Abstract
The Brucella abortus virB operon, encoding a type IV secretion system (T4SS), is required for intracellular replication and persistent infection in the mouse model. The products of the first two genes of the virB operon, virB1 and virB2, are predicted to be localized at the bacterial surface, where they could potentially interact with host cells. Studies to date have focused on characterization of transposon mutations in these genes, which are expected to exert polar effects on downstream genes in the operon. In order to determine whether VirB1 and VirB2 are required for the function of the T4SS apparatus, we constructed and characterized nonpolar deletion mutations of virB1 and virB2. Both mutants were shown to be nonpolar, as demonstrated by their ability to express the downstream gene virB5 during stationary phase of growth in vitro. Both VirB1 and VirB2 were essential for intracellular replication in J774 macrophages. The nonpolar virB2 mutant was unable to cause persistent infection in the mouse model, demonstrating the essential role of VirB2 in the function of the T4SS apparatus during infection. In contrast, the nonpolar virB1 mutant persisted at wild-type levels, showing that the function of VirB1 is dispensable in the mouse model of persistent infection.
Brucella spp. are found in association with a large number of wild and domesticated animal species, where they can cause persistent infection and abortion. Zoonotic transmission of the organism to humans can lead to a chronic febrile disease known as brucellosis or Malta fever. A characteristic of both natural and zoonotic forms of the disease is extended survival of the organism in tissues of the reticuloendothelial system, such as the spleen, lymph nodes, and bone marrow.
In a mutant screen for Brucella abortus virulence factors required for survival in the murine reticuloendothelial system, the virB operon, encoding homologs of Agrobacterium tumefaciens and Bordetella pertussis type IV secretion systems (24), was found to be essential for persistence in mice (18). The importance of these genes for intracellular survival was demonstrated in tissue culture models of infection as well (12, 15, 20, 24, 30). These genes were subsequently found to be expressed intracellularly by Brucella suis in macrophages (8), where they are required for localization of B. abortus to an intracellular niche that is associated with the endoplasmic reticulum (9), the same location where B. abortus was earlier shown to replicate in nonprofessional phagocytes (13, 14, 27) and in the ruminant placenta (2, 23). For B. abortus, the virB genes have been implicated in the initial interactions between the bacterium and the host cell during entry into macrophages (33).
Based on studies performed with A. tumefaciens, both VirB1 and VirB2 are predicted to be accessible on the surface of B. abortus. VirB2 is predicted to form a pilus-like structure, and VirB1 is predicted to have two domains—a lytic transglycosylase and a second domain that is released to the extracellular medium and remains associated with the bacterial surface (4, 10, 22). These findings for A. tumefaciens suggested that both VirB1 and VirB2 could mediate interactions of B. abortus with host cells.
Evidence for a lytic transglycosylase function for VirB1 from Brucella spp. was provided by Höppner et al., who showed that B. suis virB1 could complement an A. tumefaciens virB1 mutant for tumor formation and for extracellular localization of VirB2 and that the complementation required the lytic transglycosylase active site (19). A polar virB1 mutation in B. abortus, as well as transposon mutations in Brucella melitensis virB2 and in the virB1-B2 intergenic region of B. melitensis and B. abortus, has been reported to be defective for intracellular survival in vitro (12, 30, 32). However, for all of these mutant strains, the expected polar effect of the insertions on expression of the downstream genes in the virB operon did not allow any conclusions as to whether VirB1 or VirB2 are required for infection by Brucella spp. We therefore constructed and characterized defined deletion mutations of virB1 and virB2 in order to determine whether VirB1 and VirB2 are required for intracellular persistence of B. abortus in both tissue culture and in the mouse model of brucellosis.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
B. abortus strain 2308 was used throughout this study. Strains were cultured on tryptic soy agar (TSA; Difco/Becton-Dickinson, Sparks, Md.) or in tryptic soy broth (TSB) at 37°C on a rotary shaker. Bacterial inocula for infection of mice were cultured on potato infusion agar, as growth on this medium is described to prevent the appearance of spontaneous rough mutants (1). Antibiotics, when required, were added at the following concentrations: carbenicillin, 100 mg/liter; kanamycin, 100 mg/liter; chloramphenicol, 5 to 30 mg/liter. All work with live B. abortus was performed at biosafety level 3.
Strain construction and recombinant DNA techniques.
