Abstract
The 54-kDa extracellular metalloprotease ZapA is an important virulence factor of uropathogenic Proteus mirabilis. While ZapA has the ability to degrade host immunoglobulins (Igs), the dramatic attenuation of virulence in ZapA mutants suggests that this enzyme may have a broader spectrum of activity. This hypothesis was tested by in vitro assays with purified ZapA and an array of purified protein or peptide substrates. The data reveal that many proteins found in the urinary tract are substrates of ZapA proteolysis, including complement (C1q and C3), cell matrix (collagen, fibronectin, and laminin), and cytoskeletal proteins (actin and tubulin). Proteolysis of IgA and IgG was significantly enhanced by conditions that denatured the Igs. It was discovered that the antimicrobial peptides human β-defensin 1 (hBD1) and LL-37 are readily cleaved by the enzyme. To the best of our knowledge, this is the first report of a bacterial protease capable of cleaving hBD1, a component of the human renal tubule innate immune response. Proteolysis of hBD1 resulted in ca. six peptides, while proteolysis of LL-37 resulted in at least nine products. Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis of the molecular masses of the reaction products indicated that ZapA preferred no distinct peptide bond. The antimicrobial activity of hBD1 and LL-37 was significantly reduced following ZapA treatment, suggesting that proteolysis results in inactivation of these peptides. The data suggest that a function of ZapA during urinary tract infections is the proteolysis of antimicrobial peptides associated with the innate immune response.
Urinary tract infections (UTIs) are among the most common bacterial infections of any organ system, accounting for an estimated 8 million visits to physicians each year (19). Of the bacteria responsible for causing uncomplicated cystitis or acute pyelonephritis, Proteus mirabilis represents only a small percentage of cases. The importance of this bacterium, however, is underscored when the incidences of P. mirabilis infection in patients with complicated urinary tracts (those with functional or anatomic abnormalities or with chronic instrumentation) are examined. In these cases, P. mirabilis is a more frequent uropathogen and its infection may be more severe due to reoccurring infection and the production of urinary stones in the kidney(s) (8).
In developing and maintaining a UTI, P. mirabilis competes against the primary bladder defenses, which include physical barriers, such as bulk flow of urine, which rinses away nonattached or weakly attached microbes, as well as low pH and high concentration of salts, urea, and organic acids. If the bacteria bypass these defenses, a second line of host response, innate host defense, becomes involved. Innate immunity refers to antigen-nonspecific defense mechanisms that a host uses immediately or within several hours after exposure to an antigen. It includes the physical barriers, as well as an immune response that involves (among many others) the production of antimicrobial peptides.
There are about 500 known antimicrobial peptides (29), and at least two of them, human β-defensin 1 (hBD1) and hBD2, are active in the human urinary tract (5, 9). These two peptides are both members of the defensins, which is a family of 3- to 5-kDa, three-disulfide, cationic, β-sheet antimicrobial peptides that engage in host defense by directly killing microbes due to their ability to disrupt microbial membranes (10, 16). β-Defensins have up to 45 residues, a unique cysteine pairing (C1-C5, C2-C4, and C3-C6), and several lysines and arginines (16). Within the kidney, hBD1 is produced in the distal tubules, the loops of Henle, and the collecting ducts. Human urine contains 10 to 100 μg of hBD1 per liter, and the release of hBD1 rises about threefold during pyelonephritis (26). These innate immune response antimicrobial peptides offer the host an immediate, although nonspecific, defense against invading bacteria. They act in concert with specific host defenses in the form of the humoral-response-mediated immunoglobulins (Igs), IgA and IgG, which are time dependent in their formation (19).
P. mirabilis has multiple mechanisms to evade host defenses, including the extracellular metalloprotease ZapA (27, 28), also referred to as an IgA protease (24, 25). ZapA is expressed in many strains of P. mirabilis (24). ZapA expression is coordinately regulated with swarmer cell differentiation and swarming behavior (27), with ca. 30 times more zapA transcript expressed in swarmer cells than in uninduced swimmer cells. ZapA has been purified as a single 54-kDa protein, and the genes encoding the enzyme have been cloned (28). Based on its deduced amino acid sequence, ZapA is a member of the serralysin metalloprotease family of zinc metalloproteases, which require both zinc and divalent cations for maximal catalysis (6, 11, 12).
