Abstract
Nested genes are the most common form of protein-coding overlap in eukaryotic genomes. Previous studies have shown that nested genes accumulate rapidly over evolutionary time, typically via the insertion of short young duplicate genes into long introns. However, the evolutionary relationship between nested genes remains unclear. Here, I compare RNA-seq expression profiles of nested, proximal intra-chromosomal, intermediate intra-chromosomal, distant intra-chromosomal, and inter-chromosomal gene pairs in two Drosophila species. I find that expression profiles of nested genes are more divergent than those of any other class of genes, supporting the hypothesis that concurrent expression of nested genes is deleterious due to transcriptional interference. Further analysis reveals that expression profiles of derived nested genes are more divergent than those of their ancestral un-nested orthologs, which are more divergent than those of un-nested genes with similar genomic features. Thus, gene expression divergence between nested genes is likely caused by selection against nesting of genes with insufficiently divergent expression profiles, as well as by continued expression divergence after nesting. Moreover, expression divergence and sequence evolutionary rates are elevated in young nested genes and reduced in old nested genes, indicating that a burst of rapid evolution occurs after nesting. Together, these findings suggest that similarity between expression profiles of nested genes is deleterious due to transcriptional interference, and that natural selection addresses this problem both by eradicating highly deleterious nestings and by enabling rapid expression divergence of surviving nested genes, thereby quickly limiting or abolishing transcriptional interference.
Keywords: nested genes, overlapping genes, transcriptional interference, gene expression evolution
Introduction
Nearly 10% of Drosophila genes form nested structures, whereby one (“internal”) gene is contained within the intron of a second (“external”) gene (Assis et al. 2008; Lee and Chang 2013). Most nested genes are located on opposite strands and, relative to un-nested genes, internal genes tend to be small and have few short introns, whereas external genes tend to be large and have many long introns (Yu et al. 2005; Assis et al. 2008). There are a number of complex nestings in which multiple internal genes inhabit the same external gene (Yu et al. 2005; Assis et al. 2008), and even a few Russian doll-like structures, in which a gene resides within a gene that resides within another gene (Assis et al. 2008). Thus, gene nesting is an intriguing example of genomic organizational complexity, prompting evolutionary questions about how it occurs, how it is maintained, and how it impacts gene function and phenotype.
An early study of the dynamics of nested genes showed that nesting accumulates over evolutionary time in three separate animal lineages (Assis et al. 2008). Nesting often arises via the insertion of a young duplicate gene, which is typically produced by an RNA-mediated mechanism, into the intron of another gene (Assis et al. 2008). A recent study of nested genes in Drosophila showed that internal genes tend to be testis-specific and evolve at faster rates than both external and un-nested genes (Lee and Chang 2013). Testis specificity and rapid evolution are hallmarks of young duplicate genes in Drosophila, particularly those created from RNA intermediates (e.g., Thornton and Long 2002; Hahn et al. 2007; Bai et al. 2007, 2008; Chen et al. 2010; Assis and Bachtrog 2013; Assis 2014). Hence, given that internal genes often arise by RNA-mediated duplication, it is not surprising that they exhibit these properties. However, nesting introduces an additional layer of complexity, in that this unique physical association may alter the evolutionary trajectories of nested genes by limiting or enhancing their functional divergence.
Three hypotheses have been proposed to explain the evolutionary relationship between nested genes. First, nesting may confer a selective advantage if internal and external genes are co-regulated (Hurst et al. 2004; Oliver and Misteli 2005). Under this hypothesis, we expect expression profiles to be more similar between nested than between un-nested genes. Second, nesting may be selectively disadvantageous if internal and external genes interfere with one another’s transcription (Shearwin et al. 2005; Liao and Zhang 2008). Under this hypothesis, we expect expression profiles to be less similar between nested than between un-nested genes. Third, nesting may be selectively neutral if it simply occurs because long introns provide feasible “landing spots” for young duplicate genes (Assis et al. 2008). Under this hypothesis, we expect expression profiles to be equally similar between nested and un-nested gene pairs. It is important to note that all three of these hypotheses assume that nested genes have similar properties to one another, and the third hypothesis further assumes that introns compose typical chromatin environments.
Previous studies tested these hypotheses by examining correlations between expression profiles of nested genes. Assis et al. (2008) analyzed early human (Su et al. 2004) and Drosophila melanogaster (Chintapalli et al. 2007) microarray data and found that, although expression profiles of nested genes were positively correlated, this correlation was not significantly different from the correlation between expression profiles of random adjacent genes. Thus, they concluded that nesting is a selectively neutral event driven by the abundance of long introns in animal genomes. Using an updated microarray dataset for D. melanogaster (FlyAtlas, Chintapalli et al. 2007), Lee and Chang (2013) also uncovered a positive correlation between expression profiles of nested genes, but found that this correlation was weaker than the correlation between expression profiles of proximal intra-chromosomal genes, and not significantly different from the correlation between expression profiles of random intra-chromosomal genes. Hence, they concluded that nesting is selectively disadvantageous due to transcriptional interference between genes.
