Abstract
A preliminary genome sequence has been assembled for the Southern Ocean salp, Salpa thompsoni (Urochordata, Thaliacea). Despite the ecological importance of this species in Antarctic pelagic food webs and its potential role as an indicator of changing Southern Ocean ecosystems in response to climate change, no genomic resources are available for S. thompsoni or any closely related urochordate species. Using a multiple-platform, multiple-individual approach, we have produced a 318,767,936-bp genome sequence, covering >50% of the estimated 602 Mb (±173 Mb) genome size for S. thompsoni. Using a nonredundant set of predicted proteins, >50% (16,823) of sequences showed significant homology to known proteins and ∼38% (12,151) of the total protein predictions were associated with Gene Ontology functional information. We have generated 109,958 SNP variant and 9,782 indel predictions for this species, serving as a resource for future phylogenomic and population genetic studies. Comparing the salp genome to available assemblies for four other urochordates, Botryllus schlosseri, Ciona intestinalis, Ciona savignyi and Oikopleura dioica, we found that S. thompsoni shares the previously estimated rapid rates of evolution for these species. High mutation rates are thus independent of genome size, suggesting that rates of evolution >1.5 times that observed for vertebrates are a broad taxonomic characteristic of urochordates. Tests for positive selection implemented in PAML revealed a small number of genes with sites undergoing rapid evolution, including genes involved in ribosome biogenesis and metabolic and immune process that may be reflective of both adaptation to polar, planktonic environments as well as the complex life history of the salps. Finally, we performed an initial survey of small RNAs, revealing the presence of known, conserved miRNAs, as well as novel miRNA genes; unique piRNAs; and mature miRNA signatures for varying developmental stages. Collectively, these resources provide a genomic foundation supporting S. thompsoni as a model species for further examination of the exceptional rates and patterns of genomic evolution shown by urochordates. Additionally, genomic data will allow for the development of molecular indicators of key life history events and processes and afford new understandings and predictions of impacts of climate change on this key species of Antarctic pelagic ecosystems.
Keywords: Salpa thompsoni, Thaliacean genome, urochordate, miRNA, Antarctic ecosystem
Introduction
Pelagic zones of the open ocean represent one of the largest (by volume) habitats on Earth, with highly diverse and ecologically important assemblages of zooplankton, animals that drift with ocean currents, that can serve as early warning indicators of climate change (Ducklow et al. 2013). Despite their habitat size and species diversity, with (>7,000 species in 15 phyla; Wiebe et al. 2010), few marine zooplankton have been utilized as model organisms for genome-scale analyses. Today, genomic information is lacking for highly abundant zooplankton species in diverse groups that occupy pivotal roles in marine ecosystems and food web. The lack of genomic information for sister taxa and even distantly related species is a significant factor limiting progress in analysis of deep phylogenetic comparisons and the application of genomic perspectives to understanding the biology of these organisms.
A case in point are the salps, (Phylum Chordata, Subphylum Tunicata, Class Thaliacea, Order Salpida), which includes 45 species of gelatinous zooplankton. While there are no genomic data currently available for any members of this phylogenetically diverse and relatively divergent lineage (Govindarajan et al. 2011), there are a few existing resources for other tunicates. Within Tunicata, there are three extant classes, the appendicularians (also referred to as larvaceans), ascidians and thaliacians, comprising a single polytomy (fig. 1A). Within the tunicate Class Ascidiacea, a reference genome is available for three species, including Ciona intestinalis, which has achieved status as a model species for genomic studies (Holland and Gibson-Brown 2003; Takatori et al. 2004; Kawada et al. 2011) due in part to its ecological importance as a nuisance and invasive marine species, the closely related taxa Ciona savignyi (Small et al. 2007), and Botryllus schlosseri, a model species for studying the evolution of adaptive immunity and organ/tissue complexity (Voskoboynik et al. 2013). Another tunicate model species is Oikopleura dioica (Class Appendicularia) (Seo et al. 2001; Stach 2007; Yadetie et al. 2012), which has a genome characterized by a compact arrangement of genes and a small genome size (∼70 Mb). Collectively, these tunicate species have yielded new insights into genome evolution, including both lineage-specific innovations, such as horizontal acquisition of the cellulose synthase gene from bacteria and spliced-leader trans-splicing of mRNAs (Satoh et al. 2006), and a broader understanding of fundamental principles of chordate and vertebrate genome evolution. The tunicate genomes studied to date, particularly C. intestinalis and C. savignyi, lack the extensive gene duplication events of vertebrates, and have thus proven to be extremely valuable in elucidating the gene networks associated with key morphogenetic and developmental biology processes, including the formation of basic chordate tissues, the notochord, neural tube, and heart (Dehal et al. 2002).
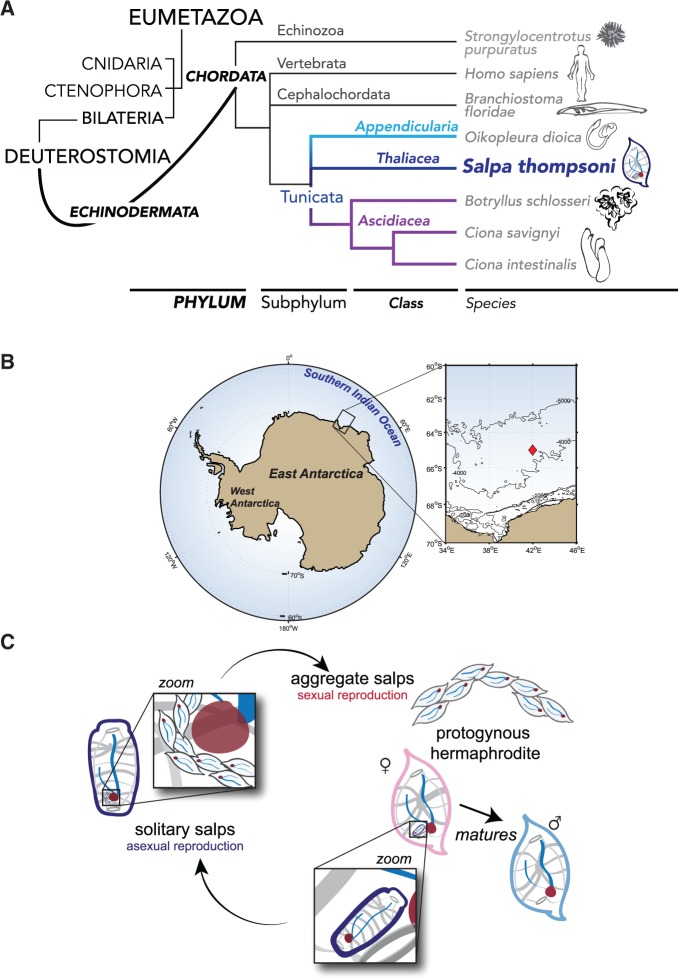
Fig. 1.—
(A) Evolutionary position of Salpa thompsoni with respect to other groups within the Subphylum Tunicata, including Ciona spp. and Botryllus schlosseri within Class Ascidiacea, Oikopleura dioica within Class Appendicularia, as well as Subphyla Cephalochrodata and Vertebrata. Overall relationships among Eumetazoa are indicated to left. (B) Map of Antarctica indicating the collection area for S. thompsoni samples (boxed inset, red diamond) in the Southern Indian Ocean. (C) Reproductive life cycle of S. thompsoni.