For construction of strain BA1143 carrying a virB4::lacZYA fusion (Table 1), a fragment corresponding to bp 1644 to 2457 of the B. abortus virB locus was amplified by PCR using primers virB-1644F and virB-2457R (Table 2) and cloned into pCR2.1 (Invitrogen). The fragment was excised with XbaI and SmaI and ligated into pFUSE (5) digested with the same enzymes to yield pBA1143. This plasmid was introduced by electroporation into B. abortus 2308 and a recombinant resistant to chloramphenicol was designated BA1143. Integration of pBA1143 into B. abortus chromosome II of BA1143 was confirmed by Southern blot (data not shown).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| B. abortus strains | ||
| 2308 | Wild type | B. Deyoe |
| BA1143 | virB4::lacZYA | This work |
| ADH1 | ΔvirB2::Km (polar) | This work |
| ADH3 | ΔvirB2 (nonpolar) | This work |
| ADH4 | ΔvirB1-12::Km | This work |
| ADH5 | ΔvirB1::Km (polar) | This work |
| ADH6 | ΔvirB1 (nonpolar) | This work |
| ADH8 | Knock-in complementation of ADH3 | This work |
| E. coli strains | ||
| DH5α | endA 1 hsdR17 (rκ− mκ−) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR (φ80 dlac Δ[lacZ]M15) | 34 |
| BL21 (DE3) | F′ompT gal [dcm] [lon] hsdSB (rB− mB−) DE3 | 31 |
| Plasmids | ||
| pBluescript KS | ColE1, bla | Stratagene |
| pCR2.1-TOPO | TA cloning vector | Invitrogen |
| pUC4-KSAC | Carries Tn903 kanamycin resistance cassette | Pharmacia |
| pAV1.4 | virB219-477 and virB1154-1562 cloned into pBluescript | This work |
| pAV1.5 | virB219-477 and virB1154-1562 separated by kanamycin resistance cassette KSAC cloned into pBluescript | This work |
| pAV2.1 | PCR products of virB988-1562 and virB1796-2458 cloned into pBluescript | This work |
| pAV2.2 | PCR products of virB988-1562 and virB1796-2458 separated by kanamycin resistance cassette cloned into pBluescript | This work |
| pAS1.1 | sacB cloned into pAV2.1 | This work |
| pAS1.2 | sacB cloned into pAV1.4 | This work |
| pMR10 | RK2, orivV + trfA, oriT, nptI, lacZα | R. M. Roop II |
| pEX18Ap | sacB | 17 |
| pHB11 | virB140-1967 cloned into pMR10 | This work |
| pFUSE | oriR6K plasmid containing promoterless lacZYA | 5 |
| pBA1143 | virB1644-2457 cloned into pFUSE | This work |
| pAVT1.4 | PCR products of B. abortus using primer pairs 98736F-99854R and 112902F-114022R separated by kanamycin resistance cassette cloned into pBluescript | This work |
TABLE 2.
Primers used in this worka
| Primer name | Sequence (restriction enzyme) | Fragment |
|---|---|---|
| virB-1644F | 5′-GCTCTAGAGCGGATTCTACCTCACCTACTGC-3′ (XbaI) | virB2-B4 |
| virB-2457R | 5′-TCCCCCGGGCCTCCTTGTCCTGCGTCTCG-3′ (SmaI) | virB2-B4 |
| virB-219F | 5′-CTCGAGTGAAAATCACCGCATACCAC-3′ (XhoI) | virB1 upstream |
| virB-477R | 5′-ATCGATGAGGACAAGGAATGGCACC-3′ (ClaI) | virB1 upstream |
| virB-1154F | 5′-ACGGCGTAGTTGTTTTCTAACCC-3′ | virB1 downstream |
| virB-1562R | 5′-TCTAGACGGTCTGCTTGCTCAATGTCTATTC-3′ (XbaI) | virB1 downstream |
| virB-988F | 5′-CTCGAGGCTGCCCCAGTAAAAAAACGAC-3′ (XhoI) | virB2 upstream |
| virB-1562R | 5′-ATCGATTCGGTCTGCTTGCTCAATGTCTAT-3′ (ClaI) | virB2 upstream |
| virB-1796F | 5′-CCATCGCCATCATCTGGTCCG-3′ | virB2 downstream |
| virB-2458R | 5′-TCTAGATCTCCTTGTCCTGCGTCTCG-3′ (XbaI) | virB2 downstream |
| KSAC707F1 | 5′-ATCGATTAACCAATTCTGATTAGA-3′ (ClaI) | Kan cassette |
| KSAC1638R | 5′-GAATTCCGTTGTGTCTCAAAATCTCTG-3′ (EcoRI) | Kan cassette |
| virB-5063F | 5′-GAATTCTACGAAGCGGTGATGAGCG-3′ (EcoRI) | GST-VirB5 fusion |
| virB-5549R | 5′-CTCGAGGTGTAGATGATGTGTTGCGTG-3′ (XhoI) | GST-VirB5 fusion |
| virB-629F | 5′-GGAATTCTCCCCCAGCAAGAAGTCG-3′ (EcoRI) | GST-VirB1 fusion |
| virB-1330R | 5′-CTCGAGTGGTGTGCGACTGCCTCCTATC-3′ (XhoI) | GST-VirB1 fusion |
| 98736F | 5′-CTCGAGGCTGCGATTTCCATTCACTCC-3′ (XhoI) | virB upstream |
| 99854R | 5′-GTCGACTCTTGCCGTCATAATAGCCTGAG-3′ (SalI) | virB upstream |
| 112902F | 5′-CTGCAGAAAGCCAACCCCTCGC-3′ (PstI) | virB downstream |
| 114022R | 5′-TCTAGACACCTGATTCCACGGAGTG-3′ (XbaI) | virB downstream |
| KSAC707F2 | 5′-GTCGACTAACCAATTCTGATTAGA-3′ (SalI) | Kan cassette |
| KSAC1638R2 | 5′-CTGCAGGTTGTGTCTCAAAATCTCTG-3′ (PstI) | Kan cassette |
| SacB-F | 5′-GAGCTCGCTGGCTTAACTATGCGGCA-3′ (XhoI) | sacB |
| SacB-R | 5′-GAGCTCTTCAAACAGGAGGGCTGGAAG-3′ (XhoI) | sacB |