Data from murine UTI experiments have demonstrated that, in the absence of ZapA, bacterial survival in the urinary tract is dramatically decreased; e.g., the rates of recovery of ZapA− P. mirabilis from the urine and bladder differ by at least 104 per ml and 103 per g, respectively, compared to the recovery of wild-type P. mirabilis from those same sites (27). This decrease is as great or greater than those for other P. mirabilis virulence factors (13), emphasizing the importance of ZapA as a virulence factor in P. mirabilis UTI. The dramatic attenuation of virulence in strains lacking ZapA suggests that ZapA may have a broad spectrum of activity associated with virulence. One hypothesis to explain the dramatic attenuation of virulence suggests that the role of ZapA in UTI extends beyond cleavage of IgA and IgG and that the principal substrates of ZapA may be other proteins found in the urinary tract.
In the present report, data are presented to show that ZapA hydrolyzes a wide range of protein and peptide substrates, including the antimicrobial peptides hBD1 and LL-37.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
P. mirabilis BB2000 is wild type for ZapA production and was used as the source of the purified enzyme. It was maintained as previously described (3, 4) in Luria-Bertani (LB) broth (22) or, when isolated colonies were required, on LSW− agar (10 g of tryptone, 5 g of yeast extract, 0.4 g of NaCl, 50 ml of glycerol, 20 g of agar/liter of distilled H2O) to phenotypically inhibit swarming (3). Escherichia coli D31 (7), a gift from Gill Diamond, New Jersey Dental School, University of Medicine and Dentistry of New Jersey, was maintained in either LB broth or on LB agar. All bacterial cultures were incubated overnight at 37°C, unless otherwise noted.
Proteins, peptides, and chemicals.
All chemicals were of the highest available purity. Protein substrates were purchased from Calbiochem (bovine serum albumin [BSA], human IgA1, IgA2, secretory IgA, and IgG), Quidel Corp. (C1q and C3), or Boehringer-Mannheim (actin, β-tubulin, collagen [type I], fibronectin, and laminin). Dynorphin A, dynorphin A fragment 1-7, and bradykinin were obtained from Bachem. hBD1 and hBD2 were obtained either as gifts from Wuyuan Lu (Institute of Human Virology, University of Maryland Biotechnology Institute) or were commercially purchased from the Peptide Institute, Inc. LL-37 was a gift from John S. Gunn (Ohio State University) or was purchased from Phoenix Pharmaceuticals, Inc. Protegrin PG-1 was a gift from John S. Gunn.
Purification of ZapA.
The ZapA protease was purified by phenyl-Sepharose hydrophobic interaction chromatography using a modification (28) of the method originally described by Loomes et al. (17). Briefly, P. mirabilis was incubated overnight at 37°C in 1-liter cultures of LB broth. Cells and debris were removed by centrifugation at 10,000 × g for 30 min at 4°C. The protease-containing supernatants were then filtered through 0.45-mm-pore-size filters (Micron Separations Inc.), and the filtrates were loaded at a rate of 1 ml/min at 4°C onto columns (2.5 by 50 cm; total bed volume, 240 ml) of phenyl-Sepharose (Pharmacia) equilibrated in 50 mM Tris-HCl (pH 8.0). The columns were washed with 10 column volumes of 50 mM Tris-HCl buffer (pH 8.0) or until unbound material was removed. Bound protease was eluted with 50 mM Tris buffer (pH 11). The pH of the fractions containing the ZapA protease was adjusted to 8.0 with HCl after collection, and the fractions were pooled. The purified protease eluted from this column was then dialyzed against 50 mM Tris (pH 8.0) and stored at −20°C in 20% glycerol until further use. The purity of the ZapA was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described by Wassif et al. (28), and averaged 98 to 99%.
Protein analyses.
Protein concentration was determined with the micro-bicinchoninic acid protein assay reagent as recommended by the manufacturer (Pierce Chemical Co.).
Proteolysis of protein substrates.
Proteolysis of protein and peptide substrates by ZapA was done with 20 nM ZapA and 1 to 125 μM substrate in a buffer consisting of 50 mM MES (morpholineethanesulfonic acid), 10 mM NaCl, 2 mM CaCl2 and 0.5 μM ZnCl2, pH 6.0, at 37°C for 1 to 16 h, depending on the substrate used. For example, proteolysis of antimicrobial peptides was measured with 100 ng of ZapA and 50 μg of the respective peptide in a reaction volume of 100 μl. Reactions were stopped by addition of EDTA to a final concentration of 20 mM, as described by Wassif et al. (28). To study the effects of denaturation of the protein substrate on hydrolysis by ZapA, a protein substrate was either incubated at 85°C or incubated in 8 M urea for 15 min prior to exposure to the enzyme. Protein substrates in 8 M urea were diluted with buffer to 0.5 M urea, a concentration of urea that does not affect ZapA hydrolysis (data not shown).
