Abstract
The mitochondrial nad1 gene of seed plants has a complex structure, including four introns in cis or trans configurations and a maturase gene (matR) hosted within the final intron. In the geranium family (Geraniaceae), however, sequencing of representative species revealed that three of the four introns, including one in a trans configuration and another that hosts matR, were lost from the nad1 gene in their common ancestor. Despite the loss of the host intron, matR has been retained as a freestanding gene in most genera of the family, indicating that this maturase has additional functions beyond the splicing of its host intron. In the common ancestor of Pelargonium, matR was transferred to the nuclear genome, where it was split into two unlinked genes that encode either its reverse transcriptase or maturase domain. Both nuclear genes are transcribed and contain predicted mitochondrial targeting signals, suggesting that they express functional proteins that are imported into mitochondria. The nuclear localization and split domain structure of matR in the Pelargonium nuclear genome offers a unique opportunity to assess the function of these two domains using transgenic approaches.
Keywords: Geraniaceae, intron splicing, matR maturase, nad1 gene, retroprocessing
Introduction
The mitochondrial genomes of angiosperms exhibit a diverse array of features that contribute to increased genomic complexity, including numerous genes and introns, a large amount of intergenic and repetitive DNA, and abundant cytidine-to-uridine (C-to-U) RNA editing (Knoop 2012; Mower et al. 2012). The assortment of these complex genomic features is highly variable among individual angiosperms. For example, the 783 kb mitogenome of Liriodendron tulipifera has 64 genes, 25 introns, and >750 edit sites (Richardson et al. 2013), whereas the 6.7 Mb mitogenome of Silene noctiflora has only 32 genes, 18 introns, and <200 edit sites (Sloan et al. 2010b, 2012). Mitochondrial genes encode for products involved either directly or indirectly in aerobic respiration, and the variation in gene content among species is caused primarily by intracellular gene transfer from the mitochondrial to the nuclear genome (Adams et al. 2002). This process is generally assumed to be RNA mediated due to the absence of introns and the conversion of most RNA editing sites to their edited state in the nuclear gene copies (Nugent and Palmer 1991; Covello and Gray 1992; Wischmann and Schuster 1995).
Plant mitochondrial introns can be classified as either group I or group II introns based on their folded structure and splicing mechanism (Michel et al. 1989; Cech et al. 1994), with nearly all angiosperm mitochondrial introns classified as the group II type. Some of these introns have evolved a split arrangement, requiring a trans-splicing event to remove the fragmented intron and reconnect the independently transcribed gene halves into a continuous, functional transcript (Malek et al. 1997; Qiu and Palmer 2004). Loss of introns, as well as RNA edit sites, from the mitochondrial genome is often assumed to occur via an RNA-mediated process termed retroprocessing, where a spliced and edited transcript is reintegrated into the genome to physically and/or functionally replace the original gene (Ran et al. 2010; Sloan et al. 2010b; Grewe et al. 2011). Horizontal transfer can also contribute to both the gain and loss of introns in angiosperms (Vaughn et al. 1995; Sanchez-Puerta et al. 2008; Hepburn et al. 2012).
The angiosperm nad1 gene epitomizes the genomic complexity of plant mitogenomes, with five exons, four introns in cis- or trans-spliced arrangements, and abundant RNA editing. In addition, embedded within the final nad1 intron [named nad1i728 based on Dombrovska and Qiu (2004) intron notation] of nearly all angiosperms is another gene (matR) that encodes a putative intron splicing factor termed a maturase, although recent survey sequencing has identified a few plant lineages in which matR is no longer in this position. In Viscum album, matR was established as a freestanding gene due to loss of the host gene nad1 (Petersen et al. 2015). The matR gene is also freestanding in several species of Geranium, presumably by translocation prior to the loss of the host intron (Park et al. 2015). In the gnetophyte Welwitschia mirabilis, matR is no longer adjacent to any nad1 exons, but it is flanked by segments of the nad1i728 intron and may still be associated with the intron through a double trans-splicing event (Guo et al. 2016). In some other species of Viscum (Petersen et al. 2015; Skippington et al. 2015) and two species within Malpighiales (Wurdack and Davis 2009), the matR gene appears to be missing completely from the mitochondrial genome. Using the extensive genomic and transcriptomic data available for many species within Geraniaceae (Weng et al. 2014; Park et al. 2015; Zhang et al. 2015; Blazier et al. 2016), we assessed the status of matR and nad1 in this family and present an evolutionary scenario to explain the unusual structural and functional diversity among species.
