Abstract
The population structure of the Bacillus cereus group (52 strains of B. anthracis, B. cereus, and B. thuringiensis) was investigated by sequencing seven gene fragments (rpoB, gyrB, pycA, mdh, mbl, mutS, and plcR). Most of the strains were classifiable into two large subgroups in six housekeeping gene trees but not in the plcR tree. In addition, several consistent clusters were identified, which were unrelated to species distinction. Moreover, interrelationships among these clusters were incongruent in each gene tree. The incongruence length difference test and split decomposition analyses also showed incongruences between genes, suggesting horizontal gene transfer. The plcR gene was observed to have characteristics that differed from those of the other genes in terms of phylogenetic topology and pattern of sequence diversity. Thus, we suggest that the evolutionary history of the PlcR regulon differs from those of the other chromosomal genes and that recombination of the plcR gene may be frequent. The homogeneity of B. anthracis, which is depicted as an independent lineage in phylogenetic trees, is suggested to be of recent origin or to be due to the narrow taxonomic definition of species.
The Bacillus cereus group, which is a subdivision of the genus Bacillus, includes the closely related species Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis (22). However, their pathogenic potentials and disease spectrums are quite different despite their genetic relatedness. B. cereus is an opportunistic pathogen and causes several types of infections in humans. It is frequently isolated as a contaminant of milk, cereals, and various other foods, and it produces an emetic toxin and one or several enterotoxins. On the other hand, B. thuringiensis is primarily an insect pathogen, and it produces intracellularly insecticidal crystal toxins of different specificities during sporulation or in the stationary phase, which is the only established difference between it and B. cereus (7). B. anthracis causes the potentially lethal disease anthrax, and it has been identified as a nonhemolytic, nonmotile, penicillin-sensitive, encapsulated bacterium. B. anthracis is important in that it is considered a potential biological weapon (26). The genes causing the lethal effect of anthrax are located on two large virulence plasmids, pXO1 and pXO2 (26).
The genomes of these three species show high levels of similarity; for example, they share almost identical 16S ribosomal DNA sequences (2), although an association of a distinct type of 16S ribosomal DNA sequence with B. anthracis was recently reported (30). Although several phenotypes (such as capsule, lack of hemolysis, lack of motility, and susceptibility to gamma phage) and biochemical tests can differentiate B. anthracis from B. cereus and B. thuringiensis (38), species delimitation is unclear. In fact, they were suggested to be one species based on a multilocus enzyme electrophoresis (MLEE) result (14) and by the presence of an S-layer on the cell surface (24). Therefore, an investigation of the population genetic structure of the B. cereus group seemed warranted as a means of understanding their evolution.
To date, the population genetic structure of the B. cereus group has been studied mainly by MLEE (12, 13, 14, 38) and by amplified fragment length polymorphism (AFLP) analysis (18, 36). All MLEE analyses, which compared the allozyme patterns of 10 to 20 housekeeping genes, showed that strains of the B. cereus group are separated into two clusters and that B. cereus and B. thuringiensis are indistinguishable on the basis of their genetic backgrounds. These approaches have contributed to our understanding of the population genetic structure of these three species of the B. cereus group. However, as indicated previously, MLEE analysis has its drawbacks compared to genetic analysis, specifically, (i) comparison of results from MLEE studies between laboratories is difficult, (ii) enzyme mobility reflects genetic background only indirectly, and (iii) variations may be limited (23). Moreover, AFLP analysis has problems associated with inaccurate DNA fragment length determination (18, 36) and of result nonportability, as for MLEE. To overcome such shortcomings in bacterial population and epidemiological studies, the multilocus sequence typing (MLST) method, which involves comparisons of the nucleotide sequences of several DNA fragments, was recently introduced (23, 33). Studies based on sequence analysis have several advantages, including improved precision, portability, and reproducibility (5). Moreover, population genetic studies using MLST have revealed the existence of clonal groupings and have been used to evaluate the extent of recombination and mutation within the same bacterial species (5, 23, 33).
In the present study, we investigated the population structure of B. anthracis, B. cereus, and B. thuringiensis by using sequences of seven protein-encoding genes, namely, rpoB (RNA polymerase β subunit), gyrB (gyrase B subunit), pycA (pyruvate carboxylase A), mdh (malate dehydrogenase), mbl (cell shape determination-like protein, mreB like), mutS (DNA mismatch repair protein MutS), and plcR (transcriptional regulator PlcR). rpoB, gyrB, and mutS have frequently been used for the phylogenetic study of bacterial pathogens (9, 11, 19, 20, 21), and pycA, mdh, and mbl were successfully used in a previous study of the B. cereus group (14). The plcR gene was selected because of its importance for virulence in the B. cereus group (1). It was hoped that the present study might determine whether B. anthracis, B. cereus, and B. thuringiensis are genetically distinct and the origin of B. anthracis.