Detection of protein substrate hydrolysis.
Proteolysis of protein and peptide substrates by purified ZapA was measured by two methods: SDS-PAGE and high-performance liquid chromatography (HPLC). SDS-PAGE was done using commercial premade 4 to 20% gradient gels (Ready Gels; Bio-Rad, Hercules, Calif.) and the system established by Laemmli (14). After electrophoresis, protein bands were stained with SYPRO Orange (Molecular Probes) according to the manufacturer's recommendation and the protein band patterns were analyzed with a Typhoon 9410 variable-mode imager (Amersham Biosciences). The loss of the substrate band or the presence of new digestion product bands after incubation with ZapA indicated that the enzyme hydrolyzed the protein.
In HPLC analyses, ZapA hydrolysis of protein and peptide substrates was assessed by measuring the peptide fragments resulting from the cleavage of the substrate with a Zorbax 300 SB-C8 rapid-resolution column (4.6 by 50 mm; 3.5-μm matrix; Agilent). To separate these small peptides, a gradient starting at 3% acetonitrile (ACN) in 0.1% trifluoroacetic acid (TFA) and progressing over 20 min to 64% ACN plus 0.8% isopropanol in 0.07% TFA was used and the separation was run at 50°C. Peptides were detected with a diode array detector at 214 nm, and the chromatograms were analyzed for peak height and area with ChemStation software (Agilent).
MALDI-TOF spectrometry.
hBD1, hBD2, LL-37, and protegrin were incubated in the presence of ZapA, and the reaction was stopped by the addition of a saturating amount of EDTA, as described earlier. This material (20 ng) was cocrystallized in α-cyano-4-hydroxycinnamic acid (10 mg/ml in 50% ACN-0.3% TFA) and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectrometry on a Voyager DE STR (Applied Biosystems, Foster City, Calif.) using Data Explorer (version 4.0). Data were acquired in linear mode, and masses were externally calibrated with a standard peptide mixture. Peptide fragments were predicted from MALDI-TOF masses with PROWL World Wide Web software (http://prowl.rockefeller.edu/).
Measurement of antibacterial activity.
Inactivation of the antimicrobial activity of hBD1 and LL-37 by ZapA hydrolysis was measured by a MIC assay based on the NCCLS guidelines (20) and modified as follows. E. coli D31 was incubated overnight in Mueller-Hinton broth (Difco) at 37°C and then diluted 100-fold in fresh broth and incubated at 37°C until the culture density reached an optical density at 600 nm of 0.2. The culture was then diluted 200-fold in 10 mM NaPO4 buffer, pH 7.4. The cells were pipetted into the wells of a 96-well microtiter dish (Costar; 3790) to which was added either (i) buffer, (ii) a dilution series of the antimicrobial peptide, or (iii) a dilution series of the antimicrobial peptide that had been incubated in the presence of ZapA (as described for proteolysis of protein substrates). E. coli D31 cells in buffer plus ZapA served as a control. Each well contained 5 × 104 CFU in 100 μl of fluid. The cells were incubated at 37°C for 2 h, after which an aliquot was diluted and the CFU were determined by spreading the inoculum on Mueller-Hinton agar and incubating it overnight at 37°C. The percentage of cells killed was determined from the mean of a triplicate series of CFU data by using the equation percent killing = (CFUcontrol − CFUdiluted peptide)/CFUcontrol× 100, where the control is 0 μg of antimicrobial peptide per ml.
RESULTS
ZapA cleaves a wide array of proteins associated with urinary tract cells.
To test the hypothesis that ZapA hydrolysis is not limited solely to IgA and IgG, an SDS-PAGE method was used to qualitatively assess the spectrum and range of substrates hydrolyzed by the metalloprotease. Figure 1 shows a representative set of gels from these analyses, in which the loss of the substrate band upon incubation with ZapA (Fig. 1, lanes with +) indicates that the protein is cleaved by the enzyme. These data show that IgA1, IgA2, and IgG (Fig. 1A), components of the cell matrix and interconnecting filaments (actin, β-tubulin, fibronectin, collagen, and laminin; Fig. 1B and C), and the complement proteins C1q and C3 (Fig. 1D) are hydrolyzed by ZapA. BSA (Fig. 1D, lanes 2 and 3), in contrast, is recalcitrant to proteolysis by ZapA under the conditions (37°C for 8 h) used.
FIG. 1.