Thus, the evolutionary relationship between nested genes remains unclear. To assess this relationship, I took an approach that addressed several limitations common to both previous studies. One limitation of these studies is the use of microarray data, which have low signal-to-noise ratios as a result of hybridization issues. This can be particularly problematic when quantifying the expression of duplicate genes, which are overrepresented among internal genes (Assis et al. 2008). For this reason, I chose to use RNA-seq data, which has less noise because sequences can be unambiguously mapped to unique regions of the genome. A second limitation of previous studies is the quantification of expression divergence via correlation coefficients, which are sensitive to measurement error (Pereira et al. 2009). In particular, if measurement error is large, correlation coefficients tend to show high divergence for genes with relatively uniform expression levels across conditions (Pereira et al. 2009). Thus, here I estimated expression divergence with Euclidian distances, which are robust to measurement error (Pereira et al. 2009). Additionally, I obtained relative expression values across tissues before computing Euclidian distances, which decreases biases introduced by experimental differences in abundance estimates across tissues and species, and is comparable with the standardization inherent to correlation coefficients (Pereira et al. 2009). A third limitation of prior studies is their relatively narrow scopes. Assis et al. (2008) only compared nested gene pairs to random adjacent gene pairs separated by any distance, and Lee and Chang (2013) compared nested gene pairs to proximal intra-chromosomal (bp) and random intra-chromosomal (bp) gene pairs. However, a number of studies have shown that there is a strong positive correlation between expression divergence and genomic distance between genes (Cohen et al. 2000; Boutanaev et al. 2002; Lercher et al. 2002, 2003; Trinklein et al. 2004; Williams and Bowles 2004; Weber and Hurst 2011). To account for this association, I separated un-nested genes into four classes based on separation distance: proximal intra-chromosomal ( bp), intermediate intra-chromosomal ( bp), distant intra-chromosomal ( bp), and inter-chromosomal (different chromosomes).
Gene expression was assessed by examination of RNA-seq data from developmental stages of D. melanogaster, as well as from six orthologous tissues of D. melanogaster and D. pseudoobscura: carcass, male head, female head, ovary, testis, and accessory gland (see Materials and Methods section for details). Using genome annotation and RNA-seq data, I identified all expressed nested, proximal intra-chromosomal, intermediate intra-chromosomal, distant intra-chromosomal, and inter-chromosomal gene pairs in D. melanogaster and D. pseudoobscura (see Materials and Methods section for details). In total, there are and expressed nested gene pairs annotated in the genomes of D. melanogaster and D. pseudoobscura, respectively (supplementary tables S1 and S2, Supplementary Material online). Of these pairs, in D. melanogaster and in D. pseudoobscura are part of complex nested structures, most of which are cases whereby multiple genes (as many as ) reside within a single gene, and a handful of which are Russian doll-like configurations. Although there appear to be more expressed nested genes and complex cases in D. melanogaster, these figures are similar to those of D. pseudoobscura ( and , respectively) when limiting D. melanogaster RNA-seq samples to those available for both species.
Whereas transcriptional interference can refer to any influence of the transcription of one gene on that of another gene (Liao and Zhang 2008), it can broadly be split into two types: interference that affects the actions of RNA polymerases and interference that influences gene splicing. One RNA polymerase may interfere with the actions of a second RNA polymerase in several ways, including by blocking its promoter site, stalling its elongation, or even knocking it off its transcript. Mis-splicing may occur when both genes contain introns, and is more likely when genes are located on the same strand (Shearwin et al. 2005). Several properties of nested genes support the hypothesis that nesting is selectively disadvantageous due to one or both types of transcriptional interference. In particular, consistent with earlier studies (Assis et al. 2008; Lee and Chang 2013), of D. melanogaster nested genes are located on opposite strands, which is a significantly greater proportion than expected by chance (, binomial test with ). A similar strand bias exists in D. pseudoobscura, for which of nested genes are found on opposite strands (, binomial test with ). Moreover, as previously reported (Lee and Chang 2013), intronless internal genes are overrepresented in same-strand () relative to opposite-strand () nestings in D. melanogaster (, Fisher’s exact test; supplementary table S3, Supplementary Material online); and this bias also exists in D. pseudoobscura, for which of same-strand internal genes and only of opposite-strand internal genes are intronless (, Fisher’s exact test; supplementary table S4, Supplementary Material online). Together, these biases indicate that selection may act to prevent mis-splicing of nested genes (Lee and Chang 2013). Further, as described in D. melanogaster (Lee and Chang 2013), internal genes tend to be more tissue-specific (median in D. melanogaster and in D. pseudoobscura), and external genes more broadly expressed (median in D. melanogaster and in D. pseudoobscura), than un-nested genes (median in both species; for all comparisons in both species, permutation tests; see Materials and Methods section for details). Expression in different tissues may reduce one or both types of transcriptional interference. Unfortunately, because most nestings involve the longest intron of the external gene, it is difficult to assess whether promoters of nested genes tend to be farther apart than expected by chance, which we might expect if interactions between their RNA polymerases causes transcriptional interference. Thus, these observations suggest that transcriptional interference may be due to mis-splicing of nested genes, although effects on RNA polymerases cannot be ruled out as a contributing mechanism.