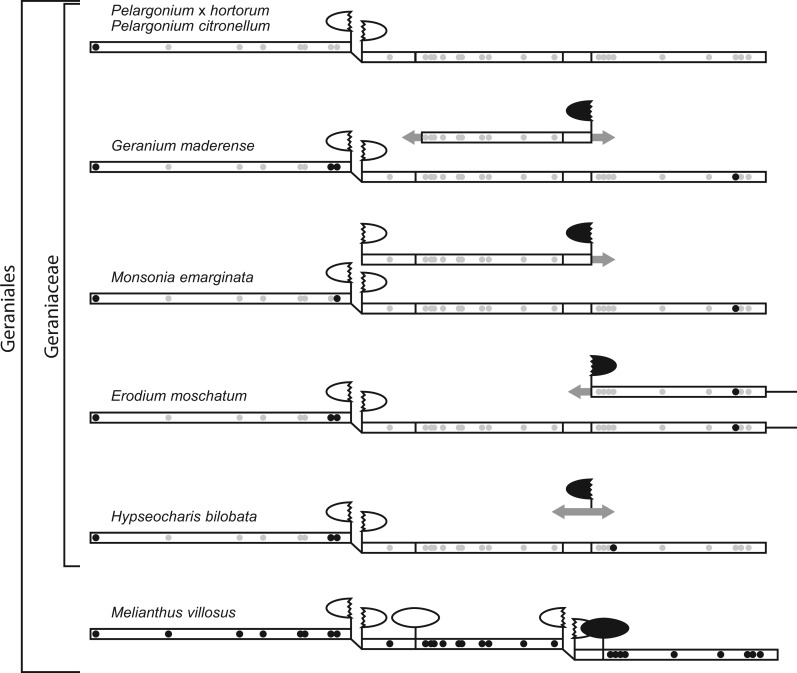
Loss of Multiple Introns from the Geraniaceae nad1 Gene
In the large majority of angiosperms, the nad1 reading frame is interrupted by four group II introns (fig. 1). The first (nad1i394) and third (nad1i669) nad1 introns require trans splicing for removal whereas the second intron (nad1i477) is removed by cis splicing. The fourth intron (nad1i728), which harbors the maturase gene matR, has evolved from a cis- to trans-spliced configuration several times in angiosperms due to genomic rearrangement occurring upstream or downstream of matR (Qiu and Palmer 2004). Thus, nad1i728 can be found as an ancestrally cis-spliced intron as observed for Melianthus villosus (fig. 1) and many other angiosperms, whereas in other species it can have a trans-spliced arrangement with matR located either in the 3′ or 5′ intron fragment (Qiu and Palmer 2004).
Fig. 1.—
Structure of mitochondrial nad1 and matR genes of Geraniales. Mitochondrial nad1 gene sequences and fragmented copies are represented by open boxes. Homologous intergenic regions are shown by a single line. Non-homologous regions are indicated by grey arrows. Introns in cis and trans configurations are represented by full and broken ellipses, respectively; a black color indicates the presence of the maturase gene matR. Filled black and grey dots indicate the presence and absence of RNA edit sites in Geraniales, respectively.
Within Geraniaceae, however, the structure of the nad1 gene has experienced a unique loss of complexity (fig. 1). Although the first intron (nad1i394) has been retained, the other three ancestral nad1 introns (nad1i477, nad1i669, and nad1i728) are absent from all examined species. In addition, whereas most sequenced angiosperms contain >20 sites of RNA editing in this gene (Rice et al. 2013) as exemplified by the 28 edit sites in Melianthus nad1, nearly all of these sites have been converted to thymidines in the corresponding positions of Geraniaceae nad1 genes. The correlated loss of introns and edit sites from Geraniaceae nad1 is consistent with retroprocessing activity. Despite these changes, the Geraniaceae nad1 gene is probably functional: it is full length and free of internal stop codons, and RNA editing of the few remaining sites improves sequence conservation to homologous genes from non-Geraniaceae species.