Qualitative measurement of the degradation of proteins associated with urinary tract epithelial cells by ZapA. A qualitative assessment of protein degradation by ZapA was made by an SDS-PAGE assay in which protein substrates by themselves and in the presence of ZapA (plus signs) were compared. The loss of a given protein band when reacted with the protease indicates that the protein is a substrate of ZapA. (A) IgA1, IgA2, and IgG. (B) Actin, tubulin, and fibronectin. (C) Collagen and laminin. (D) BSA (a negative control), IgA, C1q, and C3. All reaction mixtures were incubated at 37°C for 8 h in a reaction volume of 100 μl containing 20 nM ZapA and 100 to 125 μM respective protein substrate. Stds, standards.
Substrate conformation is critical for ZapA proteolysis.
During SDS-PAGE analyses, it was discovered that the age and condition of the protein substrate greatly affected the outcome of the hydrolysis by ZapA; e.g., older or poorly stored protein samples were more readily digested than were fresh preparations (data not shown). This observation suggests that ZapA proteolysis may be enhanced by a substrate that is partially or fully denatured and may explain the broad spectrum of substrates cleaved by ZapA in SDS-PAGE analysis (Fig. 1). This possibility was examined by a more sensitive, quantitative reverse-phase C8 HPLC assay comparing native proteins to the same substrates preexposed to either denaturant (8 M urea) or heat (85°C). The results of this analysis are shown in the chromatograms for IgA and IgG displayed in Fig. 2. While most of the protein substrates in their native conformation were efficiently digested by ZapA, both native IgA and native IgG were only marginally hydrolyzed by the enzyme. This can be seen by comparing the data in Fig. 2A (native IgA), B (IgA denatured with urea), and C (IgA denatured by heat). Some enzymatic hydrolysis of this protein is evident after 16 h of incubation with ZapA (Fig. 2A); however, heat denaturation of the substrate (Fig. 2C) significantly improved the proteolysis, resulting in nearly complete loss of the substrate by 4 h. Preincubation of IgA in urea only slightly improved proteolysis by ZapA (Fig. 2B, 16-h chromatogram). Denaturation of IgG (Fig. 2D) also significantly improved its digestion by ZapA. These data show that denaturation of some of the protein substrate, specifically IgA and IgG, improved the rate of hydrolysis by ZapA. However, the majority of substrates tested did not require denaturation to be effectively cleaved by ZapA, and hydrolysis of these proteins occurs at the same rate regardless of their conformational state.
FIG. 2.
Quantitative measurement of ZapA proteolysis of IgA and IgG by HPLC analysis. In this method, the decrease in the substrate protein peak (rightmost peak in each chromatogram; retention time of ca. 12 min) area and increases in the number and peak areas of product peaks (generally in the range of 2- to 10-min retention time) were used to assess proteolysis as a function of time of incubation and preincubation denaturation of the substrate. (A) Digestion of native IgA over time. The cascade of chromatograms from front to back is IgA without ZapA and in the presence of ZapA at 0, 0.5, 1, 2, 4, 6, and 16 h. (B) Digestion of IgA preincubated in 8 M urea. From front to back, the chromatograms are IgA without ZapA and in the presence of ZapA at 0, 0.5, 1, 2, 4, 6, and 16 h. (C) Digestion of heat-denatured IgA (preincubated at 85°C for 15 min). From front to back, the chromatograms are IgA without ZapA and in the presence of ZapA at 0, 0.5, 1, 2, 4, 6, and 16 h. (D) Digestion of IgG. From front to back, the chromatograms are IgG without ZapA (0 h), IgG plus ZapA (0 h) at 16 h and 37°C, heat-denatured IgG without ZapA, heat-denatured IgG with ZapA, urea-denatured IgG without ZapA, and urea-denatured IgG with ZapA. Reaction mixtures were incubated at 37°C for the specified times in a reaction volume of 100 μl containing 20 nM ZapA and 100 to 125 μM respective protein substrate.
Antimicrobial peptides as substrates for ZapA.
ZapA has been reported to digest peptides (1). Our results confirm these earlier reports and show that dynorphin A, a fragment of dynorphin A containing the first seven amino acids, and the β chain of insulin are substrates effectively hydrolyzed by ZapA (data not shown). In addition to these peptides, it was discovered that the antimicrobial peptide hBD1 is a substrate of ZapA and that the digestion of the peptide is time dependent (Fig. 3). These data allow an estimate of the rate of hydrolysis of this substrate to be calculated, based on the molecular masses of ZapA (53,976.8 Da) and hBD1 (3,928.6 Da), the amount of each used in the reaction, and the volume of the reaction mixture applied to the HPLC column. A standard curve comparing known hBD1 concentrations against HPLC chromatogram substrate peak areas was used to determine the amount of hBD1 in the substrate peak on the chromatograms shown in Fig. 3. This analysis indicated that ca. 0.44 μg of hBD1 was present at 0 h and that, after 16 h at 37°C, only 0.18 μg of hBD1 remained. From this, a conservative estimate suggests that 1 pmol of ZapA digests 0.6 pmol of hBD1 per h at 37°C.
