Abstract
Borrelia burgdorferi undergoes differential gene expression during transmission from its tick vector to a vertebrate host. The addition of blood to a spirochete culture at 35°C for 48 h had a dramatic effect on gene expression of this organism. Utilizing B. burgdorferi whole genome DNA arrays, we compared the transcriptomes of the spirochetes following a 2-day temperature shift with blood and without blood. Using combined data from three independent RNA isolations we demonstrated that the addition of blood led to a differential expression of 154 genes. Of these, 75 genes were upregulated, with 49 (65%) of them encoded on plasmids. Blood supplementation of cultures also resulted in the downregulation of 79 genes, where 56 (70%) were plasmid encoded. We verified our results by reverse transcriptase PCR of several genes in both flat and feeding ticks. In the 2-day experiment we observed the effect that exposure to increased temperature and blood combined had on B. burgdorferi gene expression at this crucial time when the spirochetes begin to move from the vector to a new vertebrate host. These changes, among others, coincide with the upregulation of the chemotaxis and sensing regulons, of the lp38-encoded ABC transporter, of proteases capable of remodeling the outer surface of the spirochetes, and of the recombination genes of cp32 as a transient or initial part of the stress response of the phage. These are all functions that could cause or facilitate the changes that spirochetes undergo following a blood meal in the tick.
Lyme disease is an illness caused by an infection of the spirochete Borrelia burgdorferi (5, 9). The spirochete persists in nature in a zoonotic cycle between its arthropod vector, the Ixodes scapularis tick, and vertebrates and is responsible for over 10,000 cases of Lyme disease in the United States annually (40).
The ability of the spirochete to invade and colonize two different kinds of hosts has been an area of active research. Differential gene expression of spirochetes during tick feeding is thought to facilitate the passage of the spirochete from the tick to its new vertebrate host. In the unfed tick, the spirochetes are in the lumen of the midgut, present in low numbers, and express unique antigens, including the outer surface lipoprotein A (OspA) (10, 17, 48). As the tick begins to feed, the temperature in the midgut rises, pH drops, and in response to this radically changing environment, the spirochetes undergo drastic changes in physiology. They begin to multiply dramatically and change their gene expression profile. One well-known example of changes in gene expression is the downregulation of OspA and upregulation of OspC (15, 37, 57, 58), and this switch in expression has been used to propose pathogen-vector interactions with these lipoproteins. OspA has been implicated in adherence to the midgut of the flat tick (42, 68). Likewise, there may be a role for OspC in directing the spirochetes to the salivary glands, which may in part explain its initial upregulation (23, 43). However, this molecule may be more important for vertebrate infection than for traffic within the tick (26). Many other proteins in the spirochetes undergo differential expression during this critical time (3, 25). The spirochetes are able to penetrate the midgut and enter the salivary glands of the ticks. Once in the salivary glands, the spirochetes are deposited into the skin of the vertebrate host (17, 45, 49, 65). The entire process of the spirochete migration from the midgut through the salivary glands to the new host takes approximately 2 days (47).
The genome of B. burgdorferi strain B31, containing a linear chromosome and 12 linear and 9 circular plasmids, has been completely sequenced (13, 24). The genome sequence has allowed the opportunity to study the gene expression profile of the spirochete in response to experimental changes with DNA array technology. Several recent publications described such studies utilizing differential gene expression in Borrelia grown at temperatures mimicking unfed (flat) ticks and feeding tick-mammal conditions (39, 54, 71). Another study identified genes differentially expressed when B. burgdorferi spirochetes were grown in dialysis membrane chambers transplanted into peritoneal cavities of rats, i.e., in a “host-adapted” state (8). In addition, several differentially expressed genes in flat and feeding ticks were identified utilizing decal technology (34). These studies have presented valuable data on changes in gene expression and how B. burgdorferi gene expression oscillates throughout its life cycle due to the spirochete circulating between new environments. The studies have also shown that various results can occur not just because of the technology used but also due to differences in experimental conditions.
Several factors affect differential gene expression of B. burgdorferi in vitro. Changes in temperature, pH, and growth phase can result in differential expression of several genes (11, 12, 51, 52, 66). However, the changes during tick feeding do not occur in isolation from each other. For example, increasing temperature and pH changes in the midgut occur simultaneously as a result of the incoming blood meal and its large influx of nutrients. In fact, this interdependence of environmental factors having a role in gene expression has already been documented (66). The goal of the present study was to determine the combined effect of blood influx and temperature shift on B. burgdorferi gene expression, in similarity to the changes the spirochetes encounter in feeding ticks. Using B. burgdorferi DNA whole genome arrays, we identified genes differentially expressed when B. burgdorferi encounters human blood following a 48-h temperature shift from 23 to 35°C. These changes in gene expression may be essential for the spirochete in transmission and adaptation to the vertebrate host. In addition, reverse transcriptase PCR (RT-PCR) in flat and feeding ticks was used to validate array results and to show that the timing and extent of the differential expression of many of these genes in the natural tick-vertebrate cycle are the result of the combined effects of the blood and of the increase in temperature.
MATERIALS AND METHODS
Culture conditions.
The B. burgdorferi low-passage B31 MI strain was used in all experiments. The strain contained all 21 plasmids, as confirmed by PCR using plasmid-specific primers (20). Frozen stock vials of the strain were inoculated directly into 250 ml of BSK-H medium supplemented with 6% rabbit serum (Sigma, St. Louis, Mo.) and incubated at 23°C until the cell density reached approximately 1.5 × 106 spirochetes/ml. The culture was then divided into 50-ml Falcon tubes, with 47 ml of culture placed into each tube. For blood studies, 3 ml of human blood from which the buffy coat was removed and which contained 10% 0.1 M sodium citrate was added. Control cultures received 3 ml of BSK-H medium with 10% 0.1 M sodium citrate. All cultures were incubated at 35°C for 48 h, with periodic mixing to prevent the settling of the blood. These experiments were repeated three times. Each of the three experiments yielded separate RNA preparations, which were used for three separate cDNA probe syntheses and three separate array hybridizations (see below). The pH of the cultures was measured with a 300 series pH meter (Beckman Instrument Inc., Fullerton, Ca.) prior to addition of blood, as well as at 24 and 48 h afterwards. All spirochete enumeration was performed utilizing dark-field microscopy.
RNA extraction.
Following the 48-h incubation, the cultures were centrifuged at 400 × g for 4 min to sediment the red blood cells. Spirochetes in the supernatant were then centrifuged in a new 50-ml tube for 10 min at 7,000 × g, and the pellet was resuspended in 0.4 ml of BSK-H; the remaining medium was decanted. RNA extraction was performed with Tri-Reagent LS (Molecular Research Center Inc.) according to the manufacturer's instructions. The RNA was then passed through an RNeasy spin column (QIAGEN Inc., Valencia, Ca). An aliquot of the RNA was used for A260-A280 spectrophotometry readings as well as for agarose gel electrophoresis. Three separate 35°C and 35°C-blood RNA preparations were isolated as described above and used in the array experiment.
cDNA probe synthesis.
A cDNA probe was created for hybridization to the DNA array membranes as described previously, with some modifications (2, 38). A 15-μl reaction mixture was set up which contained 5 μg of RNA and genome-directed primers (GDP) (54). Amplifications were carried out in a PTC-1152 MiniCycler (MJ Research, Waltham, Mass.) with the parameters set at 90°C for 2 min followed by a ramp to 42°C over 20 min. A final volume of 30 μl containing 1× reaction buffer (50 mM Tris-HCl, 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol, pH 8.5), 50 U of avian myeloblastosis virus RT, 333 μM dCTP, 333 μM dGTP, 333 μM dTTP (Roche, Indianapolis, Ind.), and 20 μCi of [α-33P]dATP (ICN, Costa Mesa, Calif.) was added to the reaction mixture. This reaction was incubated at 42°C for 2.5 h in a MiniCycler. Unincorporated radiolabeled nucleotide was removed from the labeled cDNA by use of a Panorama Sephadex G-25 column on the basis of the protocol of the manufacturer (Sigma-Genosys).
Membrane hybridization and array image analysis.
The B. burgdorferi genome arrays used in these experiments consisted of 1,689 PCR amplified open reading frames (ORFs) spotted in duplicate on charged nylon membranes (8, 38, 39). All details regarding construction and handling of these membranes have been extensively documented by Ojaimi et al. (38, 39). Two membranes were utilized in the experiment. Each RNA preparation was hybridized twice, once to each membrane, to account for any slight membrane variations, and the average intensity from both experiments was calculated. Membranes were neutralized in 35- by 300-mm hybridization bottles (Robbins Scientific, Sunnyvale, Calif.) in 250 ml of 2× SSPE (0.3 M NaCl, 20 mM NaH2PO4, 2 mM EDTA) for 25 min; in subsequent experiments, membranes were neutralized for 5 min. Membranes were prehybridized for 60 min in 5 ml of 65°C hybridization solution (Sigma-Genosys) containing 100 μg of salmon testes DNA (Sigma-Genosys)/ml. The entire radiolabeled cDNA sample was added to 10 ml of hybridization solution containing 100 μg of salmon testes DNA/ml, denatured in a 95°C water bath for 10 min, and then added to the membranes and allowed to hybridize for 16 to 18 h at 65°C in a Model 2000 Micro hybridization incubator (Robbins Scientific). The cDNA was removed from the bottles, and 40 ml of wash solution (0.5× SSPE, 2% sodium dodecyl sulfate) was added. Bottles were inverted 10 times at room temperature, and the wash solution was decanted. This step was repeated twice. Following the third wash, 80 ml of wash solution was added and the bottles were incubated at room temperature in the hybridization oven for 20 min. Membranes were exposed to a 35- by 43-cm PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.) for 2 to 3 days. Screens were viewed using ImageQuant 1.2 software in a Storm 860 PhosphorImager (Molecular Dynamics). Pixel densities for each spot were generated with ArrayVision software version 6.0.