Genomic resources are also proving instrumental in garnering new insights into the evolution of organismal adaptive innovations and organism–environment interactions (Franssen et al. 2014; Villarino et al. 2014), including responses to environmental variability associated with climate change (Meyer et al. 2015). Although any given species may be uniquely impacted by the dynamic range of physical and biological parameters that accompany shifts in global climate profiles, processes involved in responses to climate change at the molecular level may share common features across species, for example, in the evolution of gene networks associated with environmental stress responses. Genomic analysis is critically needed for key species that can impact entire ecosystems and are already exhibiting marked changes with climate change. One such species is the Southern Ocean salp, S. thompsoni (fig. 1B), a highly efficient filter-feeder capable of consuming significant fractions of the organisms and particulate organic matter between 1 and 1,000 μm in pelagic ecosystems (Madin and Deibel 1998). The repackaging of waste material into large, fast-sinking fecal pellets means that S. thompsoni is a substantial contributor to vertical flux of organic matter (Pakhomov 2004; Pakhomov and Froneman 2004; Phillips et al. 2009). The species also has exceptional capacity for rapid population growth—bloom formation (Alldredge and Madin 1982)—via a complex life history alternating between morphologically distinctive asexual and sexual forms (fig. 1C). The asexual form (solitary) is shaped like a cylinder and has a stolon that forms chains of aggregates, which are released into the water column when conditions are favorable, such as during austral spring and summer (Loeb and Santora 2012), and reproduce sexually. Aggregates are sequential protogynous hermaphrodites, ensuring cross-fertilization between different chains (Godeaux et al. 1998; Loeb and Santora 2012). The complex life history of S. thompsoni allows rapid responses and life cycle adjustments to environmental variability (Chiba et al. 1999). The species has shown altered patterns of distribution and abundance in Antarctic Ocean (fig. 1B) ecosystems in recent years in response to climate change (Loeb and Santora 2012; Kokubun et al. 2013) and has the potential to displace Antarctic krill, Euphausia superba, a keystone species that is the foundation for the highly productive Southern Ocean pelagic food web (Atkinson et al. 2004).
We have completed the first draft genome sequence for the third class of tunicates (Thaliacea) for S. thompsoni and, in concert with transcriptome sequencing efforts (Batta-Lona et al. 2016; submitted), have generated a new genetic resource for this urochordate species, for which genome sequence information was previously wholly lacking. These data establish the Southern Ocean salp as a new marine model species and allow renewed appreciation for genomic evolution within the Subphylum Urochordata. The application of new genomic and transcriptomic resources for this phylogenetically important species will expand our understanding of the genetics underlying its adaption to a planktonic and polar environment, and its organism–environment interactions, which are the foundation for predictions of the species’ population dynamics and responses to climate change, as well as its impacts on the Antarctic pelagic ecosystem
Material and Methods
Sample Collection
Collections of S. thompsoni were made in January 2009 during a cruise of the Japanese Research Vessel Umitaka-Maru in the Indian sector of the Southern Ocean, from Cape Town, South Africa to Fremantle, Australia. Samples were collected at Station 33 (fig. 1B) at depths of 50–100 m with a Rectangular Midwater Trawl (RMT) 1 + 8 (mesh size 0.33 mm and 4.5 mm). Immediately after collection, salps were analyzed under the microscope to confirm species identification. All specimens were categorized by life stage (solitary vs. aggregates), reproductive status (presence of gonads and embryos), and designation of maturational stages (0–5; see Daponte et al. 2001). The gut of S. thompsoni was removed by dissection to avoid DNA contamination from prey, and muscular tissue was flash-frozen with liquid nitrogen and stored at −80°C. For this analysis, four specimens (aggregates of life stage 2) were chosen; DNA was extracted following the QIAGEN (Germantown, MD) DNeasy extraction protocol (Cat. no. 69582) and RNA was purified prior to sequencing.
Genome Size Estimation
We used real-time quantitative PCR (qPCR) to estimate the genome size for S. thompsoni (Wilhelm et al. 2003). A single copy gene, Tbx1, found in C. intestinalis (Takatori et al. 2004) was identified within S. thompsoni and validated for use as a reference standard. Four specimens were chosen and their DNA was extracted in the same manner as described earlier. First, a PCR product ∼500 bp was generated, followed by development of internal primers to produce a smaller PCR product amenable to qPCR. The smaller PCR product was amplified from each of the four salp DNA samples and then quantified using the PicoGreen assay to determine a highly specific estimate of nucleotide composition. The PCR product was 1/10 serially diluted from 1×10−1, 1×10−2, to 1×10−9. A qPCR was then performed on these dilutions and PicoGreen assay estimates were used to build a standard curve showing the regression relationship of the number of cycles to the number of moles of internal primer product. This standard curve was then used to estimate the number of copies of each gene; molar amounts were then used to calculate the size of S. thompsoni genome.
Next Generation Sequencing—454 and Ion Torrent Platforms
Four different S. thompsoni specimens collected in the Indian Ocean were used for genome sequencing. Three specimens were used for library construction and subsequent sequencing using shotgun and paired-end protocols provided by the instrument manufacturer (GS FLX Titanium General Library Preparation Guide; Roche Applied Science, Branford, CT). Sequencing results for all libraries are listed in supplementary table S1, Supplementary Material online; details associated with library construction are as follows:
Shotgun DNA Library Generation and Sequencing
Salp genomic DNA was mechanically sheared into fragments and sequenced according to the Roche 454 FLX WGS protocol.
Paired-End DNA Library Generation and Sequencing
For the paired-end libraries, genomic DNA from a third specimen was sheared into ∼3 kb fragments and processed for Paired End sequencing according to the manufacturers’ protocol. When sequenced on the GS FLX, this protocol generated two, ∼100 bp tags known to be ∼3 kb apart. These paired-end reads were used to aid in the assembly of contigs into scaffolds.
Ion Torrent Proton
The fourth specimen was sequenced on two runs of the Ion TorrentTM Proton (Life Technologies, Grand Island, NY). Library construction and the emPCR were performed according to the Ion Torrent protocol associated with the kit (Ion Xpress™ Fragment Library Kit).
Filtering and Assembly
Sequencing reads generated on both the Roche 454 System and the Ion Torrent PGM were used in developing a de novo genome assembly. Initially, low quality sequencing reads were removed from inclusion in assembly efforts based on phred quality scores (Q) <30 (Q30 is defined as the probability that a given base is called incorrectly 1 in 1,000). All remaining reads were then filtered against known bacterial or algal genome sequences and any reads that specifically mapped to these genomes were excluded from all further analyses, as follows. The criterion for exclusion was if a sequencing read showed 90% similarity to a bacterial or algal genome over 90% of the read length, the read was tagged as a possible contaminant and removed from the data set. All Proton reads were error-corrected using the program Coral v1.4, with settings of mr = 2, mm = 2, and g = 3 (Salmela and Schroder 2011). The remaining data were assembled into contigs and scaffolds using the assembly program Newbler v2.8. To further improve our genome assembly, the program L-RNA Scaffolder (Xue et al. 2013), which scaffolds contigs using a BLAST-mapped transcriptome reference, was used to increase scaffold lengths. The transcriptome sequence used in the study was generated from sequencing runs for RNA-Seq from multiple individuals (N = 40), which were then normalized and assembled using the program Trinity and associated scripts (see Batta-Lona et al. 2016, submitted, for methods).
Annotation
The final genome assembly was annotated by generating de novo predictions for both repetitive elements and gene sequences. Repetitive elements were predicted using the programs RepeatModeler (Smit and Hubley 2008–2010) and RepeatMasker (Smit 1996–2010) to identify both de novo and reference-based repetitive elements, respectively. All unclassified repetitive elements were further examined using the program TEClass (Abrusán et al. 2009) for a more detailed annotation assignment. The genome sequence was then assessed for total repetitive element content using RepeatMasker and our de novo repeat annotation library. Gene and protein sequences were predicted using the Maker2 (Holt and Yandell 2011) pipeline, in which repetitive elements were masked and salp transcriptome sequences and C. intestinalis proteins for which the salp transcriptome showed at least 90% coverage were aligned to the salp genome to generate gene predictions. Subsequent iterations of the program used two de novo gene prediction programs, SNAP (Korf 2004) and Augustus (Stanke and Morgenstern 2005), to generate additional de novo gene predictions. Both SNAP and Augustus were trained for S. thompsoni using full-length protein sequences isolated from the transcriptome; these proteins were identified by comparing their predicted open-reading frame sequences to those of existing proteins in the UNIPROT database (http://www.uniprot.org/; last accessed September 19, 2016). Ultimately, both evidence-based and de novo predictions were used to cross-validate and finalize gene predictions. All predicted proteins were compressed into a nonredundant protein set using the CD-Hit algorithm with default parameters (Fu et al. 2012), and then annotated by identifying their protein identity using BLASTP (e-value = 1E-5).