FIG. 3.
HPLC chromatograms of the digestion of hBD1 by ZapA. Shown is cleavage of hBD1, part of the kidney innate defense mechanism, by the ZapA protease over time. Chromatograms are, in order from front to back, hBD1 at 0 h; hBD1 at 16 h of incubation; and hBD1 plus ZapA at 0, 1, 2, 4, 6, and 16 h of incubation. The reaction mixtures were incubated at 37°C for the specified times in a reaction volume of 100 μl containing 20 nM ZapA and 125 μM (50 μg) hBD1.
The ability of ZapA to hydrolyze three other peptides with antimicrobial activity, hBD2, LL-37, and porcine protegrin PG-1, was also measured (Fig. 4). Both LL-37 (Fig. 4C) and protegrin (Fig. 4D), in addition to hBD1 (Fig. 4A), are substrates of the enzyme, while hBD2 (Fig. 4B) was not digested under the conditions used. These data show that ZapA effectively cleaves two antimicrobial peptides, hBD1 and LL-37, and partially degrades protegrin.
FIG. 4.
ZapA proteolysis of antimicrobial peptides. Shown are HPLC chromatograms of the digestion of hBD1 (A), hBD2 (B), human LL-37 (C), and porcine protegrin (D). In each panel the foremost chromatogram represents a reaction of the antimicrobial peptide without ZapA, while the rearmost chromatogram is the same substrate in the presence of the protease. In each, 20 nM ZapA and 125 μM respective antimicrobial peptide were incubated for 8 h at 37°C.
Examination of the chromatograms from ZapA hydrolysis of hBD1, LL-37, and protegrin shown in Fig. 4 indicates that the digestion of each substrate results in a discrete set of major peaks for each peptide. Approximately six peptides are produced from hBD1, nine are produced from LL-37, and two or three are produced from digestion of protegrin. Further, the hydrolysis of these antimicrobial peptides is not significantly affected by prior denaturation (data not shown).
Location of the scissile bonds in hBD1 and LL-37.
The location of the enzyme cleavage sites on the antimicrobial peptide and the identity of the resulting peptide fragments generated upon ZapA proteolysis of hBD1 and LL-37 were determined by MALDI-TOF spectrometry. Nine fragments were detected as significantly above the baseline in the digest of hBD1 (Fig. 5). The masses of six of these fragments corresponded to one or more peptide sequences within the hBD1 sequence (Table 1). The location of these fragments within the hBD1 amino acid sequence is shown in Fig. 5.
FIG. 5.
MALDI-TOF analysis of the cleavage of hBD1 by ZapA. (A) hBD1 alone. (B) hBD1 peptide fragments after incubation with ZapA under standard conditions (Materials and Methods). Arrows under the sequence of hBD1, potential sites of cleavage and peptide fragments deduced from the MALDI-TOF mass data.
TABLE 1.
Predicted peptide fragments resulting from ZapA proteolysis of hBD1
| Massa (Da) | Predicted fragment(s)b |
|---|---|
| 542.81 | TCYR |
| 847.09 | PIFTKIQ |
| 1,060.87 | FTKIQGTCY |
| 1,285.85 | SSGGQCLYSACPI |
| 1,705.61 | FTKIQGTCYRGKAKC, DHYNCVSSGGQCLYSA |
| 1,906.98 | DHYNCVSSGGQCLYSACP |
Mass from MALDI-TOF analysis.
As described in Materials and Methods.