MATERIALS AND METHODS
Bacterial strains.
Fifty-two strains of B. cereus, B. thuringiensis, and B. anthracis were included in this study (Table 1). Sequence data for four B. anthracis strains (Ames, Florida, Kruger B, and Western NA) and for two B. cereus strains (ATCC 14579T and ATCC 10987) were retrieved from GenBank and from The Institute for Genomic Research website (www.tigr.com); their genomes have been sequenced. Five reference strains and five Korean isolates (GJ-1, GJ-2, BC, CN, and HS) of B. anthracis were provided by J. M. Kim, W. Kim, and I. J. Kim as DNA. Other strains of B. cereus and B. thuringiensis were purchased from the Korean Collection for Type Cultures (KCTC) and from the Institute of Microbiology, Seoul National University (IMSNU). One strain of Bacillus mycoides (KCCM 40260), which is usually classified as a member of the B. cereus group, was also included.
TABLE 1.
Strains used in this study
| Species and strain | Cluster | Origin |
|---|---|---|
| B. anthracis | ||
| ATCC 14185 | A | pX01+, pX02− |
| ATCC 14186 | A | pX01−, pX02− |
| ATCC 14578T | A | Vollum strain (pX01−, pX02+) |
| Sternea | A | Vaccine strain (pX01+, pX02−) |
| Pasteur no. 2 Armya | A | pX01+, pX02− |
| Floridaa | A | Isolate related to bioterror, A2012 |
| Kruger Ba | A | |
| Western NAa | A | |
| GJ-1b | A | Patient isolate, Korea |
| GJ-2b | A | From cow, Korea |
| BCb | A | From cow, Korea |
| CNb | A | From cow, Korea |
| HSb | A | From cow, Korea |
| B. cereus | ||
| ATCC 14579Ta | C-I | Dairy-isolated strain |
| ATCC 9634 (NRRL B-1530) | C-II | New York |
| IMSNU 11011 (IAM 1729) | C-II | |
| IMSNU 11012 (NCIB 9207) | C-I | |
| IMSNU 11013 (ATCC 11778) | C-I | New York |
| IMSNU 12076 | C-I | |
| IMSNU 12077 (ATCC 21366) | B-IV | From soil |
| IMSNU 12078 (ATCC 21768) | B-III | From turkey and chicken manure |
| IMSNU 12079 (KCTC 1092) | C-II | From contaminated flask |
| IMSNU 13043 (KCTC 1093) | E | |
| IMSNU 13044 (KCTC 1094) | C-II | From soil, Japan |
| IMSNU 13045 (NRRL B-569) | C-II | From contaminated flask |
| IMSNU 13046 (ATCC 21772) | B-I | From chicken and turkey manure |
| IMSNU 13047 (ATCC 12480) | C-I | From sheep rumen |
| KCTC 1012 | C-I | |
| KCTC 1014 | B-III | From turkey and chicken manure |
| IMSNU 10013 | C-II | From soil, Korea |
| IMSNU 10014 | C-II | From soil, Korea |
| IMSNU 11014 | E | From soil, Korea |
| 2127 | B-I | |
| ATCC 10987a | B-IV | Dairy-isolated strain |
| B. thuringiensis | ||
| IMSNU 12095 (KCTC 1513) | B-II | |
| IMSNU 12096 (KCTC 1514) | D-I | B. thuringiensis subsp. dendrolimus |
| IMSNU 12097 (KCTC 1515) | C-I | B. thuringiensis subsp. entomocidus |
| IMSNU 12098 (KCTC 1516) | B-II | B. thuringiensis subsp. finitimus |
| IMSNU 12099 (KCTC 1511) | C-II | B. thuringrensis subsp. indiana |
| IMSNU 10051 | D-I | From diseased insect larvae |
| IMSNU 12092 (KCTC 1512) | C-I | B. thuringiensis subsp. pakistani |
| IMSNU 12089 (ATCC 35646) | D-I | From sewage, Israel |
| KCTC 1507 (ATCC 33679) | C-II | From diseased insect larvae |
| KCTC 1509 | D-I | From sewage |
| IMSNU 11043 | D-I | |
| IMSNU 12086 (KCTC 1034) | C-I | From flour moth |
| IMSNU 12087 (KCTC 1108) | D-II | Philippine |
| IMSNU 12088 (ATCC 10792) | D-II | From flour moth |
| IMSNU 12090 (KCTC 1524) | D-I | From sewage |
| IMSNU 12091 (KCTC 1525) | D-I | From sewage |
| IMSNU 12093 (KCTC 1519) | D-II | From silkworm |
| B. mycoides KCCM 40260 (ATCC 21929) | From soil, New Guinea |
Sequence was retrieved from GenBank and www.tigr.com and used in this study.
Korean isolates.
Molecular methods.