Membranes were stripped in roller bottles, initially with 250 ml of 0.4 M NaOH followed by incubation with stripping solution (250 ml of 200 mM Tris-HCl, 0.1× SSPE, 0.2% sodium dodecyl sulfate, pH 7.0) for 30 min at 45°C each with rotation. This procedure was followed by incubation with 250 ml of stripping solution at room temperature in the hybridization oven for 30 min with moderate rotation. Membranes were exposed to the PhosphorImager screen (as described above) overnight to ensure that the stripping was successful so that membranes could be reprobed.
Statistical analysis and bioinformatics.
ArrayVision software (Imaging Research, St. Catharines, Ontario, Canada) was used to generate raw pixel density data for every ORF from each hybridized membrane. Each B. burgdorferi ORF is represented by two spots. An initial gene filter created a threshold for each gene individually on the basis of the local background of each spot. In brief, the mean density of membrane areas that did not have spotted DNA was subtracted from each spot value to yield the raw data. Only genes with raw data values ≥ 2 standard deviations (SD) above the local background were analyzed. The raw data were imported into a Microsoft Excel spreadsheet (Microsoft Corp.), where each individual spot was divided by the sum of all spots to get a percent intensity value for each individual spot. All genes with expression levels above the background from individual RNA preps were plotted by calculating their log 10 percent intensity culture with blood (designated 35B culture)/culture without blood (designated 35 culture) ratio. Correlation plots between separate RNA isolations were obtained by plotting the percent intensity 35B culture/35 culture ratio for each ORF in Microsoft Excel. Transcript abundance was calculated as described previously (39). Our final analysis consisted of the mean of the combined data from all three experiments. The data was subjected to an unpaired two-tailed t test to determine which ORFs were differentially expressed at a P value of ≤0.01. Differentially expressed genes were further analyzed for possible functions, paralogous family members, and structural motifs, domains, and regions by the use of The Institute for Genomic Research software and protein databases (http://www.tigr.org, http://smart.embl-heidelberg.de/, http://psort.nibb.ac.jp/form.html, http://www.sanger.ac.uk/Software/Pfam/, and http://us.expasy.org/sprot/).
Quantitative real-time RT-PCR.
This procedure was used for validation of whole genome DNA array experiments. One of the three RNA preparations utilized in the array study was also used for RT-PCR. For cDNA synthesis, 2 μg of total RNA was used in an RT reaction with GDP as described previously. The resulting cDNA sample was diluted 1:50 for use in the RT-PCR. Three replications were performed for each gene. LightCycler-FastStart DNA Master SYBR I Green (Roche) reaction mix was used in the experimental procedure with the LightCycler (Roche) on the basis of the manufacturer's protocol. Cycle parameters were as follows: preincubation, 1 cycle at 95°C for 5 min; amplification (quantification), 40 cycles of 95°C for 15 s, 52°C for 6 s, and 72°C for 6 s; melting curve, 95°C for 0 s, 65°C for 15 s, and 72°C for 0 s; and cooling, 1 cycle of 40°C for 30 s. 16S rRNA and flaB were used as internal controls for all data. Oligonucleotide primers for RT-PCR were designed using MacVector software (Kodak International Biotechnologies, Inc., New Haven, Conn.).
RT-PCR of Borrelia-infected ticks.
Infected I. scapularis nymphs were placed on the neck of temporarily restrained uninfected mice. The nymphs were allowed to feed for 72 h, after which they were removed and homogenized. Total RNA was isolated using Tri-Reagent LS (MRC Inc.) according to the manufacturer's instructions. RNA was treated with 10 U of DNase I (Roche) for 1 h at 37°C followed by purification by RNeasy kit spin columns (QIAGEN). Total RNA (5 μg) was reverse transcribed to cDNA by the use of avian myeloblastosis virus RT (Roche) and GDP. PCR consisting of 1 cycle at 94°C for 5 min and 40 cycles of 94°C for 20 s, 52°C for 30 s, and 72°C for 30 s was performed. A PCR of an RNA sample without RT was always run to ensure that no DNA contamination was present. For flat ticks, total tick RNA was isolated from adult ticks collected on Shelter Island, N.Y., and treated in an identical manner.
Western blot analysis.
Minislab gels were cast with a 10-well comb, and samples (12 μl/well) were applied and run for 2 h at 20 mA/gel. Proteins were electroblotted onto nitrocellulose, blocked in 2% casein, and incubated for 1 h each in primary antibody (1/1,000 dilution of murine anti-OspC immunoglobulin G) and secondary antibody (1/1,000 dilution of a 0.2 mg/ml stock of alkaline phosphatase conjugated to goat anti-mouse immunoglobulin G [Fisher, Pittsburgh, PA]) and reacted with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (Kirkegaard & Perry, Gaithersburg, Md.).
RESULTS AND DISCUSSION
Experimental design.
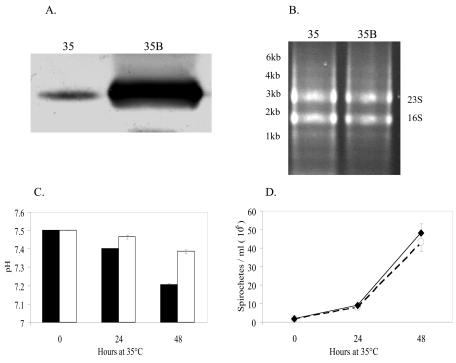
B. burgdorferi spirochetes are present in very low numbers in flat ticks, but there is a dramatic increase in spirochete numbers during tick feeding (17, 48, 49). To mimic the conditions found in tick midguts in vitro, spirochetes were grown at 23°C to a very low concentration prior to shifting them to 35°C. Once the spirochetes reached the concentration of 1.5 × 106/ml, the 23°C culture was split and then temperature shifted to 35°C either with or without blood and incubated for 48 h. The time interval was chosen because the highest number of spirochetes in feeding ticks expressing OspC was found after 48 h (57). In addition, it is around this time that passage of infectious spirochetes from ticks to animals is most likely to occur (46, 47). In our experiments, OspC protein levels vastly increased following a 2-day incubation with blood at 35°C (Fig. 1A).
FIG. 1.
Comparisons between 35B and 35 cultures. (A) Western blot of OspC from spirochetes grown in control cultures and in cultures supplemented with blood after 48 h of incubation. (B) Agarose gel electrophoresis of RNA obtained from cultures grown at 35°C and at 35°C with blood for 2 days. A total of three separate RNA isolations were performed; the figure shows a representative 16S and 23S rRNA electrophoretic pattern of one isolation. Molecular mass markers are shown on the left. (C) pH changes in blood (black bars) and control (white bars) cultures of B. burgdorferi at 24 and 48 h. (D) Growth curve of B. burgdorferi in blood (solid line) and control (dashed line) cultures of B. burgdorferi at 24 and 48 h.
Two DNA array studies have been published describing differential gene expression of B. burgdorferi at temperatures that would be expected in flat and feeding ticks (39, 54). In those experiments, the spirochetes were initially grown at 23°C, diluted, and shifted to either 35°C (39) or 37°C (54). The comparison was then made between the gene expression profiles at 23°C and at the elevated temperatures. These studies, while uncovering temperature-regulated genes, may have missed genes expressed transiently during the first 2 days of blood feeding. More importantly, they would not uncover genes differentially expressed due to factors in the incoming blood meal. For the array results described here, we analyzed the B. burgdorferi transcriptome changes that occurred after a 48-h temperature shift to 35°C of spirochetes in 35B culture compared to results for spirochetes temperature shifted in 35 culture. In a 2-day experiment we observed the effect that exposure to increased temperature and blood combined had on B. burgdorferi gene expression at this crucial time, when the spirochetes begin to move from the vector to a new vertebrate host.
A representative pattern of the RNA generated under these experimental conditions showed the 16S and 23S typical of prokaryotic samples and none of the 18S and 28S rRNA that could be derived from the human blood (Fig. 1B). The mean pH of the blood cultures was 7.20 compared to that of the control at 7.38 after 48 h of incubation (Fig. 1C). This drop in pH is less than the 6.8 reported for feeding ticks but may have contributed to some of the differences reported here (66). The addition of blood did not result in growth inhibition of the spirochetes in the time frame of the experiments (Fig. 1D).
Different RNA isolations produced reproducible array results.
