Abstract
The evolution of new organs is difficult to study because most vertebrate organs evolved only once, more than 500 million years ago. An ideal model for understanding complex organ evolution is the placenta, a structure that is present in live bearing reptiles and mammals (amniotes), which has evolved independently more than 115 times. Using transcriptomics, we characterized the uterine gene expression patterns through the reproductive cycle of a viviparous skink lizard, Pseudemoia entrecasteauxii. Then we compare these patterns with the patterns of gene expression from two oviparous skinks Lampropholis guichenoti and Lerista bougainvillii. While thousands of genes are differentially expressed between pregnant and non-pregnant uterine tissue in the viviparous skink, few differentially expressed genes were identified between gravid and non-gravid oviparous skinks. This finding suggests that in P. entrecasteauxii, a pregnant-specific gene expression profile has evolved, allowing for the evolution of pregnancy-specific innovations in the uterus. We find substantial gene expression differences between the uterus of the chorioallantoic and the yolk sac placenta in P. entrecasteauxii, suggesting these placental regions are specialized for different placental functions. In particular, the chorioallantoic placenta is likely a major site of nutrient transport by membrane-bound transport proteins, while the yolk sac placenta also likely transports nutrients but via apocrine secretions. We discuss how the evolution of transcription factor networks is likely to underpin the evolution of the new transcriptional states in the uterine tissue of viviparous reptiles.
Keywords: viviparity, placenta, RNA-seq, lizard, convergent evolution, Pseudemoia
I.ntroduction
Understanding how complex structures such as organs evolve is a question fundamental to evolutionary and organismal biology (Schwenk et al. 2009). Complex traits offer an intriguing puzzle to evolutionary biologists because they require changes to several tissues that must be underpinned by multiple genetic changes (Gregory 2008). Comparisons of organs that have evolved convergently in different lineages provide a unique opportunity to identify the genetic changes that have led to the evolution of complex organs (Stern 2013).
Complex organs evolve by modifications to pre-existing structures (Shubin et al. 2009). The evolution of new functions in pre-existing tissues occurs by three broad processes: the recruitment of genes expressed elsewhere in the organism (gene expression recruitment), the co-option of expressed genes so that they now support novel physiological functions (gene co-option), and the introduction of new genes (typically introduced by viruses, retro-transposons, or gene duplications) (True and Carroll 2002; Cross et al. 2003; Long et al. 2003; Zhang 2003; Weake and Workman 2010).
A placenta is an organ formed by the apposition of maternal and embryonic tissue to exchange materials between mother and embryo during development (Mossman 1937). Placentae provide an ideal model for understanding organ evolution as they have evolved many times independently, have evolved relatively recently in some lineages, and they involve complex functions that require changes in the expression of suites of genes that cannot be caused by mutations in just a few genes (Reznick et al. 2002; Brandley et al. 2012; Hou et al. 2012; Griffith 2015). Placentae are necessary in all live bearing (viviparous) reptiles and mammals (amniotes) because they facilitate the exchange of respiratory gases and water between mother and embryo—physiological functions that would otherwise occur across the eggshell in a laid egg. The placenta has evolved independently in at least 115 amniote lineages, with all but one of these transitions occurring in lizards and snakes (Blackburn 2014; Griffith et al. 2015). While a recent analysis suggested that the number of independent origins of viviparity may be controversial, there is a consilience of data that counters any suggestion that there have been a high number of reversions from a viviparous to oviparous reproductive mode (Griffith et al. 2015; Shine 2015; Wright et al. 2015). The presentation of these data led the lead author of the original analyses to concede that the data support the hypothesis that there have been frequent transitions to viviparity in squamates (Pyron 2015). In viviparous squamates, a vast majority of placentae are simple structures with mainly gas and water exchange functions, and the embryo relies on yolk reserves for nutrition, much like an embryo of an oviparous species. However, placentotrophy (embryonic nutrition provided via a placenta during development) has evolved only seven times in viviparous amniote vertebrates: once in therian mammals and six times in lizards (Murphy and Thompson 2011; Blackburn 2014; Metallinou et al. 2016). That placentotrophy has evolved so few times in the more than 115 viviparous amniote lineages, suggesting that mechanisms of placental nutrient transport are either relatively difficult to evolve (i.e. require multiple complex steps) or placentotrophy has not been selected for in many of the viviparous lineages of squamates due to it not being adaptive in most environments (Murphy and Thompson 2011).
In all viviparous amniote lineages, placentae are derived from three tissues, the uterus of the mother, and the embryo’s chorioallantoic and yolk sac membranes. However, differing placental invasiveness has resulted in differences in the tissues responsible for nutrient transport from mother to embryo. In reptiles and humans, the tissues responsible for nutrient uptake during pregnancy are not homologous. Humans exhibit highly invasive hemochorial placentation, which has resulted in embryonic trophoblast invading through multiple layers of maternal cells, to be bathed in maternal blood (Wildman et al. 2006). As fetal tissue has direct access to maternal blood, the embryonic cells of the trophoblast are directly responsible for nutrient uptake (Fowden et al. 2006; Lager and Powell 2012). Most reptiles such as the Australian southern grass skink (Pseudemoia entrecasteauxii) exhibit non-invasive epitheliochorial placentation, so nutrients must be transported from the maternal blood to the embryo via the maternal uterine epithelium (Adams et al. 2005; Griffith, Van Dyke, et al. 2013). Although the tissues that facilitate nutrient uptake from the mother differ between species, the function of these different tissues is fundamentally the same. In the skink, nutrients are absorbed from the maternal blood by the uterine endothelium, transported across several layers of uterine cells, and then released into the uterine lumen by the uterine epithelium. In humans, nutrients are absorbed from the maternal blood by the trophoblast epithelium, are transported across several layers of trophoblast tissue, and then released into the embryonic blood supply by placental endothelial cells. By examining the convergent evolution of placental transport of nutrients in non-homologous tissues, we can test if the convergent evolution of a complex trait (placentotrophy) is constrained in the absence of shared developmental histories.
In most viviparous squamates, two distinct embryonic tissues come into contact with the uterus, the chorioallantois, and yolk sac membrane. When this contact between maternal and embryonic tissue persists through pregnancy, two separate placentae form, the chorioallantoic placenta, which forms at the embryonic pole, and the yolk sac placenta which sits at the ab-embryonic pole (Murphy and Thompson 2011). In the matrotrophic skink, P. entrecasteauxii, the chorioallantoic and yolk sac placentae form with distinct cellular morphologies, which likely support different physiological functions at these sites (Van Dyke et al. 2014). For example, the yolk sac placenta has been suggested to support the exchange of macro-nutrients including lipids, while the chorioallantoic placenta supports the exchange of respiratory gasses and some ions (Adams et al. 2005; Griffith, Ujvari, et al. 2013, Van Dyke et al. 2015).
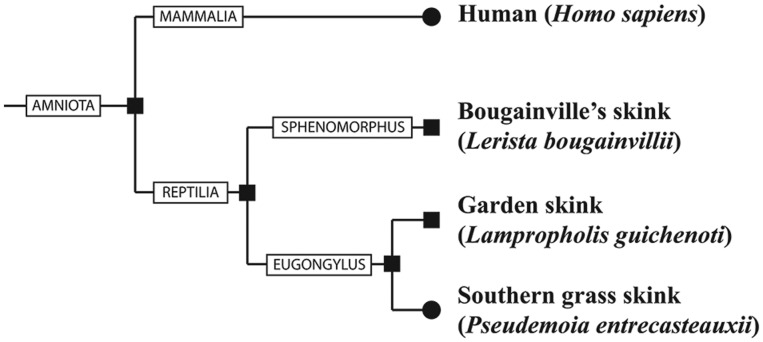
To investigate the evolution of placentation in a matrotrophic skink, we compare patterns of gene expression from the uterus of the chorioallantoic and yolk sac placenta, in pregnant P. entrecasteauxii, with the uterus of non-pregnant females. To determine whether the expression patterns in this skink are associated with viviparity rather than simply patterns shared with the ancestral oviparous squamate condition, we compared the P. entrecasteauxii transcriptomes to those from gravid and non-gravid uterine transcriptomes of two related oviparous skinks Lampropholis guichenoti and Lerista bougainvillii. While a viviparous population of Le. bougainvillii exist, our study focused on the oviparous population located in the Burra Region of South Australia. These oviparous taxa were selected to allow us to infer the ancestral state of the skink taxa examined, with Lampropholis and Pseudemoia both belonging to the Eugongylus group skinks, while Lerista represents an outgroup in the Sphenomorphus group skinks (fig. 1; Skinner et al. 2011; Brandley et al. 2015). Finally, we statistically compare the amino acid transporter gene expression in skinks and humans to test whether they have convergently evolved placental amino acid transport by the use of the same genes during pregnancy.
Fig. 1.—
Phylogenetic relationship between species discussed in this study. Viviparous taxa are indicated by circles and oviparous taxa by squares at branch tips and nodes. Lizards examined in this study are members of either the Eugongylus group or the Sphenomorphous group skinks.
