Abstract
CD44 polymorphisms have been previously associated with cancer risk. However, the results between independent studies were inconsistent. Here, a meta-analysis was performed to systematically evaluate associations between CD44 polymorphisms and cancer susceptibility. A comprehensive literature search conducted in PubMed, Embase, and Web of Science databases through August 10, 2016 yielded 11 eligible publications consisting of 5,788 cancer patients and 5,852 controls. Overall, odds ratios (OR) calculated with 95% confidence intervals (CI) identified a significant association between CD44 polymorphism rs13347 and cancer susceptibility under all genetic models. Additionally, the minor allele of polymorphism rs11821102 was associated with a decreased susceptibility to cancer in allele contrast, dominant, and heterozygous models, while no significant association was identified for polymorphisms rs10836347, rs713330, or rs1425802. Subgroup analysis by ethnicity revealed rs13347 was significantly associated with cancer susceptibility for Chinese but not for Indians. Linkage disequilibrium (LD) between different polymorphisms varied across diverse ethnic populations. In conclusion, the results indicate that CD44 polymorphism rs13347 acts as a risk factor for cancer, especially in Chinese, while the minor allele of polymorphism rs11821102 may be associated with a decreased susceptibility to cancer. Nevertheless, further studies on a larger population covering different ethnicities are warranted.
CD44 is a multistructural and multifunctional transmembrane glycoprotein, which functions as a receptor for hyaluronan and many other extracellular matrix components, as well as a coreceptor for growth factors and cytokines1. Numerous studies have demonstrated that CD44 is involved in many crucial cellular processes, including cell survival, proliferation, differentiation, adhesion, and migration1,2.
Increasing evidence indicates that CD44 has a role in cancer. First, CD44 is found to be widely expressed in various cancers, and its expression correlates with poor prognosis1,3,4,5. In functional studies, specific targeted knockdown of CD44 has been shown to impede cancer progression6. Second, CD44 is a common cancer stem cell (CSC) marker. CSCs are a selected population of tumor cells that display many features of embryonic or tissue stem cells, and likewise, possess the capacity for self-renewal and differentiation. CD44 is in fact a critical player in the regulation of properties of the CSCs, such as self-renewal, tumor initiation, metastasis, and chemo- or radio-resistance1,7. CD44 contributes in part as a downstream target of Wnt, a gene involved in the maintenance of the CSC phenotype, and has been shown to be essential for Wnt-induced tumor progression in cancers8. The interaction between CD44 and hyaluronan furthermore promotes protein kinase C activation which leads to phosphorylation and translocation to the nucleus of NANOG, a transcription factor also involved in the maintenance of stem cell characteristics9. Finally, CD44 has a critical role in epithelial-mesenchymal transition (EMT). EMT is a tightly regulated and highly conserved cellular process in which a cell type changes from an epithelial to a mesenchymal phenotype6,10. In cancer, EMT is involved in the acquisition of the stemness of epithelial tumor cells, which confers an invasive phenotype onto cells that may underlie tumor recurrence and metastasis11,12.
Based on these roles in cancer development, molecular mechanisms contributing to the regulation of CD44 and/or function have been under intensive investigation. Recent studies have examined the association of specific single nucleotide polymorphisms (SNPs) in the CD44 gene with cancer risk but the significance of these findings remains unclear. Jiang et al. first discovered that the CD44 rs13347 polymorphism might affect breast cancer development and prognosis by increasing CD44 expression in the Chinese population13. Several new studies performed on Chinese also revealed a similar positive correlation with statistical power in acute myeloid leukemia, nasopharyngeal carcinoma, and colorectal cancer14,15,16,17. However, three other studies on Chinese, failed to demonstrate any statistically significant associations18,19,20,21. Furthermore, two studies conducted on Indians demonstrated that no significant association existed between CD44 rs13347 polymorphism and risks for gallbladder or breast cancers22,23,24. Here, a comprehensive meta-analysis was performed to derive a more precise estimation of the relationship between CD44 polymorphisms and the susceptibility to cancer. Eligible studies included diverse forms of cancer and two ethnicities, Chinese and Indian. These results have potential implications for the basis of cancer development as well as for functional consequences of allele specific expression of CD44.
Materials and Methods
Search strategy
PubMed, Embase, and Web of Science databases were searched for studies reporting association of CD44 polymorphisms with cancer risk in any type of cancer performed up to the date of August 10, 2016. Gene-specific terms (CD44) were combined with polymorphism- (polymorphism or polymorphisms or variation or variations or variant or variants or mutation or mutations or genotype or genotypes) and disease-specific terms (cancer or cancers or tumor or tumors or neoplasm or neoplasms). The search was performed by the method of free-text word combined with the Medical Subject Headings (MeSH) and included the following search terms: “Neoplasms”, “Antigens, CD44” and “Polymorphism, Single Nucleotide”. A thorough review of all referenced materials within retrieved studies was also performed in order to identify additional potentially eligible studies.