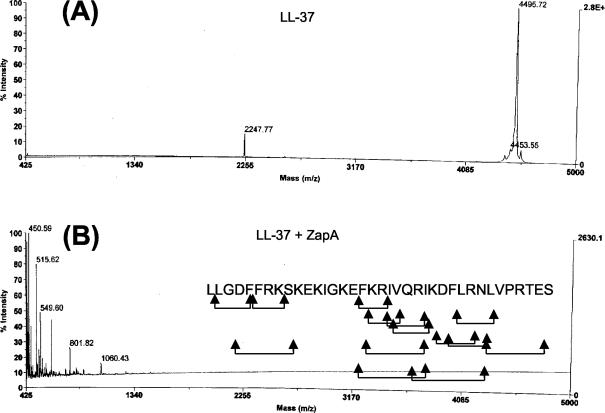
Similarly, MALDI-TOF spectrometry detected the masses of five peptide fragments from the digest of LL-37 as significantly above the baseline (Fig. 6). It is apparent from looking at the data in Fig. 6 that many more fragments may have been produced in this digest, but their masses were not statistically significant. The predicted peptide fragments from LL-37 that correspond to the masses detected are listed in Table 2, and the possible scissile bonds within LL-37 are indicated in Fig. 6. As is apparent for hBD1, ZapA shows no clear preference for specific amino acid residues, except that Arg at the P1 position was noted at four sites in the combined data sets. Many of the potential cleavage sites on LL-37 occur within a domain, EKIGKEFKRIVQRIKDFLRNLV, that has been shown to possess antimicrobial activity (23), suggesting that hydrolysis of LL-37 by ZapA may result in a loss or decrease in antimicrobial activity of the peptide.
FIG. 6.
MALDI-TOF analysis of the cleavage of LL-37 by ZapA. (A) LL-37 alone. (B) LL-37 peptide fragments after incubation with ZapA. Arrows under the sequence of LL-37, potential sites of cleavage and peptide fragments deduced from the MALDI-TOF mass data.
TABLE 2.
Predicted peptide fragments resulting from ZapA proteolysis of LL-37
| Massa (Da) | Predicted fragmentsb |
|---|---|
| 450.59 | LGDF, FRK, FKR |
| 515.62 | KRIV, IVQR, VQRI, LRNL |
| 549.60 | DFLR, FLRN |
| 801.82 | DFFRKS, KRIVQR, LVPRTES |
| 1,060.43 | FKRIVQRI, RIKDFLRN |
Mass from MALDI-TOF analysis.
As described in Materials and Methods.
ZapA proteolysis of hBD1 and LL-37 decreases the antibacterial activity of the peptide.
The effect of proteolysis on the antibacterial activity of hBD1 and LL-37 was measured by a MIC assay with E. coli D31, a strain that is well characterized and that is frequently used in testing antimicrobial peptide activity (2). By this method, the 50% lethal dose (LD50), as measured by the percent killing, for hBD1 was determined to be 1.68 μM or 6.6 μg/ml (Fig. 7A). Proteolysis of hBD1 by ZapA significantly increased the LD50 of this antimicrobial peptide to >13 μM (or 50 μg/ml), at least a sevenfold reduction in activity. Hydrolysis of LL-37 also showed marked effects on the activity of this peptide (Fig. 7B). The mean LD50 after ZapA hydrolysis was 3.16 μM (12.4 μg/ml), compared to an LD50 of 0.1 μM (0.4 μg/ml) prior to proteolysis, a value that agrees well with other reports (23). This represents at least a 30-fold decrease in antibacterial activity. These results demonstrate that ZapA proteolysis has a negative effect on the antibacterial activity of hBD1 and LL-37.
FIG. 7.
ZapA proteolysis of hBD1 and LL-37 reduces the antimicrobial activity of hBD1 and LL-37. The antimicrobial activity of each peptide was determined with E. coli D31, as described in Materials and Methods. (A) Antimicrobial activity, as LD50, of hBD1 prior to digestion (squares) and after digestion with ZapA (circles). (B) Antimicrobial activity, as LD50, of LL-37 prior to digestion (squares) and after digestion with ZapA (circles). The reaction mixtures were incubated at 37°C for 8 h.
DISCUSSION
The results demonstrate that the ZapA metalloprotease of P. mirabilis is capable of cleaving a variety of proteins and peptide species, including peptides with antibacterial activity. This activity of the ZapA metalloprotease is noteworthy; as it is generally thought that antibacterial peptides are resistant to proteolytic degradation by virtue of their structure and disulfide bonds (21). We believe that this may be the first report of a bacterial enzyme that hydrolyzes hBD1, an important antibacterial peptide and component of the human urinary tract innate immune system (26), and the second report of bacterial proteolysis of LL-37 (23). Schmidtchen et al. (23) reported that a 50-kDa extracellular protease obtained from the extracellular spent media of P. mirabilis digested LL-37 in an in vitro assay (23). Based on the present data and other reports (27, 28), it is very likely that the 50-kDa extracellular enzyme identified by Schmidtchen et al. (23) is ZapA.