Materials and methods
Animal Collection
All animal work was conducted with University of Sydney Animal Ethics approval. Gravid female P. entrecasteauxii were collected in Kanangra Boyd National Park, NSW, Australia, in November 2011. Pseudemoia entrecasteauxii is a highly seasonal breeder and reproductive stage can be approximated by the time of year (Murphy et al. 2006). Once lizards had reached the appropriate period of the reproductive cycle, they were euthanized by injection with 0.1 mL of sodium pentobarbital (6 mg/mL). Non-pregnant lizards were collected during pregnancy and housed for an additional 2 months after giving birth before processing. Pregnant females were euthanized at developmental stage 40, the final stage of the Dufaure and Hubert (1961) staging scheme. The egg chamber of the uterus in gravid females was excised and uterus was cut along the boundary of the chorioallantoic membrane and yolk sac, resulting in two hemispheric portions of uterine tissue. This made it possible to collect uterine tissue of the chorioallantoic placenta (n = 3) and of the yolk sac placenta (n = 3) separately. In P. entrecasteauxii and most viviparous squamates, there is little physical attachment between the uterus and the extra-embryonic membranes, so the uterus can be cleanly separated from embryonic tissue (Biazik et al. 2010, 2012). In non-gravid females (n = 2), egg chambers constituting stretched regions of the uterus were excised individually (Girling 2002). After excision, uterine tissues were fixed in RNA later (24 h) and stored at −80 °C.
We collected gravid (n = 3) and non-gravid (n = 3) oviparous Le. bougainvillii from the Burra Region of South Australia in December 2014 and January 2015. It is not possible to identify if oviparous lizards were post-reproductive, as they were collected from the field. Lizards were euthanized, uteri dissected immediately, and tissues were fixed and stored as above. We collected gravid (n = 3) La. guichenoti from the Sydney region in 2009–2014. Non-gravid La. guichenotti samples (n = 3) were obtained by maintaining gravid females in the lab until they had deposited eggs, and then harvesting tissues after an additional 2 month period in captivity. For both Le. bougainvillii and La. guichenoti, eggs were shelled and at developmental stage 25–30. Lizards were returned to the University, euthanized, and uteri collected. Tissues were fixed and stored as above.
Transcriptome Sequencing, Assembly, and Annotation
There was no pooling of biological replicates, each sample was sequenced separately. Tissue was macerated using a mechanical homogenizer in 600 μL of Buffer RLT (QIAGEN) then homogenized using a QIAshredder spin column (QIAGEN). Total RNA was extracted using the RNeasy Mini Kit (QIAGEN). Extracted RNA was treated with Amplification Grade DNase 1 (Sigma-Aldrich). RNA quality was measured on the Agilent 2000 Bioanalyzer (Agilent Technologies) and was only used for transcriptome analysis if the RIN was greater than 8. Sequencing libraries were generated in house using the TruSeq RNA Sample Preperation kit (Illumina Inc.). All P. entrecasteauxii, and four La. guichenoti samples (two non-gravid and two gravid) were sequenced on the HiSeq2000 (Illumina Inc.). All Le. bougainvillii and two La. guichenoti (one non-gravid and one gravid) had their sequencing libraries prepared by the Ramaciotti Centre using the TruSeq RNA Sample Preperation kit (Illumina Inc.) and were sequenced on the HISeq2500 (Illumina Inc).
For P. entrecasteauxii, raw transcriptome reads were aligned to the de-novo assembled transcriptome for this species (Griffith, Brandley, Belov, et al. 2016). For the oviparous skink species, transcriptomes were assembled de-novo from the data presented in this study, using the same transcriptome assembly approach that was used for P. entrecasteauxii. De-novo transcriptome assembly was performed with ABySS (Simpson et al. 2009). Assemblies were made for each sample individually using a kmer of 39 and 59. To build reference transcriptomes for each species, we combined the assemblies from each sample and then removed contigs smaller than 100 bp and redundant contigs using CD-HIT-EST (Huang et al. 2010) with default options. Contigs in the assembled transcriptomes were annotated by aligning to complete proteomes of Anolis carolinensis (Ensemble build 79), Gallus gallus (Ensembl build 82), and Homo sapiens (Ensembl build 82) with BlastX with an e-value of 10−5. The alignment rate of the raw reads to the assembled transcriptome was > 90% for all samples.
Differential Gene Expression Analysis
Expression of each contig in the reference transcriptome was measured for each biological sample by aligning the raw reads of each sequenced transcriptome against the species specific reference transcriptome with Bowtie2 (version 2.0.0-beta7) (Langmead and Salzberg 2012). Counts from contigs that aligned to the same gene were summed, and only counts for contigs that could be identified were used in statistical analyses.
For oviparous taxa, differential gene expression between gravid and non-gravid uterine tissue was compared in pairwise comparisons with DESeq (Anders and Huber 2010). For P. entrecasteauxii differential gene expression between uterine tissue of non-pregnant uterine tissue, uterine tissue of the chorioallantoic and yolk-sac placentae were compared with each other in three pairwise comparisons using DESeq (Anders and Huber 2010).
A false discovery threshold of 0.05 was used for all downstream analyses. Transcriptome data were validated by reverse transcription quantitative PCR on a subset of genes. To ensure the observed gene expression patterns were real, we confirmed the expression of five genes (AQP3, CTSA, SLC22A5, and RARRES1) that showed differential expression between sample groups in P. entrecasteauxii (see supplementary methods, Supplementary Material online, for more details). All genes that were identified as significantly differentially expressed in the transcriptomes were also significantly differentially expressed when measured by qPCR (supplementary material fig. S1, Supplementary Material online).
Gene-ontology (GO) annotation was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resources 6.7 (Huang et al. 2009). Gene symbol lists for significantly up and down regulated genes when comparing the chorioallantoic and yolk sac placenta of P. entrecasteauxii were entered and converted to DAVID ID’s. Genes were annotated with GO terms corresponding to biological processes (BP-FAT).
Further Statistical Analyses
To visualize the data, we performed principle component analyses (PCA) on the expression of several gene sets from our study. The PCAs were performed on the square root TPMs and data were transformed to equally weigh each gene by subtracting the mean expression of each gene from the individual expression of that gene in each sample. PCAs were performed using the stats package in R. Principle component analysis was performed on the expression of all genes in all species in this study, the expression of amino acid transport protein genes in all species in this study, and the expression of transcription factors in uterine tissue of P. entrecasteauxii. We limited our PCA analysis of transcription factor expression to P. entrecasteauxii as comparisons with the other species largely show a species level gene expression signal. Our list of known transcription factors was taken from Ravasi et al. (2010).
We calculated the probability of utilizing as many or more overlapping amino acid transport protein genes in the placentae of skinks and humans. To assess this probability, we made two assumptions; (a) co-option events were independent and (b) co-option events were random. We used a hypergeometric test to identify if the observed overlap in genes used in placental tissues of skinks and humans during pregnancy likely arose by chance. If a P value of less than 0.05 was observed, we rejected the assumptions of our analysis suggesting either co-option events are not independent or non-random.
Results
Transcriptome Sequencing and Assembly
For Pseudemoia entrecasteauxii, we sequenced a mean of 5.04 × 107± 2.8 × 106 101 bp paired end reads per sample. Our assembled transcriptome contained 367,722 contigs with a mean contig length of 465 bp. Alignment of these contigs to reference proteomes identified 88,204 contigs. These contigs map to 16,942 unique genes.
For Lampropholis guichenoti, we sequenced a mean of 4.66 × 107± 4.1 × 106 101 bp paired end reads per sample. Our assembled transcriptome contained 427,110 contigs with a mean contig length of 284 bp. Alignment of these contigs to reference proteomes identified 71,021contigs, which map to 15,492 unique genes.
For Lerista bougainvillii, we sequenced a mean of 6.91 × 107± 1.8 × 106 101 bp paired end reads per sample for Le. bougainvillii. Our assembled transcriptome contained 230,011 contigs with a mean contig length of 541 bp. Alignment of these contigs to reference proteomes identified 54,886 contigs. These contigs map to 14,521 unique genes.