Inclusion criteria and exclusion criteria
Inclusion criteria for eligible studies were the following: (1) case-control or cohort study design; (2) sufficient data for the evaluation of CD44 SNPs in cancer risk; (3) articles published in English; (4) studies performed on humans; and (5) in the case of multiple publications from the same study group, the most complete and recent results were used.
The exclusion criteria were defined as the following: (1) abstracts, reviews and animal studies; (2) useless reported data such as genotype number or frequency was not included; and (3) articles published in languages other than English.
TagSNPs selection
Previous studies performed bioinformatics analysis with Haploview software 4.2 to analyze the haplotype block based on the Chinese Han Beijing population data of HapMap (HapMap Data Rel 27 Phase II + III, February 2009, on NCBI B36 assembly, dbSNP b126). Six tagSNPs were found to cover all the potential functional common SNPs (MAF > 0.05) in the CD44 gene: rs8193, rs11821102, rs10836347, and rs13347 in the 3′UTR, rs1425802 in the promoter and rs9666607 in exon region. CD44 rs8193 and rs13347 were in high linkage disequilibrium (LD) (D’ = 1.0, r2 = 0.527), so the selection of rs13347 was sufficient to represent the two SNPs. In addition, due to the difficulty in genotyping rs9666607 using MALDI-TOF, the polymorphism rs713330, which is in complete LD with rs9666607 (D’ = 1.0, r2 = 1.0) was chosen to replace it13,15,17.
Data extraction
Two reviewers (Q.C.Q. and J.W.W.) independently selected studies and extracted the following data from each study: first author’s surname, publication year, ethnicity, cancer types, numbers of cases and controls, and genotype distributions of cases and controls. The results were compared and disagreement was resolved through discussion until consensus was reached. Study design was stratified into population-based studies and hospital-based studies.
Quality assessment
The Newcastle-Ottawa Scale and Agency for Healthcare Research and Quality (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp, maximum score = 9 points) was used to evaluate the methodological quality, which scored studies by the selection of patients, the comparability of the groups, and the quality of the sampling process. Studies awarded a score of 0–3, 4–6, or 7–9 were considered as low-, moderate-, or high-quality studies, respectively.
Statistical analysis
The meta-analysis assessed associations between polymorphisms and cancer risk under allele contrast, dominant, recessive, homozygous, and heterozygous models. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to estimate the strength of associations. Heterogeneity among the included studies was evaluated using the Chi square-based Q statistic. The fixed-effect (Mantel-Haenszel method) or random-effects model (DerSimonian-Laird method) was used to calculate pooled effect estimates in the presence (P < 0.05) or absence (P > 0.05) of heterogeneity. Sensitivity analysis was conducted by sequentially excluding one study at a time. The Begg’s test and the Egger’s test were performed to assess publication bias. If any possible bias was observed, the trim and fill method was used to identify and adjust for those studies. Data analysis was carried out using Stata software, version 11.0 (Stata Corporation; College Station, TX, USA). P-values < 0.05 were considered to be statistically significant.
Linkage disequilibrium (LD) analysis across populations
We extracted data from the 1000 genomes Project Phase III (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/) regarding the CD44 polymorphisms evaluated in the current study. Briefly, populations enrolled in the project including African ancestry in Southwest USA (ASW), Utah residents with Northern and Western European ancestry (CEU), Han Chinese in Beijing, China (CHB), Southern Han Chinese, China (CHS), Finnish in Finland (FIN), British in England and Scotland (GBR), Gujarati Indians in Houston, Texas (GIH), Indian Telugu in the UK (ITU), Japanese in Tokyo, Japan (JPT), and Toscani in Italy (TSI). Haploview software 4.2 was used to perform the analysis, and linkage disequilibrium (LD) was evaluated by D’ and r2 statistics in each of the above-mentioned populations25.
Results
Study characteristics
The literature search yielded 15 studies that were carefully reviewed based on the inclusion criteria13,14,15,16,17,18,19,20,21,22,23,24,26,27,28. Among these articles, the following studies were excluded: 2 studies that did not investigate the association between cancer risk and the polymorphism of interest27,28; 1 study that did not offer sufficient raw data26; and 1 study in which the data was repeated in a second publication with a larger population conducted by the same study group22,24. Thus, 11 publications were eligible for the meta-analysis for a total of 5,788 cancer patients and 5,852 controls and 9 different cancer types. The main characteristics of the eligible studies are listed in Table 1. All studies scored a value of 7 or above (high-quality) as determined in the Newcastle-Ottawa Scale, and individuals therein were of Chinese or Indian descent.
