Abstract
LINE-Alu-VNTR-Alu-like (LAVA) elements comprise a family of non-autonomous, composite, non-LTR retrotransposons specific to gibbons and may have played a role in the evolution of this lineage. A full-length LAVA element consists of portions of repeats found in most primate genomes: CT-rich, Alu-like, and VNTR regions from the SVA retrotransposon, and portions of the AluSz and L1ME5 elements. To evaluate whether the gibbon genome currently harbors functional LAVA elements capable of mobilization by the endogenous LINE-1 (L1) protein machinery and which LAVA components are important for retrotransposition, we established a trans-mobilization assay in HeLa cells. Specifically, we tested if a full-length member of the older LAVA subfamily C that was isolated from the gibbon genome and named LAVAC, or its components, can be mobilized in the presence of the human L1 protein machinery. We show that L1 proteins mobilize the LAVAC element at frequencies exceeding processed pseudogene formation and human SVAE retrotransposition by > 100-fold and ≥3-fold, respectively. We find that only the SVA-derived portions confer activity, and truncation of the 3′ L1ME5 portion increases retrotransposition rates by at least 100%. Tagged de novo insertions integrated into intronic regions in cell culture, recapitulating findings in the gibbon genome. Finally, we present alternative models for the rise of the LAVA retrotransposon in the gibbon lineage.
Keywords: gibbon, retrotransposon, LAVA, LINE-1, trans-mobilization assay
Introduction
Repetitive and repeat-derived sequences have been estimated to comprise 66–69% of the human genome (Lander et al. 2001; de Koning et al. 2011), with similar proportions being annotated in other non-human primate genomes (The Chimpanzee Sequencing and Analysis Consortium 2005; Gibbs et al. 2007; Locke et al. 2011). The majority of these sequences are non-long terminal repeat (LTR) retrotransposons, which include the LINE-1 (L1) and Alu families, as well as the composite SVA (SINE-R-VNTR-Alu) family (Cordaux and Batzer 2009). In addition to being the largest contributors of genetic variation during species evolution, insertions of these elements have been linked to human diseases, including cancer (Callinan and Batzer 2006; Belancio et al. 2009; Belancio et al. 2010; Konkel and Batzer 2010; Hancks and Kazazian 2012; Kaer and Speek 2013). Further, the high sequence similarity between copies within each family can result in large-scale structural changes by inducing non-homologous allelic recombination (NAHR) events that can result in duplications, deletions, and inversions of the intervening sequence, including genes and other functional sequences, hence resulting in a disease phenotype (Han et al. 2007b; Belancio et al. 2008; Han et al. 2008; Lee et al. 2008; Belancio et al. 2010).
LINE-Alu-VNTR-Alu-like (LAVA) elements are a newly discovered family of non-autonomous, composite, non-LTR retrotransposons that can be classified in 22 subfamilies which originated and expanded exclusively in one group of primates: the gibbons, or small apes (Carbone et al. 2012; Carbone et al. 2014) (supplementary fig. S1, Supplementary Material online). Gibbons are part of the superfamily Hominoidea and shared a common ancestor with hominids (humans and other great apes) around 17 million years ago (mya) (Carbone et al. 2014). Although evolutionarily close to the hominids, gibbons have some very distinct genomic traits, including highly divergent karyotypes as a consequence of numerous chromosomal rearrangements that occurred during a relatively short period of time. As a consequence, each of the four gibbon genera (Nomascus, Hylobates, Hoolock, and Symphalangus) carries a different karyotype with chromosome numbers ranging from 38 to 52 (Muller et al. 2003) although the genera diverged from each other as recently as about 5 mya (Veeramah et al. 2015). The recent analysis of the first gibbon genome assembly uncovered evidence that the appearance of the LAVA element might have been associated with the accelerated karyotype evolution in the gibbon lineage through the disruption of genes linked to chromosome segregation (Carbone et al. 2014). Finally, one of the gibbon genera, the Hoolock, displays a remarkable expansion of the LAVA element detectable by fluorescent in situ hybridization in almost all centromeres (Carbone et al. 2012), further highlighting a link between the LAVA element and the karyotype variability observed in gibbons. Altogether, these observations suggest that the LAVA element was an evolutionary novelty that may have greatly impacted the evolution of the gibbon lineage through several different avenues.
The LAVA element can be described as a “composite-of-composites” because, together with fragments from other repeats, it includes a portion from the hominoid-specific SVA element, a composite non-LTR retrotransposon itself (fig. 1A). Specifically, it consists of the CT-rich, Alu-like, and VNTR (variable number tandem repeat) regions of the SVA element, a portion of an AluSz element, and a portion of an L1ME5 element. Two small spacer sequences, derived from a unique genomic locus in the genome of the anthropoid common ancestor, separate SVA and AluSz components (28-bp U1 sequence) and AluSz and L1ME5 modules (103-bp U2 sequence) (fig. 1A). Given their unique location in the other primate genomes, these spacers allow the identification of the putative progenitor locus for the 3′ end of the LAVA element. Just such a locus containing the U1-AluSz-U2-L1ME5 sequence can be found on gibbon chromosome 1a, human chromosome 9, and the orthologous loci in all other great apes, old world monkeys (i.e., macaque), and marmoset genomes. The steps leading to the origin of the LAVA element, however, are still not entirely clear.
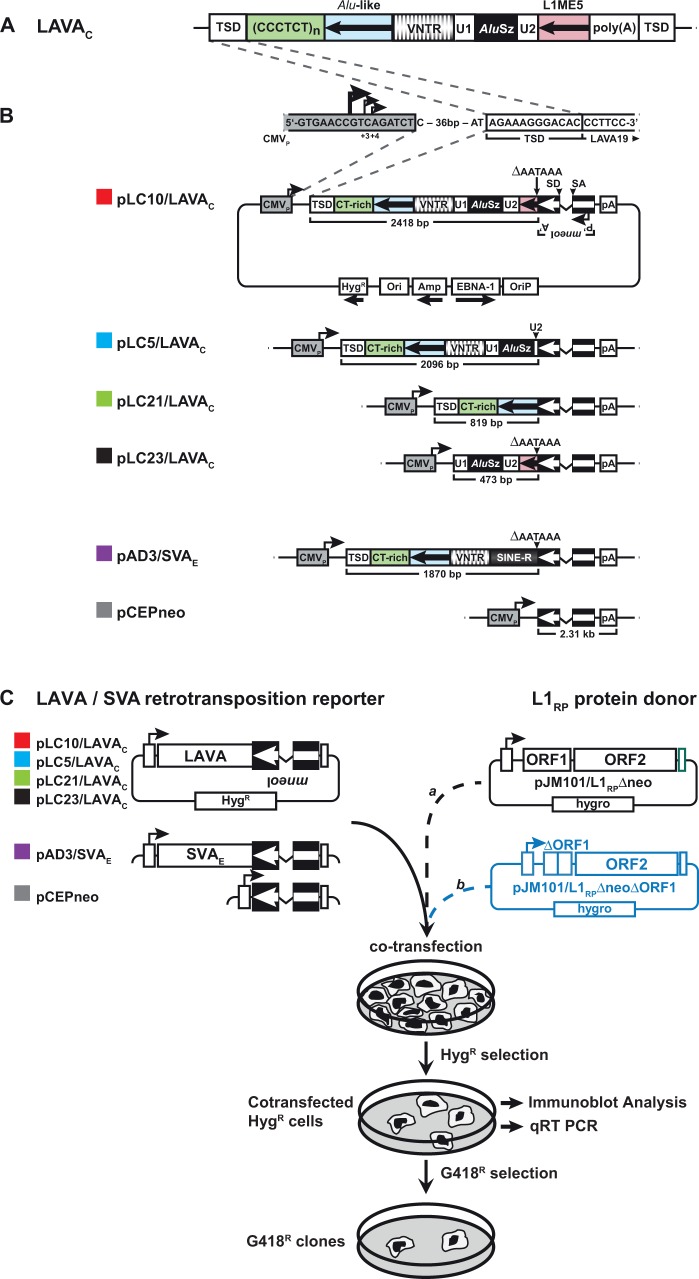
Fig. 1.—
Structures of LAVAC retrotransposition reporter plasmids and rationale of the LAVA trans-mobilization assay. (A) Organization of the non-autonomous non-LTR retrotransposon LAVA in the gibbon genome. The SVA-derived module consists of CT-rich (green), Alu-like (light blue), and VNTR (banded) region. It is separated from the AluSz-derived module (black) by Unique Region 1 (U1). The L1ME5-derived module (pink) is separated from the AluSz module by Unique Region 2 (U2). Black arrows in boxed Alu-like region and L1ME5 module indicate antisense orientation. Poly(A), poly(A) stretch; TSD, target site duplication. (B) Schematics of the retrotransposition reporter plasmid pLC10/LAVAC carrying the full-length LAVAC element, the expression cassettes of the reporter plasmids pLC5/LAVAC, pLC21/LAVAC, pLC23/LAVAC expressing truncated LAVAC versions, and pAD3/SVAE (Raiz et al. 2012). pCEPneo is used to measure processed pseudogene formation frequency. Each of the LAVA and SVA reporter elements and the processed pseudogene formation cassette were tagged with the indicator gene mneoI, and set under transcriptional control of the human CMV immediate early enhancer/promoter (CMVP). Splice donor (SD) and splice acceptor (SA) sites of the oppositely oriented γ-globin intron are indicated. mneoI is flanked by an SV40 promoter (P’) and polyadenylation signal (A’).Transcripts starting from CMVP driving LAVA mneoI, SVA mneoI or pCEP mneoI transcription, can splice the intron, but contain an antisense copy of the neoR gene. G418 resistant (G418R) colonies accrue only if this transcript is reverse transcribed, integrated into chromosomal DNA, and expressed from its own promoter P’. LAVA or SVA sequences were inserted between CMVP and the mneoI cassette. pLC5/LAVAC differs from pLC10/LAVAC exclusively in the absence of the L1ME5 and U2 modules covering 322 bp. pLC21/LAVAC and pLC23/LAVAC encode the 5′-terminal 819 bp of LAVAC covering TSD, CT-rich and Alu-like region, and the 3′-terminal 473 bp covering U1, AluSz and L1ME5 modules, respectively. pAD3/SVAE (Raiz et al. 2012) serves as positive control construct for trans-mobilization. Transcriptional termination signals at the 3′ ends of the L1ME5 (pLC10/LAVAC, pLC23/LAVAC) and SINE-R (pAD3/SVAE) modules were deleted from the LAVA and SVA reporter cassettes (ΔAATAAA) to ensure transcriptional read-through into the mneoI cassette and polyadenylation at the pCEP4-encoded SV40 polyadenylation signal (pA). pCEPneo is distinguished from the remaining presented reporter constructs by the absence of any LAVA or SVA sequence. CMVP sequences are highlighted in grey. CMVP major and minor transcription start sites (Isomura et al. 2008) are indicated by arrows. TSD, target site duplication; CT-rich, Alu-like, VNTR (Variable number of tandem repeats), U1, AluSz, U2 and L1ME5 represent repeat modules, LAVA is composed of; pA, poly(A) tail. HygR, hygromycin resistance gene serving as selectable marker for eukaryotic cells. (C) Design of the experimental approach to test for trans-mobilization of the mneoI-tagged LAVAC element by the human L1 protein machinery. LAVA or SVA retrotransposition reporter plasmids, or pCEPneo were each co-transfected with L1 protein donor plasmid pJM101/L1RPΔneo or pJM101/L1RPΔneoΔORF1 (blue) into HeLa-HA cells that were subsequently selected for hygromycin resistance for 12 days. HygR cells were assayed for retrotransposition events by selecting for 9–12 days for G418R HeLa colonies.
