Abstract
The numbers of host-adapted Borrelia burgdorferi (HAB) organisms in rabbit skin were assessed by real-time PCR over the first 3 weeks of infection. Maximal numbers were found at day 11, while spirochete numbers decreased by more than 30-fold by day 21. The antigenic composition of HAB in skin biopsy samples was determined by use of a procedure termed hydrophobic antigen tissue Triton extraction. Immune sera from rabbits, sera from chronically infected mice, and monospecific antiserum to the antigenic variation protein, VlsE, were used to probe parallel two-dimensional immunoblots representing each time point. Individual proteins were identified using either specific antisera or by matching protein spots to mass spectrometry-identified protein spots from in vitro-cultivated Borrelia. There were significant changes in the relative expression of a variety of known and previously unrecognized HAB antigens during the 21-day period. OspC and the outer membrane proteins OspA and OspB were prominent at the earliest time point, day 5, when the antigenic variation protein VlsE was barely detected. OspA and OspB were not detected after day 5. OspC was not detected after day 9. VlsE was the most prominent antigen from day 7 through day 21. BmpA, ErpN, ErpP, LA7, OppA-2, DbpA, and an unidentified 15-kDa protein were also detected from day 7 through day 21. Immunoblot analysis using monospecific anti-VlsE revealed the presence of prominent distinct VlsE lower forms in HAB at days 9, 11, and 14; however, these lower forms were no longer detected at day 21. This marked diminution in VlsE lower forms paralleled the clearance of the spirochete from skin.
There have been recent advances in understanding the molecular adaptations of the Lyme disease spirochete, Borrelia burgdorferi, relevant to tick transmission and to survival in mice. The spirochetal surface proteins that are downregulated or upregulated in response to tick feeding, OspA/B and OspC, have been a major focus of such work (7, 35). Recent gene knockouts have addressed the roles of OspA/B and OspC in tick and mouse infections. OspA/B are essential for colonization of the tick midgut but are not necessary for infection of mice (44). Pal et al. found that without OspC, B. burgdorferi strain N40 was unable to invade the salivary glands (32). In contrast, Grimm et al. found that ospC knockouts of strain B31-A3 invaded the salivary glands; however, this knockout strain could not infect either immunocompetent or SCID mice upon direct injection or by tick bite (14). Both groups directly visualized spirochetal OspC expression in salivary glands by using confocal microscopy, and the meaning of their divergent findings is unclear at this time.
Most information about changes in B. burgdorferi gene expression during infection of mice is based upon detection of specific transcripts, not upon direct visualization in tissue, presumably due to the small numbers of spirochetes present. Recent studies have given discordant results regarding detection of ospA/B and ospC transcripts. Liang et al. have shown by reverse transcription-PCR that ospC is downregulated after 2 weeks in immunocompetent mice, following the appearance of OspC antibodies (25). Spirochetes infecting SCID mice expressed ospC as judged by reverse transcription-PCR (25). A microarray analysis of 137 lipoprotein genes revealed that many additional genes in addition to ospC became downregulated in immunocompetent mice by several weeks after infection, but not in SCID mice (26). ospB transcripts, but not ospA transcripts, were also detected throughout this period. However, Hodzic et al. recently reported that ospC transcripts were readily detected in immunocompetent C3H mice throughout an 8-week course of infection and that ospA transcripts were also detected (19). Transcription of the antigenic variation lipoprotein gene, vlsE, and of the decorin binding protein gene, dbpA, was not reduced in normal mice during 33 days of infection (26).
We have taken a different approach to assess B. burgdorferi gene expression during infection of murine tissues. Using a method termed HATTREX (hydrophobic antigen tissue Triton extraction), the constellation of hydrophobic protein antigens present during infection of SCID mice was visualized by immunoblotting two-dimensional (2D) gels (6). Using the SCID mouse, we showed the presence of several outer membrane proteins, including very high levels of the antigenic variation protein, VlsE. Smaller forms of VlsE detected in infected tissues were also very prominent in mouse ear and joint tissues. OspC was readily detected in SCID mouse tissues.
In the rabbit model of Lyme disease, B. burgdorferi infection is naturally cleared, with resulting immunity to reinfection (11). In mice, the immune response does not result in clearance of the infection, and chronic infection is established. The rabbit model therefore presents an opportunity to examine the spirochete and the host immune response during the ontogeny of protective immunity. This study presents real-time PCR-based data that indicate that the numbers of host-adapted Borrelia (HAB) in rabbit skin become maximal by 2 weeks after infection but then fall logarithmically in the following week. We used HATTREX to identify hydrophobic proteins of HAB in rabbit skin expressed at intervals ranging from 5 to 21 days after infection.
MATERIALS AND METHODS
Bacterial strains.
Virulent B. burgdorferi sensu stricto strain B31 was isolated from infected rabbit skin and grown at 34°C in BSKII supplemented with 6% normal rabbit serum as described previously (10). Low-passage (≤3) virulent B31 was used in all the experiments. Strain ME3-2 is a B31 clonal isolate grown from SCID mouse blood and is lacking vlsE (data not shown).
Infection of rabbits with B. burgdorferi.
Male New Zealand White rabbits (Irish Farms, Norco, California) were inoculated by needle injection intradermally with 104 virulent in vitro-cultivated borrelia (IVCB) (strain B31). Two animals were sacrificed at days 5, 7, 9, 11, 14, and 21 after inoculation. The skin from the back was placed in Ziploc freezer bags and frozen in dry ice-ethanol.
Real-time PCR.