Three RNA isolations generated from three separate experiments were used to probe the arrays. To compare the RNA isolations, 35B culture/35 culture percent intensity ratios of all the ORFs were plotted, and correlation coefficients between all isolations were calculated (Fig. 2). Furthermore, the log-transformed percent intensity ratio was calculated for each experiment and plotted to identify genes differentially expressed from each RNA isolation as well as to identify genes whose expression levels differed between different RNA isolations (Fig. 3). The array results from the three RNA isolations showed a high degree of correlation to one another (Fig. 2 and 3). This allowed us to pool and analyze the data by using the mean values for all three RNA isolations. The final analysis only included genes with significant change in expression (P ≤ 0.01) and with at least twofold differential ratios.
FIG. 2.
Scatter plots of gene expression derived from three separate RNA isolations. Panel A compares RNA isolation 1 to isolation 3, panel B compares RNA isolation 2 to isolation 3, and panel C compares RNA isolation 1 to isolation 2. The 35B culture/35 culture percent intensity ratio from each RNA isolation was calculated and plotted against the ratios from the two other RNA isolations. The correlation coefficient r value is provided for each plot.
FIG. 3.
Comparison of the three individual RNA isolations-hybridizations. Each individual spot represents one log 10 35B culture/35 culture percent intensity ratio of a single ORF with expression 2 SD above the background. Panels A to C represent individual cDNA results from experiments 1 to 3, while panel D represents the plot of all three combined RNA isolations. The dashed lines represent two SD above and below the means of all the plotted ORFs.
Incubation of B. burgdorferi with blood for 48 h leads to a gene expression profile that is different from the profiles obtained by temperature shifts alone.
The combined data from our three separate RNA preparations yielded 154 genes that were differentially expressed, as determined by the established statistical criteria (Table 1 and Table 2). Of these, 75 genes were upregulated, with 49 (65%) of them encoded on plasmids (Table 1). Likewise, the genes with the highest fold increases were also plasmid encoded (Table 1). Blood supplementation of cultures resulted in the downregulation of 79 genes, of which 56 (70%) were plasmid encoded (Table 2). As B. burgdorferi contains many paralogs in its genome, it was expected that many genes sharing high sequence similarity and belonging to the same paralogous gene family would exhibit similar expression characteristics on the array, although it is unknown whether all of the paralogs are identically regulated or even expressed (8, 39).
TABLE 1.
Mean fold upregulation of genes on the cDNA array by use of three separate RNA isolations
| Upregulation (fold) | Gene description | Replicon | Gene family |
|---|---|---|---|
| 45.0 | BBA72 hypothetical protein | lp54 | |
| 38.2 | BB0175 conserved hypothetical protein | Chromosome | |
| 33.6 | BBB19 outer surface protein C (OspC) | cp26 | |
| 31.2 | BBA66 antigen, P35, putative | lp54 | 54 |
| 24.2 | BBA73 antigen, P35, putative | lp54 | 54 |
| 19.7 | BBA24 decorin binding protein A (DbpA) | lp54 | 74 |
| 17.8 | BBA65 hypothetical protein | lp54 | 54 |
| 16.7 | BBA64 antigen, P35 | lp54 | 54 |
| 15.4 | BBE31 antigen, P35, putative | lp25 | 60 |
| 12.5 | BBC12 phage exonuclease | cp9 | 165 |
| 12.3 | BBJ25 hypothetical protein | lp38 | |
| 10.7 | BBA71 hypothetical protein | lp54 | 54 |
| 9.9 | BBJ46 hypothetical protein | lp38 | |
| 9.5 | BB0771 RNA polymerase sigma factor (RpoS) | Chromosome | 89 |
| 8.4 | BB0565 purine-binding chemotaxis protein (CheW-2) | Chromosome | 33 |
| 8.3 | BBC05 phage ssb binding protein (ERF) | cp9 | 161 |
| 7.8 | BBL36 phage exonuclease | cp32-8 | 165 |
| 7.6 | BB0418 hypothetical protein | Chromosome | |
| 6.9 | BB0844 hypothetical protein | Chromosome | 32 |
| 6.9 | BBQ43 phage exonuclease | lp56 | 165 |
| 6.5 | BBA05 antigen, S1 | lp54 | |
| 6.5 | BBO36 phage exonuclease | cp32-7 | 165 |
| 6.4 | BBP35 phage exonuclease | cp32-1 | 165 |
| 6.3 | BBP28 lipoprotein | cp32-1 | 113 |
| 6.1 | BBS38 phage exonuclease | cp32-3 | 165 |
| 6.0 | BB0681 methyl-accepting chemotaxis protein (Mcp-5) | Chromosome | 13 |
| 5.8 | BB0680 methyl-accepting chemotaxis protein (Mcp-4) | Chromosome | 13 |
| 5.5 | BBJ28 hypothetical protein | lp38 | |
| 5.4 | BBM28 Mlp lipoprotein | cp32-6 | 113 |
| 5.3 | BBQ74 conserved hypothetical protein, pseudogene | lp56 | 60 |
| 5.2 | BB0253 ATP-dependent protease LA (Lon-1) | Chromosome | 22 |
| 5.1 | BBN35 conserved hypothetical protein | cp32-9 | 165 |
| 4.8 | BBJ27 ABC transporter, permease protein | lp38 | |
| 4.7 | BB0400 hypothetical protein | Chromosome | |
| 4.5 | BBR36 phage exonuclease | cp32-4 | 165 |
| 4.5 | BB0385 basic membrane protein D (BmpD) | Chromosome | 36 |
| 4.2 | BBC11 phage integrase | cp9 | 96 |
| 3.8 | BBR28 Mlp lipoprotein | cp32-4 | 113 |
| 3.7 | BB0567 chemotaxis histidine kinase (CheA-1) | Chromosome | 134 |
| 3.7 | BBS30 Mlp lipoprotein | cp32-3 | 113 |
| 3.6 | BBB09 hypothetical protein | cp26 | |
| 3.5 | BB0776 hypothetical protein | Chromosome | |
| 3.3 | BB0782 nicotinate-nucleotide adenylyltransferase (NADD) | Chromosome | |
| 3.2 | BBL28 Mlp lipoprotein | cp32-8 | 113 |
| 3.2 | BB0671 chemotaxis operon protein (CheX) | Chromosome | |
| 3.1 | BBM29 phage Ssb binding protein (ERF) | cp32-6 | 161 |
| 3.0 | BBO28 Mlp lipoprotein | cp32-7 | 113 |
| 2.9 | BBL29 phage Ssb binding protein (ERF) | cp32-8 | 161 |
| 2.8 | BBQ37 phage Ssb binding protein (ERF) | lp56 | 161 |
| 2.7 | BBJ43 hypothetical protein | lp38 | 90 |
| 2.7 | BB0548 DNA polymerase I (PolA) | Chromosome | |
| 2.7 | BBS22 conserved hypothetical protein | cp32-3 | 142 |
| 2.6 | BB0232 HbbU protein | Chromosome | |
| 2.6 | BBJ45 hypothetical protein | lp38 | 59 |
| 2.5 | BB0279 flagellar protein (FliL) | Chromosome | |
| 2.5 | BBR29 phage Ssb binding protein (ERF) | cp32-4 | 161 |
| 2.5 | BBN28 Mlp lipoprotein | cp32-9 | 113 |
| 2.4 | BB0297 Smg protein | Chromosome | |
| 2.4 | BB0231 conserved hypothetical protein | Chromosome | 125 |
| 2.4 | BBP29 phage Ssb binding protein (ERF) | cp32-1 | 161 |
| 2.4 | BBO29 phage Ssb binding protein (ERF) | cp32-7 | 161 |
| 2.3 | BBM35 phage exonuclease | cp32-6 | 165 |
| 2.3 | BB0843 C4-dicarboxylate transporter protein | Chromosome | |
| 2.3 | BBS31 phage Ssb binding protein (ERF) | cp32-3 | 161 |
| 2.3 | BBK32 immunogenic protein P35 | lp36 | |
| 2.3 | BBA04 antigen, S2 | lp54 | 44 |
| 2.2 | BB0313 rRNA methylase (FtsJ) | Chromosome | |
| 2.2 | BBR44 DNA binding protein, putative | cp32-4 | 115 |
| 2.2 | BB0689 hypothetical protein | Chromosome | |
| 2.2 | BB0017 ABC transporter, permease | Chromosome | |
| 2.2 | BBJ31 hypothetical protein | lp38 | 59 |
| 2.1 | BB0443 SpoIIIJ-associated protein (Jag) | Chromosome | |
| 2.1 | BB0728 NADH oxidase, water-forming (Nox) | Chromosome | |
| 2.1 | BB0647 ferric uptake regulation protein (Fur) | Chromosome | |
| 2.1 | BBL38 phage integrase | cp32-8 | 96 |
TABLE 2.