Establishment of matR as a Freestanding Gene in Most Geraniaceae Species
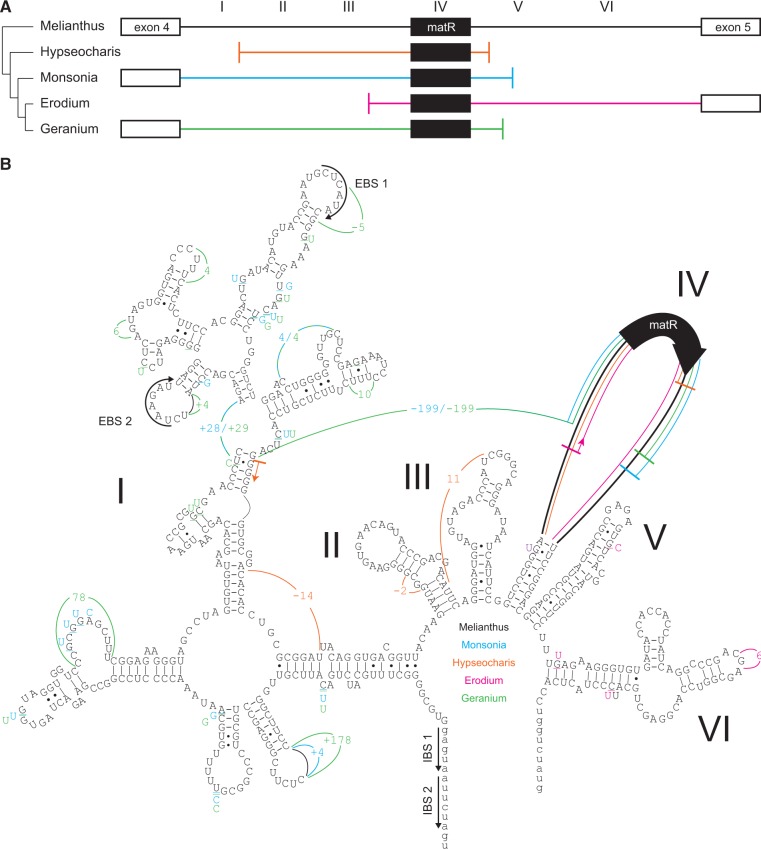
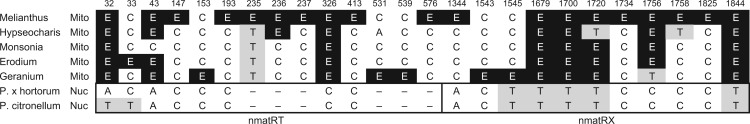
In addition to the presence of a putatively functional nad1 gene, most Geraniaceae species contain another partial nad1 sequence at a distinct genomic position (fig. 1). Unlike the putatively functional nad1 copies, however, which lack the nad1i728 intron and the associated matR gene, these partial nad1 sequences include the matR gene and parts of the nad1i728 intron (fig. 2A). The flanking intron sequences show clear signs of degradation based on the numerous nucleotide substitutions, insertions, and deletions that disrupt the predicted secondary structure (fig. 2B). In contrast, the matR sequence is presumably functional because it is full length, free of internal stop codons, and transcribed based on detectable reads in the RNAseq library. The RNAseq reads also revealed 25 positions that are edited in at least one of the seven Geraniales matR sequences, of which eight positions (32, 43, 326, 1679, 1700, 1720, 1756, and 1844) are edited in most species (fig. 3). These data indicate that matR functions as a freestanding gene in most Geraniaceae species.
Fig. 2.—
Structural degradation of the nad1i728 intron but retention of matR in Geraniaceae. (A) The diagram depicts the degree of degradation of nad1i728 in selected Geraniaceae species. (B) The secondary structure of the functional cis-spliced nad1i728 intron of M. villosus (in black) was determined by a manual comparison with existing group II introns models (Michel and Ferat 1995; Qin and Pyle 1998; Toor, et al. 2001). The intron folds into a typical group II intron secondary structure, with a central core and six branched domains (I–VI). Colored intron regions represent changes in the secondary structure of the degraded introns in G. maderense (in green), E. moschatum (in magenta), H. biloba (in orange), and M. emarginata (in blue): numbers, positive numbers, and negative numbers show sequence replacements, insertions, and deletions, respectively. The position of the maturase matR gene in domain IV is indicated by a bold black arrow. Exon binding sites (EBS) within domain I of the intron sequence (capital letters) and intron binding sites (IBS) in the adjacent exon sequence (small letters) are highlighted by a thin black arrow.