Genome Statistics
Gene Ontology terms were assigned to the S. thompsoni genome with the BLAST2GO B2G4 v2.5 pipeline using default parameter settings (Conesa et al. 2005) and protein family and domain information using InterProScan v.5.6 (e-value = 1E-5). Enzyme codes and KEGG pathway membership were all done in BLAST2GO. All final annotations were further summarized in GO SLIM (http://geneontology.org/page/go-slim-and-subset-guide; last accessed September 19, 2016).
Evolutionary Rate Estimation and Positive Selection Analyses
Evolutionary rates of urochordate genes were examined using an analysis pipeline that identified orthologs, generated multiple sequence alignments (MSAs), filtered out MSAs with ambiguities, estimated genetic distances between among members of orthologous protein groups, and tested hypotheses of positive selection in S. thompsoni on these genes. Using MAKER, predicted transcript sequences for S. thompsoni and the other “official” transcript sequences for four other species of urochordates with genome sequences (Oikopleura diocia, Botryllus schlosseri, Ciona savignyi, and Ciona intestinalis) and the cephalochordate Branchiostoma floridae, orthologs were predicted among these six species using reciprocal best-BLAST hit comparisons (TBLASTX). Once orthologs were identified, all genes lacking a predicted ortholog from any of our test species, or identified using a high-scoring segment pair (HSP) alignment region of <200 bp long, were discarded. Using an in-house Python script, the remaining orthologs were passed through a series of analysis steps. Groups of orthologs were first reconstructed in the same strand and aligned using the codon-guided multiple sequence alignment (MSA) algorithm MACSE v 0.9b1 (Ranwez et al. 2011). Aligned sequences (MSAs) were cleaned using trimAl (Capella-Gutiérrez et al. 2009) to remove all gaps both from within and at the ends of the aligned sequences. MACSE includes the convenient feature of assessing frameshift and stop codon issues associated with multiple sequence alignment. Thus, in order to avoid confounding alignment problems related to poor data quality, low scoring MSAs and true pseudogenized gene sequences, all of which would contribute to false positives in subsequent PAML analyses, this feature was leveraged to identify and remove from further analysis any MSA with either a frameshift ambiguity or base ambiguity.
The remaining MSAs were then used to calculated estimates of ortholog protein distances of all urochordates as compared with the common ancestor, B. floridae, and tested for evidence of positive selection in rapid codon evolutionary rates in S. thompsoni using the branch-sites models implemented in the program PAML (Yang 2007). To calculate amino acid distances among orthologs, MSAs were used to calculate Jones–Taylor–Thornton (JTT) model distance using the program Fprotdist (see Berná et al. 2012 for methods). For positive selection analyses, classification of sites having significant evidence for being under positive selection required a significantly better fit of the branch-sites alternative model of positive selection over the null model [implemented as described in the PAML manual with a χ2 test (P value < 0.05) for significant improvements in maximum likelihood model fit] and identification using the Bayes empirical Bayes (BEB) method (P value >0.95). All analyses were done with S. thompsoni defined as the phylogenetic “foreground” and the four urochordates and B. floridae defined as the “background.”
Small RNA Analyses
Small RNA libraries were constructed using the Illumina Truseq Small RNA library preparation kit, without modifications with a gel selection targeting 9–45 nt. Post-sequencing on an Illumina Hiseq 2000, small RNA data sets were adaptor trimmed, and quality filtered using the Fastx toolkit on Galaxy. After adaptor trimming, data sets were size filtered to 18–24 nt for miRNA and 28–32 nt for piRNA. All genome and transcriptome alignments were performed using Bowtie 1, calling the best alignment (−k 1) and varying the number of mismatches allowed (−v). Repeat content of the piRNA pools was assessed using RepeatMasker. The piRNA pools were masked to both the chordate and de novo salp models and the output results were combined to generate the final estimates. Novel miRNA target prediction on the transcriptome was performed using miRanda, requiring strict alignment in the seed region. miRNA gene folding predictions were performed using RNAfold (Lorenz et al. 2011) implemented in Geneious 8.1.
Results and Discussion
For our assembly, we sequenced over 11 GB of DNA, yielding ∼20× genome coverage. The resulting salp genome assembly, Salp1.0, is 318,767,936 bp, covers >50% of the genome (table 1) based on a genome size estimate for this species of 602 Mb (±173 Mb) that was determined using a quantitative approach independent of a genome assembly. Salp1.0 genome sequence contains 478,293 contigs, indicating a somewhat fragmented genome assembly (N50 = 934). Approximately 33% of the assembled salp genome is predicted to be interspersed repetitive elements, demonstrating a higher repetitive element content than observed in many other urochordates with smaller genomes. For example, the genome of C. intestinalis and O. dioica are estimated to contain <20% interspersed repetitive elements (Chalopin et al. 2015). The majority of the repetitive elements identified in S. thompsoni are DNA elements, “unknown” elements, or long interspersed nuclear elements (LINEs) (table 2). The observed dearth of both short interspersed nuclear elements (SINEs) and long terminal repeats (LTRs) in the salp further demonstrates the uniqueness of the composition of this genome compared with other urochordates such as C. intestinalis and O. dioica (Chalopin et al. 2015). In addition to repetitive elements identified in the assembled contigs, 37.5% (40,834,207) of all reads sequenced were excluded from the final draft genome assembly as they were identified by Newbler as being too repetitive for accurate contig assignment. The ∼1:3 ratio of unassembled reads-to-assembled reads was consistently observed across different sequencing platforms and library preparation methods, indicating that an additional 37% of the genome is likely repetitive in nature. Combining these discarded reads unrepresented in predicted repeats with predicted interspersed repetitive element content based on assembled contigs, we estimate ∼60–70% of the salp genome may be repetitive in nature.
Table 1.
General Assembly Information for Genomic and Transcriptomic Assemblies for Salpa thompsoni
| Genome | |
|---|---|
| Number of individuals | 4 |
| Total amount of 454 sequence | 1,052,340,564 |
| Total amount of ion proton sequence | 10,774,831,593 |
| Number of contigs (or predicted transcripts) | 478,293 |
| N50 contig size (bp) | 934 |
| Total length of contigs (bp) | 318,767,936 |
| Average depth of coverage per base | 28.6 |
| Total number of bases sequenced (Mb) | 11,825 |
| Number of predicted genes (strictly ab initio predicted genes) | 5,467 (26,415) |
| Number of BLAST hits of predicted genes/transcipts | 16,823 |
| Trinity de novo predicted transcripts | 217,849 |
| Total coverage | ∼20× |
Table 2.
Repetitive Element Counts and Base-Pair Coverage for the Salp Genome
| Type of element | Count | Total base pairs | Percent of genome |
|---|---|---|---|
| SINEs | 45,324 | 2,935,478 | 0.92 |
| ALUs | 39 | 3,429 | 0 |
| LINEs | 136,014 | 14,682,786 | 4.61 |
| LINE1 | 230 | 32,570 | 0.01 |
| LINE2 | 6,222 | 818,015 | 0.26 |
| L3/CR1 | 13,793 | 1,676,529 | 0.53 |
| LTR elements | 113,301 | 8,988,469 | 2.82 |
| ERV_class I | 53 | 5,784 | 0 |
| DNA elements | 649,419 | 56,060,847 | 17.59 |
| hAT-Charlie | 8,128 | 889,239 | 0.28 |
| TcMar-Tigger | 215 | 30,504 | 0.01 |
| Unclassified | 280,392 | 20,865,094 | 6.55 |
| Total interspersed repeats | 103,532,674 | 32.48 | |
| Small RNAs | 511 | 49,308 | 0.02 |
| Satellites | 404 | 72,054 | 0.02 |
| Simple repeats | 64,031 | 2,454,620 | 0.77 |
| Low complexity | 10,916 | 502,724 | 0.16 |
| Total repeat content | 105,389,055 | 33.45 | |
While this is a seemingly high repeat content estimate for a relatively small eukaryotic genome, our estimates match those recently described for another urochordate with a similarly sized genome, the ascidian Botryllus schlosseri (genome size ∼725 Mb with 65% repetitive content) (Voskoboynik et al. 2013). Thus, the observed high repetitive element content for the S. thompsoni genome is likely a key contributing factor to the fragmented assembly and our inability to generate longer scaffolds (Salzberg et al. 2012). In an attempt to further define the content of the collected unassembled reads, these reads were fragmented into 50-mer bins and the resulting 50-mer sequences were searched against the complete repbase repeat library. Only 15% of the reads could be associated with a known reference sequence using our 50-mer approach; the majority of reads with a matching reference are associated with rDNA sequence (∼12% of all unassembled reads and ∼80% of those read sequences with a 50-mer match to a known reference, or ∼4.4% of the entire salp genome) (supplementary table S2, Supplementary Material online). This finding indicates that numerous rDNA repeats, which also present significant assembly challenges in many other eukaryotic genome assemblies (e.g., human; Eichler et al. 2004; Chaisson et al. 2015), may also contribute to our difficulties in building longer scaffolds in our genome assembly, even more so if rDNA sequences are distributed broadly in the salp genome.
