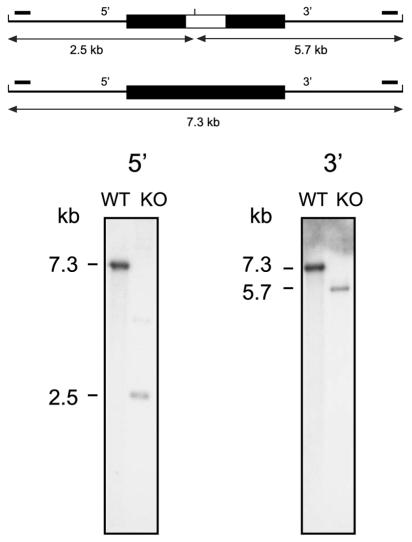
The targeting construct was electroporated into M. tuberculosis strain 1424, a streptomycin-resistant derivative of H37Rv (2). Both one-step and two-step selections were performed by streptomycin selection as previously described (12, 13). Colonies obtained conforming to the desired phenotype were further screened by PCR (data not shown) and Southern analyses (Fig. 1) to identify genuine double-crossover recombinants. In addition, the genotype of the isolate selected for further study was further confirmed with microarrays (data not shown).
FIG. 1.
Identification of an Rv1931c mutant by Southern hybridization. At the top a schematic representation of the gene locus is shown in which the upper part shows the arrangement in the mutant and the lower part shows that in the wild type. The cloned region is shown as a thick black line, and the introduced kanamycin resistance gene in the mutant is shown as a white box within this. The locations of the hybridizing probes (small solid bars) and the expected sizes of the hybridizing fragments for each of the 5′ and 3′ Southern blots are indicated. Genomic DNAs isolated from the mutant strain (KO) and the parental wild-type (WT) strain were digested with ClaI, transferred to nylon membrane, and hybridized to radioactively labeled DNA probes. The sizes of the hybridizing bands were determined from the migration distances of the DNA molecular size markers λ-HindIII+EcoRI and λ-HindIII.
In a preliminary experiment to assess the effects of the mutation obtained on M. tuberculosis pathogenesis, the mutant strain was compared with wild-type strain H37Rv in a mouse model of infection. Groups of BALB/c mice were infected intravenously with approximately 5 × 105 cells of each strain, and the progress of the infection was assessed by determining organ CFU counts at various time points. Mice infected with the wild-type strain developed clinical symptoms of disease between days 69 and 133 postinfection and had to be sacrificed. In contrast, all mice infected with ΔRv1931c were still surviving at day 133 postinfection, and CFU counts from both lungs and spleens of mice infected with this mutant were approximately 1 logarithm lower that those obtained following infection with the wild type, with counts from spleens decreasing after 69 days (data not shown), although growth under normal culture conditions in vitro was indistinguishable from that of the wild type.
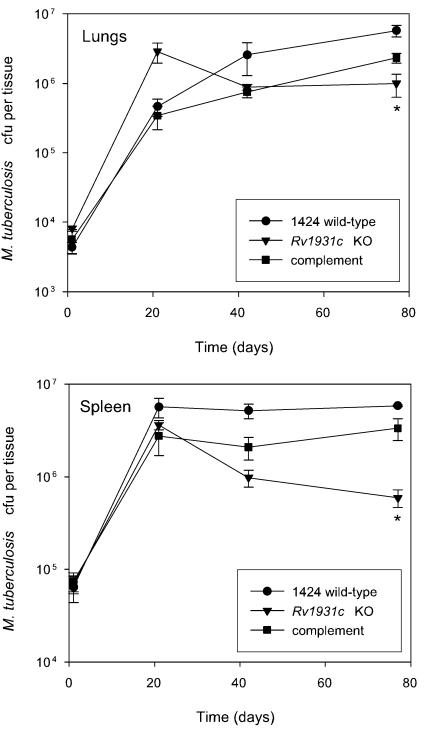
To confirm that the attenuation seen in the initial experiment with ΔRv1931c was due to the introduced mutation, it was necessary to both complement the mutation with the cloned Rv1931c gene and compare the progress of infection caused by these strains with that of the isogenic wild-type strain, 1424. Therefore, a 968-bp fragment of DNA containing the entire Rv1931c coding sequence plus 160 bp of the upstream sequence was cloned into mycobacterial integrating vector pKP201. This plasmid carries a hygromycin resistance gene and the attP site but lacks the integrase, which must be supplied in trans for integration to occur; this function is provided by cotransformation with pBluescriptint (16), a plasmid that fails to replicate in mycobacteria, resulting in increased stability of the clone. The insert in the complementing clone, pCF51, was sequenced to confirm that no mutations had been introduced. Following introduction of pCF51 into the ΔRv1931c strain, RNA was isolated and comparative expression analysis with microarrays revealed that the inserted gene was expressed (data not shown). A second mouse infection experiment was conducted as before but with strains 1424 (wild type), ΔRv1931c, and ΔRv1931c::pCF51. The growth of wild-type strain 1424 was similar to that seen previously with H37Rv (11), although a direct comparison was not made. Although it appeared that the mutant strain grew better than the wild type initially, we believe this to be an artifact as the difference between the mutant and the wild type at day 21 is not significant by t test (P > 0.05) and in the preliminary experiment described above this was not observed. Overall a level of attenuation similar to that seen in the previous experiment was observed with the ΔRv1931c mutant, whereas with the complemented strain the bacterial loads in the lungs and spleen resembled those obtained with the wild-type strain (Fig. 2), confirming that the phenotype seen was due to the disruption of the Rv1931c gene.