Differential Gene Expression in the Viviparous Skink Placenta
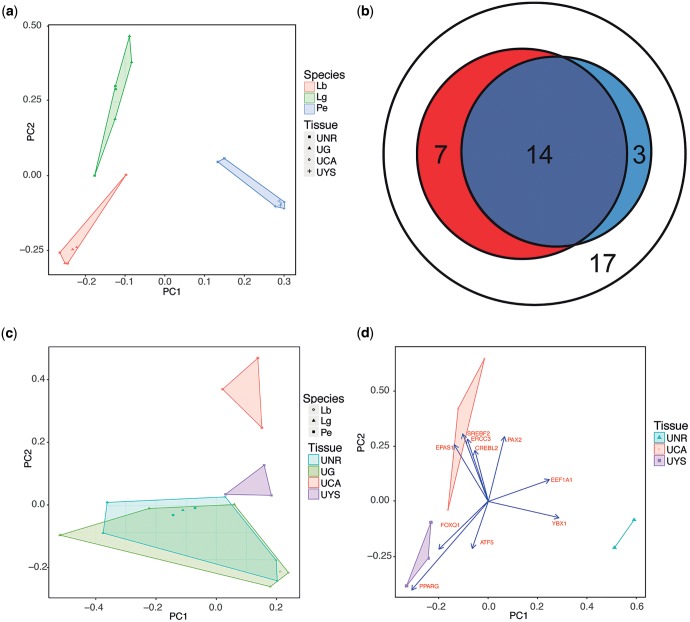
Principle component analysis on all shared genes showed three distinct species-specific clusters. (fig. 2a). In the clusters of both oviparous taxa, there is no separation of non-gravid and gravid uterine transcriptomes, but there is separation of pregnant from non-pregnant transcriptomes in P. entrecasteauxii along principle component 1. The species-specific clustering of transcriptome sequences is consistent with concerted transcriptome evolution, which has been predicted and observed in other studies (Liang et al. 2016).
Fig. 2.—
Comparisons of placental gene expression between different species and placental region. (a) PCA of square root TPM for all shared genes in the uterine tissue of the oviparous skinks Le. bougainvillii (Lb) and La. guichenoti (Lg), and the viviparous skink P. entrecasteauxii. Uterine tissue is from either non-reproductive (UNR) or gravid (UG) females, or from the chorioallantoic (UCA) or yolk sac (UYS) placenta of late pregnant females. Samples form discrete clusters based on the species of origin. (b) Amino acid transporter gene use in the placentae of reptiles and mammals. There is an overlap in the use of orthologous amino acid transport protein genes by human trophoblast (red) and grass skink uterine tissue (blue) during pregnancy. 41 orthologous amino acid transport proteins are shared by both lineages, 21 are utilized in human placenta, while 17 are used in grass skinks. About 17 genes are not utilized in the placental tissues of either species (white). Lineages show a significantly non-random use of genes (P = 0.003, hypergeometric test). (c) PCA of square root TPM for amino acid transporter gene expression in the uterine tissue of oviparous and viviparous skinks. In P. entrecasteauxii, uterine tissue from each placental tissue clusters with each other based on amino acid transporter expression, and there is an overlap in the expression profiles of gravid oviparous uterine tissue and non-pregnant uterine samples from all species. (d) PCA of square root TPM for transcription factors expressed in the uterine tissue of P. entrecasteauxii. Three discrete clusters form one for each placental region, and one for non-pregnant tissue. The data are overlaid with the loadings for the top 10 genes that contribute to the distance between samples in these principle components.
Uterine tissues from the chorioallantoic and yolk sac placenta of P. entrecasteauxii have vastly different expression profiles when compared with non-reproductive tissue. The uterus of the chorioallantoic placenta has 2,555 differentially expressed genes compared with non-pregnant uterine tissue (all of which had more than a 2-fold difference in gene expression, supplementary table S1, Supplementary Material online). The uterus of the yolk sac placenta has 3,861 differentially expressed genes compared to non-pregnant uterine tissue (3,823 of which had more than a 2-fold difference in gene expression, supplementary table S2, Supplementary Material online). Differential gene expression from the uterus of the chorioallantoic and yolk sac placenta identified 282 differentially expressed genes (all of which had more than a 2-fold difference in gene expression, supplementary table S3, Supplementary Material online).
Gene ontology (GO) analysis on these differentially expressed genes suggests the uterus is regionally specialized to support different functions associate with pregnancy (fig. 3). Gene ontology analysis on genes that were significantly more highly expressed in the chorioallantoic placenta, compared to the yolk sac placenta identified eight clusters of genes which were associated with significantly over-represented GO terms (table 1). Four of these clusters are specifically related to nutrient transport processes, and three related to steroid hormone metabolism and transports. Gene ontology analysis on genes that were significantly more highly expressed in the yolk sac placenta, compared with the chorioallantoic placenta identified 10 clusters of genes which were associated with significantly over-represented GO terms (table 2). Four of these clusters specifically related to nutrient transport processes, and three related to steroid hormone metabolism and transport.
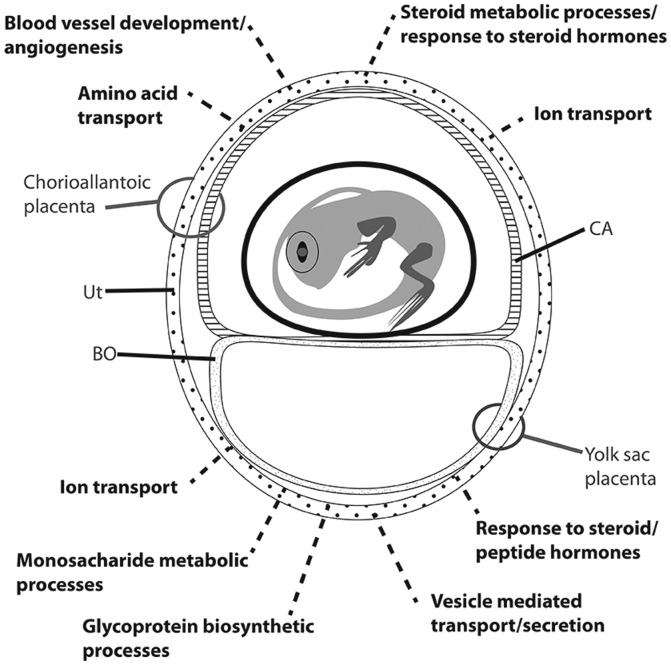
Fig. 3.—
Inferred processes occurring in the two placentae of the southern grass skink. Stylized figure (modified from Stewart and Thompson 2003) outlining the processes (dashed lines) occurring in the uterine tissue of each placenta in grass skinks as identified by functional annotation analysis of differentially expressed genes between chorioallantoic and yolk sac placental samples. Ut, uterus; BO, bilaminar omphalopleure; CA, chorioallantois.
Table 1.
Clustering of Significantly Over-represented GO Terms from a List of Genes Significantly More Highly Expressed in the Chorioallantoic Placenta Compared the Yolk Sac Placenta in P. entrecasteauxii
| GO term (biological processes) | P valuea | Benjamini corrected P value | Fold enrichment | Gene symbols in cluster |
|---|---|---|---|---|
| Organic acid transport (Cluster 1) | Enrichment Score: 4.22 | |||
| GO:0015837∼amine transport | 0.0000 | 0.00 | 17.37 | SLC38A3, AQP9, SLC6A2, SLC6A4, SLC7A8, SLC3A2, SLC22A3, SLC7A5, PDZK1, CRYM |
| GO:0046942∼carboxylic acid transport | 0.0000 | 0.00 | 11.15 | SLC38A3, SLC16A1, AQP9, AQP8, SLC7A8, SLC3A2, SLC7A5, PDZK1 |
| GO:0015849∼organic acid transport | 0.0000 | 0.00 | 11.08 | SLC38A3, SLC16A1, AQP9, AQP8, SLC7A8, SLC3A2, SLC7A5, PDZK1 |
| GO:0015804∼neutral amino acid transport | 0.0001 | 0.02 | 39.04 | SLC38A3, SLC7A8, SLC3A2, SLC7A5 |
| GO:0006865∼amino acid transport | 0.0009 | 0.10 | 11.39 | SLC38A3, SLC7A8, SLC3A2, SLC7A5, PDZK1 |
| GO:0015807∼L-amino acid transport | 0.0054 | 0.26 | 26.74 | SLC38A3, SLC7A8, SLC7A5 |
| GO:0015718∼monocarboxylic acid transport | 0.024 | 0.58 | 12.30 | SLC16A1, AQP9, AQP8 |
| Neuron-related transport (Cluster 2) | Enrichment Score: 2.36 | |||
| GO:0015844∼monoamine transport | 0.0001 | 0.01 | 48.23 | SLC6A2, SLC6A4, SLC22A3, CRYM |
| GO:0019226∼transmission of nerve impulse | 0.0014 | 0.13 | 4.69 | UNC119, CLDN19, SLC6A2, GALR3, SLC6A4, LPAR3, SLC22A3, VIPR1 |