Genomic DNA from rabbit skin samples (5-mm skin punches at sites of erythema migrans [EM]) and from IVCB were prepared using the Easy-DNA kit from Invitrogen (Carlsbad, Calif.) according to the manufacturer's instructions. Primers and probe were selected for the flagellin gene (GenBank accession no. AE001126) for use in quantitation of B. burgdorferi strain B31 in infected tissues. The upstream primer for flaB corresponds to the region from bases 579 to 602 (TGTTGCAAATCTTTTCTCTGGTGA) of the open reading frame. The downstream primer corresponds to the region from bases 635 to 656 (CCTTCCTGTTGAACACCCTCTT). The probe corresponds to the region from bases 609 to 631 (TCAAACTGCTCAGGCTGCACCGG). The collagenase 1 precursor gene (MMP-1) (exon 2) was selected for rabbit tissue quantitation (GenBank accession no. M17820). The forward primer for MMP-1 corresponds to the region from base 4220 to base 4237 (5′-CCGTCTACCCTGGGTGCC-3′) of the open reading frame. The reverse primer corresponds to the region from base 4274 to base 4296 (5′-ATGGATTTCCTTGCTTGATTCTG-3′). The probe corresponds to the region from base 4243 to base 4270 (5′-TGTGCAGACCACAGGAGCACTTGACAAC-3′). Probes were labeled with 6-carboxyfluorescein at the 5′ end and 6-carboxy-N,N,N′,N′-teramethylrhodamine at the 3′ end. Primers and probes were purchased from QIAGEN. Real-time PCR was performed as previously described (6). Control rabbit DNA was obtained from Seegene (Seoul, Korea).
Extraction of hydrophobic HAB antigens from B31-infected skin (HATTREX).
One gram of infected rabbit skin at the site of inoculation was homogenized in a PowerGen 125 tissue grinder (Fisher) in 10 mM Tris (pH 8.0), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (approximately 50 mg of tissue/ml). HATTREX was performed as previously described (6). For immobilized pH gradient-2D electrophoresis (IPG-2DE) analysis, detergent-phase samples were precipitated by addition of 3 volumes of a 10% trichloroacetic acid (TCA) in −20°C acetone solution and incubated at −20°C overnight. The precipitate was pelleted by centrifugation at 16,000 × g for 30 min at 4°C, and the pellet was washed once with −20°C acetone. The pellet was then air dried and resuspended in 7 M urea, 2 M thiourea, and 1% ASB-14 (4, 30).
IPG-2DE.
Protein samples were analyzed by 2DE using the IPGPhor-2D system from Amersham Pharmacia Biotech. Acetone pellets were resuspended in 7 M urea, 2 M thiourea, and 1% ASB-14. Dithiothreitol was added to 30 mM prior to loading onto the first dimension (immobilized pH gradient), along with the appropriate pH (3 to 10) range IPG buffer to 0.5%. Samples were included directly in the rehydration buffer and allowed to swell the immobilized pH gradient strip overnight. The isoelectric focusing (IEF) parameters were as follows: (i) 1 min at a 500-V gradient, (ii) 1.5 h at a 4,000-V gradient, (iii) 8,000 V for 40,000 V · h, with a 50-μA-per-strip maximum setting at 20°C. The completed first-dimension strip was separated on a conventional sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (24).
Immunoblot analysis.
The proteins were transferred to polyvinylidene difluoride membranes and stained with amido black. The membranes were probed with either immune rabbit serum (IRS; 1:1,000), anti-VlsE (α-VlsE; 1:2,000), or chronic mouse serum (CMS; 1:1,000), all diluted as indicated in Tween-phosphate-buffered saline (0.05% Tween 20) and 5% nonfat dry milk. For CMS, horseradish-linked sheep anti-mouse secondary antibody (Amersham Biosciences) was used at 1:15,000 for secondary antibody. For all others, horseradish-linked donkey anti-rabbit antibody (Amersham Biosciences) was used at 1:15,000. Visualization was performed using the Supersignal West Dura extended-duration substrate from Pierce and the Alpha Innotech Fluorchem 8000 imager. Band densitometry was performed using the Fluorchem imaging software (version 2.0). α-VlsE was a gift of Steven Norris, University of Texas at Houston (unpublished data). IRS was obtained from rabbits with complete infection-derived immunity to reinfection (10, 11, 37). CMS was obtained from four C3H/Hej mice infected for 4 months with strain B31.
IPG-2DE protein gel analysis and spot identification.
IPG-2DE gels were stained with SYPRO Ruby according to the manufacturer's instructions (Molecular Probes, Eugene, Oreg.). Proteins were visualized using the Alpha Innotech Fluorchem 8000 imager with UV transillumination at 302 nm. In-gel trypsin digestion was performed as described by Shevchenko et al. (38) with modifications. Spots were excised and dehydrated with 200 μl of acetonitrile for 30 min. The liquid was removed, and the acrylamide pellet was dried in a SpeedVac (Thermo Savant) and then rehydrated with 100 μl of 100 mM NH4HCO3 for 30 min. The pellet was dehydrated and dried again as indicated above and then rehydrated in 100 μl of 50 mM NH4HCO3-5 mM CaCl2 containing 12.5 ng of trypsin (Promega, Madison, Wis.)/μl for 45 min on ice. The liquid was removed, and 10 μl of the above buffer minus the trypsin was added. The samples were then incubated overnight at 37°C. The pellet was then washed with 25 μl of 20 mM NH4HCO3. The peptide fragments were extracted as follows: 50 μl of a 5% formic acid-50% acetonitrile solution was added to each sample and incubated at room temperature for 20 min. The solution was retained in a separate tube. This was repeated two more times, and the extracts were pooled for each sample. A final extraction was performed using 50 μl of acetonitrile for 20 min.