The primers used for amplification and sequencing are shown in Table 2 with the lengths of the sequenced fragments. Total DNA extraction, PCR amplification, and direct sequencing of PCR products were performed as described previously (20, 21). Briefly, total DNA for PCR was extracted by using the bead beater-phenol extraction method. Amplifications were performed with a GeneAmp PCR system 9700 (Perkin-Elmer) as follows: denaturation at 95°C for 5 min; then 30 cycles of 95°C for 30 s, 45 to 55°C for 30 s, and 72°C for 1 min; and then a final extension at 72°C for 5 min. PCR products were purified by using a QIAEX II gel extraction kit (Qiagen, Hilden, Germany) for sequencing. Sequences were directly determined with an Applied Biosystems model 377 automated sequencer and a BigDye terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom).
TABLE 2.
Gene fragments and primers used for amplification and sequencing
| Gene (length [bp] of sequenced fragment | Primer | Sequence |
|---|---|---|
| rpoB (318) | BA-rpoBF | 5′-GAC GAT CAT YTW GGA AAC CG-3′ |
| BA-rpoBR | 5′-GGN GTY TCR ATY GGA CAC AT-3′ | |
| gyrB (300) | BA-gyrBF | 5′-AAA ACA ACC RAT TCA TGA AG-3′ |
| BA-gyrBR | 5′-TCG CTT CAC TAT TYC CAA GT-3′ | |
| pycA (379) | BA-pycAF | 5′-CCT AAA CAT ATA GAA GTN CAA-3′ |
| BA-pycAR | 5′-TTT TTC CTG TAT CCG GCA TG-3′ | |
| mdh (278) | BA-mdhF | 5′-CCA TAT CGT CAC CGT GTC C-3′ |
| BA-mdhR | 5′-TTG TTG TGA TTA CAG CAG GT-3′ | |
| mbl (360) | BA-mblF | 5′-CCA AGC GGT AAC ATG GTT G-3′ |
| BA-mblR | 5′-CCT GTT AGA ATA ACA CCG C-3′ | |
| mutS (367) | BA-mutSF | 5′-GCT GAA ACG TGT ACA TTY TT-3′ |
| BA-mutSR | 5′-TTA ATT ACA GGA CCG AAC ATG-3′ | |
| plcR (424) | BA-plcRF | 5′-AAA AAG GAA GAA TAT CAT C-3′ |
| BA-plcRR | 5′-ATG CAT CTT CAA TCT CTG-3′ |
Sequence analyses.
Raw sequences were concatenated and analyzed by using EditSeq and MegAlign programs (DNASTAR, Madison, Wis.). Amino acid sequences were deduced by using the MegAlign program. Phylogenetic trees based on the concatenated sequences of six genes and the plcR sequence were constructed by using the neighbor-joining method in PAUP* (35), using the maximum-likelihood distance option. The gene tree obtained from the concatenated six housekeeping gene sequences was rooted by using B. mycoides as an outgroup, and the plcR gene tree was rooted by using the midpoint-rooting option. Branch supporting values were evaluated by performing 1,000 bootstrap replications. The seven individual gene data sets were compared statistically for incongruence by using the incongruence length difference (ILD) test (or the partition homogeneity test) (8, 10), which was implemented in PAUP* (35). In addition, to investigate the effects of recombination on the evolution of the six genes (except plcR), for the 16 randomly selected strains used in this study, a split decomposition tree was generated by using the SplitsTree 3.1 program (16). This enabled the detection of conflicting phylogenies, suggestive of recombination.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study were submitted to GenBank. The corresponding accession numbers of rpoB, gyrB, pycA, mdh, mbl, mutS, and plcR are AY169510 to AY169541 and AY265467 to AY265738.
RESULTS
Sequence diversity.
To investigate the population structure of the B. cereus group, we obtained the 278- to 424-bp fragment sequences of seven genes from 14 strains of B. anthracis, 21 strains of B. cereus, and 17 strains of B. thuringiensis. When each unique sequence was assigned a different allele number, each gene was found to contain from 15 to 22 alleles (Table 3). The maximum sequence diversity of nucleotides ranged from 4.40% (rpoB) to 19.34% (plcR). Ratios of sites showing nucleotide and amino acid differences varied from 7.86% (rpoB) to 32.55% (plcR) and from 1.00% (gyrB) to 34.04% (plcR), respectively (Table 3). The 14 strains of B. anthracis possessed identical alleles for the four gene fragments (rpoB, gyrB, pycA, and mdh). mbl of strain Kruger B, mutS of strains GJ-1 and GJ-2, and plcR of strain ATCC 14186 showed a single base substitution. The G+C ratio was highest for rpoB (41.82%) and lowest for plcR (30.47%) (Table 3). In regard to signature nucleotides, i.e., those discriminating B. anthracis from B. cereus, B. thuringiensis, and B. mycoides, the rpoB, pycA, and mutS sequences contained one and the plcR sequences contained four.