| GO:0007268∼synaptic transmission | 0.0030 | 0.22 | 4.81 | UNC119, SLC6A2, GALR3, SLC6A4, LPAR3, SLC22A3, VIPR1 |
| GO:0006836∼neurotransmitter transport | 0.0074 | 0.29 | 9.88 | SLC6A2, SLC6A4, SLC22A3, SLC6A17 |
| GO:0007267∼cell-cell signaling | 0.025 | 0.57 | 2.73 | UNC119, SLC6A2, GALR3, SLC6A4, LPAR3, SLC22A3, VIPR1, IHH |
| Lipid metabolism (Cluster 3) | Enrichment Score: 2.09 | |||
| GO:0008202∼steroid metabolic process | 0.0004 | 0.05 | 7.10 | SDR42E2, NPC1, OSBPL3, HSD17B2, SDR42E1, SORL1, NPC1L1 |
| GO:0006869∼lipid transport | 0.033 | 0.63 | 5.65 | NPC1, OSBPL3, SORL1, NPC1L1 |
| GO:0010876∼lipid localization | 0.040 | 0.68 | 5.22 | NPC1, OSBPL3, SORL1, NPC1L1 |
| Ion transport (Cluster 4) | Enrichment Score: 1.95 | |||
| GO:0006811∼ion transport | 0.0034 | 0.21 | 2.94 | SLC12A7, SLC38A3, SLC16A1, SLC20A2, SLC9A3, CLIC6, SLC3A2, SLC22A3, ABCC4, SLC13A4, PDZK1 |
| GO:0006814∼sodium ion transport | 0.0035 | 0.20 | 7.88 | SLC12A7, SLC38A3, SLC20A2, SLC9A3, SLC13A4 |
| GO:0006820∼anion transport | 0.0049 | 0.25 | 7.17 | SLC12A7, SLC16A1, SLC20A2, CLIC6, SLC13A4 |
| GO:0055085∼transmembrane transport | 0.0057 | 0.25 | 3.24 | SLC12A7, SLC16A1, SLC25A31, AQP9, SLC9A3, SLC22A3, ABCC4, SLC13A4, PDZK1 |
| GO:0015698∼inorganic anion transport | 0.010 | 0.34 | 8.82 | SLC12A7, SLC20A2, CLIC6, SLC13A4 |
| GO:0006812∼cation transport | 0.016 | 0.48 | 2.97 | SLC12A7, SLC38A3, SLC20A2, SLC9A3, SLC3A2, SLC22A3, SLC13A4, PDZK1 |
| Steroid metabolism (Cluster 5) | Enrichment Score: 1.65 | |||
| GO:0008202∼steroid metabolic process | 0.0004 | 0.05 | 7.10 | SDR42E2, NPC1, OSBPL3, HSD17B2, SDR42E1, SORL1, NPC1L1 |
| GO:0006694∼steroid biosynthetic process | 0.0079 | 0.29 | 9.65 | SDR42E2, HSD17B2, SDR42E1, NPC1L1 |
| Cell homeostasis (Cluster 6) | Enrichment Score: 1.07 | |||
| GO:0042592∼homeostatic process | 0.0029 | 0.23 | 3.00 | SLC12A7, NPC1, GCLC, AQP9, FGGY, SLC9A3, VEGFA, TRHR, SLC7A8, LPAR3, NPC1L1 |
| GO:0048878∼chemical homeostasis | 0.0030 | 0.20 | 3.60 | NPC1, GCLC, AQP9, SLC9A3, VEGFA, TRHR, SLC7A8, LPAR3, NPC1L1 |
| Response to steroid hormones (Cluster 7) | Enrichment Score: 0.99 | |||
| GO:0006790∼sulfur metabolic process | 0.018 | 0.49 | 7.13 | GCLC, GPX4, SULF1, GHR |
| GO:0032355∼response to estradiol stimulus | 0.028 | 0.59 | 11.39 | GPX4, GHR, IHH |
| Endocytosis (Cluster 8) | Enrichment Score: 0.90 | |||
| GO:0006898∼receptor-mediated endocytosis | 0.027 | 0.59 | 11.60 | SLC9A3, SORL1, GHR |
aFor brevity, only go terms with P< 0.05 are displayed.
Table 2.
Clustering of Significantly Over-represented GO Terms from a List of Genes Significantly More Highly Expressed in the Yolk Sac Placenta Compared the Chorioallantoic Placenta in P. entrecasteauxii
| GO term (biological processes) | P valuea | Benjamini corrected P value | Fold enrichment | Gene symbols in cluster |
|---|---|---|---|---|
| Metabolic processing (Cluster 1) | Enrichment Score: 2.16 | |||
| GO:0006041∼glucosamine metabolic process | 0.0055 | 0.60 | 26.35 | PGM3, GNE, CHST4 |
| GO:0006044∼N-acetylglucosamine metabolic process | 0.0055 | 0.60 | 26.35 | PGM3, GNE, CHST4 |
| GO:0005996∼monosaccharide metabolic process | 0.0082 | 0.59 | 4.75 | PGM3, GNE, ALDOB, GFPT2, HK2, CHST4 |
| GO:0006040∼amino sugar metabolic process | 0.0086 | 0.57 | 21.08 | PGM3, GNE, CHST4 |
| Regulation of transport (Cluster 2) | Enrichment Score: 1.64 | |||
| GO:0051048∼negative regulation of secretion | 0.0036 | 0.51 | 12.78 | FAM3D, ERBB3, EDN2, INHA |
| GO:0048511∼rhythmic process | 0.0058 | 0.56 | 6.86 | EGR2, ERBB3, EDN2, TIMP4, INHA |
| GO:0051051∼negative regulation of transport | 0.0070 | 0.58 | 6.51 | PTGS2, FAM3D, ERBB3, EDN2, INHA |
| GO:0046888∼negative regulation of hormone secretion | 0.0093 | 0.53 | 20.27 | FAM3D, EDN2, INHA |
| GO:0051046∼regulation of secretion | 0.027 | 0.77 | 4.35 | UNC13D, FAM3D, ERBB3, EDN2, INHA |
| GO:0051050∼positive regulation of transport | 0.037 | 0.80 | 3.94 | UNC13D, ERBB3, EDN2, P2RY1, INHA |
| organic acid transport (Cluster 3) | Enrichment Score: 1.36 | |||
| GO:0015718∼monocarboxylic acid transport | 0.032 | 0.78 | 10.54 | SLC16A3, PPARG, ANXA1 |
| GO:0046942∼carboxylic acid transport | 0.0498 | 0.87 | 4.78 | SLC16A3, PPARG, ANXA1, SLC22A5 |
| response to hormones (Cluster 4) | Enrichment Score: 1.25 | |||
| GO:0042493∼response to drug | 0.0013 | 0.35 | 5.69 | PTGS2, ERBB3, PPARG, TIMP4, SLC22A5, SLC46A2, MVP |
| GO:0048511∼rhythmic process | 0.0058 | 0.56 | 6.86 | EGR2, ERBB3, EDN2, TIMP4, INHA |
| GO:0043434∼response to peptide hormone stimulus | 0.011 | 0.57 | 5.70 | EGR2, ERBB3, ALDOB, PPARG, TIMP4 |
| Regulation of nerve activity (Cluster 5) | Enrichment Score: 1.23 | |||
| GO:0044057∼regulation of system process | 0.0017 | 0.34 | 4.55 | SRI, EGR2, PTGS2, EDN2, EPHX2, BHLHE40, INHA, SLC22A5 |
| Carbohydrate metabolism (Cluster 6) | Enrichment Score: 1.15 | |||
| GO:0005996∼monosaccharide metabolic process | 0.0082 | 0.59 | 4.75 | PGM3, GNE, ALDOB, GFPT2, HK2, CHST4 |
| GO:0016051∼carbohydrate biosynthetic process | 0.022 | 0.75 | 6.57 | PGM3, GNE, ALDOB, GFPT2 |
| Cell homeostasis (Cluster 7) | Enrichment Score: 1.11 | |||
| GO:0044057∼regulation of system process | 0.0017 | 0.34 | 4.55 | SRI, EGR2, PTGS2, EDN2, EPHX2, BHLHE40, INHA, SLC22A5 |
| GO:0042592∼homeostatic process | 0.0090 | 0.55 | 2.57 | SRI, SLC26A4, EGR2, EDN2, PPARG, SLC9A2, EPHX2, CLDN1, HEPH, INHA, SLC22A5 |
| GO:0048878∼chemical homeostasis | 0.025 | 0.76 | 2.75 | SRI, SLC26A4, EGR2, EDN2, PPARG, SLC9A2, EPHX2, CLDN1 |
| GO:0050801∼ion homeostasis | 0.027 | 0.76 | 3.01 | SRI, SLC26A4, EGR2, EDN2, SLC9A2, EPHX2, CLDN1 |
| Fatty acid metabolism (Cluster 8) | Enrichment Score: 1.04 | |||
| GO:0008217∼regulation of blood pressure | 0.019 | 0.71 | 7.03 | PTGS2, EDN2, PPARG, EPHX2 |
| GO:0033559∼unsaturated fatty acid metabolic process | 0.033 | 0.78 | 10.33 | PTGS2, EDN2, EPHX2 |
| Inflammation response (Cluster 9) | Enrichment Score: 1.02 | |||
| GO:0009611∼response to wounding | 0.0291 | 0.76 | 2.65 | IRAK2, UNC13D, ERBB3, P2RY1, ANXA1, EPHX2, CHST4, GRHL3 |
| Regulation of transport (Cluster 17) | Enrichment Score: 0.5159817951223741 | |||
| GO:0051050∼positive regulation of transport | 0.037 | 0.79721324 | 3.939199814 | UNC13D, ERBB3, EDN2, P2RY1, INHA |
aFor brevity, only go terms with P< 0.05 are displayed.