For construction of plasmids to generate marked and unmarked deletions of virB1, fragments upstream and downstream of virB1 were amplified by PCR using primer pairs virB-219F-virB-477R and virB-1154F-virB-1562R. The resulting fragments were cloned into pCR2.1-TOPO. The fragment virB219-477 was excised with XhoI and ClaI and cloned into pBluescript KS. The fragment virB1154-1562 was excised with EcoRI and XbaI and cloned into pBluescript KS containing virB219-477, generating plasmid pAV1.4 (Table 1). The Tn903 kanamycin resistance gene was amplified from pUC4-KSAC (Pharmacia) with primers KSAC707F1 and KSAC1638R, digested with EcoRI and ClaI, and cloned into EcoRI/ClaI-digested pAV1.4 to yield pAV1.5 containing a deleted copy of virB1 replaced with the Tn903 kanamycin resistance gene. Plasmid pAS1.2, containing an unmarked deletion of virB1 and the counterselectable marker sacB, was constructed by amplification of the sacB gene from pEX18Ap (17) using primers SacB-F and SacB-R. The product was digested with XhoI and ligated into XhoI-digested pAV1.4.
For construction of plasmids to generate marked and unmarked deletions of virB2, fragments upstream and downstream of virB2 were amplified by PCR using primer pairs virB-988F-virB-1562R and virB-1796F-virB-2458R. The resulting fragments were cloned into pCR2.1-TOPO. The fragment virB988-1562 was excised with XhoI and ClaI and cloned into pBluescript KS. The fragment virB1796-2458 was excised with EcoRI and XbaI and cloned into pBluescript KS containing virB988-1562, generating plasmid pAV2.1 (Table 1). The Tn903 kanamycin resistance gene was amplified from pUC4-KSAC (Pharmacia) with primers KSAC707F1 and KSAC1638R, digested with EcoRI and ClaI, and cloned into EcoRI/ClaI-digested pAV2.1 to yield pAV2.2 containing a deleted copy of virB2 replaced with the Tn903 kanamycin resistance gene. Plasmid pAS1.1, containing an unmarked deletion of virB2 and the counterselectable marker sacB, was constructed by amplification of the sacB gene from pEX18Ap using primers SacB-F and SacB-R. The product was digested with XhoI and ligated into XhoI-digested pAV2.1.
For construction of a deletion encompassing the entire B. abortus virB operon, plasmid pAVT1.4 was generated as follows: fragments upstream and downstream of the virB operon were amplified using the primer pairs 98736F-99854R (upstream) and 112902F-114022R (downstream), designed using the preliminary B. suis genome sequence at The Institute for Genomic Research. The fragments were cloned in pCR2.1-TOPO and then excised with XhoI/SalI (upstream) and PstI/XbaI (downstream). The fragments were cloned sequentially into pBluescript KS digested with the same enzymes. The Tn903 kanamycin resistance gene was cloned with PstI/SalI ends and inserted between the upstream and downstream fragments to generate pAVT1.4. This plasmid was introduced into B. abortus strain 2308 by electroporation, and a kanamycin-resistant, carbenicillin-sensitive recombinant was selected. The resulting strain, ADH4, contains a deletion from 167 bp upstream of virB1 to 1,133 bp downstream of virB12 and was confirmed by Southern blot analysis to lack the virB1-B12 genes (data not shown). The plasmids constructed in this work are listed in Table 1, and the primers used in the construction of the plasmids are listed in Table 2. Plasmid DNA was isolated using ion-exchange columns from QIAGEN. Standard methods were used for Southern blotting, PCR, restriction endonuclease analyses, and ligation and transformation of plasmid DNA into E. coli (3). PCR products were cloned into pCR2.1-TOPO using a TOPO-TA cloning kit (Invitrogen).
Cell lines.
The mouse macrophage-like cell line J774 (28) was cultured in Dulbecco's modified Eagle's medium (Gibco, Rockville, Md.) supplemented with 10% heat-inactivated fetal bovine serum, 1% nonessential amino acids, and 1 mM glutamine (DMEMsup). Since this cell line can lose the ability to generate an oxidative burst after passage in culture (B. Bloom, personal communication), we routinely tested J774 cultures for generation of oxygen radicals via the hexose monophosphate shunt (oxidative burst) after induction with phorbol myristate acetate (Sigma) as described elsewhere (11).