Table 1. Characteristics of studies on association between CD44 single nucleotide polymorphisms and cancers.
| Single Nucleotide Polymorphisms | Author | Year | Ethnicity | Cancers | Genotyping Method | Quality Score | Cases | Controls | Source of Controls | P for HWE in controls | Cases |
Controls |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | |||||||||||
| rs13347 | Yadava | 2015 | Indian | GBC | TaqMan, ARMS-PCR, PCR-RFLP | 9 | 610 | 250 | PB | 0.62 | 378 | 201 | 31 | 162 | 80 | 8 |
| rs13347 | Wu | 2015 | Chinese | CRC | MALDI-TOF mass spectrometry | 9 | 946 | 989 | PB | 0.28 | 416 | 441 | 89 | 578 | 348 | 63 |
| rs13347 | Weng | 2015 | Chinese | TCC | TaqMan | 8 | 275 | 275 | HB | 0.15 | 138 | 111 | 26 | 143 | 117 | 15 |
| rs13347 | Liu | 2015 | Chinese | NSCLC | Taqman | 8 | 234 | 468 | HB | 0.82 | 179 | 51 | 4 | 337 | 121 | 10 |
| rs13347 | Sharmaa | 2014 | Indian | GBC | TaqMan, PCR-RFLP | 9 | 405 | 200 | PB | 0.57 | 293 | 104 | 8 | 154 | 42 | 4 |
| rs13347 | Lou | 2014 | Chinese | NPC | ABI 3730xl sequencing platform | 8 | 287 | 507 | HB | 0.91 | 104 | 126 | 42 | 288 | 174 | 27 |
| rs13347 | Chou-1b | 2014 | Chinese | OSCC | TaqMan | 7 | 599 | 561 | HB | 0.92 | 287 | 262 | 50 | 295 | 223 | 43 |
| rs13347 | Chou-2b | 2014 | Chinese | HCC | TaqMan | 7 | 203 | 561 | HB | 0.92 | 110 | 72 | 21 | 295 | 223 | 43 |
| rs13347 | Xiao | 2013 | Chinese | NPC | MALDI-TOF mass spectrometry | 9 | 906 | 943 | PB | 0.64 | 386 | 418 | 102 | 606 | 297 | 40 |
| rs13347 | Wu | 2013 | Chinese | AML | MALDI-TOF mass spectrometry | 9 | 421 | 461 | PB | 0.34 | 163 | 196 | 62 | 254 | 171 | 36 |
| rs13347 | Tulsyan | 2013 | Indian | BC | ABI 7500 Real Time PCR system | 8 | 258 | 241 | HB | 0.58 | 191 | 60 | 7 | 178 | 57 | 6 |
| rs13347 | Jiang | 2012 | Chinese | BC | MALDI-TOF mass spectrometry | 9 | 1049 | 1157 | PB | 0.84 | 451 | 484 | 114 | 654 | 430 | 73 |
| rs11821102 | Wu | 2015 | Chinese | CRC | MALDI-TOF mass spectrometry | 9 | 946 | 989 | PB | <0.001 | 815 | 119 | 12 | 843 | 131 | 15 |
| rs11821102 | Weng | 2015 | Chinese | TCC | TaqMan | 8 | 275 | 275 | HB | 0.92 | 234 | 39 | 2 | 222 | 50 | 3 |
| rs11821102 | Lou | 2014 | Chinese | NPC | ABI 3730xl sequencing platform | 8 | 287 | 507 | HB | 0.35 | 252 | 27 | 1 | 439 | 54 | 3 |
| rs11821102 | Chou-1b | 2014 | Chinese | OSCC | TaqMan | 7 | 599 | 561 | HB | 0.28 | 531 | 63 | 5 | 481 | 75 | 5 |
| rs11821102 | Chou-2b | 2014 | Chinese | HCC | TaqMan | 7 | 203 | 561 | HB | 0.28 | 173 | 29 | 1 | 481 | 75 | 5 |
| rs11821102 | Xiao | 2013 | Chinese | NPC | MALDI-TOF mass spectrometry | 9 | 906 | 943 | PB | 0.14 | 796 | 100 | 10 | 805 | 129 | 9 |
| rs11821102 | Wu | 2013 | Chinese | AML | MALDI-TOF mass spectrometry | 9 | 421 | 461 | PB | 0.28 | 370 | 50 | 1 | 398 | 59 | 4 |
| rs11821102 | Jiang | 2012 | Chinese | BC | MALDI-TOF mass spectrometry | 9 | 1049 | 1157 | PB | 0.22 | 912 | 125 | 12 | 997 | 151 | 9 |
| rs10836347 | Wu | 2015 | Chinese | CRC | MALDI-TOF mass spectrometry | 9 | 946 | 989 | PB | 0.10 | 821 | 120 | 5 | 851 | 129 | 9 |
| rs10836347 | Lou | 2014 | Chinese | NPC | ABI 3730xl sequencing platform | 8 | 287 | 507 | HB | 0.85 | 249 | 27 | 2 | 438 | 55 | 2 |
| rs10836347 | Chou-1b | 2014 | Chinese | OSCC | TaqMan | 7 | 599 | 561 | HB | 0.15 | 522 | 73 | 4 | 487 | 69 | 5 |
| rs10836347 | Chou-2b | 2014 | Chinese | HCC | TaqMan | 7 | 203 | 561 | HB | 0.15 | 180 | 23 | 0 | 487 | 69 | 5 |