Fig. 3.—
Nucleotide content at positions of RNA editing in matR. All positions with an edited site in at least one species are shown. Edited cytidines are marked as “E” and shaded with a black background. Thymidines are shaded grey. All other nucleotides are unshaded.
Migration of matR into the Nuclear Genome of Pelargonium
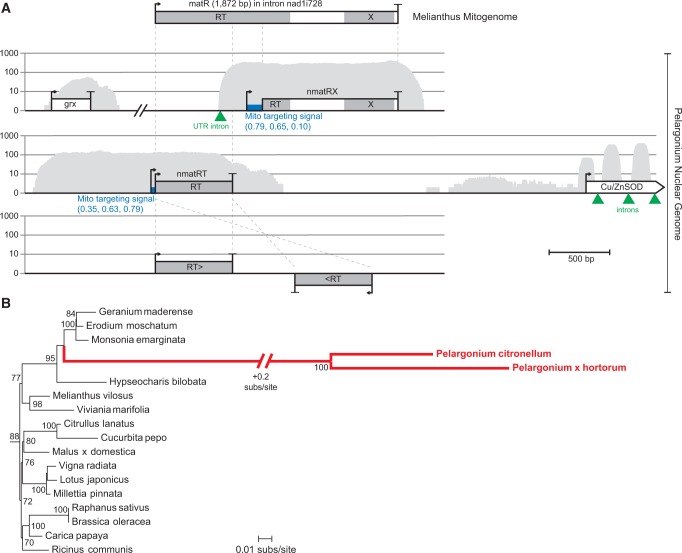
Because of the deep Illumina sequencing performed here, the matR gene was easily detectable in the draft mitochondrial assemblies of most Geraniaceae species. In contrast, matR was not detected in the mitochondrial assemblies of P. x hortorum and P. citronellum (fig. 1). Instead, a homolog was recovered in the assemblies of the nuclear genome and transcriptome (fig. 4A). In both Pelargonium species, these nuclear matR sequences (annotated as nmatR) were split into two distinct genes termed nmatRT, encoding the subdomains II to IV of the reverse transcriptase (RT) domain, and nmatRX, encoding subdomains V to VII of the RT domain and the entire maturase (X) domain. A glutaredoxin (grx) gene was identified upstream of nmatRX and a copper/zinc superoxide dismutase (CuZnSOD) gene was identified downstream of nmatRT, demonstrating that both nmatR genes are located in the nuclear genome. Furthermore, both nmatR genes are transcribed, and the nuclear intron residing in the 5′ UTR of nmatRX is properly spliced, indicating that both nmatR genes are functionally expressed nuclear genes. Mitochondrial targeting signals are predicted for both genes (fig. 4A), providing evidence that their protein products are localized to mitochondria. In contrast, two additional copies of nmatRT (found in a tail-to-tail arrangement on a different scaffold) had no transcriptional activity and may not be functional.
Fig. 4.—
Nuclear-encoded nmatR genes in Pelargonium. (A) The mitochondrial matR gene in the nad1 intron nad1i728 in M. villosus is shown at top, and homologous matR sequences in contigs from the Pelargonium nuclear genome are shown below. Start and stop codons are indicated by the arrow and bar, respectively. Grey shading shows conserved reverse transcriptase (RT) and maturase (X) domains. The thick blue bar following the first potential start codon of the nmatR sequences defines the location of the acquired mitochondrial targeting sequence; targeting prediction scores for MitoProt, TargetP, and Predotar are shown in blue in parentheses below the target sequence. The gene expression pattern of the nuclear contigs is mapped in grey. The location of nuclear spliceosomal introns are marked with a green triangle. All gene arrangements were drawn to scale. (B) Excerpt of a phylogenetic tree of the mitochondrial matR gene of angiosperms (thin black branches) and the nuclear nmatR paralogs of Pelargonium (thick red branches). All bootstrap values >70% are shown at respective nodes in the tree. The full phylogenetic tree is presented in supplementary figure S1, Supplementary Material online.