The Effects of Gravidity on Uterine Gene Expression in Oviparous Taxa
In contrast to P. entrecasteauxii, oviparous skinks do not exhibit strong changes in uterine gene expression during gravidity. Lerista bougainvillii exhibits only two differentially expressed genes (Klotho and an uncharacterized protein homologous to ENSACAG00000029408) between gravid and non-gravid states (supplementary table S4, Supplementary Material online). No genes were differentially expressed between gravid and non-gravid uterus in La. guichenoti (supplementary table S5, Supplementary Material online).
Convergent Use of Amino Acid Transport Proteins in Independent Placentotrophic Lineages
We tested for non-random use of amino acid transport protein genes between human trophoblast tissue (Carter 2012) and uterine placental tissues from the southern grass skink. It is important to note that the genes identified as used in P. entrecasteauxii were identified because they are differentially expressed through pregnancy in our study, while those used in the trophoblast, have been identified by multiple studies from both RNA and protein analysis, from multiple stages of trophoblast development, and were summarized by Carter (2012). The human trophoblast and skink uterus are non-homologous tissues, and, therefore, any similarities in genes used must rely on independent processes that led to the evolution of these tissues in this context. We found a non-random use of amino acid transport protein genes between southern grass skink uterus and human trophoblast tissues (P = 0.003, fig. 2b). Therefore, placental protein transport has evolved using the same genes in both skinks and humans due to non-random use of amino acid transport proteins.
Discussion
The Origin of Pregnancy Specific Functions in the Uterus
Gravidity in oviparous taxa results in no substantial change in gene expression, suggesting that the ancestral condition for skinks is to have a consistent transcriptome profile throughout the reproductive cycle. This is in stark contrast to P. entrecasteauxii where approximately a quarter of the identified genes show differential expression between pregnant and non-pregnant states. This finding is consistent with another viviparous skink Chalcides ocellatus, which also has substantial differences in gene expression between pregnant and non-pregnant states (Brandley et al. 2012). Our results suggest that the evolution of pregnancy in P. entrecasteauxii has involved the evolution of a mechanism for the uterine tissue to respond to pregnancy by substantially changing gene expression in these tissues. By modulating uterine gene expression into two character states (non-pregnant and pregnant), it is easier for new pregnancy associated innovations (e.g. nutrient transport) to evolve, because uterine expression of any gene during pregnancy is not constrained by the disadvantage of its expression outside of pregnancy. We propose that the evolution of pregnancy in P. entrecasteauxii has involved the evolution of a new character state for uterine tissue.
One caveat of our study is that the uterine tissue from gravid oviparous females is at an early developmental stage whereas those of the viviparous females are at a late developmental stage. This means that some of the gene expression changes observed in viviparous females may not simply be due to pregnancy, but rather because of the presence of a large embryo inside the uterus. Future work is needed to examine whether differential gene expression is observed at early stages of pregnancy in P. entrecasteauxii, which would allow for a direct comparison with the oviparous taxa. However, we might not expect changes to occur at this early stage, because there is little maternal fetal interaction or change to uterine morphology until after embryonic stage 30 (the stage at which most oviparous squamate eggs are laid).
We acknowledge the presence of a large embryo is likely to impact on maternal gene expression, as ancestrally embryos, and their extra-embryonic membranes produce a range of hormones at the maternal–embryo interface (Griffith, Brandley, Whittington, et al. 2016). In fact, we expect that the apposition of maternal and fetal-tissue to be a novel source of gene expression regulators, and may be an important mechanism by which novel placental gene regulatory processes evolve. However, the presence of a late stage embryo is not sufficient to induce the morphological changes and functional innovations of the uterus seen in P. entrecasteauxii, as these are absent in most viviparous squamates. It is not clear from our data whether differential gene expression associated with a gravid/non-gravid state is associated with the evolution of viviparity or with the evolution of complex placental functions such as nutrient transport. More research is needed in species in which viviparity has evolved more recently, to identify if this modularization of uterine functions occurs concurrently with the evolution of viviparity, or with the evolution of complex placental functions.
Placentation in Pseudemoia Is Underpinned by Complex Changes in Gene Regulation
In most viviparous squamates, placentation is relatively simple, with minimal structural re-modeling to uterine tissue during pregnancy (Thompson and Speake 2006). In contrast, eutherian pregnancy is characterized by discretely specialized regions that perform different functions, including nutrient transport and gas exchange (Enders and Carter 2004). Like eutherian mammals, P. entrecasteauxii has morphologically distinct regions which it has been suggested to support discrete pregnancy associated functions (Adams et al. 2005; Griffith, Ujvari, et al. 2013). We found that uterine tissue of the chorioallantoic and yolk sac placentae exhibit substantial gene expression differences. Gene ontology analysis of these different regions supports the notion that these discrete placental regions support different pregnancy associated functions (summarized in fig. 3). Our data are the first to show that reptile placentae can be underpinned by regionalized differences in gene expression, that likely support the diversity of placental functions observed in this lizard, including lipid, protein, and ion transport, gas exchange, and structural remodeling of the uterus.
Gene ontology analysis of differentially expressed genes suggests that the chorioallantoic placenta transports nutrients to the embryo via membrane bound nutrient transport proteins, while the yolk sac placenta transports packaged vesicles to embryos containing lipids and other nutrients (fig. 3), as previously suggested (Adams et al. 2005; Griffith, Ujvari, et al. 2013). Differences in gene expression show that functional specializations of these tissues are underpinned by complex changes in gene regulation, on par with the complex mechanisms of placental function in eutherians (Sood et al. 2006). The morphological and gene expression changes necessary for placental nutrient transport may explain why placentotrophy has evolved so few times in amniote vertebrates (Thompson and Speake 2006).
Transport in the Chorioallantoic Placenta
The chorioallantoic placenta has been assumed to play a role in gas exchange and the transport of some organic and inorganic solutes (Stewart et al. 2006; Biazik et al. 2009; Herbert et al. 2010). Our gene ontology (GO) analysis identified that the chorioallantoic placenta contains membrane bound transport proteins which may support the transport of a range of molecules, including amino acids, carboxylic acids, sulfates, inorganic ions, and water (table 1). The large number and diversity of nutrient transport proteins expressed in the chorioallantoic placenta is strong evidence that the area transports a large range of nutrients to developing embryos.
Nutrients that are transported to embryos must first be brought to the placenta by uterine vasculature. The expression of genes involved in blood vessel development may provide a mechanism for the uterine vascular proliferation seen in this and other viviparous species (table 1; Adams et al. 2005). Vascular endothelial growth factor A (VEGFA) is a highly potent angiogenic factor that has been found in reptile uterine tissue (Ferrara et al. 2003; Murphy et al. 2010; Whittington, Grau, Murphy, et al. 2015). VEGFA is up-regulated during pregnancy in the viviparous skink Chalcides ocellatus (Brandley et al. 2012) and we found that VEGFA exhibits greater uterine expression in the chorioallantoic placenta than the yolk sac placenta (table 1). Increased VEGF expression in the chorioallantoic placenta likely supports both increased nutrient penetration into the uterus, and increased potential for gas exchange between mother and embryo.
Given that amino acid transport proteins represent the largest and most over-represented group of genes in the list of genes upregulated in the chorioallantoic placenta, this region is likely to be a major site of protein processing and transport to embryos. This finding is at odds with conclusions drawn from Itonaga et al. (2012), where they suggested the correlation between leucine transport rates through pregnancy and vascularization of the yolk sac splanchnopleure implicated the yolk sac placenta as the site of amino acid transport. However, the patterns of leucine transport presented by Itonaga et al. (2012) are also correlated with development of the placentome (a tightly folded region of the chorioallantoic placenta with large uterine secretory epithelial cells) in the chorioallantoic placenta (Stewart and Thompson 1996). The embryonic tissues in the placentome are capable of taking up organic molecules, including dextran, suggesting any maternally secreted molecules could be absorbed by the embryo (Stewart et al. 2006). Combined, these data support the placentome as a major site of amino acid/protein transport to embryos, further highlighting the similarities of reptile placentomes with the placentomes found in the cotyledonary placentae of ruminants (Fowden et al. 2006; Wildman et al. 2006).
The evolution of amino acid transport across the placenta is an essential step in the evolution of substantial matrotrophy, as proteins are a vital and substantial component of egg yolk. To understand the evolution of amino acid transport we compared amino acid transport protein gene expression across skinks in this study (table 3). While differential expression of amino acid transporters is observed in the matrotrophic skink P. entrecasteauxii only, all differentially expressed genes are expressed to some degree in the non-pregnant state, and in the uterus of oviparous species. Therefore, our results suggest that the evolution of amino acid transport has not required recruiting the expression of genes expressed elsewhere in the organism, but rather by the evolution of a mechanism for the uterus to respond to pregnancy by inducing a pregnancy specific state of gene regulation. When comparing amino acid transporter expression in skink transcriptome data, we found that oviparous transcriptomes both gravid and non-gravid clustered with non-gravid P. entrecasteauxii; however, uterine tissue from the chorioallantoi and yolk sac placentae formed two discrete clusters (fig. 2c).