FIG. 2.
Phenotype of the ΔRv1931c mutant and complemented strains in a mouse model of infection. BALB/c mice were inoculated intravenously with approximately 5 × 105 CFU of each strain. The survival and multiplication of the M. tuberculosis strains in the lungs and the spleen were determined by CFU counts and are shown for the 1424 wild-type (•), ΔRv1931c (▾), and Rv1931c-complemented (▪) strains. The results for each time point are the means of CFU determinations performed on organs from three mice, and the error bars show the standard deviations. The asterisk indicates that the result is statistically significantly different from that of the wild type by the two-tailed Student t test for groups of unequal variance (P < 0.01), as well as by single-factor analysis of variance (P < 0.01). KO, knockout.
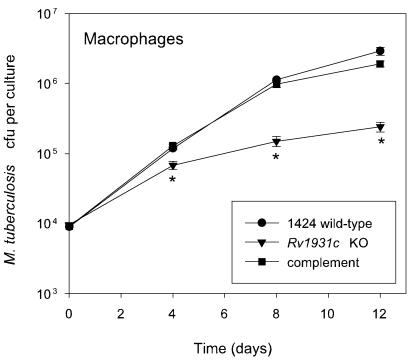
To further assess the phenotype of the ΔRv1931c mutant strain, strains 1424 (wild-type), ΔRv1931c, and ΔRv1931c::pCF51 were used to infect mouse bone marrow-derived macrophages in vitro and the numbers of CFU were determined at various time points following infection. Bone marrow cells were extracted from the hind leg bones of 6- to 8-week-old female BALB/c mice and cultured in Iscove's modified Dulbecco's medium supplemented with 5% heat-inactivated fetal calf serum, 2 mM glutamine, 80 μM 2-mercatoethanol, and 10% supernatant of L929 cells to provide macrophage colony-stimulating factor in 12-well plates for 5 days. Following this, cells were infected for 6 h at a multiplicity of infection of one bacterium to two macrophages with each strain. The medium was removed, the cells were washed with prewarmed medium, and fresh medium was added. At selected time points samples were lysed with 2% saponin and viable bacterial counts were determined by plating serial dilutions on Middlebrook 7H11 agar plates. Again the ΔRv1931c mutant exhibited a reduced ability to survive and replicate in comparison with the wild type, yielding CFU counts approximately 1 logarithm lower that those obtained following infection with the wild type after 8 to 12 days, while the complemented strain was indistinguishable from the wild type (Fig. 3).
FIG. 3.
Phenotype of the ΔRv1931c mutant and complemented strains following infection of mouse bone marrow-derived macrophages in vitro. Macrophages were isolated from BALB/c mice as described in the text and infected at a multiplicity of infection of one bacterium to two macrophages with each strain. The survival and multiplication of the M. tuberculosis strains were determined by CFU counts and are shown for the 1424 wild-type (•), ΔRv1931c (▾), and Rv1931c-complemented (▪) strains. The experiment was performed twice with similar results; the results of a representative experiment are shown. The results for each time point are the means of CFU determinations performed on triplicate infections, and the error bars show the standard deviations. The asterisk indicates that the result is statistically significantly different from that of the wild type by the two-tailed Student t test for groups of unequal variance (P < 0.01), as well as by single-factor analysis of variance (P < 0.01).