For macrophage killing assays, 24-well microtiter plates were seeded with macrophages at a concentration of 1 × 105 to 2 × 105 cells/well in 0.5 ml of DMEMsup and incubated overnight at 37°C in 5% CO2. For preparation of the inoculum, B. abortus strains were grown for 24 h and then diluted in DMEMsup, and about 5 × 107 bacteria in 0.01 ml of DMEMsup was added to each well of macrophages. The microtiter plates were centrifuged at 250 × g for 5 min at room temperature in order to synchronize infection. Cells were incubated for 20 min at 37°C in 5% CO2, free bacteria were removed by three washes with phosphate-buffered saline (PBS), and the zero time point was taken as described below. DMEMsup plus 50 μg of gentamicin per ml was added to the wells, and the cells were incubated at 37°C in 5% CO2. After 1 h, the DMEMsup plus 50 μg of gentamicin/ml was replaced with medium containing 25 μg of gentamicin/ml. Wells were sampled at 0 and 24 or 48 h after infection by aspirating the medium, lysing the macrophages with 0.5 ml of 0.5% Tween 20, and rinsing each well with 0.5 ml of PBS. Viable bacteria were quantified by dilution in sterile PBS and plating on TSA. All experiments were performed independently in triplicate at least three times, and the standard error for each time point was calculated. The statistical significances of differences observed were analyzed by analysis of variance (ANOVA) and Tukey's honestly significant difference (HSD) test (25).
Infection of mice.
Female BALB/c ByJ mice were obtained from the Jackson Laboratory (Bar Harbor, Maine) and used at 6 to 10 weeks of age. For infection experiments, groups of four or five mice were inoculated intraperitoneally with 0.2 ml of PBS containing 1 × 105 to 5 × 105 CFU of B. abortus. Infected mice were held in microisolator cages in a biosafety level 3 facility. At the appropriate time points, the mice were euthanized by CO2 asphyxiation, and the spleens were collected aseptically at necropsy. The spleens were homogenized in 3 ml of PBS, and serial dilutions of the homogenate were plated on TSA containing antibiotics, as appropriate, for enumeration of CFU. The statistical significances of differences observed were analyzed by ANOVA followed by Tukey's HSD test. All animal experiments were approved by the Texas A&M University Laboratory Animal Care and Use Committee and conducted in accordance with institutional guidelines.
Generation of polyclonal antisera to VirB1 and VirB5.
virB1 and virB5 genes were PCR amplified from B. abortus by use of the primer sets described in Table 2, cloned into pCR2.1 (Invitrogen), and subsequently cloned in frame with glutathione S-transferase (GST) in pGEX4T-1 (Pharmacia) using the enzymes EcoRI and BamHI. GST fusion proteins were overexpressed and purified according to standard protocols (3) and used to raise polyclonal antisera in rabbits at the Texas A&M Laboratory Animal Research Resource facility. Nonspecific antibodies to Escherichia coli proteins and GST were depleted from sera by incubation with killed E. coli Top10F′(pGEX4T-1) in which overexpression of GST had previously been induced with isopropyl-β-d-thiogalactopyranoside.
Detection of VirB proteins.
B. abortus cultures inoculated in TSB to a starting optical density at 600 nm of 0.01 were incubated at 37°C with shaking at 200 rpm for 36 h. Bacteria were pelleted and resuspended in 1× Laemmli sample buffer at a concentration of 1010 CFU/ml and heated at 100°C for 5 min, and the total protein from 108 CFU (0.01 ml) was loaded into each well for separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (21). Proteins were transferred to polyvinylidene difluoride membranes by electroblotting and were detected using polyclonal rabbit sera specific for VirB1 and VirB5, and goat anti-rabbit immunoglobulin G was conjugated to horseradish peroxidase. Horseradish peroxidase activity was detected with a chemiluminescent substrate (NEN).
RESULTS
Construction of marked and unmarked deletion mutations of virB1 and virB2.
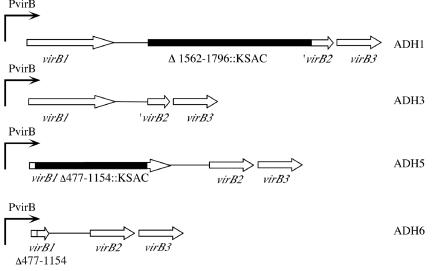
In order to assess whether VirB1 and VirB2 are essential components of the B. abortus type IV secretion system (T4SS), we constructed marked and unmarked deletions in virB1 and virB2 by a two-step method. In the first step, the deletion mutation was marked with a kanamycin resistance cassette. In the second step, the kanamycin resistance determinant was deleted using sucrose counterselection. This strategy has the advantage over a one-step sucrose selection strategy that in each step gain or loss of antibiotic resistance can be monitored, facilitating screening for the desired recombinants from the background of sucrose-resistant, nonrecombinant colonies. For the first step in the mutation construction, plasmids pAV1.5, containing part of the virB locus, in which virB1 was replaced by the kanamycin resistance cassette KSAC, and pAV2.2, in which virB2 was replaced by KSAC, were constructed (see Materials and Methods for details). The resulting constructs pAV1.5 (pBluescriptKS-virB1::Km) and pAV2.2 (pBluescriptKS-virB2::Km) were introduced into B. abortus 2308 by electroporation (Table 1). Transformants were screened for resistance to kanamycin and sensitivity to carbenicillin (the resistance determinant of pBluescript KS) to identify recombinants in which the allelic exchange had occurred. Kanamycin-resistant and carbenicillin-sensitive recombinants were tested for deletion of the virB1 and virB2 genes, respectively, by Southern blot analysis using probes containing the virB1 and virB2 genes (data not shown). The resulting marked deletion strains were designated ΔvirB2::Km (ADH1) and ΔvirB1::Km (ADH5). To delete the kanamycin resistance cassettes in these two strains, plasmids pAS1.1 and pAS1.2, carrying unmarked deletions of virB1 and virB2 and the sacB gene, were constructed and introduced into ΔvirB2::Km (ADH1) and ΔvirB1::Km (ADH5). Recombinants in which pAS1.1 and pAS1.2 had integrated into the virB locus were selected on carbenicillin and then subjected to sucrose counterselection in TSB containing sucrose. Sucrose-resistant recombinants were screened for loss of both the kanamycin resistance cassette and the sacB gene by screening for sensitivity to kanamycin and carbenicillin. Deletion of ΔvirB1::Km and ΔvirB2::Km was again confirmed by Southern blot analysis using probes containing virB1 and virB2 (data not shown). The resulting unmarked virB1 and virB2 deletion mutations (Fig. 1) were designated ΔvirB2 (ADH3) and ΔvirB1 (ADH6). The strains and plasmids constructed in this work are described in Table 1.