If arrival of the internal gene is detrimental to the biological function of the external gene, as is expected under transcriptional interference, one would expect more deleterious phenotypes to be associated with external genes than with un-nested genes that have similar genomic properties. To test this hypothesis, I examined known phenotypes associated with alleles of external genes in D. melanogaster (see Materials and Methods section for details). There are D. melanogaster external genes, and contain alleles that have known phenotypes, of which are viable, are sterile, and are lethal. For comparison, I obtained random samples of un-nested genes, for which each gene in a particular sample was matched to a D. melanogaster external gene by chromosome, expression breadth, and highest expressed tissue (see Materials and Methods section for details). All random samples contained at least genes associated with a viable phenotype. However, only three samples had at least genes associated with a sterile phenotype, and none of the samples had at least genes associated with a lethal phenotype (the greatest was genes). Hence, this analysis supports the hypothesis that gene nesting interferes with the biological function of the external gene.
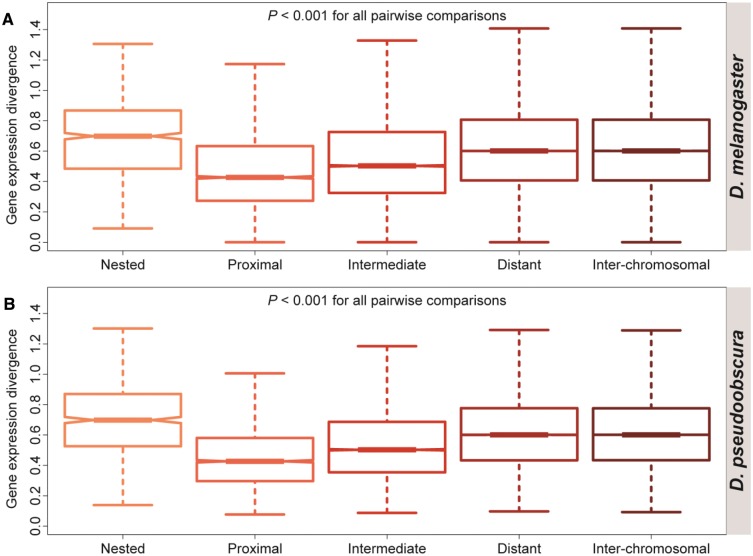
To further probe the relationship between nested gene functions in Drosophila, I compared Euclidian distances between expression profiles of nested gene pairs to those between expression profiles of proximal intra-chromosomal, intermediate intra-chromosomal, distant intra-chromosomal, and inter-chromosomal gene pairs (fig. 1). As expected (Cohen et al. 2000; Boutanaev et al. 2002; Lercher et al. 2002, 2003; Trinklein et al. 2004; Williams and Bowles 2004; Weber and Hurst 2011), divergence between expression profiles of intra-chromosomal genes pairs increases with distance—proximal pairs are most similar, intermediate pairs are less similar, and distant pairs are least similar—and expression profiles of inter-chromosomal pairs are more divergent than those of intra-chromosomal pairs ( for all comparisons in both species, permutation tests). However, although nested genes are physically the closest in distance, their gene expression profiles are more divergent than those of genes in any other class ( for all comparisons in both species, permutation tests). This pattern persists even after removal of same-strand nested genes ( for all comparisons in both species, permutation tests) and pairs within complex nested structures ( for all comparisons in both species, permutation tests). Thus, the exceptional divergence between expression profiles of nested genes provides additional support for the hypothesis that nesting of similarly expressed genes is selectively disadvantageous due to transcriptional interference.
Fig. 1.—
Expression divergence between pairs of nested, proximal intra-chromosomal, intermediate intra-chromosomal, distant intra-chromosomal, and inter-chromosomal genes. Distributions of Euclidian distances between expression profiles of pairs are depicted for each class of genes in (A) D. melanogaster and (B) D. pseudoobscura. Boxes represent first, second (median), and third quartiles of distributions, notches indicate confidence intervals of medians, and whiskers denote outliers (smallest and largest points within 1.5 times the interquartile range, respectively). Significant differences between groups were assessed with permutation tests (see Materials and Methods section for details).
Expression divergence of nested genes may occur via two mechanisms. First, natural selection may disfavor nesting of similarly expressed genes, resulting in the removal of nested genes with insufficiently divergent expression profiles. If this is the only mechanism underlying the expression divergence of nested genes, expression divergence of gene pairs before nesting should be greater than expected given their genomic properties, and should remain the same after nesting. Second, the expression profiles of genes may diverge after nesting. If this is the only mechanism underlying expression divergence of nested genes, expression divergence of gene pairs before nesting should be equal to expected divergence given their genomic properties, and should be elevated after nesting.