TABLE 3.
Allelic diversities and G+C ratios of genes used in this studya
| Gene fragment | No. of alleles | No. of variable sites/total (% of variable sites)
|
Maximum sequence divergence (%) | G+C ratio (%) | |
|---|---|---|---|---|---|
| Nucleotide | Amino acid | ||||
| rpoB | 16 | 25/318 (7.86) | 2/106 (1.89) | 4.40 | 41.82 |
| gyrB | 17 | 52/300 (17.33) | 1/100 (1.00) | 11.33 | 34.42 |
| pycA | 17 | 52/379 (13.72) | 10/127 (7.87) | 8.97 | 36.63 |
| mdh | 15 | 22/278 (7.91) | 7/93 (7.53) | 5.04 | 38.85 |
| mbl | 22 | 34/360 (9.44) | 7/120 (5.83) | 6.39 | 39.23 |
| mutS | 15 | 46/367 (12.53) | 4/123 (3.25) | 7.08 | 36.53 |
| plcR | 20 | 138/424 (32.55) | 48/141 (34.04) | 19.34 | 30.47 |
B. mycoides was excluded in this analysis.
Phylogenetic relationships in rpoB, gyrB, pycA, mdh, mbl, and mutS gene trees.
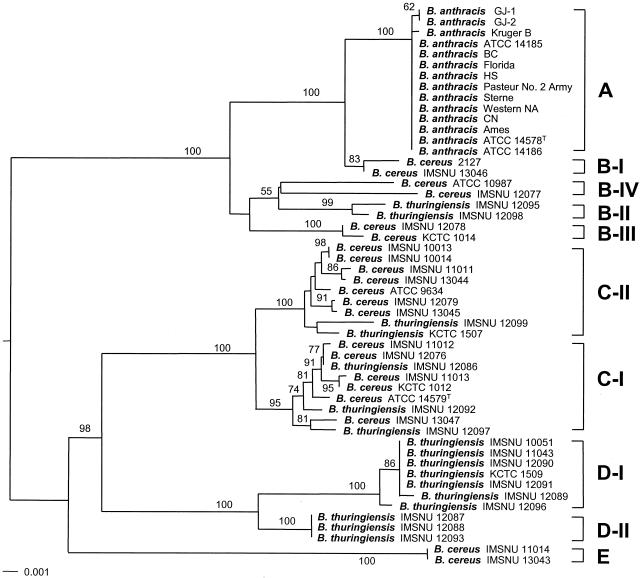
The neighbor-joining tree was inferred from the concatenated nucleotide sequences of six housekeeping gene fragments (rpoB, gyrB, pycA, mdh, mbl, and mutS) (Fig. 1). Five clusters, designated A to E, were observed in the housekeeping gene tree. Most strains of the B. cereus group were subdivided into two large subgroups, one consisting of clusters A and B and the other consisting of clusters C and D. When the six housekeeping genes were analyzed independently, the gene trees consistently showed congruent clustering overall, although some of the interrelationships between clusters were inconsistent (data not shown).
FIG. 1.
Neighbor-joining trees based on the concatenated sequences of six chromosomal genes (52 strains of the B. cereus group). B. mycoides KCCM 46260 was used as an outgroup. Clusters are indicated at the right. The branch lengths are proportional to nucleotide changes. Branches supported by more than 50% in the bootstrap analysis (1,000 replications) are indicated.
Fourteen B. anthracis strains formed a distinct cluster, cluster A, in all independent and concatenated gene trees. The pycA genes of B. cereus 2127 and IMSNU 13046 and the mdh genes of B. cereus 2127 and IMSNU 13046 and of B. thuringiensis IMSNU 12095 and IMSNU 12098 have sequences identical to those of the B. anthracis strains and clustered with them in each gene tree (data not shown). Although cluster B-I (B. cereus 2127 and IMSNU 13046) was a sister group of the B. anthracis cluster in the concatenated six-housekeeping-gene tree (Fig. 1), clusters closest to B. anthracis were gene tree dependent.
Unlike those of B. anthracis, strains of B. cereus and B. thuringiensis were not separated into distinct clusters in any tree. While clusters B-I, B-III, and E consisted of only B. cereus strains, clusters B-II, D-I, and D-II contained only B. thuringiensis strains. However, strains of both B. cereus and B. thuringiensis were mixed in clusters C-I and C-II. Cluster E, which consisted of two B. cereus strains (IMSNU 11014 and IMSNU 13043), was a minor cluster with a distinct position (Fig. 1).
plcR gene.