Analysis of tryptic peptide sequence tags by μLC-MS/MS.
Samples were analyzed by microliquid chromatography-tandem mass spectrometry (μLC-MS/MS) with data-dependent acquisition (LCQ-DECA; ThermoFinnigan, San Jose, Calif.) after dissolution in 5 μl of 70% (vol/vol) acetic acid. A reverse-phase column (200 μm by 10 cm; PLRP/S 5 μm, 300 Å; Michrom Biosciences, San Jose, Calif.) was equilibrated for 10 min at 1.5 μl/min with 95% solution A, 5% solution B (buffer A, 0.1% formic acid in water; buffer B, 0.1% formic acid in acetonitrile) prior to sample injection. A linear gradient was initiated 10 min after sample injection, ramping to 60% solution A, 40% solution B after 50 min and 20% solution A, 80% solution B after 65 min. Column eluent was directed to a coated glass electrospray emitter (TaperTip, TT150-50-50-CE-5; New Objective) at 3.3 kV for ionization without nebulizer gas. The mass spectrometer was operated in triple-play mode with a survey scan (m/z 400 to 1,500), data-dependent zoom scan, and MS/MS with exclusion of singly charged ions. Individual sequencing experiments were matched to a custom B. burgdorferi sequence database NC_001318 (downloaded from The Institute for Genomic Research [www.tigr.org]) using Sequest software (ThermoFinnigan) (2, 12). The search was run under the no-enzyme mode to identify nontryptic peptides. The results of Sequest searches were carefully scrutinized. MS/MS spectra of doubly charged ions with cross-correlation scores (Xcorr) greater than 2.8 and triply charged ions with scores over 3.2 were examined manually. Some noisy spectra were discarded despite high Xcorr scores. Nontryptic peptide returns were retained only if the data looked to be of an especially high signal-to-noise ratio. Where two or more peptides were matched reliably, a strong hit was reported. Where a single good-quality peptide hit was returned, a potential hit was reported. The mass spectral data were interpreted without knowledge of the pI or Mr predicted from the 2D analysis.
RESULTS
Quantitation of B. burgdorferi in infected rabbit skin.
To study the antigenic profile during the early course of the rabbit model of Lyme disease, we injected intradermally 104 low-passage B31 strain B. burgdorferi cells into the backs of New Zealand White rabbits, and the animals were sacrificed at days 5, 7, 9, 11, 14, and 21 postinfection (two rabbits per time point). All animals developed typical EM lesions approximately 7 days postinfection at the injection site (data not shown). Skin biopsies were taken at the site of EM for each time point, and real-time PCR was used to determine the numbers of spirochetes present. As shown in Table 1, B. burgdorferi organisms were present at all the time points tested. A marked increase in numbers of HAB occurred between days 5 and 7. The number of HAB then plateaued over the next 7 days. By day 21, there was a 33-fold drop in HAB number. Day 5 revealed approximately 100 organisms per microgram of rabbit DNA at the site of EM, which then increased dramatically to 74,000 organisms by day 7. A similar number, 50,000, was found at day 9, followed by an increase to 370,000 by day 11. The numbers dropped off to 100,000 at day 14, and by day 21 only 10,000 Borrelia organisms were detected. Over a period of 6 days (days 5 to 11), the number of Borrelia organisms increased by over 3,300-fold, with the largest increase occurring between days 5 and 7 (670-fold). From days 11 to 21, the number of Borrelia organisms dropped by 33-fold, corresponding with the ability of the rabbit host to clear the organisms (10). These numbers are based on the estimate of one copy of the chromosome per cell (31).
TABLE 1.
Numbers of B. burgdorferi B31 in rabbit skin determined by real-time PCRb
| Day | No. of Borrelia/μg of rabbit DNAa | No. of Borrelia/5-mm plugc |
|---|---|---|
| 5 | 1.1 × 102 ± 2.5 × 101 | 2.8 × 103 |
| 7 | 7.4 × 104 ± 6.7 × 104 | 1.0 × 107 |
| 9 | 4.6 × 104 ± 8.5 × 103 | 2.5 × 106 |
| 11 | 3.7 × 105 ± 1.9 × 105 | 7.8 × 106 |
| 14 | 9.4 × 104 ± 4.0 × 104 | 1.6 × 107 |
| 21 | 1.1 × 104 ± 4.7 × 103 | 6.8 × 105 |
Mean ± standard error.
Two separate animals were used for each time point.
Based upon total DNA extracted from a 5-mm skin plug.
Hydrophobic proteins and antigens of IVCB.