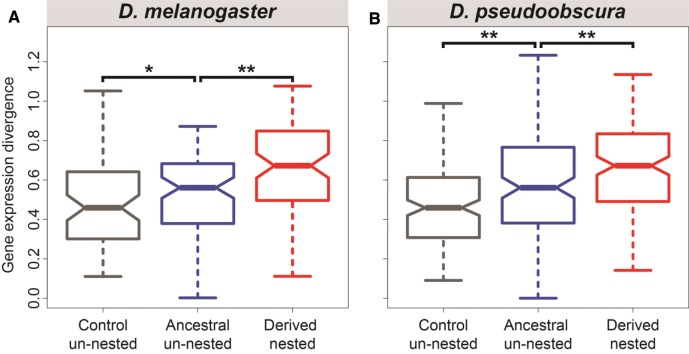
To disentangle these mechanisms, I identified pairs of genes ( in simple nested structures) that underwent nesting in the D. melanogaster lineage (supplementary table S5, Supplementary Material online), and pairs of genes ( in simple nested structures) that underwent nesting in the D. pseudoobscura lineage (supplementary table S6, Supplementary Material online), after the divergence of these lineages approximately million years ago (Beckenbach et al. 1993; see Materials and Methods section for details). These derived nested genes in one species and their ancestral un-nested orthologs in the other species were used as proxies for genes after and before nesting, respectively. Of the ancestral un-nested gene pairs, were located on the same chromosome, of which were on opposite strands, with a median separation distance of 10,770 bp. Thus, to represent expected expression divergence of genes with similar genomic properties, I obtained equal-sized random samples of un-nested (control) pairs that were matched to ancestral un-nested pairs by chromosome, orientation, and distance (see Materials and Methods section for details). Comparison of expression divergence between pairs of control un-nested, ancestral un-nested, and derived nested genes uncovered an interesting trend (fig. 2). In both lineages, expression divergence is greater between ancestral un-nested than between control un-nested pairs ( for D. melanogaster, for D. pseudoobscura, permutation tests), suggesting that expression divergence between genes prior to nesting is greater than expected given their genomic properties. However, expression divergence is even greater between derived nested pairs in both lineages ( for D. melanogaster and D. pseudoobscura, permutation tests), indicating that expression divergence also increases after nesting. Thus, these findings support both hypothesized mechanisms. First, nestings between genes with insufficiently divergent expression profiles are removed by natural selection; and second, expression profiles of nested genes that are not removed by selection diverge even further.
Fig. 2.—
Expression divergence between control un-nested, ancestral un-nested, and derived nested pairs. Distributions of Euclidian distances between expression profiles of gene pairs in each class, where derived nested pairs refer to those that underwent nesting in the lineage of (A) D. melanogaster or (B) D. pseudoobscura after their divergence. Boxes represent first, second (median), and third quartiles of distributions, notches indicate confidence intervals of medians, and whiskers denote outliers (smallest and largest points within 1.5 times the interquartile range, respectively).Significant differences between groups were assessed with permutation tests (see Materials and Methods section for details). Asterisks indicate (*) and (**).
Although control un-nested pairs were matched closely to ancestral un-nested pairs, it is important to emphasize that there are additional genomic properties, such as chromatin environment, that were not accounted for and may also influence gene expression and evolution. In particular, a recent study showed that open chromatin can promote genomic rearrangements (Berthelot et al. 2015). Thus, it is possible that chromatin environment may bias the mutational aspect of nesting, resulting in preferential nesting of genes with divergent gene expression profiles. Under this scenario, chromatin environment may by itself produce the difference observed between control un-nested and ancestral un-nested pairs in fig. 2. However, the bias toward opposite-strand nesting and overrepresentation of intronless internal genes in same-strand nestings both specifically point to selection disfavoring nesting of genes that interfere with one another’s transcription. Moreover, the overrepresentation of deleterious phenotypes associated with external genes also implicates transcriptional interference in nested gene evolution. Finally, chromatin environment cannot explain expression divergence that occurs after nesting (fig. 2). Thus, although chromatin environment may influence nested gene evolution to some degree, all of these findings together point to a major role of natural selection against transcriptional interference.