During the analysis and annotation of the gibbon genome assembly, all 1097 full-length LAVA elements were retrieved and classified into 22 subfamilies (Carbone et al. 2014) based on the presence/absence of diagnostic mutations (supplementary fig. S1, Supplementary Material online). These 22 subfamilies were further classified into six larger subfamilies designated LAVA_A through LAVA_F. The age of LAVA copies of the oldest subfamilies (LAVA_A1 and LAVA_A2) was estimated to be about 17–18 my, and therefore very close to the inferred date of divergence of the small ape and great ape lineages (Carbone et al. 2014). This observation suggests that the LAVA element was present from the very beginning of the gibbon lineage and the rise of LAVA might actually be one of the events involved in gibbon divergence. Subfamily LAVA_B consists of a large and diverse expansion of ten small and midsized subfamilies that evolved between 12 and 18 mya. One early branch of this LAVA_B expansion, LAVA_B1G, led to the lineage of the remaining middle-aged and younger LAVA subfamilies. LAVA_C and LAVA_D consist of three and two subfamilies aged 16–18 and 13–15 my, respectively, whereas LAVA_E and LAVA_F contain the most recently evolved subfamilies. The single large LAVA_E subfamily and the three LAVA_F subfamilies are estimated to have evolved between 10–11 mya and 6–9 mya, respectively (Carbone et al. 2014). Analyses of the evolutionary history of the LAVA elements identified in the gibbon reference genome revealed that about 50% of LAVA insertions have been fixed in all gibbon species, indicating that this retrotransposon was active before the split of the four gibbon genera about 5 mya (Carbone et al. 2014; Veeramah et al. 2015). Additionally, the discovery of polymorphic insertions specific to only some individuals of one species (Nomascus leucogenys) (Carbone et al. 2014) implies that LAVA activity was ongoing, at least until recently, and suggests the possibility that some source elements could be currently active.
Endogenous LAVA insertions display all the hallmarks of L1-mediated retrotransposition by target-primed reverse transcription (TPRT) (Carbone et al. 2014), including a consensus target sequence closely matching the human L1 endonuclease (EN) cleavage motif (5′-TTTT/AA-3′), target site duplications (TSDs) flanking each insertion, variable length poly(A) tails following putative polyadenylation signals, and the existence of 5′-truncated LAVA insertions (Szak et al. 2002; Ostertag et al. 2003). It has been demonstrated experimentally that both Alu and SVA elements, which lack any protein-coding capacity, accomplish their retrotransposition by hijacking the L1-encoded protein machinery in trans (Dewannieux et al. 2003; Raiz et al. 2012). In order to test the hypothesis that the gibbon genome harbours functional LAVA elements that can be trans-mobilized by the L1-encoded protein machinery, we established a LAVA retrotransposition reporter assay in HeLa cells.
Taking advantage of the modular structure of full-length LAVA elements, we set out to test the retrotransposition competence of its components relative to the full-length element in the presence of human L1 proteins. To this end, we picked one (GL397285: 9072637-9074632 in Nleu1.5) of the 207 members of the LAVA subfamily LAVA_C4B (supplementary fig. S1, Supplementary Material online) from the gibbon genome (Carbone et al. 2014). Here we show that this full-length LAVA element (hereafter referred to as LAVAC) is able to mobilize in human cells at frequencies exceeding those of a human SVAE retrotransposon. Data demonstrate that the SVA-derived portion makes the LAVAC element a preferred substrate for trans-mobilization by the human L1 protein machinery. Surprisingly, removal of the L1ME5-derived portion increased the mobilization rate of LAVAC, indicating that the 3′-truncated element is mobilized more efficiently than the full-length LAVAC. We also found that mneoI-tagged LAVA de novo insertions integrate into intronic regions of human genes in cell culture experiments recapitulating endogenous LAVA insertion preferences in the gibbon genome. Our results are in agreement with the inferred evolutionary origin of the LAVA element and indicate that there are genomic LAVA elements that are still “hot and flowing”, explaining why numerous insertions are polymorphic within the gibbon lineage.
Materials and Methods
Selection and Isolation of the LAVA Element to Be Used for Trans-Mobilization Assays
The full-length LAVAC element studied in this work was recently identified within the insert of the BAC clone CHORI 271-458C4 (Carbone et al. 2014) of the Nomascus leucogenys genome library (CHORI 271, http://bacpac.chori.org/library.php?id=228; last accessed September 21, 2016) that was also used for the gibbon genome assembly (Nleu1.0). A glycerol stock of CHORI271-458C4 was obtained from BACPAC Resources (http://bacpac.chori.org/; last accessed September 21, 2016). Using the publicly available BAC sequence (AC202765.2) as a template, we designed primers (GS_LAVA_19_F and GS_LAVA_19_R, supplementary table S1, Supplementary Material online) flanking LAVAC and performed long-range PCR on isolated BAC DNA using standard protocols (Carbone et al. 2014). The resulting 2418-bp PCR product harboring the full-length LAVAC element was isolated after separation in a 1% agarose gel, and sub-cloned in the pDRIVE vector (Qiagen) leading to the plasmid pDRIVE.LAVAC. Subsequently, primer walking was carried out to validate the isolated LAVAC element in pDRIVE.LAVAC by sequence analysis (supplementary fig. S2, Supplementary Material online).
LAVAC-Specific Filled Site/Empty Site PCR
Genomic DNA was isolated from blood of 11 individuals of the genus Nomascus leucogenys, three individuals of the genus N. gabriellae, and one individual of each of the genera Hoolock leuconedys, Hylobates moloch, and Symphalangus syndactylus applying the PureGene DNA Isolation Kit (Qiagen) according to the manufacturer’s instructions. To test for presence/absence of LAVAC, 25 μl PCR reactions were made each containing 50 ng genomic DNA, 1X Pfu Turbo Buffer, 0.32 μM GS_LAVA19_F primer, 0.32 μM GS_LAVA19_R primer (supplementary table S1, Supplementary Material online), 0.3 μM dNTPs, 1 U Platinum Taq (Invitrogen), 0.1 U Pfu Turbo Cx, and water. Cycling parameters for the reactions were: 95 °C for 3 min, 30 cycles of 95 °C for 30 s, 61 °C for 30 s, 72 °C for 8 min, and a final extension at 72 °C for 10 min. Samples in order of loading on the 1% agarose gel presented in supplementary figure S3, Supplementary Material online, are from N. leucogenys individuals Vok, Asia, China, Nancy, Johannes, Ricky, Bobby, B09007, Melouprey, Gibson, and Khao, N. gabriellae individuals Victor, China, Enick, and H. leuconedys, H. moloch, and S. syndactylus individuals Drew, Khusus, and Monty, respectively.
Retrotransposition Reporter Constructs and L1 Protein Donor Plasmids
The 2418-bp, full-length LAVAC element was isolated by PCR amplification from pDRIVE.LAVAC using primers GS.LAVA1 and GS.LAVA2 (supplementary table S1 and fig. S2, Supplementary Material online). LAVAC fragments of 2,096 bp, 819 bp and 473 bp including different portions of the LAVAC element (fig. 1A), were isolated by PCR using primer combinations GS.LAVA1/GS.LAVA3, GS.LAVA1/GS.LAVA4, and GS.LAVA2/GS.LAVA5 (supplementary table S1 and fig. S2, Supplementary Material online), respectively. Each of the four different PCR products was cloned into the pGEM-T Easy vector (Promega), sequenced, and the obtained sequences were compared with the LAVAC sequence of pDRIVE-LAVAC in order to check for PCR artifacts (data not shown). After validation of the correct nucleotide sequences of the PCR fragments, each of them was cloned as KpnI/NheI fragment into the KpnI/NheI digested pCEPneo plasmid (Raiz et al. 2012) as KpnI and NheI restriction sites are part of primers GS.LAVA1, GS.LAVA5 and GS.LAVA2, GS.LAVA3, GS.LAVA4, respectively (supplementary table S1, Supplementary Material online). Using this strategy, each of the four PCR products was inserted between CMVP and the mneoI indicator cassette of pCEPneo via KpnI/NheI, yielding pLC10/LAVAC (2418-bp fragment), pLC5/LAVAC (2096-bp fragment), pLC21/LAVAC (819-bp fragment), and pLC23/LAVAC (473-bp fragment) (fig. 1B). The construct pJM101/L1RPΔneoΔORF1 (L1RPΔORF1) was generated by introducing a 330-bp in-frame deletion in L1 ORF1 of pJM101/L1RPΔneo (Wei et al. 2001; Raiz et al. 2012) (fig. 1C).