Despite the challenges to assembly extensive rDNA duplicates throughout the salp genome present, such increases in copy number may provide insight into the adaptive evolution of this polar species and a key source of functional genetic variation in salps that shares similarities to genetic mechanisms found in humans. Variation among human individuals in rDNA copy numbers have been shown to positively correlate with gene expression patterns related to regulatory processes, particularly with regard to its interactions with mitochondrial function (Gibbons et al. 2014). Salps appear to be unique among the Urochordates in their enrichment of rDNA sequences whose possible role in interacting with mitochondria presents an intriguing opportunity to explore the impact of rDNA dosage on adaptive evolution. Interestingly, evidence from other polar species indicates that an increase in mitochondrial density per cell is a means by which some species compensate for the reduced aerobic capacity of extremely cold polar environments (Johnston et al. 1998). In humans, rDNA dosage is negatively correlated with mitochondrial number (Gibbons et al. 2014), and thus the increase in rDNA sequences in salps may likewise act as an alternative metabolic compensatory mechanism to dealing with cold environments. Moreover, increases in rDNA sequences may also have a broader functional role by affecting epigenetic patterns through the modulation of genome-wide gene expression (Paredes et al. 2011). Thus, further efforts to delineate patterns of rDNA variation both within and among salp and other polar species could provide insight into mechanisms by which repetitive variation contributes to polar adaptation.
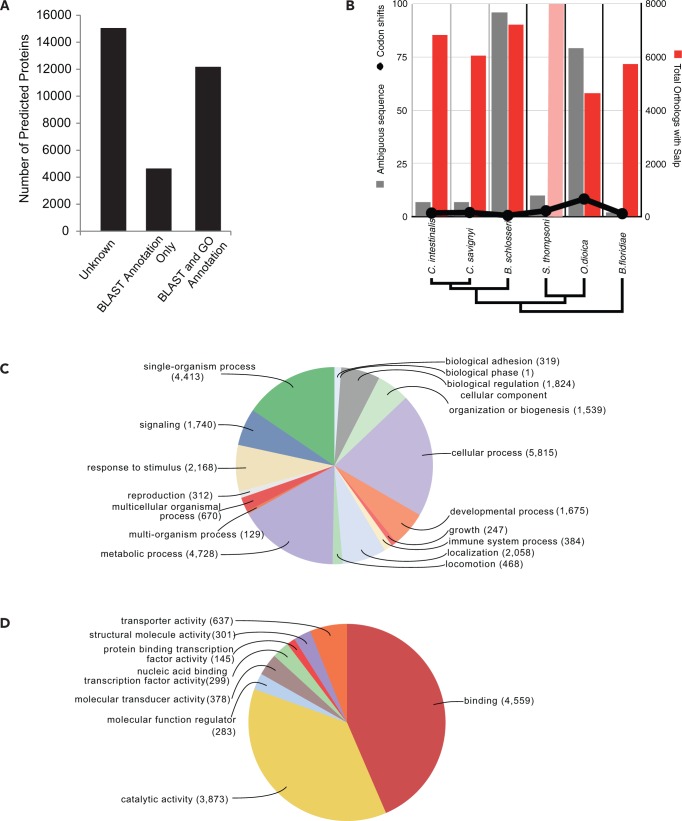
Despite fragmentation issues, the Salp1.0 genome sequence, in concert with a recent reference transcriptome assembly (Batta-Lona et al. 2016; submitted) and protein orthologs from other urochordates with well-annotated genomes (e.g., Ciona spp.), was sufficient to build initial gene models for this species. While the gene density per Mb was on the lower end of the range when compared with other urochordate genome assemblies, our estimates are comparable in quality to many other draft genome assemblies (table 3). Overall, 5,467 genes (average AED score = 0.18) were predicted and annotated, with support from multiple lines of evidence, and we identified a further 26,415 ab initio, nonredundant genes. Using this combined, nonredundant set of predicted genes, we successfully identified >50% (16,823) of these predicted gene sequences via BLASTP to the NCBI NR database and ∼38% (12,151) of the total protein predictions were associated with Gene Ontology functional information (fig. 2A).
Table 3.
Estimated Percentages of CEGMA Core Eukaryotic Genes Sequenced Either Completely or Partial in Each Urochorate Species Genome (Gray Rows) As Well As Other Select Species
| Species | Complete (%) | Partial (%) | Total (%) | Estimated genome size (Mb) | Number of predicted genes per Mb |
|---|---|---|---|---|---|
| Ciona intestinalis | 87.90 | 6.45 | 94.35 | 160 | 108 |
| Ciona savignyi | 83.06 | 12.50 | 95.56 | 190 | 106 |
| Oikopleura dioica | 85.48 | 6.05 | 91.53 | 70 | 257 |
| Botryllus schlosseri | 78.63 | 16.53 | 95.16 | 600 | 52 |
| Salpa thompsoni | 35.48 | 40.73 | 76.21 | 602 | 53 |
| Nematostella vectensis | 76.61 | 21.78 | 98.39 | 450 | 61 |
| Trichoplax adhaerens | 97.58 | 0.40 | 97.98 | 98 | 117 |
| Ixodes scaluparis | 37.50 | 35.48 | 72.98 | 2,100 | 10 |
| Strongylocentrotus purpuratus | 49.60 | 39.11 | 88.71 | 800 | 29 |
| Bombyx mori | 16.53 | 4.44 | 20.97 | 530 | 21 |
Fig. 2.—
Salpa thompsoni gene annotations. (A) Annotation status of S. thompsoni predicted proteins delineated by validation method. (B) The number of ambiguous sequences, codon shifts (left) and total orthologues (right) with S. thompsoni are shown for each species (bottom). Phylogenetic relationships for each species are indicated. (C and D) Pie chart summary of level 2 gene ontology terms associated with biological process (C) and molecular function (D) categories for all annotated Annotation status of S. thompsoni predicted proteins.
Comparisons to CEGMA (Core Eukaryotic Genes Mapping Approach) genes showed a recovery of 75% of the 248 highly conserved core eukaryotic orthologs within Salp1.0 (table 3), however a large percentage of these sequences (40.73%) were either incomplete representations of coding regions or truncated matches to reference sequences. These gene predictions indicate that while we have built a reference genome that contains a large majority of the functional gene regions, it will need additional finishing to establish sufficient assembly N50 values in order to predict full-length coding sequences for all proteins. Moreover, a comparison of the Salp1.0 assembly to the C. intestinalis protein database further supports our finding that predicted protein sequences are likely not complete; only 11.24% of 13,186 BLAST hits to this database carried a protein query that aligned to >80% of the protein subject match from the database. Thus, while our assembly is a broad representation of the entire genome, building larger scaffolds and complete gene ORFs requires genome finishing to improve gene prediction estimates and coverage.
In addition to the identification of predicted proteins, we identified useful evolutionary and population-level markers for this key indicator species. Using reciprocal best-BLAST, we identified 2,199 of the orthologous protein sequences in S. thompsoni that are present in all four other urochordates for which there are currently genome sequences (Dehal et al. 2002; Small et al. 2007; Denoeud et al. 2010; Voskoboynik et al. 2013), as well as 1,898 orthologs that are shared among these Urochordates (including S. thompsoni) and the highly divergent cephalochordate Branchiostoma floridae (Putnam et al. 2008) (fig. 1A). In pairwise comparisons with Salp1.0, there were 7,207 orthologs with B. schlosseri, 6,839 orthologs with C. intestinalis, 6,056 orthologs with C. savignyi, 4,650 orthologs with O. dioica, and 5,742 orthologs with B. floridae (fig. 2B). The overall lower number of identified orthologous groups of genes shared across all five species compared with the higher number of orthologs we observed in pairwise comparisons with other urochordate species is likely due to a difficulty in establishing orthology for rapidly evolving protein sequences across highly divergent taxa, as has been previously described in Urochordates (Berná et al. 2012). Additionally, because our genome sequencing strategy included DNA from multiple samples, we are able to generate predictions for 109,958 SNP and 9,782 indel variable sites for S. thompsoni. These resources will serve as a valuable reference for future phylogenomic and population genetic studies.