| rs10836347 | Xiao | 2013 | Chinese | NPC | MALDI-TOF mass spectrometry | 9 | 906 | 943 | PB | 0.31 | 785 | 118 | 3 | 792 | 147 | 4 |
| rs10836347 | Wu | 2013 | Chinese | AML | MALDI-TOF mass spectrometry | 9 | 421 | 461 | PB | 0.93 | 364 | 55 | 2 | 404 | 55 | 2 |
| rs10836347 | Jiang | 2012 | Chinese | BC | MALDI-TOF mass spectrometry | 9 | 1049 | 1157 | PB | 0.97 | 906 | 139 | 4 | 995 | 156 | 6 |
| rs713330 | Weng | 2015 | Chinese | TCC | TaqMan | 8 | 275 | 275 | HB | 0.87 | 231 | 42 | 2 | 223 | 49 | 3 |
| rs713330 | Chou-1b | 2014 | Chinese | OSCC | TaqMan | 7 | 599 | 561 | HB | 0.09 | 507 | 88 | 4 | 467 | 86 | 8 |
| rs713330 | Chou-2b | 2014 | Chinese | HCC | TaqMan | 7 | 203 | 561 | HB | 0.09 | 167 | 36 | 0 | 467 | 86 | 8 |
| rs713330 | Xiao | 2013 | Chinese | NPC | MALDI-TOF mass spectrometry | 9 | 906 | 943 | PB | 0.74 | 732 | 164 | 10 | 751 | 180 | 12 |
| rs713330 | Wu | 2013 | Chinese | AML | MALDI-TOF mass spectrometry | 9 | 421 | 461 | PB | 0.39 | 341 | 74 | 6 | 371 | 87 | 3 |
| rs713330 | Jiang | 2012 | Chinese | BC | MALDI-TOF mass spectrometry | 9 | 1049 | 1157 | PB | 0.39 | 865 | 172 | 12 | 950 | 194 | 13 |
| rs1425802 | Weng | 2015 | Chinese | TCC | TaqMan | 8 | 275 | 275 | HB | 0.001 | 105 | 121 | 49 | 99 | 109 | 67 |
| rs1425802 | Chou-1b | 2014 | Chinese | OSCC | TaqMan | 7 | 599 | 561 | HB | < 0.001 | 197 | 249 | 153 | 194 | 235 | 132 |
| rs1425802 | Chou-2b | 2014 | Chinese | HCC | TaqMan | 7 | 203 | 561 | HB | < 0.001 | 70 | 75 | 58 | 194 | 235 | 132 |
| rs1425802 | Xiao | 2013 | Chinese | NPC | MALDI-TOF mass spectrometry | 9 | 906 | 943 | PB | 0.11 | 270 | 450 | 186 | 299 | 442 | 202 |
| rs1425802 | Wu | 2013 | Chinese | AML | MALDI-TOF mass spectrometry | 9 | 421 | 461 | PB | 0.08 | 126 | 204 | 91 | 122 | 248 | 91 |
| rs1425802 | Jiang | 2012 | Chinese | BC | MALDI-TOF mass spectrometry | 9 | 1049 | 1157 | PB | 0.55 | 316 | 513 | 220 | 353 | 563 | 241 |
PB: population-based. HB: hospital-based. PCR: polymerase chain reaction. RFLP: restriction fragment length polymorphism. ARMS: amplification refractory mutation system. HWE: Hardy-Weinberg equilibrium. GBC: gallbladder cancer. CRC: colorectal cancer. TCC: transitional cell carcinoma of urinary bladder. NSCLC: non-small cell lung cancer. NPC: nasopharyngeal carcinoma. OSCC: oral squamous cell carcinoma. HCC: hepatocellular carcinoma. AML: acute myeloid leukemia. BC: breast cancer.
aContaining repeated data.
bContaining the same population of controls.
Two publications that contained the same population of controls from the same research group were considered as one case-control study18,19. The following is the breakdown of the number of studies, including cancer types, cases and controls, that met our eligibility criteria for each polymorphism evaluated: rs13347 polymorphism, 10 case-control studies including 9 different cancer types with 5,788 cases and 5,852 controls; rs11821102 polymorphism, 7 case-control studies including 7 different cancer types with 4,686 cases and 4,893 controls; rs10836347 polymorphism, 6 case-control studies including 6 different cancer types with 4,411 cases and 4,618 controls; rs713330 polymorphism, 5 case-control studies including 6 different cancer types with 3,453 cases and 3,397 controls; and rs1425802 polymorphism, 5 case-control studies including 6 different cancer types with 3,453 cases and 3,397 controls (Table 1).