To infer the origin of the Pelargonium nmatR genes, phylogenetic analysis was used to determine their relationship to mitochondrial matR sequences from a diversity of other angiosperms (fig. 4B; supplementary fig. S1, Supplementary Material online). The two nmatR sequences group together with 100% bootstrap support, and they cluster within the Geraniaceae clade of mitochondrial matR genes with strong (95%) bootstrap support. Because P. citronellum and P. x hortorum span the full taxonomic diversity of Pelargonium (Weng et al. 2012), this phylogenetic result indicates that the nmatR genes were probably derived by intracellular transfer of a mitochondrial matR sequence into the nuclear genome of the common ancestor of Pelargonium. The extreme sequence divergence for the Pelargonium nmatR sequences probably results from the elevated substitution rates known to affect Pelargonium mitochondrial genes (Parkinson et al. 2005; Mower et al. 2007) and the generally higher rates of nuclear substitution relative to typical mitochondrial genomes (Wolfe et al. 1987; Drouin et al. 2008).
Similar to the mitochondrial matR sequences, the nuclear nmatR genes are more conserved within the domain regions relative to the rest of the protein sequence (supplementary fig.S2, Supplementary Material online). This pattern exists in both halves of the split gene, nmatRT and nmatRX, and indicates that the gene remained functional after the transfer into the nuclear genome. Many of the edited positions in the Geraniaceae mitochondrial matR transcripts have been converted to a thymidine in the Pelargonium nmatRT and nmatRX genes (fig. 3), suggesting that transfer to the nucleus involved a partially edited RNA intermediate. Because C-to-U RNA editing is not known to exist for nuclear genes we predict that the lack of conversion of some edit sites does not inhibit activity of the two nmatR gene products.
Discussion
The assemblies of the nad1 gene from representative Geraniaceae species revealed an unusual pattern of evolution, including (1) the first demonstrated loss of a trans-configured intron (nad1i669) from a functional gene, (2) the loss of the fourth nad1 intron (nad1i728) that hosts the matR gene, (3) the establishment of matR as a freestanding gene within the mitogenome, and (4) the first reported transfer of matR to the nuclear genome. All five nad introns in trans configurations have been lost from several mistletoe mitogenomes (Petersen et al. 2015; Skippington et al. 2015), but in these cases the intron losses were due to loss of the nad genes themselves. PCR results suggested that nad1i728 was lost from two species in Malpighiales (Wurdack and Davis 2009), but this intron is not trans-spliced in Malpighiales (Qiu and Palmer 2004; Rivarola et al. 2011; Kersten et al. 2016). To the best of our knowledge, the absence of nad1i669 in Geraniaceae represents the first reported loss of a trans-configured intron from any genome. Thus, although trans splicing appears to be a strong barrier to intron loss in eukaryotes, this result demonstrates that it is not an absolute barrier.
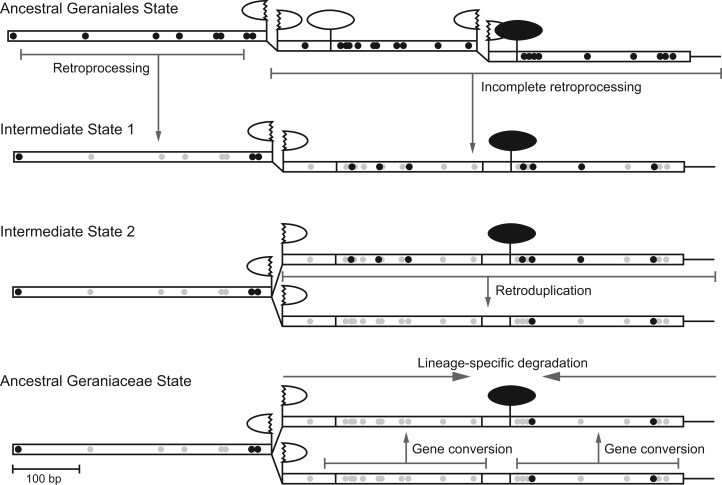
Multiple genomic changes are necessary to explain the loss of introns from nad1 and the origin of a freestanding matR gene in Geraniaceae, and one possible evolutionary scenario is shown in figure 5. The shared loss of most, but not all, introns and edit sites in Geraniaceae could be attributed to incomplete retroprocessing, in which a segment of the nad1 gene was gene converted by a transcript that was only partially edited and spliced. Most other examples of retroprocessing also involve the removal of a subset of introns and edit sites (Ran et al. 2010; Sloan et al. 2010b; Grewe et al. 2011). A second coexisting locus, lacking the nad1i728 intron and almost all edit sites, may have been created by a retroduplication event, involving the genomic integration of a partially spliced and edited transcript into a new genomic locus, eliminating the nad1i728 intron and additional edit sites from one of the loci. We then propose that the nad1 locus lacking the nad1i728 intron became the functional component of the gene. At the other locus, once the matR gene was established as a freestanding gene, the splicing of nad1i728 became unnecessary, and the intron sequence underwent lineage-specific degradation from its 5′ and 3′ ends. Finally, ongoing gene conversion, which has been documented for homologous sequences in plant mitogenomes (Hao and Palmer 2009; Hao et al. 2010; Mower et al. 2010; Sloan et al. 2010a), could explain the 100% sequence identity of the nad1 exon sequences shared between the two loci. More generally, the newly translocated matR gene described here provides a rare glimpse into the mechanism of gene translocation.