Mean fold downregulation of genes on the cDNA array by use of three separate RNA isolations
| Downregulation (fold) | Gene description | Replicon | Gene family |
|---|---|---|---|
| −2.0 | BBS33 conserved hypothetical protein | cp32-3 | 57 |
| −2.0 | BBO01 hypothetical protein | cp32-7 | 146 |
| −2.0 | BB0562 hypothetical protein | Chromosome | 35 |
| −2.0 | BBG18 hypothetical protein | lp28-2 | |
| −2.0 | BB0335 ABC transporter, ATP-binding protein (OppF) | Chromosome | 4 |
| −2.0 | BBK22 conserved hypothetical protein | lp36 | 50 |
| −2.0 | BB116 hypothetical protein | lp28-4 | 60 |
| −2.0 | BBR31 conserved hypothetical protein | cp32-4 | 57 |
| −2.0 | BBB10 conserved hypothetical protein | cp26 | 62 |
| −2.1 | BBG06 conserved hypothetical protein | lp28-2 | 57 |
| −2.1 | BBQ51 hypothetical protein, authentic frameshift | lp56 | 146 |
| −2.1 | BB0367 hypothetical protein | Chromosome | |
| −2.1 | BBF10 hypothetical protein | lp28-1 | 70 |
| −2.1 | BBR34 conserved hypothetical protein | cp32-4 | 49 |
| −2.1 | BB119 conserved hypothetical protein | lp28-4 | 57 |
| −2.1 | BBS01 hypothetical protein | cp32-3 | 146 |
| −2.1 | BBH29 conserved hypothetical protein | lp28-3 | 49 |
| −2.1 | BBH14 hypothetical protein | lp28-3 | |
| −2.1 | BB107 hypothetical protein | lp28-4 | |
| −2.2 | BBH12 hypothetical protein | lp28-3 | |
| −2.2 | BBJ08 hypothetical protein | lp38 | 12 |
| −2.2 | BBF24 plasmid partition protein, putative | lp28-1 | 32 |
| −2.2 | BB0242 hypothetical protein | Chromosome | |
| −2.2 | BB0365 lipoprotein LA7 | Chromosome | |
| −2.2 | BBD18 hypothetical protein | lp17 | |
| −2.2 | BBK23 hypothetical protein | lp36 | 62 |
| −2.2 | BB0261 hypothetical protein | Chromosome | |
| −2.3 | BBO30 conserved hypothetical protein | cp32-7 | 57 |
| −2.3 | BB0396 ribosomal protein L33 (RpmG) | Chromosome | |
| −2.3 | BB0256 ribosomal protein S21 (RpsU) | Chromosome | |
| −2.3 | BBK01 hypothetical protein | lp36 | 12 |
| −2.3 | BBM33 conserved hypothetical protein | cp32-6 | 49 |
| −2.3 | BBL01 hypothetical protein | cp32-8 | 146 |
| −2.3 | BB0034 hypothetical protein | Chromosome | 48 |
| −2.3 | BB0631 hypothetical protein | Chromosome | |
| −2.3 | BBP30 conserved hypothetical protein | cp32-1 | 57 |
| −2.4 | BB0324 hypothetical protein | Chromosome | |
| −2.4 | BB0083 hypothetical protein | Chromosome | |
| −2.4 | BB0425 hypothetical protein | Chromosome | |
| −2.4 | BBH37 hypothetical protein | lp28-3 | 12 |
| −2.4 | BB0428 hypothetical protein | Chromosome | |
| −2.5 | BB0403 hypothetical protein | Chromosome | |
| −2.5 | BBA42 conserved hypothetical protein | lp54 | 150 |
| −2.6 | BBD05.1 conserved hypothetical protein, pseudogene | lp17 | 57 |
| −2.6 | BB0356 hypothetical protein | Chromosome | |
| −2.6 | BBJ19 conserved hypothetical protein | lp38 | 62 |
| −2.6 | BB0785 stage V sporulation protein G | Chromosome | |
| −2.6 | BBH09.1 conserved hypothetical protein, pseudogene | lp28-3 | 95 |
| −2.7 | BBA52 outer membrane protein | lp54 | |
| −2.8 | BBG04 hypothetical protein | lp28-2 | |
| −2.8 | BBH18.1 conserved hypothetical protein, pseudogene | lp28-3 | 65 |
| −2.8 | BBD14 conserved hypothetical protein | lp17 | 62 |
| −2.9 | BBA23 conserved hypothetical protein | lp54 | 144 |
| −2.9 | BBA74 outer membrane porin (Oms28) | lp54 | 171 |
| −2.9 | BBE30 hypothetical protein | lp25 | 49 |
| −2.9 | BBH26 hypothetical protein | lp28-3 | 62 |
| −3.0 | BB0049 hypothetical protein | Chromosome | |
| −3.2 | BBA60 surface lipoprotein P27 | lp54 | |
| −3.3 | BBF32 recombination cassettes Vls2-Vls16, authentic frameshift | lp28-1 | 170 |
| −3.3 | BBG15 hypothetical protein | lp28-2 | |
| −3.6 | BBA59 lipoprotein | lp54 | |
| −3.7 | BBJ41 antigen, P35, putative | lp38 | 54 |
| −3.8 | BB136 antigen, P35, putative | lp28-4 | 54 |
| −3.8 | BBA62 lipoprotein | lp54 | |
| −3.8 | BBH16 hypothetical protein | lp28-3 | |
| −3.8 | BBI39 hypothetical protein | lp28-4 | 54 |
| −4.0 | BBH20 outer membrane porin, authentic frameshift (Oms28) | lp28-3 | 171 |
| −4.0 | BBI13 conserved hypothetical protein | lp28-4 | |
| −4.0 | BBA63 hypothetical protein | lp54 | |
| −4.1 | BBA03 outer membrane protein | lp54 | |
| −4.1 | BBD13 hypothetical protein | lp17 | |
| −4.3 | BB0519 grpE protein (GrpE) | Chromosome | |
| −4.5 | BB103 hypothetical protein | lp28-4 | |
| −4.5 | BB0229 ribosomal protein L31 (RpmE) | Chromosome | |
| −4.6 | BB0395 preprotein translocase subunit (SecE) | Chromosome | |
| −5.9 | BB102 conserved hypothetical protein | lp28-4 | 84 |
| −6.7 | BBJ09 outer surface protein D (OspD) | lp38 | |
| −7.2 | BB0187 hypothetical protein | Chromosome | |
| −8.1 | BB0273 flagellar biosynthesis protein (FliR) | Chromosome |
B. burgdorferi B31 encodes approximately 150 putative lipoproteins (13). On our array, a total of 13 lipoproteins, seven belonging to the mlp paralogous gene family, were upregulated whereas 10 were downregulated.
Several genes described in the present study were shown to be differentially expressed in array studies by Revel et al. and Ojaimi et al. (39, 54). Some concordance would be expected, given that this study utilized a temperature shift, a variable also present in both previous studies. However, out of 154 differentially expressed ORFs in this study, only 36 (23%) were previously shown to be temperature regulated by Ojaimi et al. (39) whereas 31 (20%) were temperature and pH regulated in the experiments by Revel et al. (54). Of further note, only 14 (9%) ORFs differentially regulated in the present study were reported for both previous temperature shift array experiments. Concordance between all three studies was seen only within our top eight upregulated genes.
Blood and time in culture are important variables for gene expression. The spirochetes grown at 35°C for 2 days did not exhibit a gene expression profile that was similar to those obtained in previous temperature shift arrays in which time in culture was longer than our 48 h (39, 54). A case in point is the change in expression of ospC. While the spirochetes had not yet upregulated ospC in a 35°C culture after 48 h, the addition of blood at the same temperature had a dramatic effect on gene expression. ospC was highly upregulated, with the third highest fold increase of all the differentially expressed genes. In contrast, ospC is expressed in culture at 23°C at a very low level; after a two-day temperature shift to 35°C, its expression was not dramatically increased and it was the 107th most highly expressed gene on the array at 35°C (Table 3). However, examination of expression level data from the 35B culture demonstrated that ospC was the most expressed gene, containing more than double the transcript level of the second highest gene (Table 3). The vast upregulation and expression of ospC further shows that it is important for the spirochetes within the first 48 h of tick feeding.
TABLE 3.
List of the ten most highly expressed genes at 35B according to transcript abundance array dataa
| Culture 35B results
|
Gene description | Culture 35 results
|
||
|---|---|---|---|---|
| Rank | Expression level | Rank | Expression level | |
| 1 | 92.26 | BBB19 outer surface protein C | 106 | 2.74 |
| 2 | 43.30 | BB0355 transcription factor, putative | 3 | 42.69 |
| 3 | 36.83 | BB0232 HbbU protein | 19 | 14.09 |
| 4 | 32.94 | BB0162 hypothetical protein | 1 | 57.42 |
| 5 | 30.26 | BB0294 flagellar basal-body rod protein (FlgB) | 4 | 28.81 |
| 6 | 23.55 | BB0426 hypothetical protein | 12 | 20.78 |
| 7 | 20.59 | BB0477 ribosomal protein S10 (RpsJ) | 7 | 25.80 |
| 8 | 18.50 | BB0231 conserved hypothetical protein | 35 | 7.61 |
| 9 | 18.46 | BB0440 ribosomal protein L34 (RpmH) | 5 | 26.40 |
| 10 | 18.33 | BB0425 hypothetical protein | 2 | 44.20 |
For comparison the expression at 35 is also provided along with the gene's rank.
Despite these differences, there are some important areas of agreement in all three studies. ospC was present as one of the top five most upregulated genes in every study. Besides ospC, several family 54 genes encoded on lp54 had the highest fold induction, implicating these genes as important in transmission of the spirochetes from vector to vertebrate host. mlp paralogs were also upregulated in all three studies. Finally, none of the genes encoded on plasmids lp5 and lp21 were differentially regulated on any arrays, including ours.