Quantitative synthesis
The results of association of CD44 SNPs with a general risk for cancer including data for all individuals are shown in Table 2. Significant association between CD44 polymorphism rs13347 and cancer susceptibility was observed under all genetic models (Fig. 1): allele contrast (T vs. C, OR = 1.391, 95% CI = 1.172–1.650, P < 0.001); dominant model (TT+CT vs. CC, OR = 1.462, 95% CI = 1.176–1.818, P = 0.001); recessive model (TT vs. CT+CC, OR = 1.810, 95% CI = 1.440–2.275, P < 0.001); homozygous model (TT vs. CC, OR = 2.122, 95% CI = 1.576–2.857, P < 0.001); and heterozygous model (CT vs. CC, OR = 1.389, 95% CI = 1.133–1.702, P = 0.002). Additionally, polymorphism rs11821102 was significantly associated with cancer susceptibility in the allele contrast (G vs. A, OR = 0.881, 95% CI = 0.789–0.983, P = 0.024), dominant (GG+GA vs. AA, OR = 0.868, 95% CI = 0.771–0.977, P = 0.019), and heterozygous models (GA vs. AA, OR = 0.864, 95% CI = 0.765–0.976, P = 0.019) (Fig. 2). However, no significant association was identified for polymorphisms rs10836347, rs713330, or rs1425802 and overall cancer susceptibility (Table 2).
Table 2. ORs and 95% CI for cancers and CD44 single nucleotide polymorphisms under different genetic models.
| Genetic models | N | OR [95% CI] | P (OR) | Model (method) | I-square (%) | P (H) | P (Begg) | P (Egger) |
|---|---|---|---|---|---|---|---|---|
| rs13347 | ||||||||
| Allele contrast | 10 | 1.391 [1.172–1.650] | <0.001 | R (D-L) | 86.6 | <0.001 | 0.210 | 0.117 |
| Dominant model | 10 | 1.462 [1.176–1.818] | 0.001 | R (D-L) | 86.9 | <0.001 | 0.210 | 0.081 |
| Recessive model | 10 | 1.810 [1.440–2.275] | <0.001 | R (D-L) | 52.1 | 0.027 | 0.371 | 0.563 |
| Homozygous model | 10 | 2.122 [1.576–2.857] | <0.001 | R (D-L) | 69.9 | <0.001 | 0.210 | 0.374 |
| Heterozygous model | 10 | 1.389 [1.133–1.702] | 0.002 | R (D-L) | 83.5 | <0.001 | 0.283 | 0.053 |
| rs11821102 | ||||||||
| Allele contrast | 7 | 0.881 [0.789–0.983] | 0.024 | F (M-H) | 0.0 | 0.942 | 0.368 | 0.074 |
| Dominant model | 7 | 0.868 [0.771–0.977] | 0.019 | F (M-H) | 0.0 | 0.958 | 0.230 | 0.192 |
| Recessive model | 7 | 0.944 [0.625–1.427] | 0.785 | F (M-H) | 0.0 | 0.821 | 0.133 | 0.088 |
| Homozygous model | 7 | 0.927 [0.613–1.401] | 0.720 | F (M-H) | 0.0 | 0.822 | 0.133 | 0.081 |
| Heterozygous model | 7 | 0.864 [0.765–0.976] | 0.019 | F (M-H) | 0.0 | 0.958 | 0.548 | 0.459 |
| rs10836347 | ||||||||
| Allele contrast | 6 | 0.923 [0.822–1.035] | 0.172 | F (M-H) | 0.0 | 0.865 | 1.000 | 0.423 |
| Dominant model | 6 | 0.927 [0.821–1.047] | 0.223 | F (M-H) | 0.0 | 0.843 | 1.000 | 0.571 |
| Recessive model | 6 | 0.733 [0.413–1.303] | 0.290 | F (M-H) | 0.0 | 0.935 | 0.024 | 0.017 |
| Homozygous model | 6 | 0.728 [0.410–1.294] | 0.279 | F (M-H) | 0.0 | 0.936 | 0.024 | 0.017 |
| Heterozygous model | 6 | 0.936 [0.827–1.060] | 0.299 | F (M-H) | 0.0 | 0.823 | 1.000 | 0.719 |
| rs713330 | ||||||||
| Allele contrast | 5 | 0.942 [0.841–1.056] | 0.307 | F (M-H) | 0.0 | 0.919 | 0.462 | 0.374 |
| Dominant model | 5 | 0.942 [0.833–1.066] | 0.344 | F (M-H) | 0.0 | 0.970 | 0.462 | 0.193 |
| Recessive model | 5 | 0.871 [0.550–1.379] | 0.555 | F (M-H) | 4.7 | 0.380 | 0.806 | 0.885 |
| Homozygous model | 5 | 0.863 [0.544–1.367] | 0.529 | F (M-H) | 3.7 | 0.386 | 0.806 | 0.873 |
| Heterozygous model | 5 | 0.948 [0.835–1.076] | 0.406 | F (M-H) | 0.0 | 0.968 | 0.221 | 0.217 |
| rs1425802 | ||||||||
| Allele contrast | 5 | 1.007 [0.941–1.077] | 0.849 | F (M-H) | 0.0 | 0.485 | 0.462 | 0.188 |
| Dominant model | 5 | 1.013 [0.914–1.122] | 0.811 | F (M-H) | 0.0 | 0.627 | 0.462 | 0.139 |
| Recessive model | 5 | 1.003 [0.894–1.126] | 0.954 | F (M-H) | 28.1 | 0.234 | 0.806 | 0.456 |
| Homozygous model | 5 | 1.009 [0.883–1.152] | 0.900 | F (M-H) | 0.0 | 0.459 | 0.221 | 0.173 |
| Heterozygous model | 5 | 1.014 [0.909–1.132] | 0.801 | F (M-H) | 0.0 | 0.503 | 0.806 | 0.429 |
OR: odds ratio. CI: confidence intervals. N: number of included studies. R: random-effects model. D-L: DerSimonian-Laird method. F: fixed-effect model. M-H: Mantel-Haenszel method. P (H): P for heterogeneity. P-values < 0.05 were considered as statistically significant and are highlighted in bold font in the table.