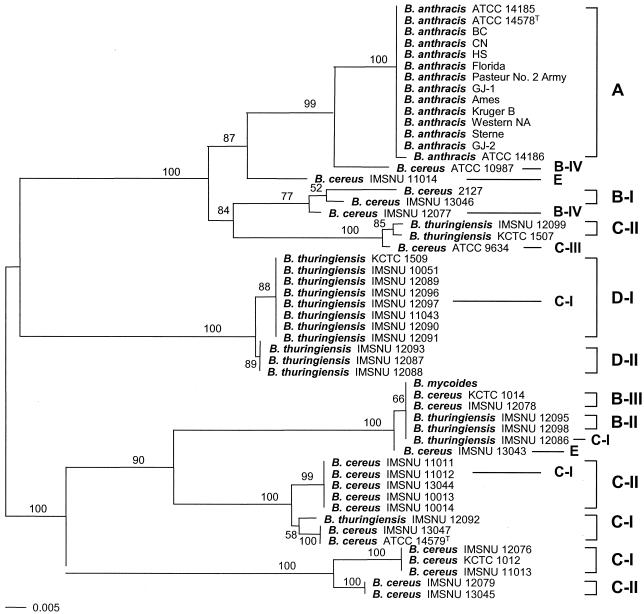
plcR had the highest sequence diversity in terms of both the ratio of variable sites and maximum sequence divergence (Table 3). Phylogenetic relationships in the plcR gene tree (Fig. 2) differed markedly from the concatenated gene tree of the six housekeeping genes (Fig. 1). B. anthracis strains were also homogeneous in plcR, as in the other independent gene trees, although B. anthracis ATCC 14186 had one nucleotide sequence that differed from those of the other B. anthracis strains. As indicated previously (1), the sequence of the plcR gene of B. anthracis strains indicates that expression of the gene would result in synthesis of a truncated PlcR protein. Specifically, the codon 214, for glutamic acid (GAA or GAG), in B. cereus and B. thuringiensis is changed to a termination codon (TAA) in B. anthracis due to single nucleotide substitution (G→T). The closest strain to the B. anthracis clade was B. cereus ATCC 10987, which had a sequence similarity of 96.9 to 97.2%.
FIG. 2.
The plcR tree of the B. cereus group, based on its nucleotide sequences. This tree was constructed by the neighbor-joining method. B. mycoides, which was used as an outgroup in Fig. 1, was not used as an outgroup because it was not distinguished from the ingroup strains. Thus, the midpoint rooting method was applied to root this tree. Clusters in the chromosomal gene tree (Fig. 1) are indicated on the right. Branches supported by more than 50% in the bootstrap analysis (1,000 replications) are indicated.
Clusters observed in the other six independent gene trees did not preserve their groupings in the plcR gene tree, except for the B-I, D-I, and D-II clusters. B. thuringiensis IMSNU 12097 of cluster C-I in the other gene trees was included in cluster D-I (Fig. 2). B. mycoides, which was clearly differentiated from other species in the other genes, had the same plcR sequence as two B. cereus strains (KCTC 1014 and IMSNU 12078) of cluster B-III and three B. thuringiensis strains of clusters B-II (IMSNU 12095 and IMSNU 12098) and C-I (IMSNU 12086). Cluster E, which had identical sequences in all six housekeeping genes, constituted a distinct clade in each gene tree (data not shown). However, two strains of cluster E showed a remarkable plcR sequence divergence (16.5% dissimilarity) and did not form a single clade in the plcR tree. B. cereus IMSNU 11014 clustered with B. anthracis and B. cereus ATCC 10987, whereas B. cereus IMSNU 13043 was very closely related to the clade containing B. mycoides. Strains of clusters C-I and C-II did not form any distinct clade and were dispersed in the plcR tree. For example, B. thuringiensis IMSNU 12097 merged into cluster D-I, B. thuringiensis IMSNU 12086 was included in the clade containing B. mycoides, and B. cereus IMSNU 11012 had a plcR sequence that was identical to those of four strains of cluster C-II (Fig. 2).
Incongruence tests and split decomposition analysis.
In order to investigate incongruences between genes, 21 pairwise ILD comparisons were performed. Of the 21 combined data sets, only four combinations (rpoB-mdh, rpoB-mbl, mdh-mbl, and mdh-mutS) coalesced; the P values of these four cases were greater than 0.05 (Table 4), which is the threshold for congruence (8). Sequence data sets of rpoB, mdh, and mbl may have been congruent with each other, based on the ILD test. In addition, the consistency of the mutS and mdh sequence data sets was also supported by the ILD test (P = 0.79). The other pairs could not be combined; i.e., they were incongruent (P < 0.05) (4, 9).
TABLE 4.