Quantitative and tick RT-PCR results validate genome array results.
To validate the array results, we utilized quantitative RT-PCR analysis of a select group of genes. Primers were generated for genes that underwent significant differential expression on the array. We also randomly chose several genes which did not undergo significant changes on the array. One of the RNA isolations hybridized to the arrays was also utilized for RT-PCR, and then the results from the two techniques were compared. The quantitative data were normalized to two internal controls, flaB and 16S rRNA. The quantitative RT-PCR data was concordant with the array results, as all the genes showed patterns of expression similar to the array data patterns (Table 4). Array fold differences are usually underestimated; it is the inherent feature of the technology, likely as a result of cross-hybridization (21). Quantitative RT-PCR validation usually shows that fold differences are several magnitudes larger than indicated by array studies. Also, genes with very high or low levels of expression often have reduced correlation between the two methods. To further determine whether our array results of in vitro gene expression correlate with gene expression in spirochetes within ticks, we selected several genes differentially expressed on the DNA array and tested them for their expression in either the flat or feeding tick by RT-PCR. The results further confirmed our genomic array data (Table 5). The array experiment was able to identify genes upregulated during feeding or genes preferentially expressed in flat ticks. These experiments showed that our in vitro array experiments closely mimic the in vivo tick feeding environment.
TABLE 4.
Comparison of cDNA array and quantitative RT-PCR results for selected genesa
| Gene | Forward and reverse primer (5′-3′) | Several induction (severalfold)
|
||
|---|---|---|---|---|
| cDNA array | 16S RNA | flaB | ||
| BBA72 | GCATAAGGAGAGTGTTTTGAC and AACTTCTTGGTAGGTGCTTC | 18.2 | 15.5 | 18.6 |
| BBB19 | GAGGTTGAAGCGTTGCTGTC and TCCATTGTGATTATTTTCGGTATC | 15.11 | 51 | 61.2 |
| BBA24 | AGATAAAAAAACGGGTAGTGGG and GCTTCCTCTTCTATTGCTATTACG | 12.66 | 13.7 | 16.44 |
| BB0175 | CGTATGAGGATTCTGATGATGC and CCTTTTTGTTTACCTTGTCTCCC | 13.7 | 1.79 | 2.15 |
| BBJ46 | TCTTTACTATTGGGGCTTTGTG and TTGTAGACTGTTGTTTATGGATTTG | 6.62 | 6.33 | 7.59 |
| BBJ25 | CCAAAATGGGTCAAGACAGTC and GCGGACAATACTAAAAATAAATCAG | 5.25 | 34.97 | 41.96 |
| BB0565 | ATAATAGGGGCAAAATCGTTCC and CCAATCTTAGGAGCATCATCAATAC | 5.47 | 1.79 | 2.15 |
| BBC05 | GAAAACCCTAAGGATGAACTTGC and TAGAACTTGTTGCTCCCCCAC | 4.86 | 16.53 | 19.84 |
| BBM35 | CTGGAGTTGAGTTTGATTGG and GGCTTTTCTCTTTTTTTATTTTCG | 1.58 | 1.11 | 1.33 |
| BB0647 | AACTTGGAGAAAACTGGGAAAC and TGGGACATTATTGTTATTATGGG | 1.42 | −1.26 | 1 |
| BB0147 | GCTTCTGATGATGCTGCTGG and ATGTGCCGTTACCTGATTGAACTGCC | 1.29 | 1.2 | 1 |
| BB0377 | CATACAAAACTCAACCCTGGC and CGGTCCAAACTTCATTATTTCTAAG | 1.17 | 1.46 | 1.75 |
| BB0690 | TTGCCCTCAATGGAAAGC and ATCGTGACAAAAACAATCGC | −1.17 | −2.26 | −1.88 |
| BBA15 | AAGAGCAGACGGAACCAGACTTG and AACCACCAATGTTGTTTTTTCAGC | −1.29 | −1.53 | −1.27 |
| BBA74 | GCTGTTTCTGTTGCTGGTGAAG and AACTTTTTGAGCCTCTTGAACTG | −1.44 | −2.43 | −2.02 |
| BBD18 | GGGGGCATAAAAGGAAC and CACCGGTTTGCATATTAAC | −1.28 | −12.68 | −10.56 |
| BBH37 | CGTAGAATCTGCGGTGTCTTTAG and CCTCAGTAGAAGGGAAAAAATCCTC | −1.65 | −6.43 | −5.35 |
| BBI39 | TGAAAAAGAAGCCGAAAAGTTG and GTAAGCGTCAATGGTTGCG | −2.52 | −6.74 | −5.61 |
| BBA62 | TATTATTTGTTGCTTGCGAAACTAC and GCTTCATTGATTTGGTATTTTTGTC | −3.31 | −10 | −8.33 |
| BBJ09 | GGCAAATAAAGTTGTAGAAGCG and TTTCAGCAGAATCAGAATAGTCAG | −3.55 | −18.58 | −15.48 |
Forward and reverse primers for each of the selected genes are provided, each with its respective fold changes. The quantitative RT-PCR assay results were normalized to 16S rRNA and flaB, utilized as internal controls.
TABLE 5.
RT-PCR results of expression assays of select B. burgdorferi genes from flat and feeding ticks
| Gene | RT-PCRa
|
cDNA array induction (severalfold) | |
|---|---|---|---|
| Flat tricks | Feeding ticks | ||
| BB0647 | − | + | 2.08 |
| BB0690 | + | − | −1.57 |
| BBA15 | + | + | −1.73 |
| BBA64 | − | + | 16.68 |
| BBA66 | − | + | 31.22 |
| BBA72 | − | + | 45.06 |
| BBA74 | + | + | −2.93 |
| BBA62 | + | − | −3.80 |
| BBD18 | + | − | −2.22 |
| BBB19 | − | + | 33.60 |
| BBJ25 | − | + | 12.33 |
| BBH37 | + | + | −2.41 |
A plus sign indicates an amplified transcripts, while a minus sign indicates lack of amplified transcript. Each gene's severalfold up- or downregulation on the cDNA array is shown on the right for comparison. Note that a downregulated gene on the array may be expressed in both flat and feeding ticks. It's likely that such a gene is preferentially expressed in the flat tricks, but the transcript can still be detected in feeding ticks, as spirochete populations within feeding ticks are heterogeneous.
lp28 genes are downregulated in blood cultures.
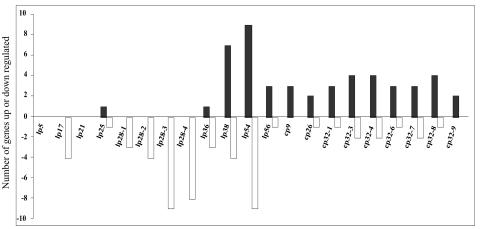
Strain B31 MI contains four lp28 plasmids designated lp28-1, lp28-2, lp28-3, and lp28-4. A total of 24 genes from all lp28 plasmids were downregulated (Fig. 4). These results strongly suggest that the downregulated genes from lp28 plasmids are required for maintenance of spirochetes in flat ticks. That three of the plasmids, lp28-2, -3, and -4, had no effect on dissemination of B. burgdorferi within mice would also support a role for these genes in the flat tick (31, 50). However, lp28-1 appears to be important in vertebrate infection, as the loss of this plasmid leads to loss of infectivity in mice (31, 50). One suggested reason why lp28-1 may be essential to infectivity is that it encodes the vlsE locus, which undergoes recombination during mammalian infection and may aid in immune evasion (69, 70).
FIG. 4.
Differential (more than twofold) expression from all 21 circular (cp) and linear (lp) plasmids of B. burgdorferi on the cDNA array. The y axis represents the number of genes up- or downregulated in spirochetes cultured with blood compared to the results for spirochetes cultured without blood. Black bars indicate the numbers of genes that were upregulated; white bars indicate the numbers of genes that were downregulated.
All but one of the downregulated lp28 genes are undescribed hypothetical proteins. The one exception is BBI16, designated VraA antigen, which encodes a lipoprotein with 21 consecutive 9-amino-acid repeat sequences near its amino terminus (30). Database searches did not provide any hints for possible functions for any of the downregulated lp28 genes. Along these lines, four genes from lp17 were also downregulated and none were upregulated, again suggesting a role for this plasmid in flat ticks similar to that of lp28 genes.
Differential expression of genes on lp54 plasmid.
The lp54 plasmid, with nine upregulated and nine downregulated genes, had the highest number of differentially expressed genes of all the plasmids (Fig. 4). These results underscore the importance of this plasmid in the life cycle of B. burgdorferi. In fact, six of the eight genes with the largest fold increase on the cDNA array were encoded on lp54. Several of these highly upregulated genes belong to paralogous gene family 54. Family 54 contains 14 members, 8 encoded at one end of the lp54 plasmid. Five genes in this family, BBA64, BBA65, BBA66, BBA71, and BBA73, were upregulated more than 10-fold on our array (Fig. 5). All but BBA71 encode putative lipoproteins. We tested for the expression of BBA64 and BBA66 in ticks, and both were expressed in feeding ticks only (Table 5). Since the genes of this family represented some of the most active genes on our array, it is likely that they are functional in the passage of Borrelia from tick to vertebrate.
FIG. 5.