LAVA Retrotransposition Reporter Assays and Statistical Analyses
HeLa-HA cells (Hulme et al. 2007) were cultured in DMEM High Glucose (Biochrom AG, Berlin, Germany) supplemented with 10% FCS (Biowest, Nuaillé, France), 100 µg/ml streptomycin and 100 U/ml penicillin. To perform retrotransposition reporter assays, 1.8 × 106 cells were plated on T75-flasks. Plated cells were co-transfected with 3 µg of each of the four LAVAC retrotransposition reporter plasmids, the SVA reporter plasmid, or pCEPneo and 3 µg of an L1 protein donor construct (pJM101/L1RPΔneo, pJM101/L1RPΔneoΔORF1; fig. 1C) or pCEP4 (Thermo Fisher Scientific) using FUGENE HD reagent (Promega) according to the manufacturer’s instructions. The L1 cis-retrotransposition rate observed after co-transfection of 3 µg of the L1RP-encoding pJM101/L1RP reporter construct with 3 µg pCEP4, served as positive control and was defined as 100% retrotransposition frequency. Each co-transfection was performed in biological triplicates using three independent DNA preparations of each plasmid, and each biological replicate was executed on a different day. Each co-transfection performed on the same day was done in quadruplicate. In each case, three co-transfections of the same biological replicate were used to quantify retrotransposition rates of the LAVA, SVA and L1 reporter elements and pseudogene formation rates of the pCEPneo construct. The fourth co-transfection was used to isolate cell lysates and total RNA in order to analyze expression of L1 proteins expressed from the L1 donor plasmids and transcription of retrotransposition reporter cassettes, respectively. Consequently, the arithmetic mean of the retrotransposition frequency of each analyzed reporter element was calculated from nine co-transfection experiments (n = 9). Starting 24 h post-transfection, cells were subjected to hygromycin (200 μg/ml, Invitrogen) selection for 12 days. After trypsinization and re-seeding, cells were selected for L1-mediated retrotransposition events in medium containing 400 μg/ml G418 (Invitrogen). After 11–12 days of selection, G418R colonies were either fixed and stained with Giemsa (Merck) to quantify retrotransposition events as described previously (Moran et al. 1996), or individual G418R colonies were isolated and expanded to characterize individual LAVA de novo retrotransposition events.
To gain countable results for retrotransposition in cis, cells co-transfected with pJM101/L1RP and pCEP4 were trypsinized after 12 days of hygromycin selection. Obtained cells were diluted 1:10 and 1:100, and diluted cells were reseeded in T75 flasks. 24 h later, cells were selected for L1 retrotransposition events in medium containing 400 μg/ml G418. Statistical evaluation was performed by means of an Analysis of Variance (ANOVA). To control the overall type I error (α = 0.05), P-values were adjusted according to Dunnett for multiple comparisons. The statistical analysis was performed with SAS®/STAT software, version 9.2 SAS system for Windows.
Immunoblot Analysis
To assess L1 ORF1p and L1 ORF2p expression, HeLa cells were co-transfected as described above and harvested after 12 days of hygromycin selection. Cells were lysed in RIPA buffer (25 mM Tris, pH 8, 137 mM NaCl, 1% glycerol, 0.5% sodium deoxycholate, 1% Nonidet P-40, 2 mM EDTA, pH 8, 0.1% SDS, and protease inhibitors), and lysates were cleared by centrifugation. Protein concentrations were quantified by applying the PierceTM BCA Protein Assay Kit (Cat.# 23225, Thermo Fisher Scientific) according to the manufacturer’s instructions. The particular amounts of protein lysates (described in the following) were boiled in Laemmli buffer, loaded on 12% polyacrylamide gels, subjected to SDS-PAGE, and electroblotted onto nitrocellulose membranes. In the case of the immunoblot analyses presented in figures 2A and 3B/left panel (αL1 ORF1p blot), 20 μg of cell lysate of each differently transfected HeLa-HA culture were loaded per lane. In the case of the gel presented in figure 3B/right panel (αL1 ORF2p blot), 40 μg of total cell lysate of each differently transfected HeLa-HA culture were loaded per lane. In the case of the immunoblot analysis presented in figures 2A and 3B/left panel, 2 μg of the cell extracts isolated from the human embryonal carcinoma cell line NTERA.2 cl.D1 (NTERA-2; ATCC#: CRL-1973; (Andrews et al. 1984)) were loaded on one lane of each gel and served as positive control for L1-ORF1p detection. 40 μg of the same NTERA-2 cell extract were loaded on one lane of the immunoblot analysis presented in figure 3B/right panel as positive control for L1 ORF2p expression. After protein transfer, membranes were blocked for 2 h at room temperature in a 10% solution of non-fat milk powder in 1× PBS-T [137 mM NaCl, 3 mM KCl, 16.5 mM Na2HPO4, 1.5 mM KH2PO4, 0.05% Tween 20 (Sigma)], washed in 1xPBS-T, and incubated overnight with the respective primary antibody at 4 °C. To detect L1 ORF1p, the polyclonal rabbit-anti-L1 ORF1p antibody #984 (Raiz et al. 2012) was used in a 1:2000 dilution in 1xPBS-T containing 5% milk powder. L1 ORF2p expression was verified using a rabbit anti-L1 ORF2p-N antibody (Goodier et al. 2004) at a 1:1000 dilution in 1× PBS, 5% milk, 0.05% Tween 20. Membranes were washed thrice in 1× PBS-T and incubated with an HRP-conjugated, secondary anti-rabbit IgG antibody (Amersham Biosciences) at a dilution of 1:30,000 in 1× PBS-T/5% milk powder for 1 h. Subsequently, the membrane was washed six times for 10 min in 1xPBS-T. ß-actin expression was detected using a monoclonal anti-ß-actin antibody (clone AC-74, Sigma-Aldrich) as primary antibody at a dilution of 1:30,000. Anti-mouse HRP-linked species-specific antibody (from sheep) at a dilution of 1:10,000 served as secondary antibody specific for anti-ß-actin. Immunocomplexes were visualized using lumino-based ECL immunoblot reagent (Amersham Biosciences).
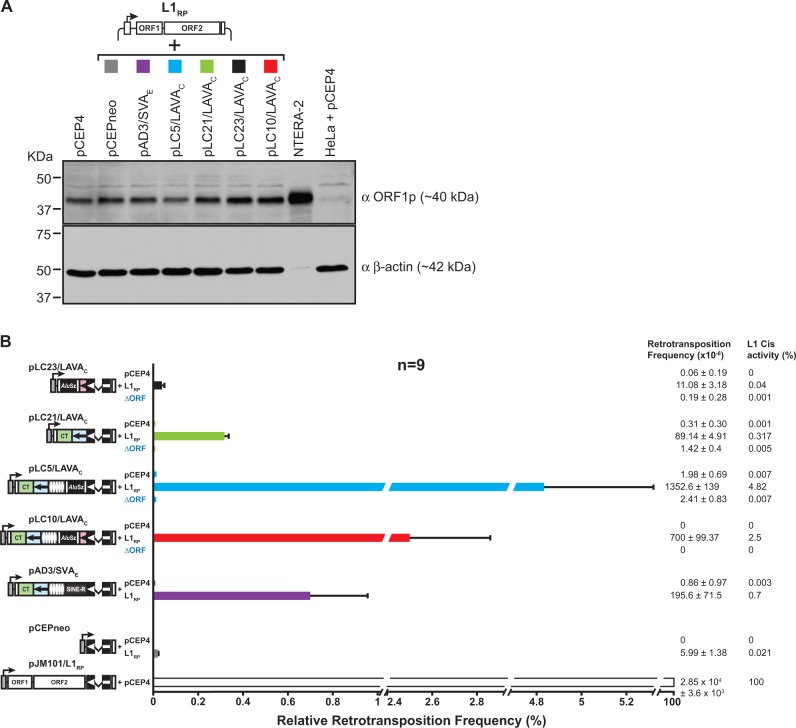
Fig. 2.—
Trans-mobilization of mneoI-tagged LAVAC reporter elements by the human L1 protein machinery requires the presence of L1 ORF1p. (A) Immunoblot analysis of L1 protein expression in HeLa-HA cells after co-transfection of the L1 protein donor plasmid pJM101/L1RPΔneo (L1RP) with the LAVAC, SVAE or pCEPneo reporter plasmids. Cell lysates were isolated 13 days after co-transfection upon completion of hygR selection and subjected to immunoblot analysis using anti-L1 ORF1p antibodies (αORF1p). About 20 µg of cell lysates from the differently transfected cells were loaded per lane. Only 2 µg of NTERA-2 cell lysates were separated on one lane of the gel. β-actin protein levels (∼42 kDa) served as loading control. Lysates from pCEP4-transfected HeLa cells (HeLa + pCEP4) and from the human embryonal carcinoma cell line NTERA-2 served as negative and positive control for L1 ORF1p detection, respectively. (B) LAVAC retrotransposition reporter assay after hygR selection for the presence of expression plasmids. LAVAC reporter plasmids pLC5/LAVAC, pLC10/LAVAC, pLC21/LAVAC, and pLC23/LAVAC, were co-transfected with the L1 protein donors pJM101/L1RPΔneo (L1RP) or pJM101/L1RPΔneoΔORF (ΔORF) or the empty vector pCEP4. Control constructs pAD3/SVAE and pCEPneo were co-transfected with either pJM101/L1RPΔneo or pCEP4. After hygR selection, G418R selection for retrotransposition events followed and retrotransposition rates were determined by counting G418R HeLa colonies. To quantify retrotransposition frequencies, each co-transfection was performed in biological triplicates with each biological replicate being executed on a separate day. Each biological replicate was conducted in technical triplicates. Absolute retrotransposition frequencies per 106 cells are listed and relative retrotransposition rates are presented as bar diagrams. Cis retrotransposition frequency of the L1 reporter element encoded by pJM101/L1RP after its co-transfection with pCEP4 was set as 100%. Each bar depicts the arithmetic mean ± SD of the relative retrotransposition frequencies that resulted from nine individual co-transfection experiments (n = 9).