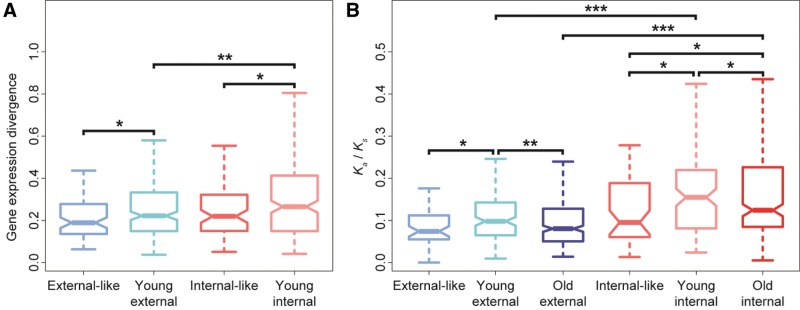
Expression divergence after nesting may occur via changes in the internal gene, external gene, or both genes. To address this question, I again examined the gene pairs that underwent nesting after the divergence of D. melanogaster and D. pseudoobscura (“young” nesting events). I computed distances between expression profiles of young external genes and their ancestral un-nested orthologs and between young internal genes and their ancestral un-nested orthologs. To represent expected expression divergence between orthologous genes in the absence of nesting, I obtained random equal-sized samples of un-nested genes that were matched to external (“external-like”) and internal (“internal-like”) genes by chromosome, expression breadth, and highest-expressed tissue (see Materials and Methods section for details). Comparison of these groups (fig. 3A) revealed that young external and internal genes experienced greater expression divergence than external- and internal-like genes, respectively ( for both comparisons, permutation tests), suggesting that expression profiles of both genes diverge more than expected after nesting. Although expression profiles of young internal genes diverged more than those of young external nested genes (, permutation test), it is important to note that this may be an unfair comparison because internal genes are also more tissue-specific and often male-biased (Lee and Chang 2013), and such genes are known to evolve rapidly (Meikeljohn et al. 2003; Zhang and Parsch 2005; Pröschel et al. 2006; Sawyer et al. 2007; Zhang et al. 2007; Assis et al. 2012) and have noisier gene expression (Díaz-Castillo 2015). Thus, a more accurate way of comparing nesting-associated expression divergence between external and internal genes is by comparing excesses in expression divergence over expectations between young external and young internal genes. Interestingly, the median expression divergence of young external genes is 1.17 times greater than the median expression divergence of external-like genes, whereas the median expression divergence of young internal genes is 1.21 times greater than the median expression divergence of internal-like genes. Although this comparison is not rigorous because it does not account for variability in expression divergence, it suggests that nesting-associated expression divergence may be marginally greater in internal than in external genes when controlling for other features that likely affect their evolution.
Fig. 3.—
Expression and sequence divergence of external and internal nested genes. (A) Distributions of Euclidian distances between expression profiles of external-like genes and their orthologs, young external genes and their un-nested orthologs, internal-like genes and their orthologs, and young internal genes and their un-nested orthologs. (B) Distributions of Ka/Ks ratios between external-like genes and their orthologs, young external genes and their un-nested orthologs, old external genes and their nested orthologs, internal-like genes and their orthologs, young internal genes and their un-nested orthologs, and old internal genes and their nested orthologs. Boxes represent first, second (median), and third quartiles of distributions, notches indicate confidence intervals of medians, and whiskers denote outliers (smallest and largest points within 1.5 times the interquartile range, respectively). Significant differences between groups were assessed with permutation tests (see Materials and Methods section for details). Asterisks indicate (*), (**), and (***).
The observation that expression divergence is greater in internal genes is consistent with their faster sequence evolution (Lee and Chang 2013). To assess whether evolutionary rate is associated with time after nesting, I identified gene pairs that underwent nesting before the divergence of D. melanogaster and D. pseudoobscura (“old” nesting events; see Materials and Methods section for details). I again included internal- and external-like genes as comparison groups to control for the effects of genomic location and tissue expression patterns on evolutionary rate. Then I compared Ka/Ks ratios among external-like genes and their orthologs, young external genes and their ancestral un-nested orthologs, old external genes and their nested orthologs, internal-like genes and their orthologs, young internal genes and their ancestral un-nested orthologs, and old internal genes and their nested orthologs (fig. 3B). Examination of Ka/Ks ratios of old nested genes revealed that old external genes are under similar levels of selective constraint as external-like genes (, permutation test), whereas old internal genes are under less constraint than internal-like genes (, permutation test), consistent with previous findings that external genes evolve slowly and internal genes evolve rapidly (Lee and Chang 2013). However, Ka/Ks ratios of young nested genes tell a different story. In particular, both young external (, permutation test) and young internal (, permutation test) genes are under less constraint than external- and internal-like genes, respectively. Moreover, Ka/Ks ratios of young internal genes are greater than those of old internal genes (, permutation test). Together, these results suggest that evolutionary rates of both internal and external genes are elevated immediately after nesting and decrease over time.
It is important to note that inferences about natural selection in this study were based solely on protein-coding sequence evolution. Although protein-coding sequences likely influence gene expression to some degree, it has become increasingly clear in recent years that regulatory sequences may play a more important role (Carroll 2005; Khaitoich et al. 2006; Whitehead and Crawford 2006; Wray 2007; Wittkopp and Kalay 2012). Additionally, most sequences under selection may lie outside of exons and function as transcriptional regulatory elements (Dermitzakis et al. 2002; Waterston et al. 2002; Cooper et al. 2004; Siepel et al. 2005; Loots 2008; Visel et al. 2009). However, because protein-coding sequences are typically under strong constraint, examining the evolution of these sequences may provide insight into the selective forces acting on genes as a whole. Thus, the elevation of evolutionary rates in protein-coding regions of nested genes suggests that these genes experience increased evolutionary rates overall.