Figure 1. Forest plot of cancer risk associated with CD44 polymorphism rs13347 using different sources of controls.
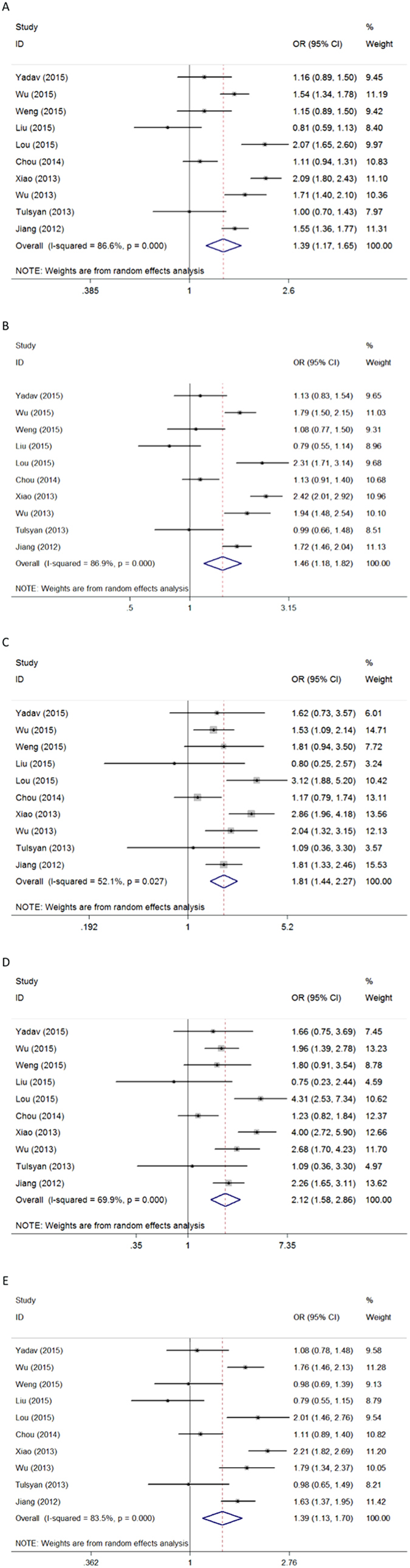
Models represented in (A) allele contrast, (B) dominant, (C) recessive, (D) homozygous, and (E) heterozygous. Each square represents a study, and the area of each square is proportional to the weight of the study. The diamond represents the summary OR and 95% CI. OR: odds ratio; CI: confidence interval.
Figure 2. Forest plot of cancer risk associated with CD44 polymorphism rs11821102 in using different sources of controls.