P values from ILD test
| Gene |
P for test witha:
|
||||||
|---|---|---|---|---|---|---|---|
| rpoB | gyrB | pycA | mdh | mbl | mutS | plcR | |
| rpo | <0.01 | <0.01 | 0.05 | 0.29 | 0.02 | <0.01 | |
| gyrB | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | ||
| pycA | 0.03 | <0.01 | <0.01 | <0.01 | |||
| mdh | 0.28 | 0.79 | <0.01 | ||||
| mbl | <0.01 | <0.01 | |||||
| mutS | <0.01 | ||||||
| plcR | |||||||
Values larger than the threshold of congruence (0.05) are in bold face.
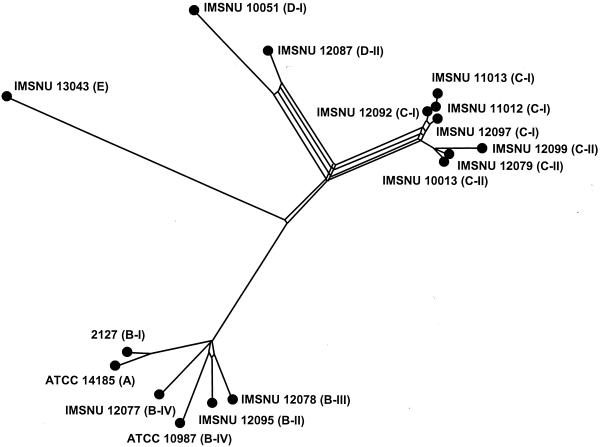
To examine how recombination between genes can affect phylogenetic relationships among strains, split decomposition analysis was performed (3). In this analysis, we used the concatenated sequences of six genes (all except plcR) of 16 randomly selected strains. Split decomposition of pairwise sequences revealed consistent results on pairwise ILD testing (data not shown). That is, no parallel evolutionary path was observed for the rpoB-mbl, mbl-mdh, and mdh-mutS comparisons, whereas the other comparisons, including rpoB-mdh, showed reticulated networks. We next analyzed the six genes together (without the plcR gene). The split graph showed evidence of a network-like evolution (Fig. 3), with a fit parameter of 0.75. Eleven distinct parallel paths were observed in the split decomposition tree.
FIG. 3.
Split graph showing the relationships between rpoB, gyrB, pycA, mdh, mbl, and mutS for 16 randomly selected strains of the B. cereus group. The split graph was generated by using SplitsTree 3.1 (16) from pairwise distances based on the maximum-likelihood model. The fit value was 0.75, indicating that the phylogenetic signal in the data is represented moderately well by the split graph. All branch lengths are drawn to scale. Cluster names are indicated in parentheses alongside the corresponding strain names.
DISCUSSION
Diversity of the B. cereus group.
Compared to the MLEE analysis (13, 14), our MLST produced a different result. Several strains that have been examined in previous population studies were also included in the present study. In previous MLEE studies (13, 14), B. cereus type strain ATCC 14579 and B. cereus ATCC 10987, the whole genomes of which have been sequenced, and B. thuringiensis subsp. kurstaki KCTC 1509, which is widely used for the preparation of biopesticides, were clustered into a narrow clade with B. anthracis strains. However, these three strains were found to be dispersed through the gene trees (Figs. 1 and 2) in our study. In other studies, several reference strains of B. cereus and B. thuringiensis were located in a single group (18, 36). This may be due to the limited number of collections of the B. cereus group strains used in the present study.
Intergenic recombination.
In the present study, comparative sequence analysis using seven chromosomal genes revealed the complicated genetic structure of the B. cereus group. Fifty-two strains of the B. cereus group were classified consistently into several clusters in all gene trees except plcR. In particular, separation into two main clusters (clusters A and B versus clusters C and D) was distinct. However, the interrelationships between clusters differed (Fig. 1).
ILD testing (Table 4) showed that most of the pairs compared were not combinable, i.e., were significantly incongruent, suggesting a relic of horizontal gene transfer or recombination (15, 17). Coupled with the ILD test, split decomposition analysis using 16 randomly selected strains showed that several recombinational events have occurred between their chromosomal loci. A split decomposition of the six genes, without the plcR gene, as shown by the different gene tree topologies (Fig. 2), affirmed substantial levels of incongruence (Fig. 3). Such results are regarded as evidence of intergenic recombination (4, 5, 34). Different topologies among gene trees suggest a relic of horizontal gene transfer or recombination (15, 17).
Evolution of the plcR gene.