From a functional perspective, the genome assembly reveals S. thompsoni to be unique from the other urochordates in terms of predicted gene ontologies, likely due at least in part to either its polar adaptations or the different life histories of the ascidians (typically characterized by larval pelagic and sessile adult stages) and appendicularians (larval only stage). Gene ontologies for S. thompsoni are enriched for Molecular Functions, such as binding and molecular function regulators, yet are underrepresented for catalytic activity, transduction and transport functions (fig. 2C). In terms of ontologies categorized as Biological Processes, S. thompsoni is enriched for a wide range of other functions including growth, reproduction, locomotion, behavior, and biological phase, the expansion of these gene ontology functional categories is consistent with the observed complex life history and free-swimming body form of this species, yet is underrepresented for Cellular Processes and Single-Organism Processes (fig. 2D). However, predicted S. thompsoni orthologs examined for evidence of rapid evolution, indicative of positive selection, highlight molecular functions and biological processes that may be specific to the adaptive history of planktonic salp, or S. thompsoni in particular.
Constrained by the ability to generate sufficiently long multiple sequence alignments to test for signals of positive selection across the four urochordate lineages and a cephalochordate outgroup lineage, 1,831 of the 1,898 orthologs identified across all five lineages were further interrogated for signals of positive selection. Of these orthologs, 192 were discarded since they contained ambiguous nucleotide calls (i.e., “N” base call) and 864 were discarded since they contained a coding frameshift in at least one of the five species. While coding frameshifts could also be the result of sequence ambiguity (ambiguous nucleotide calls were observed most frequently in B. schlosseri and O. dioica sequences), overall they were not a common factor (fig. 2B) (96, 79, 10, 7, 7, and 2 sequences in B. schlosseri, O. dioica, S. thompsoni, C. intestinalis, B. floridae, and C. savignyi, respectively). Coding frame shifts attributed to sequence mutation appeared with higher frequency across various species, but were most frequently found in O. dioica sequences (fig. 2B) (672, 231, 165, 146, 122, and 55 sequences in O. dioica, S. thompsoni, C. savignyi, C. intestinalis, B. floridae, and B. schlosseri, respectively). Collectively, these observations indicate that either the O. dioica gene predictions are of lower confidence than the other species or that O. dioica possesses proportionally more genes that have been significantly altered. Notably, despite the fragmented genome assembly, S. thompsoni gene sequence predictions are comparable to most other urochordate gene predictions for these common urochordate/cephalochordate orthologs.
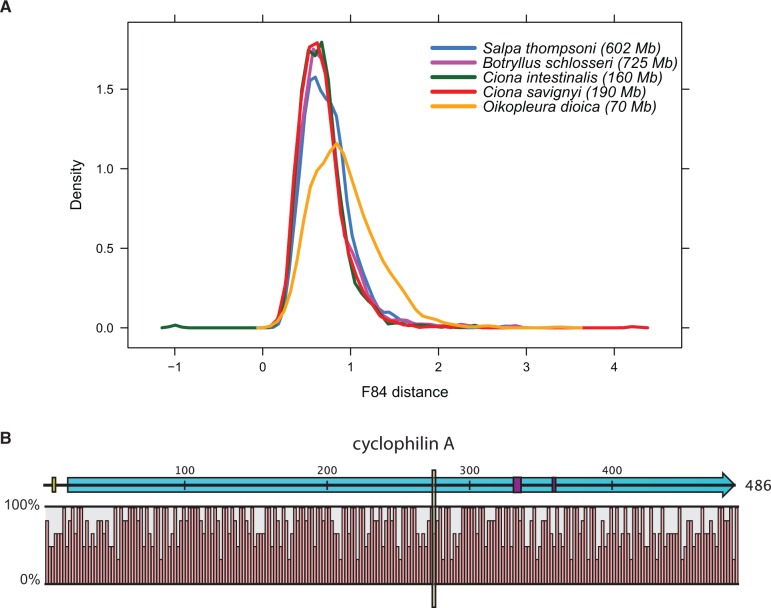
Elevated mutation rates could be a major contributing factor as to why O. dioica has a larger number of coding frame shifts than the other urochordate species. Previously, urochordate species, specifically O. dioica, have been observed to have extremely high mutation rates across their genomes compared with vertebrate species (O. dioica evolving at 3 times the rate of vertebrates; Ciona spp. evolving at 1.5 the rate of vertebrates) (Berná et al. 2009, 2012). Using the aforementioned orthologous groups, we compared the evolutionary rates among all five urochorate species to the common recent ancestor, B. floridae (fig. 3A). Rapid rates of evolution across all urochordate species were observed; O. dioica was the fastest evolving species of the five. Interestingly, S. thompsoni was observed to evolve at a rate slightly elevated compared with B. schlosseri and Ciona spp., suggesting that high mutation rates are both independent of genome size and are a broad taxonomic characteristic of the Tunicata. Given this accelerated rate of mutation in this group, salps provide a good complementary system for identifying the underlying mechanism for this phenomenon.
Fig. 3.—
(A) Kernel density plots of ortholog sequence divergence from Cephalochordate Branchiostoma floridae for each Urchordate species (see legend) reveal similar evolutionary rates among Urochordate species, with the exception of Oikopleura dioica which appears to evolve more rapidly than the others. Orthology groups (n = 967) investigated for positive selection were used in this analysis. (B) Schematic of cyclophilin A protein sequence with percent sequence conservation at each site shown for other Urochordate and Cephalochorate species analyzed. Yellow box indicates site under positive selection; purple boxes indicate metal ion-binding site; blue arrow indicates propyl isomerase functional domain.
After discarding alignments with ambiguities and reducing the number of orthologs suitable for molecular rates of evolution analysis, 894 orthologous gene groups (of the initial 1,898 gene groups) were tested for positive selection. Of these, 36 carried evidence of rapid evolution consistent with a “branch-sites” hypothesis of positive selection in S. thomponsi (supplementary table S3, Supplementary Material online). Across these 36 predicted genes within the salp, 155 sites were identified as undergoing rapid evolution; of these sites, 108 were located within known conserved protein domains, whereas 47 could not be attributed to a known protein domain and may thus be associated with previously undescribed functional domains specific to S. thompsoni and/or closely related taxa. From a functional perspective, the genes identified as experiencing positive selection were generally associated with gene ontology Biological Processes terms, such as metabolic cellular and single-organism processes, and Molecular Function terms such as binding and catalytic activity (supplementary table S3, Supplementary Material online). In addition to a variety of metabolic pathways, many genes associated with the 40S and 60S ribosomal subunit were significantly over-represented as under positive selection (supplementary table S3, Supplementary Material online). While speculative, rapid evolution in these ribosomal genes may have been facilitated by the extensive expansion of rDNA sequences in the salp genome and, may thus be a result of extensive gene duplication, contributing to the release of selective constraints traditionally experienced by these genes. Alternatively, either its nature as a zooplankton colonial species, and/or its necessity to adapt to polar environments may be driving forces behind the groups of rapidly evolving genes observed in the salp.
Metabolic processes in polar species are under a wide range of stresses related to thermal extremes and their concomitant influences on chemical rates of reaction. It has been shown that changes in temperature can have significant effects on both the activity and production of ribosomes in planktonic species, increasing the number of ribosomes in response to lower temperature (Toseland et al. 2013). Polar environments have been identified as having contributed to the rapid evolution of select gene groups: ribosomal genes (Pucciarelli et al. 2005), genes associated with cellular respiration (Welch et al. 2014), and lipid oxidation genes (Windisch et al. 2011). Within S. thompsoni, the rapid evolution of genes within each of these groups was observed (supplementary table S3, Supplementary Material online). Specifically, ribosomal genes (e.g., RPLP0, RPL10, and RPS14), genes involved in glycolysis (e.g., DLAT), neoglycogenesis (e.g., PCKG), oxidoreduction (e.g., NUOC), and lipid metabolism (e.g., ECHM) all show evidence for signals of positive selection. While this salp genome provides some suggestive patterns of gene evolution with respect to environment, more rigorous testing is needed to definitively attribute the forces driving this pattern of gene evolution to Darwinian selection specifically in this lineage rather than phylogenetic bias. As more genome sequences for polar zooplankton species and other Thaliaceans become available, this salp genome will become an important component in the disentanglement of evolutionary forces that impact polar adaptation and genome evolution.