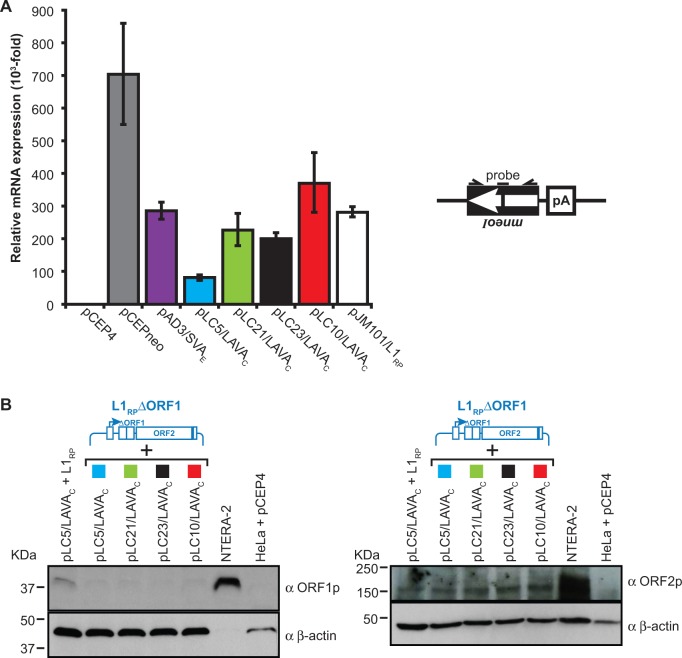
Fig. 3.—
Analysis of the expression from L1 protein donor and retrotransposition reporter plasmids. (A) qRT-PCR analyses to quantify the relative amounts of spliced transcripts expressed from the different retrotransposition reporter cassettes. Total RNA was isolated after 12 days of hygromycin selection following co-transfection of pCEPneo, pAD3/SVAE, pLC5/LAVAC, pLC10/LAVAC, pLC21/LAVAC, or pLC23/LAVAC with the L1 protein donor plasmid pJM101/L1RPΔneo and co-transfection of pJM101/L1RP with pCEP4. Each co-transfection was performed in three biological replicates. The used primer/probe combination (see “Materials and Methods” section) is specific for the spliced mneoI-cassette (black box with arrow). Real-time PCR of each biological replicate was conducted in technical triplicates. Relative amounts of mRNA expression refer to the signal obtained from total RNA from pCEP4(mock)-transfected HeLa cells which was set as 1 (pCEP4) and served as negative control. Bars depict arithmetic means ±SD of technical triplicates of three biological replicates. (B) Immunoblot analysis of L1 ORF1p (left panel) and L1 ORF2p (right panel) expression after co-transfection of the L1 protein donor plasmid pJM101/L1RPΔneoΔORF1 (L1RPΔORF1) with the LAVAC retrotransposition reporter plasmids. Whole-cell lysates were prepared 13 days after co-transfection upon completion of hygromycin selection and subjected to immunoblot analysis using antibodies against either L1 ORF1p (αORF1p) or L1 ORF2p (αORF2p). In the case of the anti-L1 ORF1p immunoblot analysis (left panel), 20 µg of the cell lysate isolated from each of the differently transfected HeLa-HA cell cultures, were loaded per lane on a 12% PAA gel. Because ORF2p (Predicted MW∼150 kDa) is expressed at a significantly lower level than ORF1p (Dai et al. 2014), 40 µg of total cell lysate from each of the differently transfected HeLa-HA cells were loaded per lane on a 6% PAA gel to perform anti-L1 ORF2p immunoblot analysis (right panel). Detectable amounts of intact L1 ORF1-encoded proteins are absent from pJM101/L1RPΔneoΔORF1-transfected HeLa cells. About 2 µg and 40 µg of the NTERA-2 cell extract were loaded on one lane of the gel used for the anti-ORF1p (left panel) Western blot analysis and the anti-ORF2p (right panel) immunoblot analysis, respectively. Lysates from pCEP4-transfected HeLa cells (HeLa + pCEP4) and from NTERA-2 cells served as negative and positive control for L1 protein detection, respectively. β-actin protein levels (∼42 kDa) were analyzed as loading control.
Analysis of LAVA De Novo Insertions
Genomic DNA from expanded G418R-HeLa colonies was isolated applying the Qiagen DNeasy® Tissue Kit according to the manufacturer’s protocol. To test for the presence of the spliced mneoI indicator cassette, a diagnostic PCR was performed using the intron-flanking primer pair GS86/GS87 (fig. 4A, supplementary table S1, Supplementary Material online). PCR cycling conditions were as follows: 3 min at 96 °C (30 s at 96 °C, 15 s at 56 °C; 2 min at 72 °C) 25 cycles, 7 min at 72 °C. To determine genomic pre- and post-integration sites of LAVA de novo insertions, we used a modified version (Kirilyuk et al. 2008) of a previously published extension primer tag selection preceding solid-phase ligation-mediated PCR (EPTS/LM-PCR) (Schmidt et al. 2001) to isolate 3′ junctions of these insertions. Products of the final PCR were separated in a 1% agarose gel, isolated from the gel using the QIAquick Gel Extraction Kit (Qiagen), and sequenced either directly or after sub-cloning into pGEM-T Easy. Obtained sequences were mapped to the human genome using the UCSC genome browser at http://genome.ucsc.edu; last accessed September 21, 2016. To characterize 5′-junctions of each LAVA de novo insertion, primers specific for the genomic sequence adjacent to the 5′ end of the de novo insertions were designed (supplementary table S2, Supplementary Material online). The second PCR primers used were designed to bind specifically to the retrotransposed LAVAC reporter cassette (LAVA19_5pSVA1 and LAVA19_5pU2A). All oligonucleotides used in this study are listed in supplementary table S1and S2, Supplementary Material online. Genomic pre-integration sites and surrounding sequences were characterized using the UCSC genome browser annotation for genes (supplementary table S3, Supplementary Material online).
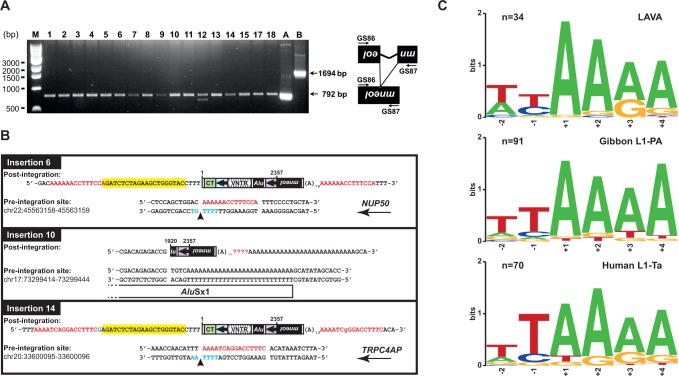
Fig. 4.—
Marked LAVAC de novo insertions have typical attributes of L1-mediated retrotransposition events. (A) Diagnostic PCR to demonstrate splicing of the artificial γ-globin intron from the mneoI indicator cassette of marked LAVAC de novo insertions. Genomic DNA was extracted from 17 G418R colonies (gDNA samples 1–15 and 17–18) that resulted from co-transfection of the LAVA reporter pLC10/LAVAC and the L1 driver construct pJM101/L1RPΔneo, and used as template for PCR using mneoI cassette-specific primers GS86 and GS87 (supplementary table S1, Supplementary Material online). Presence of the spliced and reverse-transcribed mneoI cassette resulting from retrotransposition gives rise to a 792-bp PCR product, whereas the unspliced reporter cassette which did not go through retrotransposition leads to a 1694-bp PCR product. PCR on gDNA sample #12 resulted in an additional product of ∼700 bp whose origin is unclear. PCR performed on pcDNA™ 3.1(+) (Invitrogen) served as positive control for the presence of a neoR gene lacking any intron (lane A). PCR analysis using pLC10/LAVAC as template resulted in a 1694 bp product specific for the unspliced mneoI cassette (lane B). M, 1 kb DNA ladder (New England Biolabs #N3232). (B) Structural organization of mneoI-tagged LAVAC de novo insertions. Both pre- and post-integration sites of de novo insertions 6, 10 and 14 are presented. Chromosomal nucleotide positions given for each locus indicate the sites into which the LAVAC elements inserted and refer to human reference genome hg19. Marked full-length insertions 6 and 14 cover 4692 bp and 5’-truncated insertion 10 comprises 3853 bp. The extra deoxyguanylate at the 5’-end of insertion 14 is indicated in green. The L1 EN target sequence on the bottom strand is presented in blue with the arrow indicating the L1 EN nicking site. CMVP-derived sequences are highlighted in yellow. Transcriptional directions of host genes NUP50 and TRPC4AP in which insertions 6 and 14 integrated are denoted with an arrow. TSDs could not be identified for insertion 10 because sequencing of the LAVA 3’ junction was hampered by the directly adjacent poly(A) tail which is part of a pre-existing genomic AluSx1 element. Red lettering, TSD sequences; mneoI, neomycin-phosphotransferase gene. (C) The nucleotide profile of LAVA insertion sites resembles the consensus target sequence of gibbon L1PA and human L1-Ta insertions. Target sequence logos were generated by multiple sequence alignments of genomic integration sites of LAVA insertions (top panel), gibbon L1PA (middle panel), and human L1-Ta (bottom panel) using the program WebLogo (Crooks et al. 2004). Logos for the top strand sequence cover two nucleotides of upstream and four nucleotides of downstream sequence relative to the L1 EN cleavage site on the bottom strand. Numbers on the x-axis denote nucleotide positions relative to the L1 EN nicking site. Target sequence logos were generated from genomic integration sites of 34 preexisting LAVA insertions (Carbone et al. 2012), target sequences of randomly selected L1PA insertions from the Nomascus leucogenys reference genome (n = 91), and target sequences of 70 preexisting L1-Ta insertions (Raiz et al. 2012). Integration site sequences were identified as described in the Materials and Methods section.
Sequence Logos
Using the WebLogo utility (Schneider and Stephens 1990; Crooks et al. 2004), sequence logos were generated for the genomic integration sites of LAVA elements, L1PA elements found in the gibbon genome, and L1-Ta elements reported for the human genome. The 34 LAVA target sites used in this analysis included 33 loci described in a recent report (Carbone et al. 2012) and the original LAVAC locus used to design the constructs in this study. The 70 genomic target sequences of human endogenous L1Ta elements were listed in a previous study (Raiz et al. 2012) (fig. 4C, supplementary table S4, Supplementary Material online). In order to extract genomic target sequences of L1PA elements from the gibbon genome, we first used the RepeatMasker, Version open-4.0.5 (Smit et al. unpublished) annotation of Nleu3.0 available from UCSC’s Table Browser, to select 200 full-length L1PA insertions randomly. Next, 100-bp marginal sequences consisting of ±50 bp upstream and downstream of the start and end position of each element were extracted. These pairs of marginal sequences were then aligned using Clustal Omega (Sievers et al. 2011) and the alignments were visualized in Geneious version 5.6.5 (http://www.geneious.com; last accessed September 21, 2016) (Kearse et al. 2012) and inspected to call TSDs and EN cleavage sites, if possible, for each locus. Only 91 of the 200 L1PA loci inspected showed identifiable TSDs, which allowed inference of EN cleavage sites (fig. 4C, supplementary table S4, Supplementary Material online).