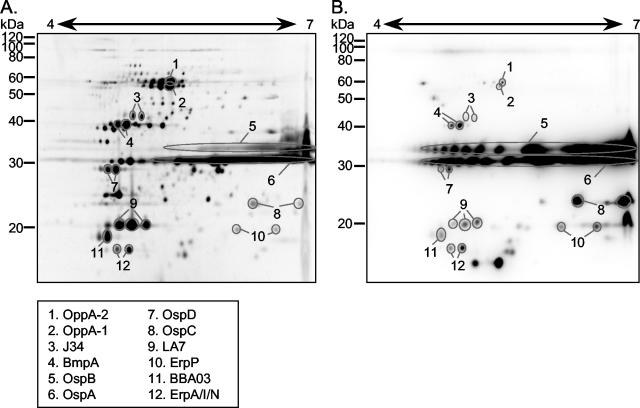
A 300-μg aliquot of Triton X-114 detergent-phase proteins from IVCB was separated by IPG-2DE and stained with SYPRO Ruby (Fig. 1A). For comparison, 0.5 μg of Triton X-114 detergent-phase proteins from IVCB was separated by IPG-2DE and immunoblotted against IRS (Fig. 1B). Most of the immunoreactive spots detected by IRS (Fig. 1B) could be matched precisely to corresponding protein spots (Fig. 1A). The protein identities indicated were all determined by μLC-MS/MS via tryptic digestion (data not shown). OspA and OspB were the dominant antigens present in IVCB and were well represented in the total protein stain. The pIs of OspA and OspB are 8.6 and 8.9, respectively. We separated our samples on a pH 4-to-7 gradient to eliminate the bulk of OspA and OspB, because the large amounts of these proteins tend to precipitate in the first dimension and cause distortion of the focusing. However, since they are so abundant, they still are prominent in the pH 4-to-7 gradient. The ABC transporter proteins OppA-1 and OppA-2 can be seen at 60 kDa (20). J34, a predicted coding frame located on lp38, is present at 42 kDa. Sequence analysis suggests that J34 is a lipoprotein, based upon the presence of a spirochetal lipobox (12, 15). BmpA, another lipoprotein found at 40 kDa, is part of a family of genes with an unknown function (8, 39, 40). OspD is readily visible at 28 kDa, as is OspC at 25 kDa. LA7, seen at 21 kDa, is a chromosomally located lipoprotein with an unknown function (13, 22, 43). A spot found at 19 kDa, BBA03, is a putative outer membrane protein and is located on lp54 (12). The two other proteins identified by MS and matched to IRS-detected spots are ErpP and ErpA/I/N. The Erp proteins, or OspE-related proteins, as they are also known, represent a large family of proteins believed to be involved in complement binding and host immune system evasion (21, 27, 29, 41, 42). ErpP can be detected at 20 kDa, while the ErpA/I/N protein is seen at 17 kDa. The protein sequences for ErpA, -I, and -N are identical and are located on cp32-1, -5, and -8, respectively (2, 42).
FIG. 1.
Identification of hydrophobic proteins and antigens of IVCB by IPG-2DE mapping. (A) Three hundred micrograms of TX-114 detergent-phase proteins from IVCB (B31) stained with SYPRO Ruby total protein stain. Indicated proteins were identified by MS. (B) Total of 107 IVCB (B31) TX-114 detergent-phase protein (∼0.7 μg) derived from 107 IVCB (B31) immunoblotted against IRS. Indicated antigens were identified by matching to results in panel A.
Antigenic composition of Borrelia during rabbit skin infection.
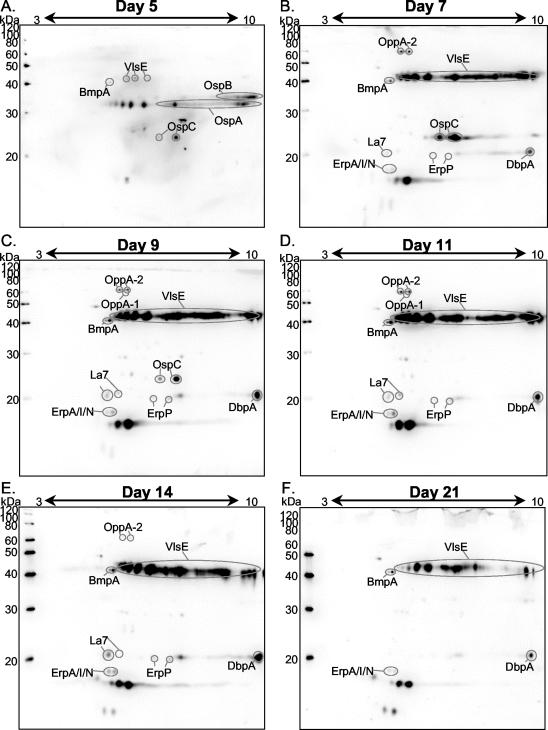
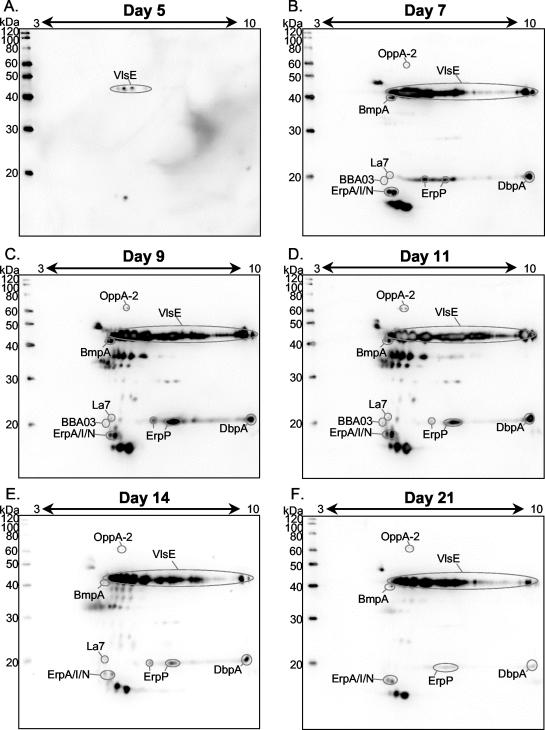
Skin samples taken at the point of inoculation at the above-described time points were prepared using the HATTREX methodology. Briefly, 1 g of skin tissue was processed and extracted with Triton X-114. The samples were precipitated with TCA-acetone and resuspended in IPG-2DE rehydration buffer. The proteins were separated by IPG-2DE and transferred to polyvinylidene difluoride membranes for Western analysis. To determine the B. burgdorferi antigenic composition at the given time points, we probed the blots