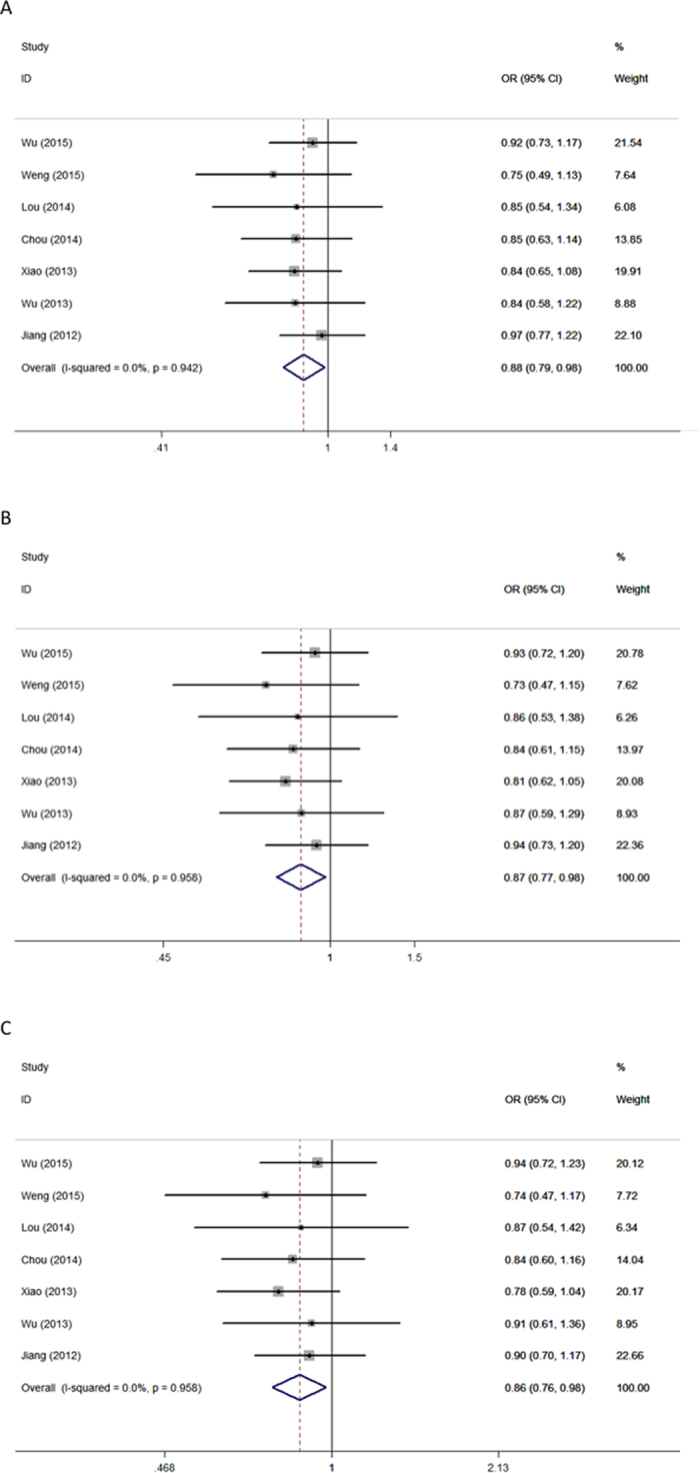
Models represented in (A) dominant and (B) heterozygous. Each square represents a study, and the area of each square is proportional to the weight of the study. The diamond represents the summary OR and 95% CI. OR: odds ratio; CI: confidence interval.
For rs13347, significant between-study heterogeneity was present in all genetic models; therefore, a random-effects model (Der-Simonian Laird) was used. For all other polymorphisms, no significant heterogeneity was found in any genetic model. A fixed-effect model (the Mantel-Haenszel method) was thus applied.
Subgroup analysis
For the rs13347 polymorphism, the studies consisted of individuals of Chinese or Indian descent. However, for other polymorphisms, individuals from all eligible studies were only of Chinese descent, and the number of studies for each cancer type was less than two. Therefore, stratified analysis on the basis of ethnicity, source of controls, and cancer type was only performed on rs13347 (Table 3).
Table 3. Subgroup analyses for CD44 rs13347 and cancers under different genetic models.
| Genetic models | N | OR [95% CI] | P (OR) | Model (method) | I-square (%) | P (H) | P (Begg) | P (Egger) |
|---|---|---|---|---|---|---|---|---|
| Allele contrast | ||||||||
| Overall | 10 | 1.391 [1.172–1.650] | <0.001 | R (D-L) | 86.6 | <0.001 | 0.210 | 0.117 |
| Chinese | 8 | 1.466 [1.218–1.765] | <0.001 | R (D-L) | 87.8 | <0.001 | — | — |
| Indian | 2 | 1.100 [0.891–1.357] | 0.376 | R (D-L) | 0.0 | 0.523 | — | — |
| PB | 5 | 1.607 [1.367–1.890] | <0.001 | R (D-L) | 78.7 | 0.001 | — | — |
| HB | 5 | 1.181 [0.869–1.605] | 0.288 | R (D-L) | 86.3 | <0.001 | — | — |
| NPC | 2 | 2.085 [1.841–2.361] | <0.001 | R (D-L) | 0.0 | 0.934 | — | — |
| BC | 2 | 1.287 [0.844–1.965] | 0.242 | R (D-L) | 80.5 | 0.024 | — | — |
| Dominant model | ||||||||
| Overall | 10 | 1.462 [1.176–1.818] | 0.001 | R (D-L) | 86.9 | <0.001 | 0.210 | 0.081 |
| Chinese | 8 | 1.571 [1.242–1.988] | <0.001 | R (D-L) | 87.7 | <0.001 | — | — |
| Indian | 2 | 1.076 [0.844–1.373] | 0.554 | R (D-L) | 0.0 | 0.610 | — | — |
| PB | 5 | 1.782 [1.451–2.189] | <0.001 | R (D-L) | 78.7 | 0.001 | — | — |
| HB | 5 | 1.182 [0.835–1.673] | 0.347 | R (D-L) | 83.7 | <0.001 | — | — |
| NPC | 2 | 2.392 [2.040–2.806] | <0.001 | R (D-L) | 0.0 | 0.802 | — | — |
| BC | 2 | 1.348 [0.786–2.311] | 0.277 | R (D-L) | 84.0 | 0.012 | — | — |
| Recessive model | ||||||||
| Overall | 10 | 1.810 [1.440–2.275] | <0.001 | R (D-L) | 52.1 | 0.027 | 0.371 | 0.563 |
| Chinese | 8 | 1.858 [1.443–2.393] | <0.001 | R (D-L) | 60.8 | 0.013 | — | — |
| Indian | 2 | 1.417 [0.745–2.697] | 0.288 | R (D-L) | 0.0 | 0.570 | — | — |