Family 54 genes differentially expressed on the cDNA array. All genes except BBA71 encode putative lipoproteins. Upregulated genes are shown in red; downregulated genes are shown in green. The gray rectangle represents a 24-bp inverted repeat located 40 bp downstream from the downregulated genes.
Five other genes in this family—BBA68, BBA69, BBI36, BBI39, and BBJ41—were downregulated (Fig. 5). BBI36, BBI39, and BBJ41 were all downregulated >3.5-fold, while BBA68 and BBA69 were both downregulated 1.8-fold. Since genes in family 54 undergo both up- and downregulation, our data agrees with the hypothesis that despite some sequence homology, the genes in this family are under the control of different regulatory mechanisms (39). Analysis of upstream and downstream regions of the downregulated family 54 genes revealed the presence of a 24-bp inverted repeat sequence, AAAGAGAAGAATTAACTTCTCTTT, which may act as a transcriptional terminator for this group of genes. This sequence is located exactly 40 bases downstream from the end of BBI36, BBI39, and BBJ41 as well as BBI38, a paralog of BBI36 that was not expressed on our array. The sequence is also located 41 bp downstream of BBA68. The repeat sequence is found only in only one other region of the genome, approximately 1 kb upstream of BBA69. It has been proposed that these five genes, along with BBI38, encode lipoproteins despite a variation in the consensus signal peptidase II sequence (13). These genes all contain an isoleucine at −1 and a threonine at −3 relative to the cysteine residue, while the rest of the family 54 genes have different residues at these positions or lack the cysteine altogether (www.tigr.org). Our findings show that the downregulated genes of family 54 all share several common characteristics which may lead to their similar regulatory patterns.
BBA74, which encodes a porin designated Oms28, was downregulated more than threefold (59). This ORF was also downregulated in both vertebrate host-adapted arrays but was shown to be upregulated in both temperature shift experiments (8, 39, 54). We found that BBA74 was expressed in flat ticks and undergoes downregulation during feeding. The downregulation of this gene in our array and in the experiments of Brooks et al. (8) suggests that some mammalian factors present in the incoming blood meal may drive the repression of BBA74. Without these factors, temperature alone seems to increase its transcription. This is the first direct evidence of a protein described as a porin in B. burgdorferi that is preferentially expressed in flat ticks and that may be instrumental in acquisition of small molecules while the spirochetes are in the nutrient-poor environment of the tick midgut.
Three lp54-encoded lipoproteins, BBA59, BBA60, and BBA62, were downregulated (at least threefold), implicating these genes as being preferentially expressed in the flat tick and downregulated during feeding. BBA62 encodes lp6.6, which has previously been shown to be downregulated during mammalian infection (32, 66). Finally, ospA and ospB were downregulated in our experiments but the decreases in expression were −1.73 and −1.75, respectively.
Upregulation of lp38-encoded ABC transporter system.
A total of 11 genes on lp38 were differentially expressed in our array (Fig. 4). Of particular interest was a cluster of three upregulated genes on lp38, BBJ25, BBJ27, and BBJ28 (12.3-, 4.8-, and 5.5-fold upregulation, respectively) (Table 1). BBJ27 is predicted to be in an operon with BBJ26. BBJ26, which was also upregulated in our array but did not pass our criteria for filtering genes, has been identified as an ATP-binding ABC transporter, the substrate of which has not yet been identified. ABC (ATP binding cassette) transporters are active transport systems. The transporters have a common global organization with three types of components: two integral membrane permease proteins, two peripheral membrane proteins that bind and hydrolyze ATP, and a periplasmic substrate binding protein. The genes for all three components frequently form an operon (64). BBJ27 contains four predicted transmembrane regions and shares homology with many other ABC transporter permeases. Both BBJ25 and BBJ28 have predicted signal peptidase cleavage sites, indicating that they may be secreted to the periplasm, where one or both may function in substrate binding. Besides this lp38-encoded system, all but two of the Borrelia ABC transporter systems are encoded on the chromosome. Notable exceptions are two known plasmid-encoded oligopeptide binding proteins. The lp38 ABC transporter system, however, is the only known B. burgdorferi system in which the genes encoding the ATP binding protein and the permease are next to each other on a plasmid. Only one other gene identified as having potential ABC transporter functions was upregulated in our array. BB0017, upregulated 2.2-fold, encodes a putative permease, but unlike other ABC transporter operons, no ATP binding protein is encoded nearby.
As shown on the basis of our DNA array data, this transporter system is not expressed or is expressed at a very low level in flat ticks and is upregulated during tick feeding. Moreover, RT-PCRs showed that BBJ25 was expressed only in feeding ticks (Table 5). Once upregulated during feeding, the system likely remains expressed throughout mammalian infection, as Brooks et al. found BBJ26 and BBJ27 to be upregulated in dialysis chambers (8). Therefore, this may be the only known transporter system utilized by Borrelia specifically during the transmission and infection process.
Differential expression of cp32 paralogous gene families.
Cp32 plasmids are members of a group of circular plasmids found in B. burgdorferi. Strain B31 MI may contain up to seven of these plasmids designated cp32-1, cp32-3, cp32-4, cp32-6, cp32-7, cp32-8, and cp32-9 (13). The cp32 plasmids are very similar to one another, and paralogs of cp32 genes are also found on cp9 plasmids and lp54 and lp56, which contain insertions of cp32. Cp32 plasmids have been implicated as prophages (13, 16, 18, 19). In our array experiments a total of 34 genes were upregulated on cp32 plasmids, all but 2 belonging to four paralogous gene families (Fig. 6). Seven paralogs of family 113 were upregulated over twofold. Family 113 encodes Mlps, which are lipoproteins that have previously been shown to undergo upregulation due to increase in temperature and/or decrease in pH and are expressed in feeding ticks but not flat ticks (25, 27, 67).
FIG. 6.
Schematic of a cp32 region which underwent significant differential expression on the cDNA array. The region depicted is from cp32-1; a similar expression pattern was observed for several cp32 paralogs. Arrows indicate whether a gene underwent upregulation or downregulation. DR stands for direct repeat regions located in front of family 161 and 165 ORFs.
Nine paralogs of family 165 and eight paralogs of family 161 were upregulated. Casjens et al. indicated that an upstream region of genes encoding members of family 161 and 165 contained identical inverted repeat sequences of approximately 180 bp (13). It was hypothesized that these genes may undergo common regulation, and our results are in agreement. Genes in family 161 encode a protein similar to ERF single-stranded binding proteins encoded by bacteriophages. Furthermore, family 165 members have been implicated as putative lambda exonuclease homologs, which are enzymes involved in recombination and repair in bacteriophages (4). Finally, two homologs of family 96 which encode a putative phage integrase were also upregulated. These results seem to indicate that an increase in temperature combined with some mammalian factors may serve as a signal to initiate a stress response leading to the upregulation of phage recombination proteins in B. burgdorferi. Furthermore, BB0548, a chromosome-encoded DNA polymerase I, was upregulated 2.71-fold. DNA polymerase I is involved in repair and recombination, and no other polymerases were differentially expressed in our experiments. What role the phage recombination machinery may serve during the infection process remains to be elucidated. The upregulation of these genes may be a transient part of a stress response of the phage, occurring early following exposure to blood and temperature. Alternatively, it may have more pronounced functions in the overall spirochete life cycle, perhaps generating genetic variability on cp32, other plasmids, and perhaps the chromosome itself.
Of the downregulated cp32 genes and their paralogs, nearly all were grouped to five families. Four of these are families 32, 49, 50, and 57. These families encode genes in a five-member locus which has been implicated on the basis of homology as functioning in plasmid maintenance and/or partitioning (Fig. 6). Members of family 32 specifically are homologous to the parA gene of Escherichia coli bacteriophage P1. ParA is an ATPase belonging to a large family of genes implicated in bacterial plasmid partitioning proteins (13, 61). On other B. burgdorferi plasmids, family 62 genes replace family 57 genes within the locus; similarly, four homologs of family 62 were downregulated more than twofold. Therefore, the downregulation of the putative maintenance locus was occurring on all the plasmids, not just cp32.
Four paralogs of family 146 were downregulated. This gene has been implicated as a bacteriophage portal protein, which allows the phage to be packaged prior to release from the cell. The downregulation of this protein would seem to indicate that the environment acts against phage release. In support of this idea, and as also noted by Ojaimi et al., very few genes found in the putative “late phage operon” (between BBP01 and BBP26 on cp32-1) were differentially expressed or even expressed at all. Ojaimi et al. (39) also noted upregulation of blyA and blyB, the putative phage holins, which we did not observe in our experiments. In fact, both failed to meet our criteria for expressed genes on our array, indicating they were either expressed at a very low level or perhaps not expressed at all.
Chromosomal genes.