Whereas both internal and external genes experience rapid sequence evolution after nesting, the effect on internal genes is stronger and longer-lasting. The tendency of internal genes to originate from duplication events, particular those that are RNA-mediated, may enable their rapid and prolonged divergence. In particular, the presence of two copies of a gene is thought to result in relaxed constraint in one copy, and recent studies have shown that this copy is often the young gene produced by the duplication event (Han et al. 2009; Assis and Bachtrog 2013; Assis 2014; Roselló and Kondrashov 2014). Moreover, young Drosophila duplicate genes tend to be testis-specific (Assis and Bachtrog 2013), and a number of studies have shown that male-biased genes experience rapid evolution (Meikeljohn et al. 2003; Zhang and Parsch 2005; Pröschel et al. 2006; Sawyer et al. 2007; Zhang et al. 2007; Assis et al. 2012). Thus, young duplicate genes may make ideal nesting partners because they are able to quickly adapt to the strict requirements of nesting configurations. This relationship may also be symbiotic, in that introns of external genes can provide ideal environmental niches for young duplicates because transcriptional interference creates “hotbeds” for rapid functional evolution of genes.
Although duplication may influence nested gene evolution, expression divergence after nesting cannot solely be attributed to the rapid evolution of young duplicate genes. For one, divergence of internal genes was compared to that of internal-like genes to control for the effects of location and tissue expression patterns on their evolution. Thus, internal genes experience greater sequence and expression divergence than expected based on other factors that may influence their evolutionary rates. Moreover, the young internal genes considered in this study do not constitute recently duplicated genes. To enable comparisons between expression profiles of genes before and after nesting, the current analysis required the presence of internal and external genes in both D. melanogaster and D. pseudoobscura. Thus, young internal genes in this study likely arose from duplication events prior to the divergence of the D. melanogaster and D. pseudoobscura lineages, and were later transposed into their nested positions. However, the strongest evidence for nesting as a source of the observed expression divergence is the sequence and expression divergence experienced by young external genes, which do not arise from gene duplication events. Therefore, although one could argue that expression divergence of internal genes is solely due to prolonged rapid evolution of young duplicate genes, there is no reason to believe that external genes should be similarly affected unless it is due to their interaction with their internal nested gene partners.
As a whole, these findings support the hypothesis that gene nesting is often deleterious due to transcriptional interference between nested genes. The observed bias toward opposite-strand nesting, overrepresentation of intronless internal genes in same-strand nestings, and elevated expression divergence of gene pairs prior to nesting all suggest that the genome’s first line of defense against transcriptional interference is to eradicate highly deleterious nestings. Most observed nestings are hence likely either slightly deleterious or selectively neutral, enabling their rise to high frequencies and fixation in the population. Nevertheless, expression divergence increases further after nesting, and sequence evolutionary rates are elevated in young nested genes and reduced in old nested genes. Thus, young nested genes may experience a burst of rapid evolution that enables their continued expression divergence, leading to the further reduction or even elimination of transcriptional interference.
Materials and Methods
Gene Expression Analysis
RNA-seq data from 30 developmental stages of D. melanogaster was obtained from Graveley et al. (2011). Paired-end RNA-seq reads from D. melanogaster carcass, male head, female head, testis, accessory gland, and ovary tissues were downloaded from the modENCODE site at http://www.modencode.org/ (last accessed October 4, 2016) (accession nos. modENCODE_4304, modENCODE_4297, modENCODE_4315, modENCODE_4299, and modENCODE_4297). Paired-end RNA-seq reads from D. pseudoobscura male carcass, female carcass, testis, accessory gland, and ovary tissues were obtained from Kaiser et al. (2011); and similar paired-end RNA-seq reads from D. pseudoobscura male head and female head were downloaded from NCBI’s sequence read archive (accession nos. SRX016182 and SRX016183). Because expression of carcass tissue was measured in a sample of mixed males and females in D. melanogaster, I used the mean expression of male and female carcass tissue as an estimate of mixed carcass expression in D. pseudoobscura. The distribution of mixed carcass expression levels in D. melanogaster was not significantly different from the distribution of mean carcass expression levels in D. pseudoobscura (, permutation test), suggesting that proportions of male and female tissues in the mixed carcass sample are approximately equal. Moreover, removal of carcass tissue from the analysis does not change any of the observed trends. Bowtie2 (Langmead et al. 2009) was used to align reads to D. melanogaster and D. pseudoobscura transcript sequences, using genome annotation files (version 6.10 for D. melanogaster and version 3.04 for D. pseudoobscura) downloaded from http://www.flybase.org; last accessed October 4, 2016. Because internal nested genes are often duplicate genes, fragments per kilobase of exon per million fragments mapped (FPKM; Trapnell et al. 2010) were calculated using eXpress, (Roberts and Pachter 2013), which uses an adaptive expectation–maximization algorithm that minimizes multi-mapping issues by continuously updating assignment probabilities for fragment mapping based on previous estimates of target sequence abundances. These values were quantile-normalized using the affy package of Bioconductor in the R software environment (R Development Core Team 2009), and tissue expression data were scaled by the median D. melanogaster FPKM so that expression levels were comparable between D. melanogaster and D. pseudoobscura. Relative expression levels across samples (developmental stages and tissues for analysis shown in figure 1, and tissues only for all remaining analyses) were used as expression profiles, and Euclidian distances between expression profiles were used to estimate expression divergence between genes. Expression breadth for each gene was estimated by its tissue specificity index τ, which ranges from 0 to 1, where low values indicate broad expression and high values indicate tissue-specific expression (Yanai et al. 2005).