Table 3.
Mean Transcripts Per Million of Amino Acid Transport Protein Genes in Skinks from Non-Reproductive Uterine Tissue (UNR), Gravid Uterine Tissue (UG), and Pregnant Uterine Tissue from the Chorioallantoic Placenta (UCA) and Yolk Sac Placenta (UYS)
|
Lampropholis guichenoti (o) |
Lerista bougainvillii (o) |
Pseudemoia entrecasteauxii (v) |
Human (v) |
|||||
|---|---|---|---|---|---|---|---|---|
| Gene symbol | UG | UNR | UG | UNR | UCA | UYS | UNR | Trophoblast (Carter 2012)d |
| SLC1A1 | 5.0 | 7.0 | 9.8 | 24.0 | 0.3a | 0.3a | 6.6b | Prot |
| SLC1A2 | 0.6 | 0.7 | — | — | 0.4 | 0.3 | 0.7 | Prot |
| SLC1A3 | 10.6 | 5.5 | 32.5 | 16.7 | 0.0 | 0.2 | 0.4 | Prot |
| SLC1A4 | 29.3 | 7.0 | 275.8 | 132.4 | 9.6a | 22.3a | 1.1b | mRNA |
| SLC1A5 | 149.1 | 14.4 | 240.9 | 184.3 | 44.1a | 54.6a | 23.3b | Prot/mRNA |
| SLC1A6 | 40.1 | 10.0 | — | — | 43.0 | 31.0 | 30.3 | — |
| SLC1A7 | — | — | 0.1 | 14.8 | — | — | — | — |
| SLC3A1 | 1.9 | 3.0 | 18.1 | 9.0 | 1.5 | 16.2 | 2.6 | — |
| SLC3A2 | 241.5 | 109.3 | 183.8 | 209.2 | 735.3a | 182.7b | 37.0c | mRNA/Prot |
| SLC7A1 | 12.8 | 16.2 | 26.3 | 36.0 | 102.0a | 26.9a,b | 10.5b | mRNA |
| SLC7A2 | 17.0 | 7.5 | 6.3 | 15.8 | 3.0a | 1.9a | 9.4b | mRNA |
| SLC7A3 | 963.0 | 509.2 | 169.5 | 158.1 | 135.5 | 107.7 | 239.2 | — |
| SLC7A4 | 72.8 | 11.8 | 18.1 | 11.1 | 88.5a | 56.3b | 32.1b | mRNA |
| SLC7A5 | — | — | — | — | 1319.5a | 144.0b | 12.5b | mRNA/Prot |
| SLC7A6 | 50.6 | 13.1 | 113.2 | 103.9 | 28.0a,b | 48.7a | 21.8b | mRNA |
| SLC7A7 | 8.3 | 9.4 | 18.1 | 31.9 | 137.8a | 10.8b | 6.4b | mRNA |
| SLC7A8 | 52.6 | 31.5 | 83.4 | 77.5 | 264.8a | 63.0b | 40.9b | mRNA |
| SLC7A9 | 0.4 | 5.1 | 112.1 | 138.7 | 2.1 | 17.6 | 4.0 | mRNA |
| SLC7A10 | — | — | 1.8 | 6.5 | 0.0 | 0.0 | 0.1 | mRNA |
| SLC7A11 | 37.8 | 7.7 | 28.1 | 23.2 | 115.3a | 82.9a | 13.1b | — |
| SLC7A12 | — | — | — | — | — | — | — | mRNA |
| SLC7A13 | 13.0 | 13.0 | — | — | 92.4 | 7.1 | 19.1 | — |
| SLC7A14 | — | — | 16.3 | 0.6 | 0.8 | 0.6 | 1.1 | — |
| SLC16A10 | 62.7 | 122.6 | 20.6 | 35.6 | 20.2 | 29.8 | 52.7 | mRNA |
| SLC32A1 | — | — | — | — | 0.0 | 0.0 | 0.5 | — |
| SLC36A1 | 19.0 | 13.3 | 18.7 | 13.6 | 36.3a | 32.4a | 14.8b | — |
| SLC36A2 | — | — | — | — | — | — | — | — |
| SLC36A3 | — | — | — | — | — | — | — | — |
| SLC36A4 | — | — | 1.6 | 0.5 | 8.7 | 6.1 | 6.9 | — |
| SLC38A1 | 14.4 | 2.4 | 311.7 | 183.4 | 22.2 | 32.1 | 11.3 | mRNA/Prot |
| SLC38A2 | 78.5 | 63.6 | 271.2 | 297.3 | 147.9a | 211.1a | 40.5b | mRNA/Prot |
| SLC38A3 | — | — | 4.4 | 3.2 | 360.4a | 15.4b | 5.0b | — |
| SLC38A4 | 0.5 | 0.2 | — | — | 164.2a | 11.9a,b | 0.7b | mRNA/Prot |
| SLC38A5 | 15.6 | 18.6 | 42.8 | 38.0 | 28.4a,b | 57.5a | 17.6b | mRNA |
| SLC38A6 | 2.7 | 2.7 | 4.4 | 7.7 | 4.1 | 3.6 | 2.4 | — |
| SLC38A7 | 9.3 | 12.4 | 14.4 | 55.8 | 5.6 | 9.7 | 6.1 | — |
| SLC38A8 | 6.3 | 4.5 | 0.5 | 37.3 | 1.3 | 11.4 | 0.4 | — |
| SLC38A9 | 7.3 | 11.5 | 27.0 | 28.8 | 5.6 | 4.1 | 9.9 | — |
| SLC38A10 | 21.5 | 13.4 | 75.8 | 54.9 | 32.0 | 29.9 | 31.6 | — |
| SLC38A11 | — | — | 11.0 | 0.1 | 0.4 | 0.2 | 0.9 | — |
| SLC43A1 | 64.6 | 43.1 | 95.5a | 54.1b | 86.7 | 21.6 | 33.9 | — |
| SLC43A2 | — | — | — | — | — | — | — | — |
| SLC43A3 | 15.5 | 21.9 | 22.5 | 20.0 | 112.0 | 27.7 | 45.2 | — |
Note—Oviparous species are denoted with an (o), and viviparous species with a (v). Significantly differentially expressed genes are indicated by different alphabetical characters (adjusted P value < 0.05)
dEvidence for each gene’s role in human placenta comes from either measuring the expression of messenger RNA (mRNA) or protein (prot).
We observed a convergence in the amino acid transporter genes used in the uterus of P. entrecasteauxii and the human trophoblast (fig. 2b). In P. entrecasteauxii, the expression of amino acid transporters does not-appear to be the result of gene expression recruitment, but why some genes are differentially expressed during pregnancy and others are not, suggests that there has been selective co-option of some genes to support the evolution of novel placental functions. This co-option is similar to what is seen with placental hormone production, where hormones are ancestrally produced in the tissues from which the placenta is derived (Griffith, Brandley, Whittington, et al. 2016). If convergent use of amino acid transporters in skink and human placental tissues is the result of independent co-option of genes for these functions, our results also suggest that complex biological traits such as nutrient transport evolve in a predictable manner, even when they occur in tissues that are not developmentally homologous.
Transport in the Yolk Sac Placenta
Transcriptomic data support the yolk-sac placenta as a transporter of organic nutrients during pregnancy, with the second most enriched cluster of GO terms including genes involved in transport regulation and secretion, while other clusters including terms for fatty acid metabolism and organic acid transport (table 2). Unlike the chorioallantoic placenta, there are few up-regulated membrane-bound transport proteins in the yolk sac placenta, reflecting the different ways in which nutrients are transported in these different placental regions. Our results are consistent with the lipid transport model presented in Griffith, Ujvari, et al. (2013) where nutrient transport in the yolk sac placenta likely occurs via membrane bound vesicles that actively bud from the uterine surface rather than secretion of nutrients directly into the lumen. Our data identify several other genes that are likely important in this transport pathway including fatty acid binding proteins 1 and 9 (FABP1 and FABP9).
Transcriptome Evolution in the Uterus of Pregnant Lizards
The evolution of a new pregnancy-specific character state in Pseudemoia is likely to be underpinned by a different gene regulatory network that allows gene expression in the pregnant state to exist as a discrete module (Wagner et al. 2007). Modularization of the uterus during pregnancy also occurs in eutherian mammals where large gene expression changes are induced during pregnancy (Lynch et al. 2015). The reprograming of uterine tissue during pregnancy in eutherians is largely regulated by the induction of a separate gene regulatory network (Gellersen and Brosens 2003). This change facilitates decidualization of the endometrial stroma and is induced in part by forkhead box O1 (FOXO1A) and Prolactin (Lynch et al. 2009; Kin et al. 2015). A similar pattern occurs in the male brood pouch of seahorses, where the pouch develops as an outgrowth of the skin, and extensive changes in gene expression occur as a result of carrying embryos (Whittington, Griffith, Qi, et al. 2015).