with IRS. Identification of the various antigens was performed by matching the spots to the MS-identified protein spots from Fig. 1. Using this matching methodology, we identified the following B. burgdorferi antigens expressed in rabbit skin that reacted with IRS: OppA-1 and -2, BmpA, OspC, LA7, ErpP, ErpA/I/N, OspB, and OspA (Fig. 2). Additionally, by using monospecific sera, both VlsE and DbpA were identified (Fig. 3 [for VlsE] and data not shown [for DbpA]). Since the levels of OspA and OspB become very low during infection, they are not a concern for any distortion during the isoelectric focusing; therefore, pH 3-to-10 gradients were used for these experiments. VlsE was not detected by IRS in Fig. 1 due to its low level of expression in IVCB. DbpA, with an expected pI of 8.5, was also not visible in Fig. 1, since the pH range of 4 to 7 was used for matching purposes. Figure 2 shows the HAB antigens detected at days 5, 7, 9, 11, 14, and 21. At day 5, very small amounts of OspA and OspB were still present and OspC was readily detectible. VlsE and BmpA were barely detected. OspA and OspB were not detected at day 7, while several new spots were visible, including OppA-1 and -2, La7, ErpP, ErpA/I/N, and DbpA. Both OspC and VlsE increased in intensity relative to the constellation of antigens. The pattern of antigens seen at day 9 was very similar to that on day 7. OspC disappeared completely at day 11, while the previously mentioned antigens were all still visible. On day 14, OppA-1 was not detected, whereas on day 21 both OppA-1 and -2 were no longer present. Additionally, on day 21, ErpP was not detected. It appears that on day 21 all the antigens were universally decreasing; however, at this time point, the numbers of B. burgdorferi organisms present had declined, most likely due to the host immune response. The two most antigenic proteins present were VlsE and an unknown spot with a molecular mass of 15 kDa. Both antigens were very prominent from days 7 through 21, with VlsE being the most intense. The 2D pattern which VlsE displayed was similar to the pattern seen in B31-infected SCID mice when probed with IRS (6). In addition to the large spot at 15 kDa, unidentified antigens were detected at 49, 30, and 10 kDa. Both rabbits from each time point yielded similar results (data not shown). IRS from different rabbits gave consistent reactivity against B. burgdorferi antigens.
FIG. 2.
Identification of HAB antigens in rabbit skin with IRS. Fifty micrograms of proteins prepared from rabbit skin by HATTREX was separated by IPG-2DE and immunoblotted against IRS. (A) Day 5; (B) day 7; (C) day 9; (D) day 11; (E) day 14; (F) day 21. Antigens were identified by their correspondence to spots of IVCB or with monospecific sera (results not shown).
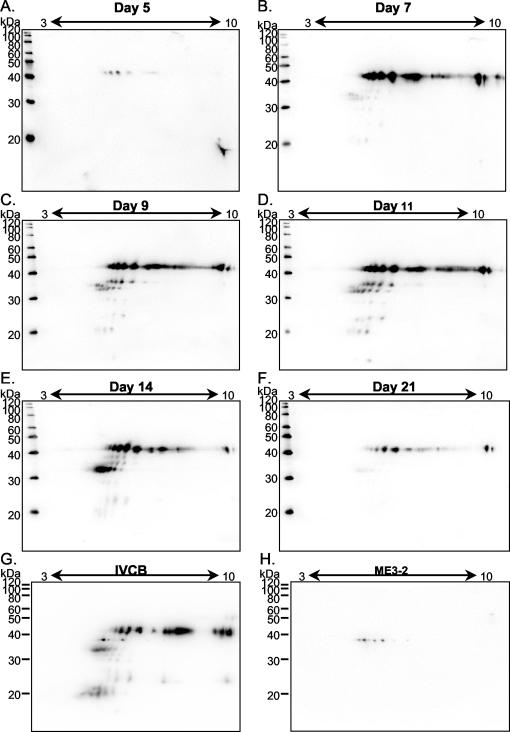
FIG. 3.
Identification of VlsE in rabbit skin. Fifty-microgram aliquots of proteins prepared from rabbit skin by HATTREX were separated by IPG-2DE and immunoblotted against α-VlsE. (A) Day 5; (B) day 7; (C) day 9; (D) day 11; (E) day 14; (F) day 21. (G) Total of 109 detergent-phase IVCB (B31, nonclonal). (H) Total of 109 detergent-phase IVCB (ME3-2, a VlsE-deficient clonal isolate [from this lab]).
VlsE forms.