Fig. 5.—
Scenario of nad1 and matR evolution in Geraniaceae. Exons, introns, and edit sites are depicted as described in figure 1. The effects of retroprocessing, retroduplication, gene conversion, and lineage-specific gene degradation, as described in the text, are shown.
The mitochondrial-to-nuclear transfer of maturase genes has occurred multiple times during land plant evolution (Mohr and Lambowitz 2003; Guo and Mower 2013), but until now, there was no evidence for a nuclear transfer of matR, the sole maturase remaining in seed plant mitogenomes. In Viscum scurruloideum, matR and nearly all introns are absent from the mitogenome, but surveys of genome sequencing data did not identify a homolog in the nucleus suggesting that MatR and all of its splicing targets have been lost from this species (Skippington et al. 2015). In some Malpighiales, PCR results suggest that matR was lost from the mitogenome, but nothing is known about a potential nuclear transfer (Wurdack and Davis 2009). The transfer of matR to the nucleus in Pelargonium was likely facilitated by several of the unusual characteristics of this genus. First, the heavy reduction in RNA editing in this genus (Parkinson et al. 2005) raises the possibility of either a DNA-mediated or an RNA-mediated transfer event (Henze and Martin 2001), whereas the presence of editing sites in mitochondrial genes of most other species precludes functional transfers occurring via DNA. A similar case has been suggested for the mitochondrial rpl5 gene in grasses, in which an ancestral retroprocessing event may have facilitated multiple instances of functional DNA-mediated gene transfer into the nuclear genome (Ong 2006). Second, if a transfer event occurs, the very high substitution rate in Pelargonium mitogenomes (Parkinson et al. 2005) would quickly degrade the mitochondrial copy, thus making the transferred nuclear copy essential.
From a functional perspective, the retention of a matR gene in Geraniaceae after loss of its host nad1i728 intron implies that MatR has functions in the mitochondrion beyond the splicing of its host intron. One possibility is that MatR facilitates the removal of other mitochondrial group II introns, and thus is still required for one or more introns remaining in Geraniaceae (supplementary table S1, Supplementary Material online). A recent review of mitochondrial splicing factors suggested an association of MatR to other introns (Brown et al. 2014) consistent with additional splicing assignments, and the plastid maturase MatK was shown in vivo to have binding activity to several chloroplast introns (Zoschke et al. 2010). It is also possible that MatR performs other essential transcriptional functions, such as RNA processing or stabilization. Genetic manipulation of the plant mitochondrial genome is not yet possible in plants, precluding any direct assessment of MatR function. In contrast, because nuclear transformation is possible in many plants, including Pelargonium (Colling et al. 2010; Garcia-Sogo et al. 2012), the nuclear location of matR in Pelargonium raises the possibility of genetic and transcriptional manipulation of this gene. Furthermore, the split nature of these nmatR genes enables the independent assessment of the functions of the RT and X domains. Thus, Pelargonium offers a unique opportunity to study matR function in plant mitochondria.
Material and Methods
Source of plant materials and procedures for nucleic acid extraction and Illumina sequencing were described previously (Weng et al. 2014; Park et al. 2015). Draft mitochondrial genomes were assembled with Velvet 1.1.06 (Zerbino and Birney 2008) using a combination of kmer (51–91) and expected coverage (20–500) values as previously described (Grewe et al. 2014; Zhu et al. 2014). For each species, the assembly with the longest average length of identifiable mitochondrial sequences (based on BlastN searches with known mitochondrial protein-coding genes from related species as query sequences) was selected for further processing (supplementary table S2, Supplementary Material online). Mitochondrial gene and intron content was identified by inspection of the BlastN search results (supplementary table S1, Supplementary Material online). For some species, the presence of two nad1 gene sequences resulted in a failure to assemble complete loci. These regions were manually corrected by aligning and inspecting individual sequence reads that cover the respective regions. The assembled matR and nad1 sequences from this study were deposited in Genbank (accession numbers KX824067–KX824107).