BB0175 was the chromosomal gene most highly upregulated under our array conditions as well as under conditions of temperature induction (39). This gene encodes a conserved protein with an undescribed function and contains a region of homology to the von Willebrand factor A (vWA) domain. This domain is associated in both prokaryotes and eukaryotes with cell-to-cell interactions, often via integrin receptors. BB0175, however, has an incomplete vWA domain, lacks two of the five conserved residues of the ligand binding Midas motif, and does not appear to be a secreted protein (www.tigr.org). One of two B. burgdorferi lon ATP proteases, BB0253, was upregulated over fivefold. In bacteria, the Lon protease functions in degradation of abnormal proteins and has also been shown to be responsible for the regulation of various pathways in many bacteria by its degradation of regulatory proteins. Recent reports focusing on Lon in Salmonella spp. showed that it can function as a regulator of genes responsible for cell invasion (62, 63). In Pseudomonas syringae, it functions as a negative regulator of type III protein secretion systems (7). Since the B. burgdorferi lon upregulation occurs at a time of vast changes in the expression of many genes, it is possible that the protease may serve similar regulatory functions in Borrelia and to degrade proteins such as OspA/B that disappear from the organisms in the midguts of the feeding ticks.
Two known regulatory proteins were upregulated in our experiment. BB0771 encodes an rpoS sigma factor. RpoS is upregulated by temperature and controls the expression of both OspC and DbpA and likely other B. burgdorferi antigens through sigma factor rpoN (28). On our array rpoS was upregulated 9.5-fold, while ospC and dbpA were upregulated 33.6- and 19.7-fold, respectively. Another upregulated regulator was BB0647, the B. burgdorferi fur homolog. Members of this family of proteins function as transcriptional regulators in response to metals. fur was upregulated 2.1-fold in our array and was expressed in feeding ticks (Table 5). It is possible that upregulation of fur occurred due to the influx of metals in the incoming blood meal. A recent report implicated fur in acting as a transcriptional activator of NapA (BB0690), a putative DNA binding protein functioning in oxidative stress response (6). Our data does not support this hypothesis, as we found that napA was downregulated on the array and was expressed in flat ticks. Our results would argue that Fur acts as a repressor of NapA.
HbbU, a B. burgdorferi HU/IHF protein family homolog, was upregulated 2.6-fold in our array. HbbU protein has been shown to bind DNA, induce a dramatic bend, and restrain negative supercoils upon binding (29). A recent study described how temperature and supercoiling affects the transcription of ospA/B as well as ospC (1). Borrelia DNA was found to be more negatively supercoiled at 23°C than in cultures grown at 35°C. Following relaxation of negative supercoils, ospA/B expression and promoter activity decreased while ospC levels increased. This implies that during tick feeding, Hbb may act in transcriptional regulation by relaxing negative supercoils, resulting in the well-described changes in ospA/B and ospC expression during feeding.
BB0782 encoding a nicotinate-nucleotide adenylyltransferase was upregulated 3.3-fold. This enzyme catalyzes the adenylation of nicotinate mononucleotide (NaMN) to nicotinic acid adenine dinucleotide (NaAD), which is then directly processed to NAD by NAD synthetase (Swiss-Prot). BB0728 was upregulated 2.1-fold, and it encodes an NADH oxidase. Its upregulation may be due to the increase in oxygen. During tick feeding, the spirochetes in the tick midgut are exposed to the oxygen in the incoming blood meal. Borrelia may upregulate NADH oxidase to allow it to deal with this increase in oxygen levels which may be toxic to the cells. A similar function has been shown in the spirochete Brachyspira, in which NADH oxidase is utilized to protect the bacteria from oxygen toxicity and therefore contributes to its pathogenicity (60).
Of the downregulated ORFs, several encode previously characterized proteins. BB0034, downregulated 2.3-fold, encodes a protein designated P13. This protein is a 13-kDa porin which is posttranslationally processed at both ends and modified by an unknown mechanism (35, 41). BB0365, encoding LA7, a lipoprotein, was downregulated 2.2-fold. This ORF was upregulated in both previous temperature shift array studies. It has been shown, however, that in similarity to results seen with OspA/B, few patients generate antibodies to this protein in early Lyme disease, lending further evidence that this gene is likely downregulated in feeding ticks (33, 53). Three genes coding for ribosomal proteins RpmG, RpsU, and RpmE were downregulated. In Anabaena and Synechocystis cyanobacteria, as well as in the soil bacterium Sinorhizobium meliloti, rpsU is preferentially expressed at colder temperatures; this gene has been suggested to be associated with adaptation to changes in the environment (36, 55, 56).
Chemotaxis-motility genes.
Several ORFs functioning in chemotaxis and motility were upregulated in our experiments. In a typical bacterial chemotaxis pathway, MCPs (methyl-accepting chemotaxis proteins) are the transmembrane chemoreceptors that regulate the autokinase activity of the histidine kinase CheA. The coupling protein CheW stabilizes the complex formed between CheA and the receptor. CheY, a response regulator, is activated by phosphorylation by CheA and then regulates flagellar activity. Adaptation enzymes CheB and CheR covalently modify specific adaptation sites on the receptor cytoplasmic domains (22, 44).
B. burgdorferi contains multiple copies of chemotaxis sensory transduction genes, encoded primarily in two operons. It has been suggested that the spirochete may preferentially express its chemotaxis genes in different habitats (14). We observed that the addition of blood had a major effect on movement, with the spirochetes exhibiting a great increase in motility. Not surprisingly, several chemotaxis genes were upregulated on our array. Two out of five MCPs encoded by B. burgdorferi were upregulated 6- and 5.8-fold, respectively; the two genes, designated mcp-4 and mcp-5, are encoded on a probable operon. cheA-1 was upregulated 3.7-fold, while cheA-2 was upregulated only 1.6-fold. cheW-2, one of the three B. burgdorferi copies of cheW, was upregulated 8.3-fold. cheX, another gene associated with chemotaxis, was upregulated 3.7-fold. No induction of cheY or cheB was observed. Our results are in full agreement with the data of Revel et al. (54), as the same chemotaxis genes were upregulated in both experiments. Since no upregulation of chemotaxis genes was reported by Ojaimi et al. (39), it's likely that lower pH or mammalian blood factors may drive the upregulation of these genes in B. burgdorferi.
Summary.
Spirochetes in the midgut of flat (unfed) ticks are metabolically active but very likely at a very low rate. Thus, genes that are expressed during this phase of the spirochete life cycle would be expected to encode proteins that are associated with the maintenance of the most elemental functions for survival in a nutrient-depleted, possibly anoxic environment with a potential wide range of temperatures. A number of genes from all four copies of lp28 are involved in maintenance of spirochetes in the midgut of flat ticks, as these genes are unanimously downregulated with the influx of blood. The same is true for some genes in lp54, including the now well-known downregulation of ospA and ospB. Without knowledge of the function of most of these plasmid genes, it is possible to speculate that genes expressed in the flat midgut may be engaged in specialized transport of simple molecules and in survival at different temperatures. In fact, in flat ticks, the lumen of the midgut is poor in residual nutrients, as nutrient storage is the function of the cells of the midgut epithelium. These cells store macromolecules (for example, undigested hemoglobin and albumin and other macromolecules) in endosomes, essentially leaving the lumen devoid of nutrients.
Although the temperature of the midgut and that of the tick itself rises upon blood feeding, this is not the only time when ticks undergo temperature shifts. Questing ticks have a constantly changing temperature that can fluctuate by several degrees within a 24-h period, at various locations within the tick microclimate, and at different seasons in their 2-year life cycle. Therefore, temperature-regulated genes would have to express differentially often and not just at the time of blood feeding. Under poor nutrient conditions, this would be unnecessarily stressful for the organisms unless accompanied, as in the blood meal, by an influx of nutrients. Some of these gene products need to change not only as a result of blood influx and its increase in temperature but also to protect the spirochete from the temperature fluctuations of the flat tick. Genes that are downregulated as the organisms come in contact with blood must be specialized enough to maintain the minimum requirements for viability.
The transformation of spirochetes as blood begins to fill the midgut and its diverticula is fueled by changes in gene expression, and these induce the marked changes in the cell cycle as the spirochetes begin to divide, the increase in motility, and in the visible changes in morphology. These changes coincide with the upregulation of the chemotaxis and sensing regulons, of the lp38-encoded ABC transporter, of proteases capable of remodeling the outer surface of the spirochetes, and of the recombination genes of cp32 as a transient or initial part of the stress response of the phage. These are all functions or occurrences that cause or facilitate the changes that spirochetes undergo following a blood meal in the tick. Most importantly, the highly active genes of unknown function that we have detected in this study may hold a lot of the answers to vector-host interactions.
In summary, we have demonstrated that the addition of blood has a dramatic effect on gene expression of B. burgdorferi. By verifying our results by tick RT-PCR, we also showed that our in vitro study was a close mimic of a feeding tick environment. When the results of our study are analyzed in combination with earlier array studies, we can obtain the picture of which genes the pathogen preferentially expresses in either the vertebrate host or the vector and may further our understanding of B. burgdorferi pathogenesis.
Acknowledgments
We thank Ira Schwartz and Caroline Ojaimi (New York Medical College) for assistance with Array Vision software and for all their efforts in the construction of the membranes used in this study. We are grateful to Darrin Akins and Chad Brooks (University of Oklahoma) for their assistance with the data analysis. We also thank Patricia Rosa (Rocky Mountain Laboratory) for the B31 MI strain.
This study was funded by a grant (AI-27044) from the National Institutes of Health.