FIG. 1.
Construction of marked and unmarked deletions of virB1 and virB2. For details, see the text. The positions of deletions in the B. abortus virB locus are numbered according to the sequence found under GenBank accession number AF226278 (30).
The ΔvirB1 and ΔvirB2 mutations do not eliminate expression of the downstream gene virB5.
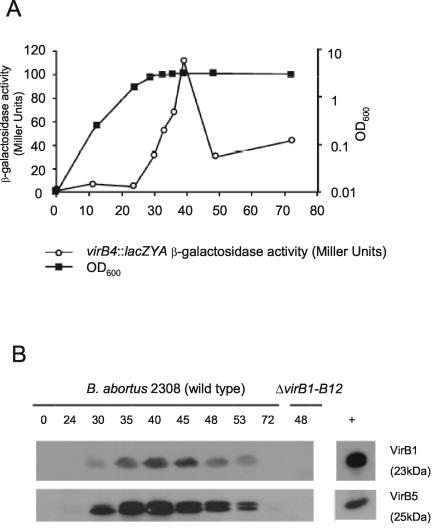
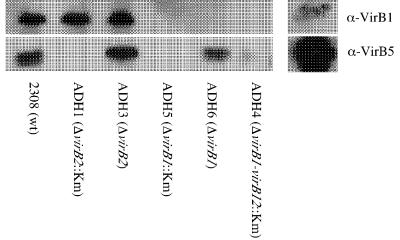
To test whether VirB1 and VirB2 are required for the function of the T4SS, it was first necessary to determine whether deletion of virB1 and virB2 affected the synthesis of proteins encoded by downstream genes. To this end, we raised polyclonal sera specific for VirB1 and VirB5, a product of a downstream gene in the virB operon, and determined in vitro conditions for synthesis of these proteins by B. abortus. Based on a previous finding that a B. abortus virB10::lacZ fusion was expressed in stationary phase (30), we performed an analysis of expression of the VirB1 and VirB5 proteins in wild-type B. abortus 2308 (Fig. 2). For this purpose, we constructed strain BA1143, containing a chromosomal virB4::lacZYA fusion, by introducing suicide vector pBA1143 into B. abortus 2308 and selecting for integration into virB4 (Tables 1 and 2). We found that virB expression peaked after entry into stationary phase and then subsequently declined. Analysis by Western blotting showed that VirB1 and VirB5 proteins could be detected starting at 30 h, with maximal levels detected between 35 and 40 h. Based on these findings, we used this time point to determine whether ΔvirB1::Km, ΔvirB1, ΔvirB2::Km, and ΔvirB2 exert polar effects on downstream virB genes. Figure 3 shows that while the marked virB1 and virB2 deletion mutations eliminate expression of the downstream gene virB5, the unmarked deletion mutations still produce VirB5, although the expression of VirB5 was reduced slightly compared to that in the other strains. We concluded from these data that deletion of virB1 in ADH6 and deletion of virB2 in ADH3 do not inhibit expression of downstream genes in the virB operon.
FIG. 2.
Expression of the virB genes during growth of B. abortus in vitro. (A) Activity of a virB4::lacZYA fusion during growth in TSB. OD600, optical density at 600 nm. (B) Western blot showing detection of VirB1 and VirB5 produced by B. abortus 2308 during growth in TSB at different times after inoculation. The positive controls shown at the right are GST-VirB1 and GST-VirB5.
FIG. 3.
Western blot showing detection of VirB1 and VirB5 produced by B. abortus mutants carrying marked or unmarked virB1 or virB2. Marked mutations exert a polar effect on virB5 expression, while VirB5 can still be detected in mutants carrying unmarked deletions. wt, wild type.