A draft nuclear genome for P. citronellum was assembled using Velvet with kmer (41) and expected coverage (20) values that were reduced (relative to the mitochondrial assembly parameters) in order to preferentially assemble the lower-depth nuclear genome. A BlastN search identified matR homologs on three contigs, with sizes of 19,729 bp (containing nmatRX), 5,204 bp (containing nmatRT), and 16,625 bp (containing pseudo-nmatRT). In plant cells, the mitogenome is typically present at a substantially higher number of copies compared with the nuclear genome (Lamppa and Bendich 1984; Draper and Hays 2000). Thus, the much lower depth of coverage for these three contigs (relative to the identified mitochondrial contigs) provides reliable evidence that these contigs are nuclear rather than mitochondrial. Additional nuclear genes were detected in these three contigs by querying them against the non-redundant protein database using BlastX, and some of the genes contain nuclear spliceosomal introns with the canonical GT and AG bases at their 5′ and 3′ splice sites, providing additional support that these contigs are from the nuclear genome. Mitochondrial targeting signals were predicted with the programs Mitoprot II (Claros and Vincens 1996), TargetP 1.1 (Emanuelsson et al. 2000), and Predotar v1.03 (Small et al. 2004). The three contig sequences were deposited in Genbank (accession numbers KX824108–KX824110).
RNAseq reads were mapped onto the matR and nad1 sequences and the nmatR nuclear contigs using Bowtie2 2.2.6 (Langmead and Salzberg 2012). Depth of sequencing coverage per position was calculated using SAMtools (Li et al. 2009). Mapping results were manually inspected to identify exon–intron splicing junctions. For the mitochondrial genes, edit sites were identified by searching for C-T mismatches detectable in at least 10% of the mapped RNA reads.
Phylogenetic analysis of matR included sequences from 39 angiosperms (supplementary table S3, Supplementary Material online). Sequences were aligned in MEGA5 (Tamura et al. 2011) and trimmed using Gblocks (Castresana 2000) in codon mode with relaxed parameters (−b2 = 20; −b4 = 5; −b5 = 20). Maximum likelihood trees were constructed with the GTR + G substitution model in RAxML (Stamatakis 2006). Bootstrap support was calculated from 100 replicates using the fast bootstrapping option.
Supplementary Material
Supplementary Figures S1–S2 and Tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We gratefully acknowledge Emily Gubbels for assistance on this project. This work was supported by the National Science Foundation (awards IOS 1027529 and MCB 1125386 to JPM). FG’s postdoc position at The Field Museum is supported in part by the Negaunee Foundation.
Literature Cited
- Adams KL, Qiu YL, Stoutemyer M, Palmer JD. 2002. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 99:9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier JC, et al. 2016. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 117:1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O. 2014. Group II intron splicing factors in plant mitochondria. Front Plant Sci. 5:35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cech TR, Damberger SH, Gutell RR. 1994. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1:273–280. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 241:779–786. [DOI] [PubMed] [Google Scholar]
- Colling J, Groenewald JH, Makunga NP. 2010. Genetic alterations for increased coumarin production lead to metabolic changes in the medicinally important Pelargonium sidoides DC (Geraniaceae). Metab Eng. 12:561–572. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. 1992. Silent mitochondrial and active nuclear genes for subunit 2 of cytochrome c oxidase (cox2) in soybean: evidence for RNA-mediated gene transfer. EMBO J. 11:3815–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrovska O, Qiu YL. 2004. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogenet Evol. 32:246–263. [DOI] [PubMed] [Google Scholar]
- Draper CK, Hays JB. 2000. Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J. 23:255–265. [DOI] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49:827–831. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Garcia-Sogo B, et al. 2012. Production of engineered long-life and male sterile Pelargonium plants. BMC Plant Biol. 12:156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, et al. 2014. Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 19, Part B:135–143. [DOI] [PubMed] [Google Scholar]
- Grewe F, et al. 2011. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 39:2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, et al. 2016. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol Biol Evol. 33:1448–1460. [DOI] [PubMed] [Google Scholar]
- Guo W, Mower JP. 2013. Evolution of plant mitochondrial intron-encoded maturases: frequent lineage-specific loss and recurrent intracellular transfer to the nucleus. J Mol Evol. 77:43–54. [DOI] [PubMed] [Google Scholar]