Quantitative Real-Time RT-PCR to Analyze Transcription of Reporter Cassettes after Cotransfection of Driver and Reporter Plasmid
To quantify mneoI-transcripts expressed from the various reporter plasmids in our assay system, plated HeLa cells were cotransfected with 3 µg of each of the four LAVAC reporter plasmids, the SVA reporter plasmid, or pCEPneo, and 3 µg of the L1 driver pJM101/L1RPΔneo, or with 3 µg pCEP4 and 3 µg of pJM101/L1RP. Hygromycin selection started 24 h post-transfection. Each co-transfection was performed in three biological replicates with each biological replicate being executed with a separate set of independent plasmid preparations. Thirteen days after co-transfection of HeLa cells, total RNA was extracted from hygromycin-selected HeLa cells using the RNeasy® Mini Kit (Qiagen) with On-Column DNase digestion following the manufacturer’s instructions. Using the SuperScript III® First-Strand Synthesis System for RT-PCR (Invitrogen) in combination with a hexamer primer, first-strand cDNA was synthesized from 0.5 µg of DNaseI-digested, total RNA according to the manufacturer’s instructions. To quantify levels of spliced transcripts expressed from the mneoI-tagged reporter elements in pAD3/SVAE, pLC5/LAVAC, pLC10/LAVAC, pLC21/LAVAC, pLC23/LAVAC, pCEPneo, and pJM101/L1RP, real-time PCR was performed as technical triplicates applying TaqMan® chemistry (Applied Biosystems) in an Applied Biosystems 7900HT Fast Real-Time PCR System base unit. We used a primer/probe combination (Neofor: 5′-GCTATTCGGCTATGACTGG-3′; Neorev: 5′-GCCACGATAGCCGCGCTGC-3′; probe: 5′-FAM-CCTCGTCCTGAAGCTCATTC-3′) specifically recognizing the spliced mneoI cassette. Spliced mneoI transcript levels were normalized against cellular 18srRNA levels. Cycling conditions were as follows: 95 °C for 15 min (initial denaturation), 95 °C for 15 s and 60 °C for 1 min (40 cycles). The software applied to analyze real-time and end point fluorescence was RQ manager 1.2. Relative quantification of RNA expression was carried out using the ΔΔCt method (Livak and Schmittgen 2001).
Results
Identification of a Potentially Active LAVA Source Element in the Nomascus leucogenys Genome
In order to investigate the competency of non-autonomous LAVA retrotransposons to be trans-mobilized by the L1 protein machinery, we chose a full-length LAVA element from the Nomascus leucogenys genome. The selected LAVA insertion was chosen among 48 elements characterized in a previous study (Carbone et al. 2012, 2014) because it does not contain any obvious indels, includes all complete LAVA-specific modules, a 3′ poly(A) tail, and is flanked by clearly identifiable TSDs covering 12 nts. A PCR-based assay over a larger panel of gibbon individuals confirmed that the same element inserted specifically in the Nomascus genus, as it can be found both in Nomascus leucogenys and its sister taxon Nomascus gabriellae, but is absent from the remaining genera (supplementary fig. S3, Supplementary Material online). This indicates that this specific insertion occurred less than 5 mya after the four gibbon genera diverged from each other but had the time to be fixed in the Nomascus genus. Consistently, comparison of its sequence to the consensus sequences identified for the 22 LAVA subfamilies (Carbone et al. 2014) indicated similarity to the older LAVA_C4B subfamily which, based on previous age analysis, emerged ∼16 to 18 mya (Carbone et al. 2014). We hence named this element LAVAC and set out to investigate whether this element, whose sequence differs from the youngest subfamilies LAVA_F and LAVA_E, is retrotransposition-competent. Given that older LAVA elements such as LAVAC represent the bulk of LAVA insertions in the gibbon genome, the finding of its activity would have significant biological implications (supplementary fig. S1, Supplementary Material online).
Engineering Functional LAVAC Reporter Elements to Be Applied in a Trans-Mobilization Assay
To investigate if the full-length LAVAC element can be mobilized in trans by the human L1 protein machinery and assess which of the LAVAC-encoded modules are relevant for such mobilization, we generated reporter plasmids expressing the full-length element (pLC10/LAVAC) and different combinations of its modules (pLC5/LAVAC, pLC21/LAVAC, and pLC23/LAVAC), respectively (fig. 1B). To this end, the full-length LAVAC and each of the three LAVAC fragments were inserted separately between the CMV promoter (CMVP) and the mneoI retrotransposition reporter cassette of the pCEPneo plasmid (Raiz et al. 2012). Each of the resulting LAVA reporter constructs was co-transfected into HeLa-HA cells (Hulme et al. 2007) with the L1 driver construct pJM101/L1RPΔneo (fig. 1C) (Wei et al. 2001) that provides the overexpressed human L1 protein machinery (fig. 2A) in the presence of mneoI-tagged LAVAC transcripts. To directly compare retrotransposition activities of the four different LAVAC-derived reporter cassettes (fig. 1B) with the recently determined trans-mobilization frequency of the human full-length SVAE element, we also co-transfected the SVAE reporter plasmid pAD3/SVAE (fig. 1B, C) (Raiz et al. 2012) with the L1 driver plasmid in HeLa-HA cells in parallel. Concurrently, we used this co-transfection with pAD3/SVAE also as positive control for the established trans-mobilization assay. Processed pseudogene formation rate was assessed by measuring the trans-mobilization frequency of the reporter cassette encoded by the empty pCEPneo plasmid (Raiz et al. 2012) in the presence of the co-transfected L1 driver plasmid (fig. 1C). As pCEPneo differs from the LAVA and SVA reporter plasmids exclusively in the absence of any retrotransposon sequence (fig. 1B) and expresses solely the mneoI cassette, it allows obtaining a rough estimate of the trans-mobilization frequency of PolII genes in our experimental setup (Esnault et al. 2000). More details on the generation of the LAVA retrotransposition reporter constructs and the LAVA retrotransposition reporter assay are outlined in the “Material and Methods” section.
Each co-transfection was performed in three biological replicates with each replicate being executed as technical triplicates (for details, see “Materials and Methods” section). After co-transfection, cells were cultivated in the presence of hygromycin to select for the presence of the expression plasmids, and subsequently selected for G418 resistance (fig. 1C). Each of the retrotransposition reporter cassettes used in this study was tagged with the selectable marker gene mneoI coding for neomycin phosphotransferase, in an antisense orientation. This combination assures that G418-resistant (G418R) cells only originate if a transcript initiated from the CMV promoter (CMVP) driving LAVAmneoI, SVAmneoI, CEPmneoI or L1mneoI expression is spliced, reverse transcribed, reintegrated into the genomic DNA, and expressed from promoter P’ (fig. 1B) (Heidmann et al. 1988; Freeman et al. 1994; Moran et al. 1996).
Retrotransposition frequencies of the different LAVAC reporter elements in the presence of the overexpressed L1 driver plasmid ranged from 0.02% to 4.7% relative to the L1RP cis-retrotransposition frequency set as 100% (fig. 2B, and supplementary figs. S4 and S5, Supplementary Material online). The absence of G418R colonies after co-transfection of LAVAC or SVAE reporter plasmids or pCEPneo with the empty episomal expression vector pCEP4 substantiates that overexpression of L1-encoded proteins is essential for the trans-mobilization of LAVAC, SVAE and for pseudogene formation and consistent with previous reports on trans-mobilization of SVA elements (Hancks et al. 2011; Raiz et al. 2012) and processed pseudogene formation (Esnault et al. 2000).
Human L1RP Proteins Trans-Mobilize Full-Length LAVAC at a Frequency Exceeding Processed Pseudogene Formation Significantly
First, we asked if L1 proteins favor the LAVAC reporter-encoded mRNA over any random RNA polymerase II transcript as substrate for trans-mobilization. In this case, trans-mobilization frequency of the LAVA reporter element should significantly exceed the frequency of processed pseudogene formation of a random PolII gene. In order to determine the processed pseudogene formation rate, we used the pCEPneo reporter plasmid. Because transcripts expressed from the CMVP of pCEPneo consist exclusively of the mneoI indicator cassette in antisense orientation and do not include any retrotransposon sequences (Raiz et al. 2012), these transcripts should be trans-mobilized as frequent as random mRNAs encoded by host PolII genes. Trans-mobilization frequencies of the canonical full-length LAVAC element expressed from pLC10/LAVAC exceeded pseudogene formation of the reverse mneoI cassette encoded by pCEPneo by ∼117-fold (fig. 2B). LAVAC reporter plasmids expressing exclusively the CT-rich and Alu-like regions (pLC21/LAVAC) or CT-rich region, Alu-like region, VNTR module, and the AluSz-derived component (pLC5/LAVAC) lead to trans-mobilization frequencies outreaching processed pseudogene formation frequency by 15- or 226-fold, respectively (fig. 2B, and supplementary figs. S4 and S5, Supplementary Material online). In contrast, trans-mobilization frequency of the LAVAC reporter element comprising only the 3′ end modules, U1, AluSz, U2, and L1ME5, (pLC23/LAVAC) does not significantly exceed processed pseudogene formation frequency of pCEPneo (fig. 2B). Reporter constructs pLC10/LAVAC (full-length) and pLC5/LAVAC (ΔL1ME5 module), whose expression results in the highest retrotransposition frequencies, are characterized by the presence of the 5′ portion of the full-length LAVAC element ranging from the CT-rich region to the U2 region (fig. 1A) suggesting that the included modules make LAVAC a preferred substrate for trans-mobilization by the L1 protein machinery. Interestingly, the pAD3/SVAE-encoded SVAE reporter element that is mobilized at a frequency exceeding pseudogene formation by ∼33-fold has CT-region, Alu-like region, and VNTR module in common with LAVAC (fig. 1A). This observation strengthens the hypothesis that the SVA-derived modules account for at least some of the preference the L1 protein machinery has for both LAVA and SVA elements, whereas LAVA-specific U1, AluSz, U2, and L1ME5 modules do not support trans-mobilization by L1 proteins.