In this study we targeted the AraC family transcriptional regulator Rv1931c for mutation in M. tuberculosis. The resulting mutant strain was attenuated in the intravenous mouse model of infection and in macrophages. This phenotype was confirmed to be due to disruption of Rv1931c by complementation. The degree of attenuation observed was intermediate within the range of phenotypes reported for M. tuberculosis mutants in other regulatory genes implicated in virulence. Mutation of the other AraC homologue characterized in this way, Rv1395, resulted in a more dramatic decrease in the bacterial load in the lungs of infected mice by 2.5 logs at 3 weeks (1), although this result was obtained with a different strain of M. tuberculosis. A greater degree of attenuation was also caused by mutation of some two-component systems such as phoP (9) and senX3 (11), although mutation of others resulted in a defect in persistence but not in initial growth as for mprA (19) or even in increased virulence as reported for devR (8). In contrast, mutation of some regulatory genes has been found to have no detectable effect on the numbers of bacteria recovered from the lungs or spleen although reduced pathology or increased host survival was observed such as, for example, with the sigma factors sigC (18) and sigH (6), as well as with whiB3 (17).
Overall, the observations presented here suggest that at least some of the genes whose expression is regulated by Rv1931c are important for the virulence of M. tuberculosis.
Acknowledgments
We thank Evangelos Stavropoulos for performing the macrophage infection experiments and for providing technical assistance with the murine infection experiments.
C.C.F. gratefully acknowledges the support of the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq-Brazil). This work was funded by the Medical Research Council (United Kingdom).
Editor: S. H. E. Kaufmann
Footnotes
In memory of the late Jo Colston and Peter Jenner, for their support and many helpful discussions.
REFERENCES
- 1.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 2.Davis, E. O., B. Springer, K. K. Gopaul, K. G. Papavinasasundaram, P. Sander, and E. C. Böttger. 2002. DNA damage induction of recA in Mycobacterium tuberculosis independently of RecA and LexA. Mol. Microbiol. 46:791-800. [DOI] [PubMed] [Google Scholar]
- 3.Durbach, S. I., B. Springer, E. E. Machowski, R. J. North, K. G. Papavinasasundaram, M. J. Colston, E. C. Bottger, and V. Mizrahi. 2003. DNA alkylation damage as a sensor of nitrosative stress in Mycobacterium tuberculosis. Infect. Immun. 71:997-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta, S., S. Jain, and A. K. Tyagi. 1999. Analysis, expression and prevalence of the Mycobacterium tuberculosis homolog of bacterial virulence regulating proteins. FEMS Microbiol. Lett. 172:137-143. [DOI] [PubMed] [Google Scholar]
- 6.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 8.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez, E., S. Samper, Y. Bordas, C. Guilhot, B. Gicquel, and C. Martin. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41:179-187. [DOI] [PubMed] [Google Scholar]
- 10.Recchi, C., B. Sclavi, J. Rauzier, B. Gicquel, and J. M. Reyrat. 2003. Mycobacterium tuberculosis Rv1395 is a class III transcriptional regulator of the AraC family involved in cytochrome P450 regulation. J. Biol. Chem. 278:33763-33773. [DOI] [PubMed] [Google Scholar]
- 11.Rickman, L., J. W. Saldanha, D. M. Hunt, D. N. Hoar, M. J. Colston, J. B. Millar, and R. S. Buxton. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem. Biophys. Res. Commun. 314:259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander, P., A. Meier, and E. C. Bottger. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 16:991-1000. [DOI] [PubMed] [Google Scholar]
- 13.Sander, P., K. G. Papavinasasundaram, T. Dick, E. Stavropoulos, K. Ellrott, B. Springer, M. J. Colston, and E. C. Bottger. 2001. Mycobacterium bovis BCG recA deletion mutant shows increased susceptibility to DNA-damaging agents but wild-type survival in a mouse infection model. Infect. Immun. 69:3562-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 15.Singh, A., S. Jain, S. Gupta, T. Das, and A. K. Tyagi. 2003. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol. Lett. 227:53-63. [DOI] [PubMed] [Google Scholar]
- 16.Springer, B., P. Sander, L. Sedlacek, K. Ellrott, and E. C. Bottger. 2001. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol. 290:669-675. [DOI] [PubMed] [Google Scholar]
- 17.Steyn, A. J., D. M. Collins, M. K. Hondalus, W. R. Jacobs, Jr., R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 19.Zahrt, T. C., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 98:12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]