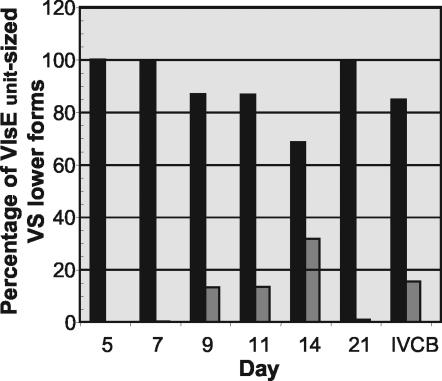
Our investigators have previously shown that IRS does not detect the VlsE lower forms (VlsElf) in B31-infected SCID mouse skin and joints (6). We probed the rabbit skin samples with α-VlsE to determine if the lower forms seen in SCID mice were also present in the rabbit model. As can be seen in Fig. 3, VlsE was readily detectable at every time point. The VlsElf that were so prominent in SCID mice were also detectable in rabbit skin. The amounts of the VlsElf increased over time in comparison with the unit-sized VlsE through day 14. However, at day 21, the VlsElf were barely detected. Using the FluorChem 8000 software (Alpha Innotech), the levels of unit-sized VlsE and lower forms were digitized and amounts were compared to each other at each time point. In Fig. 4, the total amount of VlsE at each time point was set to 100% and then divided between the unit-size and lower forms. The ratio of lower forms to the unit size increased over time and reached a maximum of 30% at day 14. By day 21, virtually all VlsE was unit sized.
FIG. 4.
Relative percentage of VlsElf to VlsE (unit size) during the first 3 weeks of rabbit skin infection.
Chronically infected mouse serum detects lower forms of VlsE.
In our previous study, sera from chronically infected mice recognized the lower forms of VlsE in addition to the unit-sized form in the SCID mouse model (6). Because rabbits clear B. burgdorferi infection while mice remain chronically infected, we probed the various time points with CMS to determine the differences between IRS and CMS in terms of antigenicity from HATTREX-derived proteins (Fig. 5). At day 5, only VlsE and the unknown 15-kDa spot were seen. This was in contrast to IRS, which also detected OspA, OspB, OspC, and BmpA. Day 7 revealed a similar pattern as with IRS, revealing OppA-2, VlsE, BmpA, LA7, ErpA/I/N, ErpP, and DbpA. However, CMS did not appear to detect OspC and OppA-1. In fact, OspC was not detected at any time point with CMS, compared with IRS. A new spot was detected around 20 kDa, which matched perfectly with BBA03 from Fig. 1. BBA03 was detected at days 7, 9, and 11. Interestingly, CMS appeared to recognize the Erp proteins better than IRS. VlsE and the 15-kDa spot were again the dominant antigens present. The same antigens were detected from day 9 to 21 with one notable exception. At days 9, 11, and 14, a large number of new spots appeared around 35 kDa. When aligned with the immunoblots from Fig. 3, these new spots matched perfectly with the VlsElf present at the same time points. These results are in agreement with our previous report, in which CMS appeared to recognize the VlsElf from infected SCID mice (6). Additionally, they highlight the largest difference between CMS and IRS. Under identical IPG-2DE conditions, CMS detected VlsElf readily and was among the dominant antigens present in infected rabbit skin, while IRS revealed the same global pattern as CMS, except for its lack of VlsElf detection. As noted earlier, rabbits can clear B. burgdorferi infection, while mice cannot.
FIG. 5.
Identification of HAB antigens in rabbit skin by CMS. Fifty-microgram aliquots of HATTREX proteins were separated by IPG-2DE and immunoblotted against CMS. (A) Day 5; (B) day 7; (C) day 9; (D) day 11; (E) day 14; (F) day 21. Proteins were identified by spot matching as described above, or with monospecific sera (results not shown).
DISCUSSION
B. burgdorferi's molecular adaptations to microenvironments in the tick have been directly demonstrated by visualizing the spirochetal surface antigens in the midgut and salivary glands (7, 32, 34, 35, 44). While it has been appreciated that B. burgdorferi undergoes further profound changes during infection of mice (45), the number of spirochetes in tissues has been small enough to preclude such direct in situ compositional analysis. Efficient methods to recover intact spirochetes from mouse tissues have not been reported. Efforts to address the surface structure of B. burgdorferi in mouse tissues have therefore relied upon detection of specific gene transcripts. Liang et al. used the presence or absence of ospC transcripts to provide evidence that OspC nonexpressors are selected during infection of immunocompetent mice as an OspC antibody response develops (25). However, Hodzic et al. recently reported that ospC transcripts were readily detected in C3H mice throughout an 8-week course of infection; far lower levels of VlsE transcripts were found, and ospA transcripts were also detected (19). It has been suggested that failure to detect certain lipoprotein transcripts as the duration of infection progresses may relate to the decreasing numbers of spirochetes present rather than to gene downregulation (19, 26). Correlation between detection of specific B. burgdorferi transcripts and the amount of protein expressed by the spirochete has not been established (19).
The HATTREX methodology we have described provides a means of extracting and concentrating hydrophobic B. burgdorferi protein antigens from infected SCID mouse tissues, including ear, heart, and joint (6). The amounts of specific antigens were assessed relative to each other. The relative overexpression of VlsE and smaller forms of VlsE (VlsElf) in mouse ear and joints was particularly striking. It is likely that the origin of VlsElf is posttranslational processing of VlsE; HATTREX affords the possibility of detecting the products of posttranslational processing if they retain their hydrophobicity (6). No evidence was found suggesting that other B. burgdorferi antigens had undergone proteolytic processing. OspC was readily detected in mouse tissues at the single time point (17 days) examined in the SCID mice, while OspA was not detected.