Quantification of overexpressed human L1 proteins by immunoblot analysis demonstrates that the observed differences in G418R colonies are not a consequence of varying amounts of overexpressed L1 proteins (fig. 2A). To evaluate if the observed differences in G418R colonies resulted from discrepancies in mRNA production or stability between the different LAVAC reporter cassettes, we tested for comparable amounts of spliced tagged LAVAC mRNAs by Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) (fig. 3A). To this end, we isolated total RNA from each of the three biological replicates of each co-transfection experiment after 12 days of hygromycin selection, applied primer/probe combinations specifically recognizing the spliced mneoI reporter cassette (Raiz et al. 2012) (see “Materials and Methods” section) and quantified the relative amounts of spliced mRNA expressed from the reporter plasmids by qRT-PCR (fig. 3A). Data show that the total quantities of spliced mneoI-carrying reporter mRNAs expressed in pAD3/SVAE-, pLC21/LAVAC-, pLC23/LAVAC-, or pJM101/L1RP-transfected cells are comparable, and reduced by only ∼22%, 39%, 44%, or 24%, respectively, relative to the reporter mRNA level expressed from pLC10/LAVAC (fig. 3A). These comparatively small differences in reporter transcript levels have only a negligible effect on the relative differences observed between trans-mobilization frequencies of the respective reporter elements (fig. 2B and supplementary fig. S5, Supplementary Material online). However, we observed mneoI–harboring reporter transcript levels expressed from pLC5/LAVAC that are significantly reduced by 55–75% relative to the transcript levels expressed from the remaining transfected LAVA, SVA, and L1 retrotransposition reporter plasmids (fig. 3A).
Considering the reduced spliced reporter transcript levels expressed in pLC5/LAVAC-transfected cells (fig. 3A), we would expect that the observed 122-fold increase in trans-mobilization frequency of the LAVAC reporter encoded by pLC5/LAVAC relative to pLC23/LAVAC (fig. 2B, supplementary fig. S5, Supplementary Material online), is even an underestimate of the relative retrotransposition frequency of the pLC5/LAVAC-encoded reporter element. The reduced level of transcripts expressed in pLC21/LAVAC- relative to pLC10/LAVAC-transfected cells (fig. 3A) has only a negligible effect on the ∼8-fold difference in trans-mobilization frequency measured between pLC21/LAVAC- and pLC10/LAVAC-transfected cells (fig. 2B). In order to distinguish the impact of variations in transfection efficiencies on mneoI-harboring reporter transcript levels from the influence of variations in reporter gene transcription rates on these transcript levels, we normalized the spliced mneoI RNA levels against RNA amounts expressed from the HygR gene located on the pCEP4 backbone (fig. 1B) of each reporter plasmid that was cotransfected before with a donor plasmid in the retrotransposition reporter assay (supplementary fig. S6, Supplementary Material online). Transcription rates of pAD3/SVAE, pLC21/LAVAC and pLC23/LAVAC differ from pLC5/LAVAC by only 45%, 36%, and 10%, respectively, and are therefore comparable (supplementary fig. S6B, Supplementary Material online). In contrast, transcription rates of the reporter cassettes of pCEPneo and pLC10/LAVAC deviate from pLC5/LAVAC more significantly by 3.1- and 2.7-fold, respectively. Whereas the comparatively high transcription rate of the pCEPneo reporter cassette can be explained by the absence of any retrotransposon sequence (fig. 1A), it is surprising that the full-length LAVAC in pLC10/LAVAC that is identical to the LAVAC element located in the gibbon genome and differs from the pLC5/LAVAC reporter exclusively in the presence of the L1ME5 module, is transcribed almost as efficiently as the pCEPneo reporter cassette (supplementary fig. S6B, Supplementary Material online). These data demonstrate that variations in individual transcription rates specific for each retrotransposition reporter cassette play a role in the observed overall differences (fig. 3A) in spliced mneoI-harboring reporter transcript levels in the variably co-transfected cells. Considering the differences in overall mneoI transcript levels between pCEPneo- and the LAVAC reporter-transfected cells, as demonstrated in figure 3A, data indicate that trans-mobilization frequencies of the reporter elements encoded by pLC23/LAVAC, pLC21/LAVAC, pLC5/LAVAC, and pLC10/LAVAC exceed processed pseudogene formation frequencies in HeLa cells by up to 7-, 46-, 1860-, and 238-fold, respectively.
L1 ORF1p Is Essential for Trans-Mobilization of LAVA Elements
Retrotransposition in cis and in trans requires reverse transcriptase (RT) activity encoded by L1 ORF2. Whereas it was shown previously that non-autonomous Alu elements require only L1 ORF2p for their mobilization in trans (Dewannieux et al. 2003), SVA retrotransposition and processed pseudogene formation were demonstrated to require the presence of both L1 ORF1p and L1 ORF2p (Esnault et al. 2000; Hancks et al. 2011; Raiz et al. 2012). In order to investigate if L1 ORF1p is also required for the trans-mobilization of LAVA elements, we co-transfected the LAVA reporter elements with the L1 protein donor pJM101/L1RPΔneoΔORF1 (fig. 1C) which differs from the L1 driver pJM101/L1RPΔneo exclusively in a 330-bp in-frame deletion in L1 ORF1 causing the expression of a mutant ORF1p that lacks ORF1p amino acid positions 99–208, and measured the resulting trans-mobilization frequency (Raiz et al. 2012). Initiation of ORF2 translation within the bicistronic RNA encoded by pJM101/L1RPΔneoΔORF1 is not perturbed. Immunoblot analysis of cell extracts isolated from the differently co-transfected HeLa cells with anti-L1 ORF1p and anti-L1 ORF2p antibodies (fig. 3B) confirmed the absence of overexpressed L1 ORF1p and the presence of comparable amounts of transiently expressed ORF2p after co-transfection of the L1 protein donor pJM101/L1RPΔneoΔORF1 with each of the four LAVAC reporter constructs. Co-expression of pJM101/L1RPΔneoΔORF1 (fig. 3B) did not result in trans-mobilization of any of the four LAVAC reporter elements (fig. 2B) although similar amounts of L1 ORF2p are expressed from both protein donor plasmids L1RP and L1RPΔORF1, indicating that L1 ORF1p expression is essential for LAVA trans mobilization.
Marked LAVAC De Novo Insertions Exhibit Hallmarks of L1-Mediated Target Primed Reverse Transcription
To confirm that mneoI-tagged LAVAC de novo retrotransposition events resulted in the observed G418R HeLa colonies, we next analyzed those colonies that followed from co-transfection of the passenger plasmid pLC10/LAVAC expressing the full-length LAVAC reporter element and the L1 driver plasmid for the presence of mneoI-tagged insertions. To this end, we isolated genomic DNA from 17 randomly chosen single expanded G418R HeLa colonies that arose from this co-transfection experiment. We first performed diagnostic PCR to specifically test for the presence of the functional neoR gene that should result from splicing of the pLC10/LAVAC-encoded mneoI cassette presuming retrotransposition. For this purpose, we used primers specifically binding to the intron-flanking mneoI sequences. As shown in figure 4A, PCR on each of the 17 genomic DNA samples generated a PCR product of ∼792 bp indicating the presence of a spliced mneoI cassette that resulted from trans-mobilization of full-length LAVAC encoded by pLC10/LAVAC.
Next, we isolated three mneoI-tagged LAVAC de novo insertions applying EPTS/LM-PCR (Schmidt et al. 2001) to analyze them for the presence of hallmarks of L1-mediated retrotransposition. Sequence analysis of pre- and post-integration sites of these pLC10/LAVAC-derived de novo insertions uncovered that insertions 6 and 14 are full-length and encompass 4,692 bp including 25 bp of CMV promoter sequence, the full length LAVAC element (2,357 bp) and the spliced mneoI reporter cassette (2,310 bp) (fig. 4B, supplementary table S2, Supplementary Material online). The 5′ ends of both full-length de novo insertions are identical and match with position 4 downstream of the transcription initiation site of the CMV promoter (Raiz et al. 2012) controlling expression of the LAVAC reporter element. Insertions 6 and 14 occurred into introns 1 and 11 of the genes NUP50 (Nucleoporin 50kDa) and TRPC4AP (Transient Receptor Potential Cation Channel, Subfamily C, Member 4 Associated Protein), respectively. Further, both retrotransposition events inserted in antisense orientation relative to the transcription direction of the host gene, mimicking the arrangement observed for the majority of intragenic LAVA elements in the gibbon genome (Carbone et al. 2014). Both de novo insertions exhibit hallmarks of L1-mediated retrotransposition such as 14-nt and 16-nt TSDs, 13-nt and 76-nt poly(A) tails, and genomic target sequences resembling the human L1 EN consensus target sequence 5′-TTTT/AA-3′ of pre-existing human L1-Ta, AluYa5, and SVAE/F insertions (Raiz et al. 2012) and gibbon L1PA and LAVA insertions (fig. 4C). The accordance of the genomic target sequences of both insertions with the human L1 EN consensus target sequence and endogenous gibbon L1 and LAVA target sequences indicates that the LAVA integration sites are determined by L1 EN activity. Also, the untemplated G nucleotide at the 5′ end of insertion 14 and the microcomplementarity at the 5′ end of insertion 6 are structural features reported previously for L1-mediated retrotransposition events (Gilbert et al. 2005; Zingler et al. 2005; Raiz et al. 2012). The intergenic 3,853 bp insertion 10 occurred into the poly(A) tail of a pre-existing AluSx1 element, is 5′-truncated and includes only the 1,543 bp of the 3′ portion of the full-length LAVAC element and the subsequent 2,310 bp of the mneoI reporter gene (fig. 4B). The extensive genomic poly(A) stretch that is located adjacent to the integration site hampered sequence analysis and identification of the 3′ junction sequence.