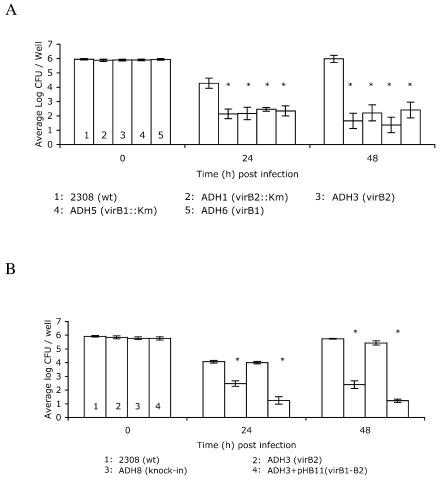
VirB1 and VirB2 are required for growth of B. abortus in macrophages.
We found previously that polar transposon mutations in the 3′ end of virB1 or in virB10 rendered B. abortus unable to grow intracellularly or persist in mice, which led us to conclude that the T4SS is essential for successful intracellular infection by B. abortus (18). To determine specifically whether VirB1 and VirB2 are essential to the function of the type IV apparatus in mediating intracellular growth, we tested whether the polar and nonpolar virB1 and virB2 mutants are able to grow within cells of the J774 macrophage-like line (Fig. 4A). Both polar and nonpolar deletions of virB1 and virB2 significantly decreased the ability of B. abortus to survive within macrophages as early as 24 h after infection, suggesting that both VirB1 and VirB2 proteins are required for intracellular survival of B. abortus in vitro. Based on data shown in Fig. 3, we knew that the unmarked deletion of virB2 did not prevent expression of downstream genes in the virB operon. However, to eliminate the possibility that passaging of B. abortus in vitro to construct the virB2 mutation could have resulted in other attenuating mutations, we complemented the virB2 mutation by two different methods. A plasmid-encoded copy of PvirB-virB1-virB2 (pHB11) was introduced into ADH3 (virB2). The second method used was “knock-in” complementation of ADH3, in which an intact copy of virB2 was reintroduced into its original location in the virB locus of ADH3, replacing the deleted allele by homologous recombination. The resulting strain was confirmed by Southern blot analysis (data not shown) to contain virB2 and was designated ADH8. As an additional control, we analyzed expression of VirB5 in this strain by Western blotting, and it was unchanged compared to ADH3 (data not shown), showing that the knock-in of virB2 did not disrupt any downstream genes. Introduction of virB2 on pHB11 did not restore the ability of ADH3 to survive in J774 cells, but knock-in complementation of virB2 restored intracellular replication of ADH3 to wild-type levels, thereby confirming that the intracellular replication defect of ADH3 was due to deletion of virB2 (Fig. 4B).
FIG. 4.
(A) Survival in J774 macrophages of B. abortus mutants carrying marked or unmarked deletions of virB1 or virB2. Data points represent the means of results from three independent experiments performed in triplicate ±standard error of the mean. (B) Complementation of intracellular survival defect of the nonpolar virB2 mutant by knock-in of virB2. Asterisks indicate significant (P < 0.05) differences in numbers of CFU of mutant strains recovered from J774 macrophages compared to the wild type (wt) by Tukey's HSD test following ANOVA.
VirB2 is required for persistence in the mouse model, but VirB1 is dispensable.
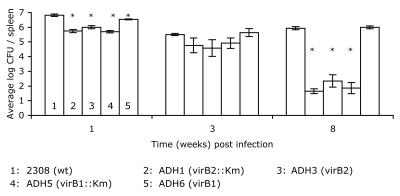
In order to determine whether VirB1 and VirB2 are required for persistence of B. abortus in vivo, we tested the abilities of both polar and nonpolar virB1 and virB2 mutants to establish infection and persist in the mouse model. Groups of 12 to 15 mice were infected with either B. abortus 2308 or one of the isogenic virB1 or virB2 mutants, and the bacterial CFU in the spleens were enumerated in groups of 4 to 5 mice at 1, 3, and 8 weeks postinoculation. By 1 week after infection, the wild-type strain, 2308, was recovered in numbers 6- to 10-fold higher than the ΔvirB2::Km, ΔvirB1::Km, and ΔvirB2 mutants and in approximately 2-fold higher numbers than the nonpolar ΔvirB1 mutant (Fig. 5). At 3 weeks after infection, the same trend was observed, with ΔvirB2::Km, ΔvirB1::Km, and ΔvirB2 mutants recovered in numbers one-third to one-eighth of those of the wild type, while the nonpolar ΔvirB1 mutant was recovered in slightly higher numbers than the wild type. At 8 weeks postinoculation, the numbers of CFU of ΔvirB2::Km, ΔvirB1::Km, and ΔvirB2 mutants recovered were 3 to 4 logs lower than those of the wild type and the nonpolar ΔvirB1 mutant. These data suggest that VirB2 is essential for survival in vivo, while VirB1 is dispensable for persistent infection in the mouse.
FIG. 5.
Persistence of polar and nonpolar virB1 and virB2 mutants in spleens of BALB/c mice. Data are the averages of results from experiments with groups of five mice ±standard error. Asterisks indicate significant (P < 0.05) differences in numbers of CFU of mutant strains recovered from mouse spleens compared to the wild type (wt) as determined by Tukey's HSD test following ANOVA.