Capitalizing on a recent companion study of population-level changes in gene expression of S. thompsoni, we were afforded the opportunity to cross-reference the genes identified as under positive selection to those identified as susceptible to changing environmental factors (Batta-Lona et al. 2016; submitted). Six genes with signal for positive selection (RPL10, EIF1A, cyclophilin A, RPS3C, RPLP0, and RPS14—supplementary table S3, Supplementary Material online) concomitantly showed differential gene expression across samples collected in two different environmental conditions, austral spring and summer, and thus demonstrated evidence for environmentally affected gene expression or reproductive lifestyle expression profiles (Batta-Lona et al. 2016; submitted). Of these six genes, only one of them carried 100% identity to its cognate transcript, whereas the other five showed < 90% similarity to their predicted protein coding transcript, and thus may represent paralogs for each gene or highly divergent alleles within the population. Our inability to definitively assign these five genes as synonymous or paralogous prompted their exclusion from further analyses. The gene identified with 100% identity as cyclophilin A is of particular interest because of its involvement in clonal reproduction, allorecognition and immune function (Liu et al. 1991; Marks 1996; Oren et al. 2013). Differential gene expression of cyclophilin A further supports its functional classification, because it is down-regulated in salps collected during the summer, when the population consisted of sexually reproducing salps, compared with the spring, when clonally reproducing salps predominated. Of the two coding sites identified as subject to positive selection within this gene, one falls outside the predicted functional domain associated with proline isomerization (Prolyl isomerase—PFAM00639), whereas the other is nested within the domain (fig. 3B). In addition to proline isomerization, this second site may also affect the binding properties of cyclophilin A, because it is proximal to—but not directly within—metal binding sites in this known functional domain (Oren et al. 2013). In addition to cyclophilin A, there are three other proteins (i.e., peroxidasin, PDCD6, OGFOD1) with possible immune functions that also show signals of positive selection (supplementary table S3, Supplementary Material online). Immune function proteins such as cyclophilin A have been shown to be key players in allogeneic interactions (Oren et al. 2013) and, thus, processes such as clonal reproduction and colonial life-styles.
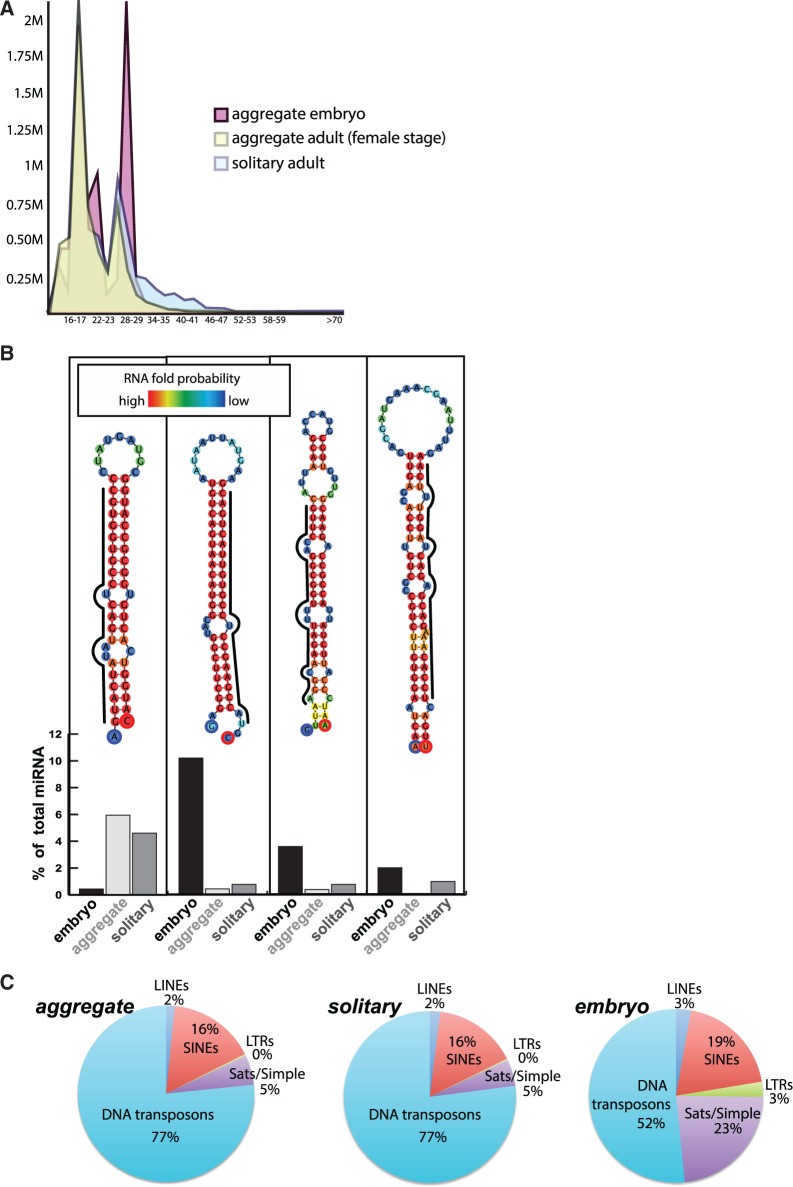
The development of a draft assembly for the salp, while complementing previous transcriptomic profiling, afforded an opportunity to explore functional classes of small RNAs shared among developmental stages and those unique to the differing reproductive cycles found in this species (fig. 1C), with specific attention to further annotating small RNA genes in the salp genome assembly. We targeted the small RNAs encompassing the micro RNAs (miRNAs) and piwi-interacting RNAs (piRNAs) from the following tissues: adult whole body asexual, adult whole body sexual female stage, and an embryo whole body that would develop into a sexually reproducing salp yet is found within an asexual parent. Post-sequencing, reads were groomed and binned by size; each sample carried a strong miRNA signal in the 22-nt size range, but there was a dramatic increase in piRNAs specific to the embryo sample (28 nt size range) (fig. 4A).
Fig. 4.—
(A) Size distribution of small RNAs post-trimming across embryo and adult solitary salp and adult female samples. (B) miRNA precursor fold predictions for each novel miRNA. Nucleotide is color coded by probability, the mature miRNA is indicated by a black line for each hairpin, and sample distribution for each is indicated. (C) Classifiable piRNA content for aggregate female stage, solitary adult and embryo samples, excluding tRNA, rRNA and unmasked (nonrepeat) sequences within the piRNA size range.
Using variable mapping parameters (see “Material and Methods” section), we mapped each data set to the Salp1.0 genome assembly and the newly assembled transcriptome (Batta-Lona et al. 2016, submitted) with a high percentage of reads mapping to each for both the miRNA and piRNA sequence pools (tables 4 and 5). Reads for each small RNA library, delineated by sample and small RNA class, were further classified using BLAST, miRANDA, miRBase, and Fold predict. Among the top 20 represented small RNAs in each miRNA pool, many miRNAs were found to have an annotated homolog in Ciona (Norden-Krichmar et al. 2007; Kozomara and Griffiths-Jones 2014) yet six miRNAs were found that lacked identity to any previously annotated small RNA from any species (supplementary table S5, Supplementary Material online). Four of these novel miRNAs were subsequently validated by using the genome scaffold to which they mapped, capturing sequence around the small RNA read that produced a putative miRNA, and further tested for miRNA precursor structures (i.e., a miRNA gene) (Lindsay et al. 2012). All novel miRNAs were confirmed as derived from a bone fide miRNA gene, with a defined hairpin structure and ORF that would produce a predicted miRNA matching the novel candidates (fig. 4B). Of these novel salp miRNAs, one is expressed predominantly in adult tissues while three are more abundant in the developing embryo.