Identification of Nested and Control Genes
Genome annotation files for D. melanogaster (version 6.10), D. pseudoobscura (version 3.04), and their outgroups D. willistoni (version 1.05), D. mojavensis (version 1.04), D. virilis (version 1.06), and D. grimshawi (version 1.3) were downloaded from FlyBase at http://www.flybase.org. There are nested gene pairs annotated in D. melanogaster, in D. pseudoobscura, in D. willistoni, in D. mojavensis, in D. virilis, and in D. grimshawi. I further filtered D. melanogaster and D. pseudoobscura nested genes to ensure that both members of each pair are expressed in at least one RNA-seq sample. In D. melanogaster, filtering resulted in expressed nested pairs when considering RNA-seq samples from developmental stages and tissues together, and 8 pairs when using only tissue data. In D. pseudoobscura, for which only tissue data was used, filtering yielded expressed nested pairs. Similar proportions of pairs were found to be in complex structures in both species (supplementary tables S1 and S2, Supplementary Material online). Chromosomal distributions of nested genes are similar to those of un-nested genes ( for both species, Fisher’s exact tests), indicating no bias in observed locations of nested genes.
Using genome annotation and RNA-seq data from all developmental stages and tissues, I identified 5,177 proximal intra-chromosomal, 21,264 intermediate intra-chromosomal, 13,544,770 distant intra-chromosomal, and 53,888,714 inter-chromosomal expressed gene pairs in D. melanogaster. Using genome annotation and tissue RNA-seq data, I identified 3,683 proximal intra-chromosomal, 15,687 intermediate intra-chromosomal, 7,162,611 distant intra-chromosomal, and 44,030,279 inter-chromosomal expressed gene pairs in D. pseudoobscura. The deficiency of expressed intra-chromosomal gene pairs in D. pseudoobscura, particularly for those that are distant, is due to division of the assembly of several chromosomes by linkage groups, rather than to a deficiency of annotated or expressed genes.
Control genes used for the analysis of phenotypes associated with external genes in D. melanogaster were obtained by closely matching each external gene to a random un-nested gene in the genome. Genes were matched by chromosome, tissue specificity index τ (), and highest-expressed tissue. Control un-nested pairs (fig. 2) were obtained by taking each ancestral un-nested pair and closely matching it to a random un-nested pair in the genome. In particular, pairs were matched by chromosome, orientation, and distance (within bp). Thus, each ancestral un-nested pair was matched to a control un-nested pair with similar genomic properties. Internal-like and external-like genes (fig. 3) were obtained by matching each individual gene to a random un-nested gene in the genome. Specifically, genes were matched by chromosome, tissue specificity index τ (), and highest-expressed tissue. Hence, each internal gene was matched directly to an internal-like gene, and each external gene to an external-like gene.
Analysis of Phenotypes Associated with D. melanogaster External Genes
A table of alleles and their known phenotypes in D. melanogaster was downloaded from FlyBase at http://www.flybase.org (2016 dataset). Following the approach of Lee and Chang (2013), phenotypes were separated into “lethal”, “sterile”, and “viable” classes based on their descriptions. A gene was considered to have a phenotype of a particular class if there was at least one allele of that gene associated with that phenotype class.
Inference of Young and Old Nesting Events
I obtained orthologs in each of the outgroup Drosophila species from the FlyBase ortholog table (2016 version 2) downloaded from http://www.flybase.org, which contains orthologs from the Drosophila 12 Genomes Consortium (2007) that were assigned by requiring sequence similarity and conserved synteny between species pairs. Nesting events that occurred in either D. melanogaster or D. pseudoobscura lineage after their divergence were inferred by parsimony, i.e., when genes were nested in one species, un-nested in the second species, and un-nested in all outgroups. To ensure that incomplete genome assembly or annotation did not bias inferences of nesting events, I required the presence of both genes in the genomes of D. melanogaster, D. pseudoobscura, and at least one outgroup species. Thus, nesting events were not inferred when one gene was simply absent ancestrally, as such inferences are prone to error. Genes inferred to have undergone nesting in either D. melanogaster or D. pseudoobscura after their divergence were designated as “young”, and genes that were nested in both species were designated as “old”.
Estimation of Sequence Evolutionary Rates
D. melanogaster and D. pseudoobscura CDS sequences were downloaded from http://www.flybase.org, and the longest transcripts of orthologous genes were aligned using MACSE (Ranwez et al. 2011), which accounts for frameshifts and stop codons. PAML (Yang 2007) was used to estimate pairwise substitution rates at synonymous (Ks) and non-synonymous (Ka) sites, as well as to obtain Ka/Ks ratios.