The antigenic variation protein, VlsE, appears to be a key virulence factor of B. burgdorferi, evidenced perhaps by the massive accumulation of VlsE and VlsElf during rabbit skin and mouse tissue infection when compared with that with IVCB. Labandeira-Rey et al. have shown that B. burgdorferi lacking the plasmid containing vlsE (lp28-1) was able to infect and disseminate in C3H mice but was cleared from the system by day 18 (in contrast to wild type, which was not cleared over the 3-week period studied) (23, 33). In contrast to this, in C3H SCID mice, the lp28-1-deficient strain was not cleared. Thus, it would seem that some gene or genes present on lp28-1 are important for persistence in an immunocompetent host (23). The obvious candidate for this function is the antigenic variation protein, VlsE. Interestingly, we have found that vlsE is upregulated in SCID mice, and it has also been shown that it undergoes antigenic variation in SCID mice (6, 46). However, McDowell et al. reported that VlsE variants do indeed possess altered antigenicity, thus providing direct evidence for antigenic variation (28). Additionally, Anguita et al. found that gamma interferon-induced signals facilitate vlsE recombination and that the rate of recombination is slowed in gamma interferon-deficient mice (1).
The rabbit model of Lyme disease offers a unique venue for addressing the experimental biology of B. burgdorferi infection. In particular, the rabbit's ability to clear infection and to develop immunity against reinfection has the potential to provide insights into the basis of protective immunity. In previous studies, our group determined that rabbit skin harbored large numbers of B. burgdorferi (36). At day 14 after needle inoculation, approximately 107 B. burgdorferi organisms were detected in a 5-mm EM skin biopsy, a number sufficient for HATTREX analysis (6, 36). We therefore decided to perform a temporal analysis of the first 3 weeks of B. burgdorferi infection in rabbit skin. As in our previous studies, we chose to infect the rabbits by needle inoculation in order to obtain large numbers of spirochetes. Although differences have been shown between needle inoculation and tick bite regarding immunization studies, our lab has found that HAB development and subsequent clearance of B. burgdorferi infection in the rabbit model are the same (3, 9, 16-18) (unpublished results). Using real-time PCR, we determined the numbers of organisms present in the skin. It is clear that the organisms thrived early on and replicated at a high rate from day 5 to day 11 (Table 1). However, after day 14, their numbers dropped off dramatically. While full clearance of infection in the rabbit model, as judged by negative cultures, was not achieved until 8 to 12 weeks, the reduction in spirochete numbers at week 3 was presumably due to clearance of the organisms by the antibody response of the host (5, 10). While the reduction in spirochete numbers could be due to dissemination rather than clearance, it is unlikely, as dissemination occurs early in the spleen, liver, and heart (24 h postinoculation) and then the organisms are no longer detected by culture after 1 week (10).
In the context of the disparate findings about detection of ospC transcripts, HATTREX-based findings regarding OspC were clear-cut (19, 25). By day 11 (Fig. 2D), OspC was no longer detected relative to the rest of the antigenic profile presented in the 2D immunoblots derived from rabbit skin. OspC was readily detected at day 17 in the infection of SCID mice (6). Thus, our findings with regard to OspC are in accord with the conclusions of Liang et al. (25). OspC is downregulated in the immunocompetent host but persistent in the SCID mouse model. The absolute extent of the downregulation of OspC is unknown, but OspC expression has clearly been dramatically reduced. As the numbers of organisms are increasing over the same time period that the OspC levels are dropping, the lack of detectable OspC is not due to low numbers of Borrelia. The failure of CMS to detect OspC even at days 5 to 9, when it is detectable by IRS, was also striking, but is of unknown significance.
OspA and OspB were detected only at day 5. Most of the other antigens detected by IRS and CMS remained at fairly consistent levels relative to each other from day 7 through day 21. This included DbpA, BmpA, ErpA/N, Erp P, and OppA-2. An unidentified very intense spot around 15 kDa was also identified at all time points.
Similar 2D profiles were observed between our group's previous studies (6) and this work. The differences detected, besides OspC, may be due to the lower numbers observed in SCID mice compared to those in infected rabbit skin. As in SCID mice, the dominant antigen detected by IRS and CMS is VlsE (6). Probing the immunoblots with α-VlsE revealed the same pattern of forms of lower molecular mass that were seen in SCID mice and also found to be present in lesser amounts in IVCB (6). The ratio of lower forms of VlsE (VlsElf) to the full-sized form initially increased over time, reaching a maximum at day 14.
At day 21, VlsElf was virtually undetectable, while unit-size VlsE remained prominent and DbpA, BmpA, ErpA/N, Erp P, and OppA-2 were also detected. The disappearance of VlsElf is the only compositional change in the spirochete that can be temporally correlated with the rapidly diminishing numbers of the spirochete in the skin. The immune response of rabbits and of chronically infected mice to VlsElf differs strikingly, as judged by immunoblots of 2D gels (Fig. 2 and 5). IRS does not bind the VlsElf, while CMS readily binds them. Given that VlsE is the dominant antigen present yet is also the main difference between the reactivity of rabbit and mouse sera, it will be interesting to determine what role, if any, the VlsElf plays in pathogenesis. Future experiments will address whether the immune response of rabbits and mice to VlsElf contributes to whether B. burgdorferi is cleared or establishes chronic infection, respectively.
Acknowledgments
This research was supported by National Institutes of Health grants AI 29733 and AI 37312 to Michael A. Lovett and James N. Miller, respectively.
We thank Evelyn Pineda for her assistance with this work.