Table 4.
Mapping Statistics for Predicted Small RNAs to the Genome (Top) and Transcriptome (Bottom) with Different Mismatched Nucleotide Frequencies Allowed (0–2) (miRNAs)
| miRNA sample | Mismatches |
||
|---|---|---|---|
| 0 | 1 | 2 | |
| Genome | |||
| Aggregate embryo | 81.18% | 89.33% | 98.30% |
| Solitary | 70.77% | 80.02% | 93.57% |
| Aggregate female | 71.79% | 82.57% | 93.86% |
| Transcriptome | |||
| Aggregate embryo | 74.77% | 81.86% | 90.51% |
| Solitary | 71.68% | 79.96% | 89.13% |
| Aggregate female | 69.60% | 79.12% | 87.31% |
Table 5.
Mapping Statistics for Predicted Small RNAs to the Genome (Top) and Transcriptome (Bottom) with Different Mismatched Nucleotide Frequencies Allowed (0–3) (piRNAs)
| piRNA sample | Mismatches |
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| Genome | ||||
| Aggregate embryo | 73.89% | 95.06% | 96.85% | 97.28% |
| Solitary | 53.36% | 78.49% | 82.29% | 83.69% |
| Aggregate female | 59.20% | 84.75% | 88.95% | 90.65% |
| Transcriptome | ||||
| Aggregate Embryo | 73.71% | 94.70% | 96.24% | 96.59% |
| Solitary | 61.37% | 86.58% | 89.13% | 89.98% |
| Aggregate Female | 63.83% | 89.10% | 92.11% | 92.99% |
A recent examination of miRNA conservation across deuterostomes revealed that increased rates of miRNA family acquisition were coincident with major morphological innovations within vertebrate lineages and that the expansion of miRNAs may have actually facilitated innovations in vertebrate development and morphological complexity (Heimberg et al. 2008). For example, bursts in the number of novel miRNA families occurred in the base of vertebrates (48 miRNA families acquired) and in the lineages leading to therian mammals (63 miRNA families acquired) and primates (414 miRNA families acquired) (reviewed in Candiani 2012). However, subsequent studies of two of the three Tunicate classes, appendicularians (O. dioica) and ascidians (C. intestinalis and C. savignyi), revealed evidence of both loss and acquisition of new miRNA families, albeit the latter at rates far lower that observed in primates (Fu et al. 2008; Dai et al. 2009; Shi et al. 2009; Hendrix et al. 2010; Candiani 2012; Hertel and Stadler 2015). These data support the hypothesis that while conserved miRNAs likely define the “robustness” of developmental programs (e.g., temporally) (Fu et al. 2008), lineage specific morphological and developmental novelty is coupled with novel miRNA acquisition across urochordates (Candiani 2012; Hertel and Stadler 2015). While the presence of embryo-specific miRNAs in salps implicates an RNA interference regulatory mechanism involved in developmental processes, the specific origin and function of these novel miRNAs do not appear conserved within other urochordates, including tunicates, and thus may represent a unique paradigm for early development and post-transcriptional gene regulation in this tunicate class of species. The scarce available small RNA scans in closely related species, however, limit our confidence in assigning species-specific function to these miRNAs rather than a function in Thaliacian-specific developmental novelty.
For our piRNA analyses, we used a novel pipeline to define the repeat family from which a particular small RNA in this class is derived. We used a combination of chordate and de novo repeat models to maximize identification of novel repeat elements in this species. We find that, in addition to the increased representation of piRNAs in the total small RNAs overall within the embryo sample (fig. 4A), we find that the composition of the total piRNA repeat types is markedly different between the embryo and the adults (fig. 4C). As with the miRNA analyses, the piRNAs are similar among adults, despite their different reproductive stage (fig. 1C).
In addition to novel miRNAs, the embryo displays a significant shift in piRNA production, in contrast to previously predicted models of piRNA processing (Brennecke et al. 2007), which are thought to be seeded from the germ line of the parental genome into the early embryo to protect against potentially deleterious repeat activity. In the case of the salp, the embryo is the product of asexual reproduction and, thus, has not received germ line material from two parents. Notably, the embryo produced by asexual reproduction will become capable of sexual reproduction upon maturity. Thus, it is possible that the piRNAs we observe are produced in the germ cells present in this early embryo, although their initial production is independent of parental contribution and thus is initiated de novo.
Conclusions
We have assembled the first draft genome sequence for Salpa thompsoni (Urochordata, Thaliacea), as well as the first estimates of genome size and gene predictions for this species. This assembly, while still in need of further finishing to reduce fragmentation and generate longer sequence scaffolds, was used to establish an extensive resource of gene, repetitive element, and small RNA gene predictions. These resources provide a significant step forward in the development of this species as a model species for studying urochordate evolution, the genetics underlying the adaptation to a polar, planktonic environment and the enhancement of molecular processes underlying organism–environment interactions that determine responses of this key species to climate change in Antarctic pelagic ecosystems. Ortholog and SNP loci identification has established significant resources for both further phylogenomic and population genetic studies. Patterns in protein divergence in S. thompsoni appear to confirm the urochordate characteristic of elevated rates of genome evolution, indicating that urochordates may be particularly responsive to selective pressures. For instance, genome size, repetitive element content and patterns of positive selection in ribosomal genes indicate that expansion of rDNA gene families is extensive in this species, and may be related to metabolic adaptations to polar environments. Additionally, the rapid evolution of the variably expressed gene cyclophilin A supports the functional importance of immune gene in urochordate biology, specifically the unusual complex life history, with both solitary and colonial stages. This rate of evolutionary change is also notable in the small RNA repertoire of this species that is characterized by both conserved and novel miRNAs implicated in embryonic development, as well as a possibly novel piRNA biogenesis mechanism. Overall, the S. thompsoni genome provides an important reference point for the continued understanding of the adaptive evolution of polar and planktonic species as well as the elucidation of the underlying biology of an important group of chordate-related organisms.
Supplementary Material
Supplementary tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgements
This work was funded by: National Science Foundation [grant number: 1044982 to A. Bucklin and R. O'Neill; and grant number: 0920088 to M. O'Neill and R. O'Neill]. We thank P.H. Wiebe (Woods Hole Oceanographic Institution) for his assistance with the manuscript. Photographs of living specimens of Salpa thompsoni were taken by L.P. Madin (Woods Hole Oceanographic Institution).