Statistical Analyses
All statistical analyses were performed in the R software environment (R Development Core Team 2009). Binomial tests were used to assess strand biases of nested genes. For each test, to represent the expected frequency of opposite-strand nested genes if orientation is random. Fisher’s exact tests were used for all comparisons of two or more categorical variables, and two-sided permutation tests for all comparisons of two numerical variables. For each permutation test, the difference between medians of the two samples, D, was calculated. Next, the two samples of sizes m and n were combined into a single dataset of size . The combined dataset was randomly split times into two samples of sizes and n. After each of these 1,000 permutations, the difference between medians of the two random samples was computed. The obtained P-value was the proportion of permutations in which the difference between medians was greater than or equal to .
Supplementary Material
Supplementary tables S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
I would like to thank three referees and the journal editors for their valuable feedback. This work was supported by the National Science Foundation [DEB-1555981].
Literature Cited
- Assis R. 2014. Drosophila duplicate genes evolve new functions on the fly. Fly 8:91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Bachtrog D. 2013. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 110:17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS, Koonin EV, Kondrashov FA. 2008. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 24:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Zhou Q, Bachtrog D. 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 4:1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Betrán E. 2008. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 9:241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Feschotte C, Betrán E. 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8:R11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckenbach AT, Wei YW, Liu H. 1993. Relationships in the Drosophila obscura species group, inferred from mitochondrial cytochrome oxidase II sequences. Mol Biol Evol. 10:619–634. [DOI] [PubMed] [Google Scholar]
- Berthelot C, Muffato M, Abecassis J, Roest Crollius H. 2015. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep. 10:1913–1924. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420: 475–478. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 26:183–186. [DOI] [PubMed] [Google Scholar]
- Cooper GM, et al. 2004. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 14:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420: 578–582. [DOI] [PubMed] [Google Scholar]
- Díaz-Castillo C. 2015. Evidence for sexual dimorphism in gene expression noise in metazoan species. PeerJ. 3:e750.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Graveley BR, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han S-G. 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. 2009. Adaptive evolution of young gene duplicates in mammals. Genome Res. 19:859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ. 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 5:299–310. [DOI] [PubMed] [Google Scholar]
- Kaiser VB, Zhou Q, Bachtrog D. 2011. Non-random gene loss from the Drosophila miranda neo-Y chromosome. Genome Biol Evol. 3: 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitoich P, et al. 2006. Evolution of primate gene expression. Nat Rev Genet. 7:693–702. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Chang H-H. 2013. The evolution and functional significance of nested gene structures in Drosophila melanosgaster. Genome Biol Evol. 5:1978–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. 2008. Coexpression of linked genes in mammalian genomes is generally disadvantageous. Mol Biol Evol. 25:1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13:238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 31:180–183. [DOI] [PubMed] [Google Scholar]
- Loots GG. 2008. Genomic identification of regulatory elements by evolutionary sequence comparison and functional analysis. Adv Genet. 61:269–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikeljohn CD, Parsch J, Ranz JM, Hartl DL. 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci U S A. 17:9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B, Misteli T. 2005. A non-random walk through the genome. Genome Biol. 6:214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V, Waxman D, Eyre-Walker A. 2009. A problem with the correlation coefficient as a measure of gene expression divergence. Genetics 183:1597–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. 2006. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics 174:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2009. R: a language and environment for statistical computing. Vienna, Austria. [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. 2011. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One 6:e22594.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselló OP, Kondrashov FA. 2014. Long-term asymmetrical acceleration of protein evolution after gene duplication. Genome Biol Evol. 6: 1949–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Parsch J, Zhang Z, Hartl DL. 2007. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proc Natl Acad Sci U S A. 104:6504–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. 2005. Transcriptional interference – a crash course. Trends Genet. 21:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. 2005. Evolutionary conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 101:6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Long M. 2002. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol Biol Evol. 19:918–925. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, et al. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14:62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. 2009. Genomic views of distant-acting enhancers. Nature 461:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. [DOI] [PubMed] [Google Scholar]
- Weber CC, Hurst LD. 2011. Support for multiple classes of local expression clusters in Drosophila melanogaster, but no evidence for gene order conservation. Genome Biol. 12:R23. Doi: 10.1186/gb-2011-12-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. 2006. Variation within and among species in gene expression: raw material for evolution. Mol Ecol. 15:1197–1211. [DOI] [PubMed] [Google Scholar]
- Williams EJB, Bowles DJ. 2004. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 14:1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 13:59–69. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21:650–659. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu P, Ma D, Xu M. 2005. Nested genes in the human genome. Genomics 86:414–422. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Parsch J. 2005. Positive correlation between evolutionary rate and recombination rate in Drosophila genes with d expression. Mol Biol Evol. 22:1945–1947. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. 2007. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.