ZapA proteolysis of hBD1 and LL-37 is also effective in reducing the bactericidal activity of these antibacterial peptides. Upon incubation for 8 h at 37°C, proteolysis of hBD1 and LL-37 by ZapA reduced the effective bactericidal action of the peptides by at least 7- and 30-fold, respectively. Thus, one of the possible modes of action of the ZapA metalloprotease may be proteolysis of innate defense molecules in the urinary tract. As the enzyme exhibits minimal substrate or amino acid site specificity, it appears that the action of ZapA is not specific or limited to antimicrobial peptides. This virulence factor may therefore provide a general “weapon of mass destruction,” rather than a highly focused “smart bomb” approach to protecting P. mirabilis against attack from the host immune system.
In considering the role of ZapA as a generalized bacterial defense against innate immunity response and host defenses, the rate of the enzyme catalysis of its substrates should be considered. Although kcat values are not immediately derived from the data presented herein, it is safe to say that (under the conditions used in the in vitro proteolysis assays) ZapA proteolysis of its substrates is not rapid. For all protein substrates examined and some peptides, e.g., hBD1, reactions of the enzyme and substrate required at least 8 h to yield >80% proteolysis of the substrate. An estimate of the rate of proteolysis for hBD1, as an example, suggests that 1 pmol of ZapA digests 0.6 pmol of hBD1 per h at 37°C, or two molecules of the enzyme are required to hydrolyze one molecule of the peptide in 60 min.
The data presented may be used to indirectly answer questions about the role of ZapA in P. mirabilis virulence and UTIs; however interpretation of the data should be tempered. In preliminary studies leading to this report, wild-type P. mirabilis and a strain defective in ZapA production (27) were used in MIC assays. Both wild-type and ZapA− mutant strains are extremely resistant to the four antimicrobial peptides tested (hBD1, hBD2, LL-37, and protegrin; data not shown). This observation is supported by the studies of McCoy et al. (18), who reported that P. mirabilis is highly resistant to the action of antimicrobial peptides (18) and probably uses multiple mechanisms to achieve this resistance. Thus, due to the high native resistance of P. mirabilis to antimicrobial peptides, a more sensitive bacterial strain, E. coli D31, was chosen for MIC analyses, which were used to answer questions about whether ZapA proteolysis reduced the bactericidal activity of hBD1 and LL-37. While the data presented in Fig. 7 clearly demonstrate the effectiveness of ZapA in reducing the bactericidal activity of hBD1 and LL-37, the significance and relative contribution of ZapA in P. mirabilis evasion of the host immune response during UTI remain open issues awaiting experimental confirmation.
Both hBD1 and hBD2 are found in the human urinary tract (9, 15, 26). The results support the conclusion that ZapA effectively cleaves hBD1, yet does not hydrolyze hBD2 (Fig. 3). This is an interesting finding and raises questions about what distinguishes the two peptides during ZapA binding and catalysis. hBD1 and hBD2 are different from one another in their amino acid sequences and sizes, yet they also contain many common characteristics, including the three disulfide bonds and an overall charge distribution characteristic of the β-defensins. Moreover, several of the putative scissile bonds found in hBD1 and LL-37 proteolytic cleavage fragments also occur in hBD2, which suggests that the enzyme distinguishes hBD1 from hBD2 based on some other molecular property or feature. Further investigation of the reasons behind this difference may uncover the molecular mechanisms used by ZapA and other serralysins to bind to and catalyze their substrate proteins and peptides.
It is safe to conclude from these results that it is inappropriate to call ZapA an IgA-degrading protease, as the enzyme can cleave a wide range of substrates of which IgA is not the best. Instead, a more plausible function for ZapA in P. mirabilis virulence and UTI is as a general proteolytic enzyme that is well suited for an opportunistic urinary tract pathogen such as P. mirabilis. As such, ZapA may aid in the acquisition of carbon and nitrogen for bacterial growth and metabolism through proteolysis of host proteins, some of which may be involved in immune system functions. This double-edged function would not only provide nutrients to the bacteria but also prolong their existence and survival during attack from the host immune defenses.
Acknowledgments
We thank Gill Diamond (UMDNJ-New Jersey Dental School) for E. coli D31, John S. Gunn (The Ohio State University), and Wuyuan Lu (Institute for Human Virology, University of Maryland Biotechnology Institute) for the generous gifts of hBD1, hBD2, LL-37, and protegrin PG-1 and the members of the Molecular and Cellular Pathogeneses of Urinary Infection PO1 group for many helpful suggestions. The assistance of David Robinson and Carole Owen in ZapA preparation and Bob Cole (AB-Mass Spectrometry Facility at Johns Hopkins School of Medicine) with the MALDI-TOF analyses is gratefully appreciated. This manuscript was greatly aided by the comments the authors received from three anonymous reviewers and the suggestions of the members of the Belas laboratory.