Supplementary Material
Literature Cited
- Abrusán G, Grundmann N, DeMester L, Makalowski W. 2009. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25:1329–1330. [DOI] [PubMed] [Google Scholar]
- Alldredge A, Madin L. 1982. Pelagic tunicates: unique herbivores in the marine plankton. Bioscience 32:655–663. [Google Scholar]
- Atkinson A, Siegel V, Pakhomov E, Rothery P. 2004. Long-term decline in krill stock and increase in salps within the Southern Ocean. Nature 432:100–103. [DOI] [PubMed] [Google Scholar]
- Batta-Lona PG, Maas AE, O’Neill RJ, Wiebe PH, Bucklin A. 2016. (submitted). Transcriptome-wide profiles of gene expression of Salpa thompsoni in relation to variation of the pelagic environment of the Southern Ocean. Polar Biol. [Google Scholar]
- Berná L, Alvarez-Valin F, D’Onofrio G. 2009. How fast is the sessile Ciona?. Comp Funct Genomics 2009:875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná L, D’Onofrio G, Alvarez-Valin F. 2012. Peculiar patterns of amino acid substitution and conservation in the fast evolving tunicate Oikopleura dioica. Mol Phylogenet Evol. 62:708–717. [DOI] [PubMed] [Google Scholar]
- Brennecke J, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103. [DOI] [PubMed] [Google Scholar]
- Candiani S. 2012. Focus on miRNAs evolution: a perspective from amphioxus. Brief Funct Genomics 11:107–117. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin D, Naville M, Plard F, Galiana D, Volff J-N. 2015. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol. 7:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Ishimaru T, Hosie GW, Wright SW. 1999. Population structure change of Salpa thompsoni from austral mid-summer to autumn. Polar Biol. 22:341–349. [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Dai Z, et al. 2009. Characterization of microRNAs in cephalochordates reveals a correlation between microRNA repertoire homology and morphological similarity in chordate evolution. Evol Dev. 11:41–49. [DOI] [PubMed] [Google Scholar]
- Daponte MC, Capitanio FL, Esnal GB. 2001. A mechanism for swarming in the tunicate Salpa thompsoni (Foxton, 1961). Antarctic Sci. 13:240–245. [Google Scholar]
- Dehal P, et al. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298:2157–2167. [DOI] [PubMed] [Google Scholar]
- Denoeud F, et al. 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330:1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducklow HW, et al. 2013. West Antarctic peninsula: an ice-dependent coastal marine ecosystem in transition. Oceanography 26:190–203. [Google Scholar]
- Eichler EE, Clark RA, She X. 2004. An assessment of the sequence gaps: unfinished business in a finished human genome. Nature Rev Genet. 5:345–354. [DOI] [PubMed] [Google Scholar]
- Franssen SU, et al. 2014. Genome-wide transcriptomic responses of the seagrasses Zostera marina and Nanozostera noltii under a simulated heatwave confirm functional types. Marine Genomics 15:65–73. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Adamski M, Thompson EM. 2008. Altered miRNA repertoire in the simplified chordate, Oikopleura dioica. Mol Biol Evol. 25:1067–1080. [DOI] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Yu S, Lemos B. 2014. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat Commun. 5:4850. [DOI] [PubMed] [Google Scholar]
- Godeaux J, Bone Q, Braconnot J-C. 1998. Anatomy of Thaliacea In: Bone Q, editor. The biology of pelagic tunicates. Oxford: Oxford University Press; p. 1–24. [Google Scholar]
- Govindarajan AF, Bucklin A, Madin LP. 2011. A molecular phylogeny of the Thaliacea. J Plankton Res. 33:843–853. [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. 2008. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 105:2946–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix D, Levine M, Shi W. 2010. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 11:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Stadler PF. 2015. The expansion of animal MicroRNA families revisited. Life (Basel) 5:905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Gibson-Brown JJ. 2003. The Ciona intestinalis genome: when the constraints are off. Bioessays 25:529–532. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston II, Calvo J, Guderley YH. 1998. Latitudinal variation in the abundance and oxidative capacities of muscle mitochondria in perciform fishes. J Exp Biol. 201(Pt 1):1–12. [DOI] [PubMed] [Google Scholar]
- Kawada T, et al. 2011. Peptidomic analysis of the central nervous system of the protochordate, Ciona intestinalis: homologs and prototypes of vertebrate peptides and novel peptides. Endocrinology 152:2416–2427. [DOI] [PubMed] [Google Scholar]
- Kokubun N, Kim J-H, Takahashi A. 2013. Proximity of krill and salps in an Antarctic coastal ecosystem: evidence from penguin-mounted cameras. Polar Biol. 36:1857–1864. [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay J, et al. 2012. Unique small RNA signatures uncovered in the tammar wallaby genome. BMC Genomics 13:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807–815. [DOI] [PubMed] [Google Scholar]
- Loeb V, Santora J. 2012. Population dynamics of Salpa thompsoni near the Antarctic Peninsula: growth rates and interannual variations in reproductive activity (1993–2009). Prog Oceanogr. 96:93–107. [Google Scholar]
- Lorenz R, et al. 2011. ViennaRNA package 2.0. Algorithms Mol Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madin LP, Deibel D. 1998. Feeding and energetics of Thaliaceans In: Bone Q, editor. The biology of pelagic tuncates. Oxford: Oxford University Press; p. 43–60. [Google Scholar]
- Marks AR. 1996. Cellular functions of immunophilins. Physiol Rev. 76:631–649. [DOI] [PubMed] [Google Scholar]
- Meyer B, et al. 2015. Pyrosequencing and de novo assembly of Antarctic krill (Euphausia superba) transcriptome to study the adaptability of krill to climate induced environmental changes. Mol Ecol Res. 15(6):1460–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden-Krichmar TM, Holtz J, Pasquinelli AE, Gaasterland T. 2007. Computational prediction and experimental validation of Ciona intestinalis microRNA genes. BMC Genomics 8:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, et al. 2013. Marine invertebrates cross phyla comparisons reveal highly conserved immune machinery. Immunobiology 218:484–495. [DOI] [PubMed] [Google Scholar]
- Pakhomov EA. 2004. Salp/krill interactions in the eastern Atlantic sector of the Southern Ocean. Deep-Sea Res II. 51:2645–2660. [Google Scholar]
- Pakhomov EA, Froneman PW. 2004. Mesozooplankton dynamics in the eastern Atlantic sector of the Southern Ocean during the austral summer 1997/1998. 2. Grazing impact. Deep-Sea Res II. 51:2617–2631. [Google Scholar]
- Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. 2011. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 7:e1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Kremer P, Madin LP. 2009. Defecation rates of the salp Salpa thompsoni and its potential contribution to vertical flux in the Southern Ocean. Marine Biol. 156:455–467. [Google Scholar]
- Pucciarelli S, Marziale F, Di Giuseppe G, Barchetta S, Miceli C. 2005. Ribosomal cold-adaptation: characterization of the genes encoding the acidic ribosomal P0 and P2 proteins from the Antarctic ciliate Euplotes focardii. Gene 360:103–110. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. http://www.nature.com/nature/journal/v453/n7198/suppinfo/nature06967_S1.html. [DOI] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. 2011. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One 6:e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L, Schroder J. 2011. Correcting errors in short reads by multiple alignments. Bioinformatics 27:1455–1461. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, et al. 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N, Kawashima T, Shoguchi E, Satou Y. 2006. Urochordate genomes. Genome Dyn. 2:198–212. [DOI] [PubMed] [Google Scholar]
- Seo H-C, et al. 2001. Miniature genome in the marine chordate Oikopleura dioica. Science 294:2506. [DOI] [PubMed] [Google Scholar]
- Shi W, Hendrix D, Levine M, Haley B. 2009. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol. 16:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K, Brudno M, Hill M, Sidow A. 2007. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 8:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2008. –2010. RepeatModeler Open-1.0. Available from: http://www.repeatmasker.org/.
- Smit AFA, Hubley R, Green P. 1996–2010. RepeatMasker Open-3.0. Available from: http://www.repeatmasker.org/.
- Stach T. 2007. Ontogeny of the appendicularian Oikopleura dioica (Tunicata, Chordata) reveals characters similar to ascidian larvae with sessile adults. Zoomorphology 126:203–214. [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori N, et al. 2004. T-box genes in the ascidian Ciona intestinalis: characterization of cDNAs and spatial expression. Dev Dynamics 230:743–753. [DOI] [PubMed] [Google Scholar]
- Toseland A, et al. 2013. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat Clim Change 3:979–984. Available from: http://www.nature.com/nclimate/journal/v3/n11/abs/nclimate1989.html-supplementary-information. [Google Scholar]
- Villarino GH, Bombarely A, Giovannoni JJ, Scanlon MJ, Mattson NS. 2014. Transcriptomic analysis of Petunia hybrida in response to salt stress using high throughput RNA sequencing. PLoS One 9:e94651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik A, et al. 2013. The genome sequence of the colonial chordate, Botryllus schlosseri. eLife 2:e00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch AJ, et al. 2014. Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol Evol. 6:433–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe P, et al. 2010. Deep-sea holozooplankton species diversity in the Sargasso Sea, Northwestern Atlantic Ocean. Deep-Sea Res II 57: 2157–2166. [Google Scholar]
- Wilhelm J, Pingoud A, Hahn M. 2003. Real-time PCR-based method for the estimation of genome sizes. Nucleic Acids Res. 31:e56e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windisch HS, Kathover R, Portner HO, Frickenhaus S, Lucassen M. 2011. Thermal acclimation in Antarctic fish: transcriptomic profiling of metabolic pathways. Am J Physiol Regul Integr Comp Physiol. 301:R1453–R1466. [DOI] [PubMed] [Google Scholar]
- Xue W, et al. 2013. L_RNA_scaffolder: scaffolding genomes with transcripts. BMC Genomics 14:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadetie F, et al. 2012. Conservation and divergence of chemical defense system in the tunicate Oikopleura dioica revealed by genome wide response to two xenobiotics. BMC Genomics 13:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.