- Hao W, Palmer JD. 2009. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc Natl Acad Sci U S A. 106:16728–16733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Richardson AO, Zheng Y, Palmer JD. 2010. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc Natl Acad Sci U S A. 107:21576–21581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn NJ, Schmidt DW, Mower JP. 2012. Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol Biol Evol. 29:3111–3120. [DOI] [PubMed] [Google Scholar]
- Henze K, Martin W. 2001. How do mitochondrial genes get into the nucleus? Trends Genet. 17:383–387. [DOI] [PubMed] [Google Scholar]
- Kersten B, et al. 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLoS One 11:e0147209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. 2012. Seed plant mitochondrial genomes: Complexity evolving. In: Bock R, Knoop V, editors. Genomics of Chloroplasts and Mitochondria. Netherlands: Springer; p. 175–200. [Google Scholar]
- Lamppa GK, Bendich AJ. 1984. Changes in mitochondrial DNA levels during development of pea (Pisum sativum L.). Planta 162:463–468. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek O, Brennicke A, Knoop V. 1997. Evolution of trans-splicing plant mitochondrial introns in pre-Permian times. Proc Natl Acad Sci U S A. 94:553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Ferat JL. 1995. Structure and activities of group II introns. Annu Rev Biochem. 64:435–461. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82:5–30. [DOI] [PubMed] [Google Scholar]
- Mohr G, Lambowitz AM. 2003. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res. 31:647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Sloan DB, Alverson AJ. 2012. Plant mitochondrial genome diversity: the genomics revolution In: Wendel JF, editor. Plant Genome Diversity. Vol. 1 Vienna: Springer; p. 123–144. [Google Scholar]
- Mower JP, et al. 2010. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 8:150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. 2007. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 7:135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent JM, Palmer JD. 1991. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66:473–481. [DOI] [PubMed] [Google Scholar]
- Ong HC. 2006. Intracellular and Horizontal Transfer of Mitochondrial Genes in Grass Evolution: Pseudogenes, Retroprocessing, and Chimeric Genes. Ph.D. Thesis. Bloomington: Indiana University Bloomington. [Google Scholar]
- Park S, et al. 2015. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol. 208:570–583. [DOI] [PubMed] [Google Scholar]
- Parkinson CL, et al. 2005. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 5:73.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Cuenca A, Moller IM, Seberg O. 2015. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci Rep. 5:17588.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol. 8:301–308. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Palmer JD. 2004. Many independent origins of trans splicing of a plant mitochondrial group II intron. J Mol Evol. 59:80–89. [DOI] [PubMed] [Google Scholar]
- Ran JH, Gao H, Wang XQ. 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol. 54:136–149. [DOI] [PubMed] [Google Scholar]
- Rice DW, et al. 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342:1468–1473. [DOI] [PubMed] [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivarola M, et al. 2011. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS One 6:e21743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. 2008. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol Biol Evol. 25:1762–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD. 2015. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci U S A. 112:E3515–E3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10:e1001241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Storchova H, Palmer JD, Taylor DR. 2010a. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. 2010b. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics 185:1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7:1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD. 1995. Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J Mol Evol. 41:563–572. [DOI] [PubMed] [Google Scholar]
- Weng ML, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31:645–659. [DOI] [PubMed] [Google Scholar]
- Weng ML, Ruhlman TA, Gibby M, Jansen RK. 2012. Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae). Mol Phylogenet Evol. 64:654–670. [DOI] [PubMed] [Google Scholar]
- Wischmann C, Schuster W. 1995. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 374:152–156. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 84:9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdack KJ, Davis CC. 2009. Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 96:1551–1570. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Sabir J, Blazier JC, Jansen RK. 2015. Coordinated rates of evolution between interacting plastid and nuclear genes in Geraniaceae. Plant Cell 27:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Jain K, Mower JP. 2014. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 31:1228–1236. [DOI] [PubMed] [Google Scholar]
- Zoschke R, et al. 2010. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A. 107:3245–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.