MALDI-TOF analyses were funded in part from National Center for Research Resources shared-instrumentation grant 1S10-RR14702. This research was supported by an award from the National Institutes of Health (DK48720).
Editor: J. B. Bliska
Publication no. 05-105 of the Center of Marine Biotechnology.
REFERENCES
- 1.Aneas, M. A., F. C. Portaro, I. Lebrun, L. Juliano, M. S. Palma, and B. L. Fernandes. 2001. ZapA, a possible virulence factor from Proteus mirabilis exhibits broad protease substrate specificity. Braz. J. Med. Biol. Res. 34:1397-1403. [DOI] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belas, R., D. Erskine, and D. Flaherty. 1991. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J. Bacteriol. 173:6279-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belas, R., D. Erskine, and D. Flaherty. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bensch, K. W., M. Raida, H. J. Magert, P. Schulz-Knappe, and W. G. Forssmann. 1995. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 368:331-335. [DOI] [PubMed] [Google Scholar]
- 6.Bode, W., F. X. Gomis-Ruth, and W. Stockler. 1993. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins.’ FEBS Lett. 331:134-140. [DOI] [PubMed] [Google Scholar]
- 7.Cole, A. M., R. O. Darouiche, D. Legarda, N. Connell, and G. Diamond. 2000. Characterization of a fish antimicrobial peptide: gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 44:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairley, K. F., N. E. Carson, R. C. Gutch, P. Leighton, A. D. Grounds, E. C. Laird, P. H. McCallum, R. L. Sleeman, and C. M. O'Keefe. 1971. Site of infection in acute urinary-tract infection in general practice. Lancet ii:615-618. [DOI] [PubMed] [Google Scholar]
- 9.Ganz, T. 2001. Defensins in the urinary tract and other tissues. J. Infect. Dis. 183(Suppl. 1):S41-S42. [DOI] [PubMed] [Google Scholar]
- 10.Ganz, T. 2002. Immunology. Versatile defensins. Science 298:977-979. [DOI] [PubMed] [Google Scholar]
- 11.Ghigo, J. M., and C. Wandersman. 1994. A carboxyl-terminal four-amino acid motif is required for secretion of the metalloprotease PrtG through the Erwinia chrysanthemi protease secretion pathway. J. Biol. Chem. 269:8979-8985. [PubMed] [Google Scholar]
- 12.Hege, T., and U. Baumann. 2001. Protease C of Erwinia chrysanthemi: the crystal structure and role of amino acids Y228 and E189. J. Mol. Biol. 314:187-193. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, D. E., R. G. Russell, C. V. Lockatell, J. C. Zulty, J. W. Warren, and H. L. Mobley. 1993. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect. Immun. 61:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann, J., M. Retz, J. Harder, M. Krams, U. Kellner, J. Hartmann, K. Hohgrawe, U. Raffenberg, M. Gerber, T. Loch, K. Weichert-Jacobsen, and M. Stockle. 18 Septembr 2002, posting date. Expression of human β-defensins 1 and 2 in kidneys with chronic bacterial infection. BMC Infect. Dis. 2:20. [Online.] http://www.biomedcentral.com/1471-2334/2/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 17.Loomes, L. M., B. W. Senior, and M. A. Kerr. 1992. Proteinases of Proteus spp.: purification, properties, and detection in urine of infected patients. Infect. Immun. 60:2267-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy, A. J., H. Liu, T. J. Falla, and J. S. Gunn. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mobley, H., and J. Warren. 1996. Urinary tract infections. ASM Press, Washington, D.C.
- 20.NCCLS. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. NCCLS, Wayne, PA.
- 21.Oren, Z., and Y. Shai. 1998. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 47:451-463. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual., 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 24.Senior, B. W., M. Albrechtsen, and M. A. Kerr. 1987. Proteus mirabilis strains of diverse type have IgA protease activity. J. Med. Microbiol. 24:175-180. [DOI] [PubMed] [Google Scholar]
- 25.Senior, B. W., M. Albrechtsen, and M. A. Kerr. 1988. A survey of IgA protease production among clinical isolates of Proteeae. J. Med. Microbiol. 25:27-31. [DOI] [PubMed] [Google Scholar]
- 26.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. McCray, Jr., and T. Ganz. 1998. Human β-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker, K. E., S. Moghaddame-Jafari, C. V. Lockatell, D. Johnson, and R. Belas. 1999. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32:825-836. [DOI] [PubMed] [Google Scholar]
- 28.Wassif, C., D. Cheek, and R. Belas. 1995. Molecular analysis of a metalloprotease from Proteus mirabilis. J. Bacteriol. 177:5790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]