Editor: D. L. Burns
REFERENCES
- 1.Anguita, J., V. Thomas, S. Samanta, R. Persinski, C. Hernanz, S. W. Barthold, and E. Fikrig. 2001. Borrelia burgdorferi-induced inflammation facilitates spirochete adaptation and variable major protein-like sequence locus recombination. J. Immunol. 167:3383-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 3.Cassatt, D. R., N. K. Patel, N. D. Ulbrandt, and M. S. Hanson. 1998. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect. Immun. 66:5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevallet, M., V. Santoni, A. Poinas, D. Rouquie, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19:1901-1909. [DOI] [PubMed] [Google Scholar]
- 5.Chong-Cerrillo, C., E. S. Shang, D. R. Blanco, M. A. Lovett, and J. N. Miller. 2001. Immunohistochemical analysis of Lyme disease in the skin of naive and infection-immune rabbits following challenge. Infect. Immun. 69:4094-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crother, T. R., C. I. Champion, X. Y. Wu, D. R. Blanco, J. N. Miller, and M. A. Lovett. 2003. Antigenic composition of Borrelia burgdorferi during infection of SCID mice. Infect. Immun. 71:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Silva, A. M., S. R. Telford III, L. R. Brunet, S. W. Barthold, and E. Fikrig. 1996. Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J. Exp. Med. 183:271-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrikova, E. Y., J. Bugrysheva, and F. C. Cabello. 2001. Two independent transcriptional units control the complex and simultaneous expression of the bmp paralogous chromosomal gene family in Borrelia burgdorferi. Mol. Microbiol. 39:370-378. [DOI] [PubMed] [Google Scholar]
- 9.Feng, S., E. Hodzic, B. Stevenson, and S. W. Barthold. 1998. Humoral immunity to Borrelia burgdorferi N40 decorin binding proteins during infection of laboratory mice. Infect. Immun. 66:2827-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foley, D. M., R. J. Gayek, J. T. Skare, E. A. Wagar, C. I. Champion, D. R. Blanco, M. A. Lovett, and J. N. Miller. 1995. Rabbit model of Lyme borreliosis: erythema migrans, infection-derived immunity, and identification of Borrelia burgdorferi proteins associated with virulence and protective immunity. J. Clin. Investig. 96:965-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley, D. M., Y. P. Wang, X. Y. Wu, D. R. Blanco, M. A. Lovett, and J. N. Miller. 1997. Acquired resistance to Borrelia burgdorferi infection in the rabbit. Comparison between outer surface protein A vaccine- and infection-derived immunity. J. Clin. Investig. 99:2030-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 13.Grewe, C., and J. H. Nuske. 1996. Immunolocalization of a 22 kDa protein (IPLA7, P22) of Borrelia burgdorferi. FEMS Microbiol. Lett. 138:215-219. [DOI] [PubMed] [Google Scholar]
- 14.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 101:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagman, K. E., X. Yang, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson, M. S., D. R. Cassatt, B. P. Guo, N. K. Patel, M. P. McCarthy, D. W. Dorward, and M. Hook. 1998. Active and passive immunity against Borrelia burgdorferi decorin binding protein A (DbpA) protects against infection. Infect. Immun. 66:2143-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodzic, E., S. Feng, K. J. Freet, and S. W. Barthold. 2003. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 71:5042-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornacki, J. A., and D. B. Oliver. 1998. Lyme disease-causing Borrelia species encode multiple lipoproteins homologous to peptide-binding proteins of ABC-type transporters. Infect. Immun. 66:4115-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement-inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697-707. [DOI] [PubMed] [Google Scholar]
- 22.Kramer, M. D., U. E. Schaible, R. Wallich, S. E. Moter, D. Petzoldt, and M. M. Simon. 1990. Characterization of Borrelia burgdorferi associated antigens by monoclonal antibodies. Immunobiology 181:357-366. [DOI] [PubMed] [Google Scholar]
- 23.Labandeira-Rey, M., J. Seshu, and J. T. Skare. 2003. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect. Immun. 71:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marconi, R. T., S. Y. Sung, C. A. Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowell, J. V., S. Y. Sung, L. T. Hu, and R. T. Marconi. 2002. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with Lyme disease spirochetes. Infect. Immun. 70:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, T. B., Y. Ma, J. H. Weis, and J. J. Weis. 1999. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J. Clin. Microbiol. 37:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang, E. S., C. I. Champion, X. Y. Wu, J. T. Skare, D. R. Blanco, J. N. Miller, and M. A. Lovett. 2000. Comparison of protection in rabbits against host-adapted and cultivated Borrelia burgdorferi following infection-derived immunity or immunization with outer membrane vesicles or outer surface protein A. Infect. Immun. 68:4189-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang, E. S., X. Y. Wu, M. A. Lovett, J. N. Miller, and D. R. Blanco. 2001. Homologous and heterologous Borrelia burgdorferi challenge of infection-derived immune rabbits using host-adapted organisms. Infect. Immun. 69:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, W. J., W. Cieplak, M. E. Schrumpf, A. G. Barbour, and T. G. Schwan. 1994. Nucleotide sequence and analysis of the gene in Borrelia burgdorferi encoding the immunogenic P39 antigen. FEMS Microbiol. Lett. 119:381-387. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, W. J., M. E. Schrumpf, and T. G. Schwan. 1990. Reactivity of human Lyme borreliosis sera with a 39-kilodalton antigen specific to Borrelia burgdorferi. J. Clin. Microbiol. 28:1329-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson, B., and K. Babb. 2002. LuxS-mediated quorum sensing in Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 70:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallich, R., M. M. Simon, H. Hofmann, S. E. Moter, U. E. Schaible, and M. D. Kramer. 1993. Molecular and immunological characterization of a novel polymorphic lipoprotein of Borrelia burgdorferi. Infect. Immun. 61:4158-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, J. R., and S. J. Norris. 1998. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect. Immun. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J. R., and S. J. Norris. 1998. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect. Immun. 66:3689-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]