Editor: D. L. Burns
REFERENCES
- 1.Alverson, J., S. F. Bundle, C. D. Sohaskey, M. C. Lybecker, and D. S. Samuels. 2003. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol. Microbiol. 48:1665-1677. [DOI] [PubMed] [Google Scholar]
- 2.Anderton, J. M., R. Tokarz, C. D. Thill, C. J. Kuhlow, C. S. Brooks, D. R. Akins, L. I. Katona, and J. L. Benach. 2004. Whole-genome DNA array analysis of the response of Borrelia burgdorferi to a bactericidal monoclonal antibody. Infect. Immun. 72:2035-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anguita, J., M. N. Hedrick, and E. Fikrig. 2003. Adaptation of Borrelia burgdorferi in the tick and the mammalian host. FEMS Microbiol. Rev. 27:493-504. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benach, J. L., E. M. Bosler, J. P. Hanrahan, J. L. Coleman, G. S. Habicht, T. F. Bast, D. J. Cameron, J. L. Ziegler, A. G. Barbour, W. Burgdorfer, R. Edelman, and R. A. Kaslow. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308:740-742. [DOI] [PubMed] [Google Scholar]
- 6.Boylan, J. A., J. E. Posey, and F. C. Gherardini. 2003. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc. Natl. Acad. Sci. USA 100:11684-11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bretz, J., L. Losada, K. Lisboa, and S. W. Hutcheson. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45:397-409. [DOI] [PubMed] [Google Scholar]
- 8.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 10.Burgdorfer, W., S. F. Hayes, and J. L. Benach. 1988. Development of Borrelia burgdorferi in ixodid tick vectors. Ann. N. Y. Acad. Sci. 539:172-179. [DOI] [PubMed] [Google Scholar]
- 11.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect Immun. 68:6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 14.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47-73. [DOI] [PubMed] [Google Scholar]
- 15.Coleman, J. L., J. A. Gebbia, J. Piesman, J. L. Degen, T. H. Bugge, and J. L. Benach. 1997. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89:1111-1119. [DOI] [PubMed] [Google Scholar]
- 16.Damman, C. J., C. H. Eggers, D. S. Samuels, and D. B. Oliver. 2000. Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J. Bacteriol. 182:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Silva, A. M., and E. Fikrig. 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am. J. Trop. Med. Hyg. 53:397-404. [DOI] [PubMed] [Google Scholar]
- 18.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 2:365-373. [PubMed] [Google Scholar]
- 19.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etienne, W., M. H. Meyer, J. Peppers, and R. A. Meyer, Jr. 2004. Comparison of mRNA gene expression by RT-PCR and DNA microarray. BioTechniques 36:618-620, 622, 624-626. [DOI] [PubMed] [Google Scholar]
- 22.Falke, J. J. 2002. Cooperativity between bacterial chemotaxis receptors. Proc. Natl. Acad. Sci. USA 99:6530-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fingerle, V., S. Rauser, B. Hammer, O. Kahl, C. Heimerl, U. Schulte-Spechtel, L. Gern, and B. Wilske. 2002. Dynamics of dissemination and outer surface protein expression of different European Borrelia burgdorferi sensu lato strains in artificially infected Ixodes ricinus nymphs. J. Clin. Microbiol. 40:1456-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 26.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 101:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobryn, K., D. Z. Naigamwalla, and G. Chaconas. 2000. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 37:145-155. [DOI] [PubMed] [Google Scholar]
- 30.Labandeira-Rey, M., E. A. Baker, and J. T. Skare. 2001. VraA (BBI16) protein of Borrelia burgdorferi is a surface-exposed antigen with a repetitive motif that confers partial protection against experimental Lyme borreliosis. Infect Immun. 69:1409-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahdenne, P., S. F. Porcella, K. E. Hagman, D. R. Akins, T. G. Popova, D. L. Cox, L. I. Katona, J. D. Radolf, and M. V. Norgard. 1997. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect. Immun. 65:412-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam, T. T., T. P. Nguyen, E. Fikrig, and R. A. Flavell. 1994. A chromosomal Borrelia burgdorferi gene encodes a 22-kilodalton lipoprotein, P22, that is serologically recognized in Lyme disease. J. Clin. Microbiol. 32:876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narasimhan, S., F. Santiago, R. A. Koski, B. Brei, J. F. Anderson, D. Fish, and E. Fikrig. 2002. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J. Bacteriol. 184:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noppa, L., Y. Ostberg, M. Lavrinovicha, and S. Bergstrom. 2001. P13, an integral membrane protein of Borrelia burgdorferi, is C-terminally processed and contains surface-exposed domains. Infect. Immun. 69:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connell, K. P., and M. F. Thomashow. 2000. Transcriptional organization and regulation of a polycistronic cold shock operon in Sinorhizobium meliloti RM1021 encoding homologs of the Escherichia coli major cold shock gene cspA and ribosomal protein gene rpsU. Appl. Environ. Microbiol. 66:392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohnishi, J., J. Piesman, and A. M. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. USA 98:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katonah, J. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358:165-177. [DOI] [PubMed] [Google Scholar]
- 39.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orloski, K. A., E. B. Hayes, G. L. Campbell, and D. T. Dennis. 2000. Surveillance for Lyme disease-United States, 1992-1998. Morb. Mortal. Wkly. Rep. 49:1-11. [PubMed] [Google Scholar]
- 41.Ostberg, Y., M. Pinne, R. Benz, P. Rosa, and S. Bergstrom. 2002. Elimination of channel-forming activity by insertional inactivation of the p13 gene in Borrelia burgdorferi. J. Bacteriol. 184:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkinson, J. S. 2003. Bacterial chemotaxis: a new player in response regulator dephosphorylation. J. Bacteriol. 185:1492-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piesman, J. 1995. Dispersal of the Lyme disease spirochete Borrelia burgdorferi to salivary glands of feeding nymphal Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 32:519-521. [DOI] [PubMed] [Google Scholar]
- 46.Piesman, J. 1993. Dynamics of Borrelia burgdorferi transmission by nymphal Ixodes dammini ticks. J. Infect. Dis. 167:1082-1085. [DOI] [PubMed] [Google Scholar]
- 47.Piesman, J., G. O. Maupin, E. G. Campos, and C. M. Happ. 1991. Duration of adult female Ixodes dammini attachment and transmission of Borrelia burgdorferi, with description of a needle aspiration isolation method. J. Infect. Dis. 163:895-897. [DOI] [PubMed] [Google Scholar]
- 48.Piesman, J., J. R. Oliver, and R. J. Sinsky. 1990. Growth kinetics of the Lyme disease spirochete (Borrelia burgdorferi) in vector ticks (Ixodes dammini). Am. J. Trop. Med. Hyg. 42:352-357. [DOI] [PubMed] [Google Scholar]
- 49.Piesman, J., B. S. Schneider, and N. S. Zeidner. 2001. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J. Clin. Microbiol. 39:4145-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramamoorthy, R., and M. T. Philipp. 1998. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect. Immun. 66:5119-5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramamoorthy, R., and D. Scholl-Meeker. 2001. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect. Immun. 69:2739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rauer, S., R. Wallich, and U. Neubert. 2001. Recombinant low-molecular-mass proteins pG and LA7 from Borrelia burgdorferi reveal low diagnostic sensitivity in an enzyme-linked immunosorbent assay. J. Clin. Microbiol. 39:2039-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato, N. 1994. A cold-regulated cyanobacterial gene cluster encodes RNA-binding protein and ribosomal protein S21. Plant Mol. Biol. 24:819-823. [DOI] [PubMed] [Google Scholar]
- 56.Sato, N., T. Tachikawa, A. Wada, and A. Tanaka. 1997. Temperature-dependent regulation of the ribosomal small-subunit protein S21 in the cyanobacterium Anabaena variabilis M3. J. Bacteriol. 179:7063-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skare, J. T., C. I. Champion, T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1996. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J. Bacteriol. 178:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanton, T. B., E. L. Rosey, M. J. Kennedy, N. S. Jensen, and B. T. Bosworth. 1999. Isolation, oxygen sensitivity, and virulence of NADH oxidase mutants of the anaerobic spirochete Brachyspira (Serpulina) hyodysenteriae, etiologic agent of swine dysentery. Appl. Environ. Microbiol. 65:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stevenson, B., W. R. Zuckert, and D. R. Akins. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol. 2:411-422. [PubMed] [Google Scholar]
- 62.Takaya, A., M. Suzuki, H. Matsui, T. Tomoyasu, H. Sashinami, A. Nakane, and T. Yamamoto. 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar Typhimurium infection of mice. Infect. Immun. 71:690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomii, K., and M. Kanehisa. 1998. A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 8:1048-1059. [DOI] [PubMed] [Google Scholar]
- 65.Wheeler, C. M., J. L. Coleman, G. S. Habicht, and J. L. Benach. 1989. Adult Ixodes dammini on rabbits: development of acute inflammation in the skin and immune responses to salivary gland, midgut, and spirochetal components. J. Infect. Dis. 159:265-273. [DOI] [PubMed] [Google Scholar]
- 66.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 67.Yang, X. F., A. Hubner, T. G. Popova, K. E. Hagman, and M. V. Norgard. 2003. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect. Immun. 71:5012-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, J. R., and S. J. Norris. 1998. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect. Immun. 66:3689-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong, J., and A. G. Barbour. 2004. Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol. Microbiol. 51:729-748. [DOI] [PubMed] [Google Scholar]