DISCUSSION
The goal of this study was to determine whether virB1 and virB2 encode proteins required for the function of the T4SS apparatus. Based on studies performed in A. tumefaciens, these two proteins are predicted to be accessible on the surface of B. abortus. VirB2 is predicted to form a pilus-like structure, and VirB1 is predicted to have two domains—a lytic transglycosylase and a second domain that is released to the extracellular medium and associated with the bacterial surface (4, 10, 22). These findings in A. tumefaciens suggested that both VirB1 and VirB2 could be involved in Brucella-host cell interactions.
In order to examine the roles of VirB1 and VirB2 in pathogenesis, it was necessary to generate nonpolar mutants in their corresponding genes, since the virB1 to virB12 genes are transcribed as an operon in B. abortus and B. suis (8, 30). We constructed two unmarked deletion mutations (ΔvirB1 and ΔvirB2) and showed that in strains containing these mutations, the downstream gene virB5 is expressed in stationary phase at levels comparable to those in the wild type (Fig. 2), which demonstrated that the ΔvirB1 and ΔvirB2 mutations did not eliminate expression of downstream genes in the virB operon. Our data showing stationary-phase expression of the B. abortus virB genes in vitro is in agreement with the work of Sieira et al. (30). Similarly, Rouot et al. were able to detect expression of VirB5 in stationary-phase B. abortus cultures grown in rich medium, but not in B. suis cultures, demonstrating that the regulation of the virB genes differs in these two Brucella species (29).
Nonpolar mutations of both virB1 and virB2 produced an intracellular survival defect in J774 macrophages that was similar to that seen for polar mutations (Fig. 3), suggesting that both VirB1 and VirB2 are required for survival in these cells. Therefore, it is likely that both of these T4SS components are required for assembly of a functional T4SS apparatus, as has been shown for A. tumefaciens (7).
Using the mouse model of persistent infection, we found that the ΔvirB1 mutation was not attenuated for persistence through 8 weeks postinoculation (Fig. 4). These data appear to be at odds with our results showing that virB1 is required for survival in J774 macrophages (Fig. 3A). Since the lytic transglycosylase consensus motif of B. suis VirB1 was required for complementation of T4SS function in an A. tumefaciens virB1 mutant (19), it is likely that this activity is also important for the function of B. abortus VirB1. One possible explanation for our finding that B. abortus VirB1 is dispensable during in vivo infection is that the predicted lytic transglycosylase activity of VirB1 can be compensated for by other proteins encoded in the Brucella genome that have similar functions. The presence of 10 additional open reading frames in the B. melitensis genome with sequence similarity to soluble or membrane-bound lytic transglycosylases appears to support this idea (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html). One or a subset of these predicted lytic transglycosylases might function more efficiently during in vivo infection than in cultured macrophages to compensate for loss of VirB1. An alternate explanation is that in J774 cells, the C-terminal product VirB1* may contribute to intracellular survival. We have observed a smaller cross-reactive band with anti-VirB1 antisera (∼12 kDa) in B. abortus lysates that is absent from virB1 mutants (data not shown), which may represent the B. abortus form of VirB1*, as it is similar to the size reported by Baron et al. for A. tumefaciens VirB1* (4). The potential role of B. abortus VirB1* in intracellular growth could be addressed by complementation of the ΔvirB1 mutant with the C-terminal domain of VirB1. These results are an example of how tissue culture models of infection may not always reflect what occurs during host-pathogen interactions in vivo, and it will be interesting to determine whether the requirement for VirB1 in the natural bovine host is more similar to that in the mouse model or the macrophage model.
Unlike the ΔvirB1 mutant, the ΔvirB2 mutant ADH3 was attenuated for persistence in mice (Fig. 5). Intracellular replication of the ΔvirB2 mutant ADH3 could be restored by complementation (Fig. 4B), suggesting that the virulence defect of ADH3 (ΔvirB2) in mice is also due to deletion of virB2. Interestingly, we were able to complement ADH3 (ΔvirB2) only by reintroducing virB2 back into chromosome II but not by expressing it from the virB promoter on low-copy-number plasmid pMR10, even though we were able to show that the complemented strain produced VirB1. Some of the possible explanations for this may be that the presence of multiple copies of virB2 or virB1 leads to elevated expression levels that are toxic to B. abortus. Overexpression of the lytic transglycosylase VirB1 could result in elevated degradation of the murein sacculus in B. abortus and lead to bacteriolysis, as has been shown for the lytic transglycosylase P19, which is involved in conjugation of plasmid R1 (6). Alternatively, multiple plasmid-encoded copies of the virB promoter in B. abortus may titrate out a repressor or activator of the virB genes and affect the regulation of the virB genes. Finally, although Berger and Christie were able to complement individual deletions of the A. tumefaciens virB genes with plasmid-encoded copies (7), it is possible that in B. abortus, VirB2 must be expressed coordinately with VirB3 or other VirB proteins in order to function correctly.