PlcR is a pleiotropic regulator of extracellular virulence factors in B. cereus and B. thuringiensis (1, 27, 31). In this study, plcR showed strikingly different characteristics from the other chromosomal genes. Ratios of variable sites and maximum sequence divergences observed for the plcR gene were much higher than those for the other genes, both at the nucleotide level and at the amino acid level (Table 3). In addition, the G+C ratio for the plcR gene was quite different from those for the other six genes (Table 3). Moreover, most of the consistent clusterings found in the other gene trees were not observed in the plcR tree. The exceptions were cluster A, including 10 B. anthracis strains, and clusters D-I and D-II of the B. thuringiensis strains. Significantly, the different tree topology and sequence characteristics indicate that the plcR gene must have evolved in a different manner. plcR sequence analysis showed that mutations were not clustered in the gene at the nucleotide and amino acid levels. In addition, no mosaic structure resulting from intragenic recombination was observed. Thus, the contradictory branching pattern in the plcR gene tree might be due to intergenic recombination of relatively large chromosomal fragments, including the plcR gene. However, according to the final annotated B. anthracis genome (www.tigr.com), more than one plcR gene was apparently revealed. Thus, the possibility that the observed heterogeneity of the plcR gene originated from several plcR homologues cannot be excluded.
Because of the presence of the plcR gene in B. anthracis and because of variable chromosomal locations of the PlcR-regulated genes, B. anthracis, B. cereus, and B. thuringiensis were inferred to have been derived from a common ancestor containing the PlcR regulon rather than having recently acquired the PlcR regulon by horizontal gene transfer (1). However, chaotic relationships in the phylogenetic tree (Fig. 2) suggest that horizontal transfer of the plcR gene occurs between clusters. Moreover, the fact that plcR showed an allele number similar to that of the other genes despite its greater sequence divergence (Table 3) indicates frequent recombination within the B. cereus group and a rapid plcR gene mutation rate.
The lack of hemolysis of B. anthracis, which is one of the principal characteristics that distinguishes it from B. cereus and B. thuringiensis, is thought to be due to the inactivation of PlcR by a nonsense mutation (1). In our study, a nonsense mutation of plcR was found only in the B. anthracis strains. Although the gene is nonfunctional, its mutation rate within B. anthracis was very low, as in the other chromosomal genes. This finding supports the suggestion that the inactivation of the PlcR regulon due to the plcR mutation must have evolved recently, i.e., after B. anthracis speciation (25).
Evolution of B. anthracis.
Our results suggest that the homogeneous nature of B. anthracis can be explained by several possibilities, i.e., the recent origin of the species, the narrow taxonomic definition of B. anthracis, or a decreased evolutionary rate of B. anthracis due to the dormant nature of spores and almost exclusive dependence of multiplication on infections in animals (37). This was also shown by AFLP analysis (36) and is supported by several findings, such as the low diversity of the 5′-vrrB region (less than 0.125% per nucleotide), a hypervariable open reading frame that includes a noncoding region (32), and of the pagA gene on the pXO1 plasmid (0.21% per nucleotide) (29), which is one of the genes on the pathogenicity island (28).
However, the origin of B. anthracis could not be elucidated with certainty by the present study. The closest cluster to B. anthracis differed from gene to gene: cluster B-I in gyrB, pycA, mdh, and mbl; cluster B-II in rpoB and mdh; and cluster B-III in mutS. In the case of the plcR gene, B. cereus ATCC 10987 was found to be the most closely related strain to the B. anthracis clade. Although no one cluster was clearly more closely related to B. anthracis, it is evident that B. anthracis shares a common ancestor with clusters B-I, B-II, and B-III, within which recombination occurs. Furthermore, the acquisition of B. anthracis-specific plasmids leading to speciation seems to be a single event, unlike that for B. thuringiensis.
Strains of B. anthracis were grouped in a distinct branch and separated from those of B. cereus and B. thuringiensis in five gene trees, but not in pycA and mdh (data not shown). This implies that B. anthracis, although closely related, represents a series of clones that have only recently diverged from B. cereus and B. thuringiensis, as inferred from the 16S-23S ribosomal DNA intergenic transcribed spacers (6). It is likely that B. anthracis started to diverge recently from one cluster of the B. cereus group and has just begun to develop sequence differences.
Acknowledgments
This work was supported by a grant from the Korea Health 21 R&D Project, Ministry of Health and Welfare, Seoul, Republic of Korea (01-PJ10-PG6-01GM03-0002), and in part by the BK21 Project for Medicine, Dentistry, and Pharmacy.