Discussion
The LAVA element represents an evolutionary novelty in the gibbon lineage. The recent annotation and analysis of the gibbon genome revealed a possible key role for this composite retrotransposon, as LAVA has been found to preferentially insert into introns of genes involved in chromosome segregation processes (Carbone et al. 2014). It has therefore been hypothesized that disruption of these genes through LAVA insertions might have been the trigger of the unusually high rate of chromosomal rearrangements that characterize gibbon species (Carbone et al. 2009). This hypothesis was substantiated by functional studies showing that LAVA insertions can alter host gene transcription by introducing a premature termination site into a given gene (Carbone et al. 2014), similarly to the gene-breaking event described for L1 elements (Han et al. 2004; Wheelan et al. 2005). For this reason, it is important to understand how this new retrotransposon has been mobilizing and if functional LAVA copies are still present in the gibbon genome. Alignments of whole-genome sequences from different gibbon species have shown that multiple LAVA insertions are polymorphic [(Carbone et al. 2014) and data not shown], suggesting that the gibbon genome harbors functional LAVA source elements that are currently retrotransposition-competent and mobilized.
Here we show that a gibbon genome-encoded, full-length LAVA element (LAVAC), a member of the older LAVA_C4B subfamily (supplementary fig. S1, Supplementary Material online), is trans-mobilized by transiently expressed human L1 proteins in HeLa cells at frequencies exceeding processed pseudogene formation rates by at least 117-fold. Similar to SVA retrotransposition (Hancks et al. 2011; Raiz et al. 2012) and processed pseudogene formation (Esnault et al. 2000), LAVA mobilization relies on the presence of both L1-encoded proteins, ORF1p and ORF2p. Analysis of genomic pre- and post-integration sites of three isolated mneoI-tagged LAVAC de novo insertions show all hallmarks of TPRT-mediated retrotransposition events, including the presence of TSDs, a 3′ poly(A) tail, and an L1 EN-specific consensus target sequence 5′-TTTT/AA-3′ (fig. 4C), all of which were also shown to be characteristic of pre-existing genomic LAVA insertions (Carbone et al. 2012, 2014) and de novo retrotransposition events launched from both human L1 and SVA reporter elements (Gilbert et al. 2002; Symer et al. 2002;Ostertag et al. 2003; Gilbert et al. 2005; Hancks et al. 2011; Raiz et al. 2012). Although the small number of isolated insertions is a limitation of this part of our study, the insertions provide evidence for the trans-mobilization of the LAVAC reporter element by the human L1-encoded protein machinery. Unfortunately, additional insertions that we attempted to isolate had occurred into highly repetitive sequences that hindered retrieval and thorough characterization of these insertions via PCR and sequence analysis. The integration events we capture with this kind of cell culture assay are merely a reflection of selection, and it is unlikely to identify de novo insertions disrupting genes, as cells with these insertions would probably not survive. Our finding that two of the three characterized LAVAC de novo insertions are full-length and occurred into introns of host genes in antisense orientation is consistent with a previous report in which six out of seven isolated tagged LAVA_F1 de novo insertions were shown to be full-length and four of these insertions occurred into introns of host genes (Ianc et al. 2014). Additionally, our results are in line with the observation that ∼70% of the endogenous LAVA insertions characterized in the gibbon genome assembly are full-length and 98.5% of the intragenic insertions are intronic (Carbone et al. 2014).
The trans-mobilization frequency of full-length LAVAC exceeds the processed pseudogene formation rate by ∼117- to 238-fold and exceeds human full-length SVAE retrotransposition frequencies by ∼3- to 3.6-fold. In fact, this LAVAC trans-mobilization frequency is within the range of the human Alu retrotransposition frequency that was reported to be 100- to 1000-fold higher than control mRNAs (Dewannieux et al. 2003). Considering that LAVAC is a member of one of the oldest LAVA sub-families aged 16–18 my, this is a noteworthy result indicating that not only members of the youngest sub-families, LAVA_E (10–11 mya) and LAVA_F (6–9 mya) are preferred substrates of the human L1 protein machinery (Ianc et al. 2014). Clearly, retrotransposition reporter assays in a homologous system in which the reporter plasmids are co-transfected with driver plasmids expressing functional gibbon-encoded LINE-1 retrotransposons into cultured gibbon cells, would allow to draw more reliable conclusions on the in vivo retrotransposition activity of LAVA elements in the gibbon genome. For instance, it was reported recently that in a similar assay established to test transposition activity of the DNA transposon piggyBat, the number of de novo transposition events that occurred in HeLa cells, was much higher than those identified in bat fibroblasts (98,816 vs. 4,264) (Mitra et al. 2013). However, because, to the best of our knowledge, there is neither any applicable gibbon cell culture system nor any isolated functional gibbon LINE-1 retrotransposon available to date, we decided to assess the LAVAC retrotransposition activity in the well-established human cell culture system. One reason for these experimental limitations is that many gibbon species are critically endangered making biological materials extremely difficult to come by. Undoubtedly, we can neither suggest the actual in vivo LAVA retrotransposition frequency based on the LAVA trans-mobilization frequencies observed in the human cell culture system, nor compare LAVAC retrotransposition frequencies with the recently reported trans-mobilization rates of LAVA_E and LAVA_F1 members (Ianc et al. 2014), due to the overall variability of the applied cell culture based assays between different laboratories. Nevertheless, it is surprising that the copy numbers of the LAVA subfamilies C, E, and F constitute only 404, 350, and 773 members, respectively. Therefore, we speculate that the number of functional and transcribed LAVA_C master copies is likely to be relatively small in the modern genome, otherwise we would observe much higher copy numbers of this subfamily.
The modular structure of the LAVA element allowed testing of different combinations of its portions in our trans-mobilization assay to identify which modules make LAVA RNAs preferred substrates for L1 proteins. We found that the SVA-derived module containing the CT-rich, Alu-like, and VNTR regions provides the competence to retrotranspose at rates beyond that of processed pseudogene formation. Recent studies investigating the structural features of SVA elements that affect their trans-mobilization frequency (Hancks et al. 2012) demonstrated that removal of the Alu-like region in the context of a full-length SVA has little to no effect, whereas removal of the CT-hexamer or the VNTR region can result in a 75% decrease in activity. This is consistent with our observation that removal of the SVA-specific fragment covering CT-rich region, Alu-like region, and VNTR module from the full-length LAVAC reporter element (pLC10/LAVAC) reduces trans-mobilization frequency by ∼98% (fig. 2B). The VNTR region is indeed likely to be crucial for both SVA and LAVA mobilization given that SVA2 elements found in rhesus macaque (Gibbs et al. 2007; Han et al. 2007a) consist exclusively of VNTRs and show signatures of L1-mediated retrotransposition (Damert et al. 2009).
Interestingly, the LAVAC reporters exhibiting the highest trans-mobilization frequencies include both the VNTR and AluSz modules. Removal of the VNTR-U1-AluSz-U2 module results in reduction of the retrotransposition frequency by at least ∼15-fold (fig. 2B and supplementary fig. S5, Supplementary Material online). It is plausible that this effect is caused by the absence of the VNTR region that has recently been demonstrated to be crucial for LAVA_F1 mobilization (Lupan et al. 2015). However, we cannot rule out the possibility that the AluSz module may also play a role in LAVA trans-mobilization as it has been shown that the presence of an AluSp module at the 3′ end of an SVAF1 source element increases its trans-mobilization frequency by ∼25-fold (Raiz et al. 2012). Our data also indicate that the presence of the 211-bp L1ME5 module reduces mobilization frequency of the full-length LAVAC element by ∼2- to 8-fold. It remains to be investigated, which function the trans-mobilization-attenuating L1ME5 module may have and why it was stably retained as part of functional endogenous LAVA elements during evolution, despite its apparent ability to attenuate LAVA mobilization rates. An exhaustive search of the gibbon genome retrieved only seven LAVA elements that lack exclusively the L1ME5 module. However, all these insertions are missing poly(A) tails suggesting that they derived from polyadenylated full-length LAVA transcripts that were 3′-truncated upon insertion by a mechanism different from TPRT.
The 3′-end portion of LAVAC comprising the modules U1, AluSz, U2, and L1ME5, is trans-mobilized at frequencies exceeding processed pseudogene formation rate by only ∼2- to 7-fold. This is consistent with our comparative data showing that only one copy of this sequence was present in the anthropoid common ancestor genome. Because this sequence was present without apparent mobility for a very long time in primate genomes, it is obviously not a preferred substrate for trans-mobilization by L1 proteins. In the gibbon genome, this sequence is found at position chr1a:39006175–39006659, and we refer to it as LAVA 3′ Progenitor Locus Allele A. We speculate that this locus is the original source of the LAVA 3′ sequence and propose two possible models for how this sequence was united with the SVA-derived module we find at the 5′ end of the full-length LAVA (fig. 5).