Pseudemoia entrecasteauxii expresses several components of the gene regulatory network for un-differentiated endometrial stromal fibroblasts, including homeobox A11 (HOXA11), CCAAT/enhancer-binding protein beta (CEBPB), and progesterone receptor (PR) (supplementary tables S1–S3, Supplementary Material online). However, none of these components is up-regulated during pregnancy, so are unlikely to support a new pregnancy-specific gene regulatory network. PCA analysis of transcription factor expression shows that there are differences in transcription factor expression between placental regions (fig. 2d), but the transcription factors that drive these differences are not ones that have been previously investigated in mammal systems. While our data cannot yet identify the presence of a pregnancy-specific gene regulatory network in a viviparous skink, it provides justification and fundamental data for identifying one.
Transposable elements have facilitated the recruitment of many genes to the uterus during the evolution of pregnancy in mammals (Lynch et al. 2015). Transposable elements are a unique source of regulatory variation, because they can insert new regulatory elements throughout the genome. Transposons occur in the genomes of most organisms and can be hyper-abundant in the genomes of squamates and birds (Feschotte 2008; Gilbert et al. 2011; Kordis 2009). Multiple transposon genes are expressed in the uterus of the oviparous and viviparous skinks in this study (supplementary tables S1–S5, Supplementary Material online). However, it is not clear whether transcriptomes have acted as sources of variation for regulatory evolution. To answer this question, we need to sequence the genomes of viviparous and related oviparous taxa, and correlate changes in gene expression between species with the insertion of transposons in the genome.
Comparisons with Other Taxa
To date, transcriptomic studies have investigated at least five independent origins of viviparity, these include therian mammals (Lynch et al. 2015), poecilid fishes (Panhuis et al. 2011), seahorses (Whittington, Griffith, Qi, et al. 2015), the lizard genus Chalcides (Brandley et al. 2012), and the lizard genus Pseudemoia (present study). In each of these lineages, various morphological changes have evolved convergently despite placentae arising from largely non-homologous structures. These convergent morphological changes include the development of regional specializations of the placenta, changes to cell morphology to perform new placental functions, increased levels of vascularization, and increases in the surface area of placental contact (Van Dyke et al. 2014; Kwan et al. 2015).
Comparison of transcriptome data between lineages for which placentae have been independently derived reveals a similar overlap in the function of genes that are correlated with pregnancy. Gene ontology analysis repeatedly identifies the same biological processes in these phylogenetically diverse taxa including, nutrient transport, gene regulation, inflammation processes, and response to stimuli (tables 1 and 2; Panhuis et al. 2011; Brandley et al. 2012; Whittington, Griffith, Qi, et al. 2015). It is not surprising that these gene ontology terms arise repeatedly as they represent the key functions that are important for successful pregnancy.
In many cases, orthologous genes have been co-opted or recruited to placental tissues. The same amino acid transport proteins are used in the placental tissues of reptiles and eutherian mammals (fig. 3b). Cystein proteases including cathepsin L are significantly up-regulated in the uterus of Pseudemoia, Chalcides, and eutherians (Song et al. 2005, 2010; Brandley et al. 2012). The transcription factor PPARG which is up-regulated in the uterus of the yolk sac placenta of Pseudemoia is also differentially expressed in the brood pouch of sea-horses, the ovary of the platy fish, and important for trophoblast growth in eutherians (Schartl et al. 2013; Whittington, Griffith, Qi, et al. 2015). There are many other examples of the same gene being use in independent placental lineages, but there are a limited number of genes in vertebrate genomes and we would expect many of the same genes to be used in new placental functions by chance alone. There is a need for a quantitative assessment of convergence at the transcriptome level between several independent origins of viviparity that consider the ancestral state of each lineage. To perform these analyses, we need more studies that include oviparous outgroups in their experimental design.
Conclusions
By examining gene expression in the uterus of gravid and non-gravid oviparous skinks, we show that ancestrally for this skink clade, gravidity does not result in substantial gene expression changes in the uterus. In contrast, the viviparous skink P. entrecasteauxii exhibits significant changes in approximately a quarter of the identified genes as a result of pregnancy. We show substantial differential gene expression between the uterus of the chorioallantoic and yolk sac placenta of P. entrecasteauxii suggesting that these regions perform different functions during pregnancy. The chorioallantoic placenta has high expression of a range of membrane-bound nutrient transport proteins that transfer a diversity of materials including amino acids, water, and inorganic ions. In contrast, the yolk sac placenta does not have greater expression of membrane bound transport proteins, but expresses genes involved in vesicle-mediated transport. These findings support morphological data that suggests that nutrient transport occurs via apocrine secretion in the yolk sac placenta and by membrane bound transport in the placentome.
Supplementary Material
Supplementary table S1–S5, figure S1 and methods are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This research was supported by a Gaylord Donnelley Postdoctoral Environmental Fellowship to O.W.G, an Australian Research Council Discovery grants to M.B.T. (DP120100649), and Discovery Early Career Research Award to M.C.B. (DE120101615). Lizards were collected under New South Wales National Parks and Wildlife License to M.B.T. (SL100401). RNA integrity analysis was carried out in the Bosch Molecular Biology Facility, and qPCR analysis was performed in the M. Olsson Lab at the University of Sydney. We thank James Van Dyke, Camilla Whittington, Fran van den Berg, Melanie Laird, and Jessica Dudley for comments on the manuscript. We thank Intersect Australia Ltd. for supercomputing resources. Raw transcriptome reads are available for download from the NCBI Sequence Read Archive (accession no. SRP040433 & SRP090404).
Literature Cited
- Adams SM, Biazik JM, Thompson MB, Murphy CR. 2005. Cyto-epitheliochorial placenta of the viviparous lizard Pseudemoia entrecasteauxii: a new placental morphotype. J Morphol. 264:264–276. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazik JM, Parker SL, Murphy CR, Thompson MB. 2012. Uterine epithelial morphology and progesterone receptors in a mifepristone-treated viviparous lizard Pseudemoia entrecasteauxii (Squamata: Scincidae) during gestation. J Exp Zool B. 318:148–158. [DOI] [PubMed] [Google Scholar]
- Biazik JM, Thompson MB, Murphy CR. 2009. Lysosomal and alkaline phosphatase activity indicate macromolecule transport across the uterine epithelium in two viviparous skinks with complex placenta. J Exp Zool. 312B:817–826. [DOI] [PubMed] [Google Scholar]
- Biazik JM, Thompson MB, Murphy CR. 2010. Desmosomes in the uterine epithelium of noninvasive skink placentae. Anat Rec. 293:502–512. [DOI] [PubMed] [Google Scholar]
- Blackburn DG. 2014. Evolution of vertebrate viviparity and specializations for fetal nutrition: a quantitative and qualitative analysis. J Morphol. 276:961–990. [DOI] [PubMed] [Google Scholar]
- Brandley M, et al. 2015. Evaluating the performance of anchored hybrid enrichment at the tips of the tree of life: a phylogenetic analysis of Australian Eugongylus group scincid lizards. BMC Evol Biol. 15:62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandley MC, Young RL, Warren DL, Thompson MB, Wagner GP. 2012. Uterine gene expression in the live-bearing lizard, Chalcides ocellatus, reveals convergence of squamate reptile and mammalian pregnancy mechanisms. Gen Biol Evol. 4:394–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AM. 2012. Evolution of placental function in mammals: the molecular basis of gas and nutrient transfer, hormone secretion, and immune responses. Physiol Rev. 92:1543–1576. [DOI] [PubMed] [Google Scholar]
- Cross JC, et al. 2003. Genes, development and evolution of the placenta. Placenta 24:123–130. [DOI] [PubMed] [Google Scholar]
- Dufaure JP, Hubert J. 1961. Table de developpement du lezard vivipare Lacerta (Zootoca) vivipara Jacquin. Arch Anat Micr Morph Exp. 50:309–328. [Google Scholar]
- Enders AC, Carter AM. 2004. What can comparative studies of placental structure tell us?–A review. Placenta 25:S3–S9. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Gerber H-P, Le Couter J. 2003. The biology of VEGF and its receptors. Nat Med. 9:669–676. [DOI] [PubMed] [Google Scholar]
- Feschote C, et al. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 9:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowden AL, Ward JW, Wooding FPB, Forhead AJ, Constancia M. 2006. Programming placental nutrient transport capacity. J Physiol. 572:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Hernandez SS, Flores-Benabib J, Smith EN, Feschotte C. 2011. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol Biol Evol. 29:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellersen B, Brosens J. 2003. Cyclic AMP and progesterone receptor cross-talk in human endometrium: a decidualizing affair. J Endocrinol. 178:357–372. [DOI] [PubMed] [Google Scholar]
- Girling JE. 2002. The reptilian oviduct: a review of structure and function and directions for future research. J Exp Zool. 293:141–170. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2008. The evolution of complex organs. Evol Educ Outreach 1:358–389. [Google Scholar]
- Griffith OW. 2015. Mechanisms of placental evolution: the genetics and physiology of pregnancy in lizards [PhD thesis]. Australia: University of Sydney.