Editor: V. J. DiRita
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Økstad, A. B. Kolstøo, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. E. Farrow, M. Dorsh, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 3.Bandelt, H. J., and A. W. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. [DOI] [PubMed] [Google Scholar]
- 4.Brown, E. W., M. L. Kotewicz, and T. A. Cebula. 2002. Detection of recombination among Salmonella enterica strains using the incongruence length difference test. Mol. Phylogenet. Evol. 24:102-120. [DOI] [PubMed] [Google Scholar]
- 5.Bygraves, J. A., R. Urwin, A. J. Fox, S. J. Gray, J. E. Russel, I. M. Feavers, and M. C. J. Maiden. 1999. Population genetic and evolutionary approaches to analysis of Neisseria meningitidis isolates belonging to the ET-5 complex. J. Bacteriol. 181:5551-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherif, A., S. Borin, A. Rizzi, H. Ouzari, A. Boudabous, and D. Daffonchio. 2003. Bacillus anthracis diverges from related clades of the Bacillus cereus group in 16S-23S ribosomal DNA intergenic transcribed spacers containing tRNA genes. Appl. Environ. Microbiol. 69:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore, N., D. R. Zeigler, J. Feitelson, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, and D. H. Dean. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, C. W. 1997. Can three incongruence tests predicts when data should be combined? Mol. Biol. Evol. 14:733-740. [DOI] [PubMed] [Google Scholar]
- 9.Denamur, E., G. Lecointre, P. Darlu, O. Tenillon, C. Acquaviva, R. Rothstein, J. Elion, F. Taddei, M. Radman, and I. Matic. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711-721. [DOI] [PubMed] [Google Scholar]
- 10.Farris, J. S., M. Källersj, A. G. Kluge, and C. Bult. 1994. Testing significance of incongruence. Cladistics 10:315-319. [Google Scholar]
- 11.Fukushima, M., K. Kakinuma, and R. Kawaguchi. 2002. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J. Clin. Microbiol. 40:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegalson, E., D. A. Caugant, M. M. Lecadet, Y. Chen, J. Mahillon, A. Lővgre, I. Hegna, K. Kvaløoy, and A. B. Kolstø. 1998. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 37:80-87. [DOI] [PubMed] [Google Scholar]
- 13.Hegalson, E., D. A. Caugant, I. Olsen, and A. B. Kolstø. 2000. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 38:1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helgason, E., L. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, E. C., R. Urwin, and M. C. J. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 16.Huson, D. H. 1998. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 17.Kalia, A., B. G. Spratt, M. C. Enright, and E. E. Besen. 2002. Influence of recombination and niche separation on the population genetic structure of the pathogen Streptococcus pyogenes. Infect. Immun. 70:1971-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, B.-J., S.-H. Lee, M.-A. Lyu, S.-J. Kim, G.-H. Bai, S.-J. Kim, G.-T. Chae, E.-C. Kim, C.-Y. Cha, and Y.-H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko, K. S., H. K. Lee, M.-Y. Park, M.-S. Park, K.-H. Lee, S.-Y. Woo, Y.-J. Yun, and Y.-H. Kook. 2002. Population genetic structure of Legionella pneumophila inferred from RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. J. Bacteriol. 184:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko, K. S., J.-M. Kim, J.-W. Kim, B. Y. Jung, W. Kim, I. J. Kim, and Y.-H. Kook. 2003. Identification of Bacillus anthracis by rpoB sequence analysis and multiplex PCR. J. Clin. Microbiol. 41:2908-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logan, N. A., and P. C. B. Turnbull. 1999. Bacillus and recently derived genera, p. 357-369. In P. R. Murray (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 23.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignot, T., B. Denis, E. Couture-Tosi, A. G. Kolstø, M. Mock, and A. Fouet. 2001. Distribution of S-layers on the surface of Bacillus cereus strains: phylogenetic origin and ecological pressure. Environ. Microbiol. 3:493-501. [DOI] [PubMed] [Google Scholar]
- 25.Mignot, T., M. Mock, D. Robichon, A. Landler, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 26.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 27.Østad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A. B. Kolstø. 1999. Sequence analysis of three Bacillus cereus loci carrying PlcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology 145:3129-3138. [DOI] [PubMed] [Google Scholar]
- 28.Pannucci, J., R. T. Okinaka, R. Sabin, and C. R. Kuske. 2002. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J. Bacteriol. 184:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price, L. B., M. Hugh-Jones, P. J. Jackson, and P. Keim. 1999. Genetic diversity in the protective antigen gene of Bacillus anthracis. J. Bacteriol. 181:2358-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacchi, C. T., A. M. Whitney, L. W. Mayer, R. Morey, A. Steigerwalt, A. Boras, R. S. Weyant, and T. Popovic. 2002. Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salamitou, S., F. Ramisse, M. Brehélin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 32.Schupp, J. M., A. M. Klevytska, G. Zinser, L. B. Price, and P. Keim. 2000. vrrB, a hypervariable open reading frame in Bacillus anthracis. J. Bacteriol. 182:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spratt, B. G. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr. Opin. Microbiol. 2:312-316. [DOI] [PubMed] [Google Scholar]
- 34.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 1999. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 36.Ticknor, L. O., A. B. Kolstøo, K. K. Hill, P. Keim, M. T. Laker, M. Tonk, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turnbull, P. C. B. 2002. Introduction: anthrax history, disease and ecology. Curr. Top. Microbiol. Immunol. 271:1-19. [DOI] [PubMed] [Google Scholar]
- 38.Vilas-Boas, G., V. Sanchis, D. Lereclus, M. V. Lemos, D. Bourguet. 2002. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 68:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]