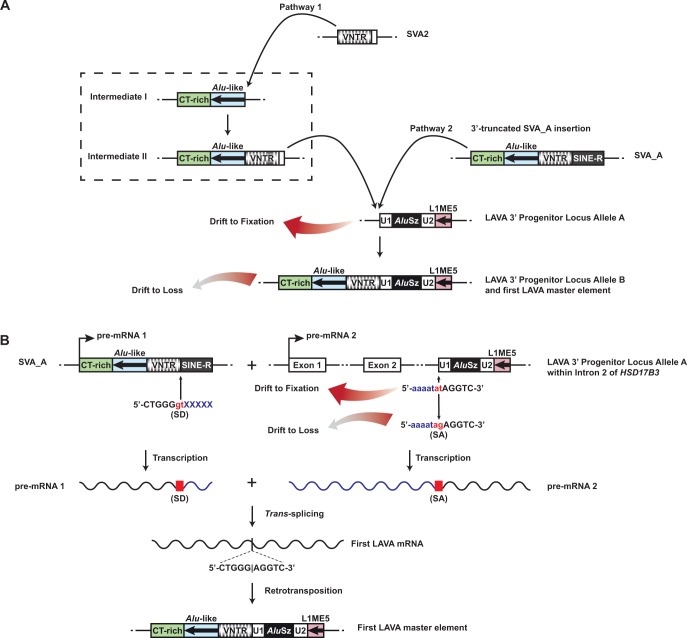
Fig. 5.—
Alternative models for the assembly process resulting in functional LAVA master copies. The putative LAVA 3′ Progenitor Locus Allele A consisting of the U1, AluSz, U2, and L1ME5 modules was present in the anthropoid common ancestor and is still present at orthologous loci in the human (chr9:99026506–99026986 in hg19) and gibbon (chr1a:39006175–39006659 in Nleu3.0) genomes, among others. (A) Model A entails the precise insertion of the CT-rich, Alu-like, and VNTR regions upstream of LAVA 3′ Progenitor Locus Allele A. The inserting sequence may have been derived from the union of an SVA2 sequence with CT-rich and Alu-like region (Pathway 1; Intermediates I and II, within the dashed box, are speculative) or from a 3′-truncated SVA_A element (Pathway 2). Because the master copy of the LAVA 3′ Progenitor Locus Allele B cannot be found at the 3′ Progenitor Locus on gibbon chromosome 1a, we speculate that this allele persisted only long enough to give rise to other master copies and was subsequently lost to genetic drift, leaving only the original allele (Allele A), which lacks the SVA-derived modules. (B) In Model B, a trans-splicing event gave rise to the first LAVA master locus. We hypothesize that a trans-splicing event occurred between a cryptic splice donor (SD) site within the 3′ end of an SVA_A mRNA (pre-mRNA 1) and a splice acceptor (SA) site immediately upstream of the chromosome 1a LAVA 3′ Progenitor Locus in the gibbon common ancestor. The mRNA from the LAVA 3′ Progenitor Locus is provided as the consequence of transcription of the gene HSD17B3, in which the LAVA 3′ Progenitor Locus is located in Intron 2 in transcriptional orientation of the gene. In all analyzed gibbon genomes, including Nleu3.0, the SA site adjacent to the LAVA 3′ Progenitor Locus does not have the necessary AG nucleotide sequence, and instead has an AT sequence. We speculate that a T-to-G point mutation at this position in an ancestral gibbon individual produced the necessary SA sequence. After giving rise to the first LAVA master element elsewhere in the genome as consequence of the trans-splicing event, this variant allele at the LAVA 3′ Progenitor Locus was subsequently lost to genetic drift.
Model A is based on the fortuitous retrotransposition of the SVA2- or SVA_A-derived sequences precisely to the 5′ end of the U1 component at LAVA 3′ Progenitor Locus Allele A (fig. 5A). The inserting sequence could have been donated either from a 3′-truncated hypothetical precursor element comprised of an SVA2 united with CT-rich and Alu-like sequences (Pathway 1) (Ianc et al. 2014) or a 3′-truncated SVA_A element (Pathway 2) (Damert et al. 2009). Insertions of either type could have resulted in the full-length LAVA element at the chromosome 1a locus (Allele B) and this could have served as the first functional LAVA master copy.
Model B is based on the fact that the LAVA 3′ Progenitor Locus is found within Intron 2 of the gene HSD17B3 and that the two nucleotides immediately adjacent to the 5′ end of the U1 component at this locus are 5′-AT-3′. We hypothesize that this 5′-AT-3′ dinucleotide may have mutated at some point to 5′-AG-3′, resulting in a cryptic splice acceptor site. A subsequent trans-splicing event could then have occurred between this splice acceptor site within an HSD17B3 transcript and a splice donor site in the 3′ end of the VNTR region of an SVA_A mRNA molecule, resulting in an mRNA molecule which, when reverse transcribed and integrated into the genome, produced the first LAVA master element (fig. 5B). We investigated whether a cis-splicing event could explain the origin of LAVA as well. However, this would require a copy of SVA upstream of the LAVA 3′ Progenitor Locus on chromosome 1a and in the same orientation. Only one such SVA copy exists upstream on this chromosome (at position chr1a: 26376513–26377921), and its VNTR sequences do not match those in the LAVA element. Additionally, this SVA copy is located ∼12.6 Mb upstream of the LAVA 3′ Progenitor Locus, a distance making cis-splicing with this locus unlikely.
In the current gibbon genome assembly (Nleu3.0), we do not find evidence of a full-length LAVA at the LAVA 3′ Progenitor Locus Allele B on chromosome 1a (Model A) nor do we find the cryptic splice acceptor sequence (Model B) (fig. 5). This suggests that under either model these variants were lost to genetic drift over time, but only after having given rise to other master copies in the genome from which the LAVA family could propagate. Because the probability of any new mutation in a population of diploid organisms being lost to genetic drift is in which is the effective population size, reversion to the ancestral sequence is a highly likely event for any new mutation (Kimura 1955). However, we cannot discount the possibility that one of the alleles that gave rise to the first master copy is present in the genome, but is obfuscated in the current assembly which, like all draft genomes, contains gaps and assembly errors.
In conclusion, we have demonstrated the ability of a full-length member of the old LAVA_C subfamily of gibbon retrotransposons to mobilize in HeLa-HA cells by recruiting the human L1 protein machinery. Our results indicate that the 5′ SVA-derived module is essential for efficient retrotransposition, likely due to the presence of the VNTR region. The AluSz module may also confer some activity, whereas the L1ME5 module appears to reduce retrotransposition efficiency. Analysis of the sequence architecture of de novo LAVAC insertions supports the conclusion that LAVA is a non-autonomous family that relies upon L1-mediated TPRT as its mechanism of propagation. Considering the recently reported trans-mobilization of members of the LAVA_E and LAVA_F1 subfamilies (Ianc et al. 2014), LAVAC represents a member of the evolutionary older LAVA_C subfamily that is the third LAVA element demonstrated to be a preferred substrate for trans-mobilization by the L1 protein machinery.
Supplementary Material
Supplementary figures S1–S6 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank John Moran for providing plasmids pJM101/L1.3, pJM101/L1RP, and pJM101/L1RPΔneo and the cell lines HeLa-HA. We are grateful to John Goodier and Haig Kazazian who provided us with the reliable αL1ORF2p-N antibody. We are indebted to Kay-Martin Hanschmann for statistical analyses. This work was supported by the Deutsche Forschungsgemeinschaft [DA 545/2-1 and SCHU 1014/8-1 to G.G.S.] and the National Institutes of Health [NCRR P51, RR000163 to LC]. Moreover, T.J.M. was supported by the National Library of Medicine of the National Institutes of Health under Award Number T15LM007088. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Literature Cited
- Andrews PW, et al. 1984. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Invest. 50:147–162. [PubMed] [Google Scholar]
- Belancio VP, Deininger PL, Roy-Engel AM. 2009. LINE dancing in the human genome: transposable elements and disease. Genome Med. 1:97.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. 2008. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res.. 18:343–358. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger PL. 2010. All y'all need to know 'bout retroelements in cancer. Semin Cancer Biol. 20:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Batzer MA. 2006. Retrotransposable elements and human disease. Genome Dyn. 1:104–115. [DOI] [PubMed] [Google Scholar]
- Carbone L, et al. 2014. Gibbon genome and the fast karyotype evolution of small apes. Nature 513:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, et al. 2012. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol. Evol. 4:648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, et al. 2009. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 5:e1000538.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, LaCava J, Taylor MS, Boeke JD. 2014. Expression and detection of LINE-1 ORF-encoded proteins. Mob. Genet. Elements 4:e29319.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damert A, et al. 2009. 5'-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 19:1992–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet.. 7:e1002384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41–48. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24:363–367. [DOI] [PubMed] [Google Scholar]
- Freeman JD, Goodchild NL, Mager DL. 1994. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques 17(46):48–49. 52. [PubMed] [Google Scholar]
- Gibbs RA, et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell Biol. 25:7780–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315–325. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr. 2004. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 13:1041–1048. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429:268–274. [DOI] [PubMed] [Google Scholar]
- Han K, et al. 2007a. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science 316:238–240. [DOI] [PubMed] [Google Scholar]
- Han K, et al. 2007b. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 3:1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, et al. 2008. L1 recombination-associated deletions generate human genomic variation. Proc. Natl. Acad. Sci. U S A. 105:19366–19371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr. 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20:3386–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr. 2012. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Mandal PK, Cheung LE, Kazazian HH., Jr. 2012. The minimal active human SVA retrotransposon requires only the 5'-hexamer and Alu-like domains. Mol. Cell Biol. 32:4718–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann T, Heidmann O, Nicolas JF. 1988. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc. Natl. Acad. Sci. U S A. 85:2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. 2007. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianc B, Ochis C, Persch R, Popescu O, Damert A. 2014. Hominoid composite non-LTR retrotransposons-variety, assembly, evolution, and structural determinants of mobilization. Mol. Biol. Evol. 31:2847–2864. [DOI] [PubMed] [Google Scholar]
- Isomura H, et al. 2008. A cis element between the TATA Box and the transcription start site of the major immediate-early promoter of human cytomegalovirus determines efficiency of viral replication. J. Virol. 82:849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaer K, Speek M. 2013. Retroelements in human disease. Gene 518:231–241. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1955. Solution of a process of random genetic drift with a continuous model. Proc. Natl. Acad. Sci. U S A. 41:144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilyuk A, et al. 2008. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 36:648–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Batzer MA. 2010. A mobile threat to genome stability: the impact of non-LTR retrotransposons upon the human genome. Semin. Cancer Biol. 20:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. 2008. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS One 3:e4047.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Locke DP, et al. 2011. Comparative and demographic analysis of orangutan genomes. Nature 469:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupan I, Bulzu P, Popescu O, Damert A. 2015. Lineage specific evolution of the VNTR composite retrotransposon central domain and its role in retrotransposition of gibbon LAVA elements. BMC Genomics 16:389.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, et al. 2013. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc. Natl. Acad. Sci. U S A. 110:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927. [DOI] [PubMed] [Google Scholar]
- Muller S, Hollatz M, Wienberg J. 2003. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum. Genet. 113:493–501. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. 2003. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 73:1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiz J, et al. 2012. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40:1666–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, et al. 2001. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 12:743–749. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer DE, et al. 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110:327–338. [DOI] [PubMed] [Google Scholar]
- Szak ST, et al. 2002. Molecular archeology of L1 insertions in the human genome. Genome Biol. 3:research0052.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah KR, et al. 2015. Examining phylogenetic relationships among gibbon genera using whole genome sequence data using an approximate bayesian computation approach. Genetics 200:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, et al. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 21:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD. 2005. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 15:1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler N, et al. 2005. Analysis of 5' junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5'-end attachment requiring microhomology-mediated end-joining. Genome Res. 15:780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.