- Griffith OW, et al. 2015. Ancestral state reconstructions require biological evidence to test evolutionary hypotheses: a case study examining the evolution of reproductive mode in squamate reptiles. J Exp Zool Part B. 324:493–503. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Brandley MC, Belov K, Thompson MB. 2016. Allelic expression of mammalian imprinted genes in a matrotrophic lizard, Pseudemoia entrecasteauxii. Dev Genes Evol. 226:79–85. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Brandley MC, Whittington CM, Belov K, Thompson MB. 2016. Comparative genomics of hormonal signaling in the chorioallantoic membrane of oviparous and viviparous amniotes. Gen Comp Endocrinol. DOI:10.1016/j.ygcen.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Ujvari B, Belov K, Thompson MB. 2013. Placental lipoprotein lipase (LPL) gene expression in a placentotrophic lizard, Pseudemoia entrecasteauxii. J Exp Zool Part B. 320:465–470. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Van Dyke JU, Thompson MB. 2013. No implantation in an extra-uterine pregnancy of a placentotrophic reptile. Placenta 34:510–511. [DOI] [PubMed] [Google Scholar]
- Herbert JF, Murphy CR, Thompson MB. 2010. Calcium ATPase localization in the uterus of two species of Pseudemoia (Lacertilia: Scincidae) with complex placentae. Herpetol Conserv Biol. 5:290–296. [Google Scholar]
- Hou Z-C, et al. 2012. Elephant transcriptome provides insights into the evolution of eutherian placentation. Genome Biol Evol. 4:713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itonaga K, Wapstra E, Jones SM. 2012. A novel pattern of placental leucine transfer during mid to late gestation in a highly placentotrophic viviparous lizard. J Exp Zool Part B. 318:308–315. [DOI] [PubMed] [Google Scholar]
- Kin K, Nnamani Mauris C, Lynch Vincent J, Michaelides E, Wagner GP. 2015. Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep. 10:1398–1409. [DOI] [PubMed] [Google Scholar]
- Kwan L, et al. 2015. An examination of the variation in maternal placentae across the genus Poeciliopsis (Poeciliidae). J Morphol. 276:707–720. [DOI] [PubMed] [Google Scholar]
- Kordis D, et al. 2009. Transposable elements in reptilian and avian (Sauropsida) genomes. Cytogenet Genom Res. 127:94–111. [DOI] [PubMed] [Google Scholar]
- Lager S, Powell TL. 2012. Regulation of nutrient transport across the placenta. J Pregnancy. 2012:179827.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Musser JM, Cloutier A, Prum R, Wagner G. 2016. Pervasive concerted evolution in gene expression shapes cell type transcriptomes. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 4:865–875. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Brayer K, Gellersen B, Wagner GP. 2009. HoxA-11 and FOXO1A cooperate to regulate decidual prolactin expression: towards inferring the core transcriptional regulators of decidual genes. PLoS One 4:e6845.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, et al. 2015. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 10:551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallinou M, et al. 2016. A single origin of extreme matrotrophy in African mabuyine skinks. Biol. Let. 12:20160430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman H. 1937. Comparative morphogenesis of the fetal membranes and accessory uterine structures. Washington (DC: ): Carnegie Institution of Washington Publication. [DOI] [PubMed] [Google Scholar]
- Murphy B, Thompson M. 2011. A review of the evolution of viviparity in squamate reptiles: the past, present and future role of molecular biology and genomics. J Comp Physiol. 181B:575–594. [DOI] [PubMed] [Google Scholar]
- Murphy BF, Belov K, Thompson MB. 2010. Evolution of viviparity and uterine angiogenesis: vascular endothelial growth factor (VEGF) in oviparous and viviparous skinks. J Exp Zool. 314B:148–156. [DOI] [PubMed] [Google Scholar]
- Murphy K, Hudson S, Shea G. 2006. Reproductive seasonality of three cold-temperate viviparous skinks from Southeastern Australia. J Herpetol. 40:454–464. [Google Scholar]
- Panhuis TM, et al. 2011. Analysis of expressed sequence tags from the placenta of the live-bearing fish poeciliopsis (Poeciliidae). 102:352–361. [DOI] [PubMed] [Google Scholar]
- Pyron RA. 2015. Advancing perspectives on parity-mode evolution. J Exp Zool Part B. 324:562–563. [DOI] [PubMed] [Google Scholar]
- Ravasi T, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick DN, Mateos M, Springer MS. 2002. Independent origins and rapid evolution of the placenta in the fish genus Poeciliopsis. Science 298:1018–1020. [DOI] [PubMed] [Google Scholar]
- Schartl M, et al. 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 45:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk K, Padilla DK, Bakken GS, Full RJ. 2009. Grand challenges in organismal biology. Integr Comp Biol. 49:7–14. [DOI] [PubMed] [Google Scholar]
- Shine R. 2015. The evolution of oviparity in squamate reptiles: an adaptationist perspective. J Exp Zool B. 324:487–492. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457:818–823. [DOI] [PubMed] [Google Scholar]
- Simpson JT, et al. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner A, Hugall AF, Hutchinson MN. 2011. Lygosomine phylogeny and the origins of Australian scincid lizards. J Biogeogr. 38:15. [Google Scholar]
- Song G, Spencer TE, Bazer FW. 2005. Cathepsins in the ovine uterus: regulation by pregnancy, progesterone, and interferon tau. Endocrinology 146:4825–4833. [DOI] [PubMed] [Google Scholar]
- Song G, et al. 2010. Cathepsin B, cathepsin L, and cystatin C in the porcine uterus and placenta: potential roles in endometrial/placental remodeling and in fluid-phase transport of proteins secreted by uterine epithelia across placental areolae. Biol Reprod. 82:854–864. [DOI] [PubMed] [Google Scholar]
- Sood R, Zehnder JL, Druzin ML, Brown PO. 2006. Gene expression patterns in human placenta. Proc Natl Acad Sci U S A. 103:5478–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2013. The genetic causes of convergent evolution. Nat Rev Genet. 14:751–764. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Thompson MB. 1996. Evolution of reptilian placentation: development of extraembryonic membranes of the Australian scincid lizards, Bassiana duperreyi (oviparous) and Pseudemoia entrecasteauxii (viviparous). J Morphol. 227:349–370. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Thompson MB. 2003. Evolutionary transformations of the fetal membranes of viviparous reptiles: a case study of two lineages. J Exp Zool Part A 299A:13–32. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Thompson MB, Attaway MB, Herbert JF, Murphy CR. 2006. Uptake of dextran-FITC by epithelial cells of the chorioallantoic placentome and the omphalopleure of the placentotrophic lizard, Pseudemoia entrecasteauxii. J Exp Zool. 305A:883–889. [DOI] [PubMed] [Google Scholar]
- Thompson MB, Speake BK. 2006. A review of the evolution of viviparity in lizards: structure, function and physiology of the placenta. J Comp Physiol. 176B:179–189. [DOI] [PubMed] [Google Scholar]
- True JR, Carroll SB. 2002. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol. 18:53–80. [DOI] [PubMed] [Google Scholar]
- Van Dyke JU, Brandley MC, Thompson MB. 2014. The evolution of viviparity: molecular and genomic data from squamate reptiles advance understanding of live birth in amniotes. Reproduction 147:R15–R26. [DOI] [PubMed] [Google Scholar]
- Van Dyke JU, Lindsay LA, Murphy CR, Thompson MB. 2015. Carbonic anhydrase II is found in the placenta of a viviparous, matrotrophic lizard and likely facilitates embryo–maternal CO2 transport. J Exp Zool Part B. 324:636–646. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM. 2007. The road to modularity. Nat Rev Genet.. 8:921–931. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. 2010. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 11:426–437. [DOI] [PubMed] [Google Scholar]
- Whittington CM, Grau GE, Murphy CR, Thompson MB. 2015. Unusual angiogenic factor plays a role in lizard pregnancy but is not unique to viviparity. J Exp Zool Part B. 342:152–158. [DOI] [PubMed] [Google Scholar]
- Whittington CM, Griffith OW, Qi W, Thompson MB, Wilson AB. 2015. Seahorse brood pouch transcriptome reveals common genes associated with vertebrate pregnancy. Mol Biol Evol. 32:3114–3131. [DOI] [PubMed] [Google Scholar]
- Wildman DE, et al. 2006. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci U S A. 103:3203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AM, Lyons KM, Brandley MC, Hillis DM. 2015. Which came first: the lizard or the egg? Robustness in phylogenetic reconstruction of ancestral states. J Exp Zool B Mol Dev Evol. 324:504–516. [DOI] [PubMed] [Google Scholar]
- Zhang JZ. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.