Our findings are consistent with an essential role for VirB2 in the assembly of the T4SS apparatus. In A. tumefaciens, VirB2 was shown to form a pilus structure that could be visualized by electron microscopy (16, 22). We have not reproducibly observed pili on the surface of B. abortus expressing the virB genes in laboratory media, but it is possible that pilus assembly occurs in vivo, or that the B. abortus T4SS has a different structure than its counterpart in Agrobacterium. Further experimentation will be required to determine how VirB2 interacts with other VirB proteins and secreted effectors to assemble a functional T4SS apparatus in B. abortus.
Acknowledgments
We thank Hortensia García Rolán and Christelle Roux for careful reading of the manuscript. We also thank R. M. Roop for providing plasmid pMR10 and H. P. Schweizer for providing plasmid pEX18Ap.
Work in R.T.'s laboratory is supported by NIH/NIAID award AI50553.
Editor: D. L. Burns
REFERENCES
- 1.Alton, G. G., L. M. Jones, and D. E. Pietz. 1975. Laboratory techniques in brucellosis, 2nd ed. World Health Organization, Geneva, Switzerland. [PubMed]
- 2.Anderson, T. D., and N. F. Cheville. 1986. Ultrastructural morphometric analysis of Brucella abortus-infected trophoblasts in experimental placentitis. Bacterial replication occurs in rough endoplasmic reticulum. Am. J. Pathol. 124:226-237. [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. J. Wiley & Sons, New York, N.Y.
- 4.Baron, C., M. Llosa, S. Zhou, and P. C. Zambryski. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J. Bacteriol. 179:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler, A. J., R. M. Tsolis, A. W. M. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentheiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183:3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chumakov, M. I., and I. V. Kurbanova. 1998. Localization of the protein VirB1 involved in contact formation during conjugation among Agrobacterium cells. FEMS Microbiol. Lett. 168:297-301. [DOI] [PubMed] [Google Scholar]
- 11.Damiani, G., C. Kiyotaki, W. Soeller, M. Sasada, J. Peisach, and B. R. Bloom. 1980. Macrophage variants in oxygen metabolism. J. Exp. Med. 152:808-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 13.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Entry and intracellular localization of Brucella spp. in Vero cells: fluorescence and electron microscopy. Vet. Pathol. 27:317-328. [DOI] [PubMed] [Google Scholar]
- 14.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58:2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulongne, V., G. Bourg, C. Cazevieille, S. Michaux-Charachon, and D. O'Callaghan. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 68:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullner, K. J., J. L. Lara, and E. W. Nester. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273:1107-1109. [DOI] [PubMed] [Google Scholar]
- 17.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 18.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höppner, C., Z. Liu, N. Domke, A. N. Binns, and C. Baron. 2004. VirB1 orthologs from Brucella suis and pKM101 complement defects of the lytic transglycosylase required for efficient type IV secretion from Agrobacterium tumefaciens. J. Bacteriol. 186:1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S., M. Watarai, Y. Kondo, J. Erdenebaatar, S. Makino, and T. Shirahata. 2003. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect. Immun. 71:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lai, E.-M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meador, V. P., and B. L. Deyoe. 1989. Intracellular localization of Brucella abortus in bovine placenta. Vet. Pathol. 26:513-515. [DOI] [PubMed] [Google Scholar]
- 24.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 25.Ott, R. L., and M. Longnecker. 2000. An introduction to statistical methods and data analysis, 5th edition. Duxbury, Pacific Grove, Calif.
- 26.Paulsen, I., R. Seshadri, K. Nelson, J. Eisen, J. Heidelberg, T. Read, R. Dodson, L. Umayan, L. Brinkac, M. Beanan, S. Daugherty, R. Deboy, A. Durkin, J. Kolonay, R. Madupu, W. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. van Aken, S. Riedmuller, H. Tettelin, S. Gill, O. White, S. Salzberg, D. Hoover, L. Lindler, S. Halling, S. Boyle, and C. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pizarro-Cerdá, J., S. Méresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goñi, E. Moreno, and J.-P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralph, P., and I. Nakoinz. 1975. Phagocytosis and cytolysis by a macrophage tumour and its cloned cell line. Nature 257:393-394. [DOI] [PubMed] [Google Scholar]
- 29.Rouot, B., M.-T. Alvarez-Martinez, C. Marius, P. Mentanteau, L. Guilloteau, R.-A. Boigegrain, R. Zumbihl, D. O'Callaghan, N. Domke, and C. Baron. 2003. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71:1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sieira, R., D. J. Comerci, D. O. Sanchez, and R. A. Ugalde. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182:4849-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Y.-H., A. B. den Hartigh, R. D. L. Santos, L. G. Adams, and R. M. Tsolis. 2002. virB-Mediated survival of Brucella abortus in mice and macrophages is independent of a functional inducible nitric oxide synthase or NADPH oxidase in macrophages. Infect. Immun. 70:4826-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watarai, M., S. Makino, Y. Fujii, K. Okamoto, and T. Shirahata. 2002. Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication. Cell. Microbiol. 4:341-355. [DOI] [PubMed] [Google Scholar]
- 34.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]