| PB | 5 | 1.945 [1.545–2.447] | <0.001 | R (D-L) | 38.4 | 0.165 | — | — |
| HB | 5 | 1.562 [0.948–2.573] | 0.080 | R (D-L) | 63.7 | 0.026 | — | — |
| NPC | 2 | 2.954 [2.181–4.001] | <0.001 | R (D-L) | 0.0 | 0.787 | — | — |
| BC | 2 | 1.746 [1.300–2.346] | <0.001 | R (D-L) | 0.0 | 0.388 | — | — |
| Homozygous model | ||||||||
| Overall | 10 | 2.122 [1.576–2.857] | <0.001 | R (D-L) | 69.9 | <0.001 | 0.210 | 0.374 |
| Chinese | 8 | 2.248 [1.624–3.110] | <0.001 | R (D-L) | 74.6 | <0.001 | — | — |
| Indian | 2 | 1.437 [0.752–2.748] | 0.273 | R (D-L) | 0.0 | 0.543 | — | — |
| PB | 5 | 2.489 [1.876–3.301] | <0.001 | R (D-L) | 55.7 | 0.061 | — | — |
| HB | 5 | 1.642 [0.877–3.074] | 0.121 | R (D-L) | 75.9 | 0.002 | — | — |
| NPC | 2 | 4.106 [3.002–5.617] | <0.001 | R (D-L) | 0.0 | 0.827 | — | — |
| BC | 2 | 1.917 [1.050–3.501] | 0.034 | R (D-L) | 35.6 | 0.213 | — | — |
| Heterozygous model | ||||||||
| Overall | 10 | 1.389 [1.133–1.702] | 0.002 | R (D-L) | 83.5 | <0.001 | 0.283 | 0.053 |
| Chinese | 8 | 1.482 [1.190–1.846] | <0.001 | R (D-L) | 84.3 | <0.001 | — | — |
| Indian | 2 | 1.040 [0.808–1.339] | 0.758 | R (D-L) | 0.0 | 0.727 | — | — |
| PB | 5 | 1.686 [1.390–2.044] | <0.001 | R (D-L) | 73.4 | 0.005 | — | — |
| HB | 5 | 1.124 [0.833–1.518] | 0.445 | R (D-L) | 76.1 | 0.002 | — | — |
| NPC | 2 | 2.152 [1.821–2.544] | <0.001 | R (D-L) | 0.0 | 0.613 | — | — |
| BC | 2 | 1.312 [0.801–2.150] | 0.281 | R (D-L) | 79.5 | 0.027 | — | — |
OR: odds ratio. CI: confidence intervals. N: number of included studies. R: random-effects model. D-L: DerSimonian-Laird method. P (H): P for heterogeneity. PB: population-based. HB: hospital-based. NPC: nasopharyngeal carcinoma. BC: breast cancer. P-values < 0.05 were considered as statistically significant and are highlighted in bold font in the table.
In an ethnicity subgroup analysis, polymorphism rs13347 was significantly associated with cancer susceptibility for Chinese: allele contrast (T vs. C, OR = 1.466, 95% CI = 1.218–1.765, P < 0.001); dominant model (TT+CT vs. CC, OR = 1.571, 95% CI = 1.242–1.988, P < 0.001); recessive model (TT vs. CT+CC, OR = 1.858, 95% CI = 1.443–2.393, P < 0.001); homozygous model (TT vs. CC, OR = 2.248, 95% CI = 1.624–3.110, P < 0.001); and heterozygous model (CT vs. CC, OR = 1.482, 95% CI = 1.190–1.846, P < 0.001). No associations were observed between the CD44 polymorphisms examined and Indians. The magnitude of association in hospital-based studies was significantly weakened for all genetic models. However, the magnitude of association in population-based studies was not significantly changed: allele contrast (T vs. C, OR = 1.607, 95% CI = 1.367–1.890, P < 0.001); dominant model (TT+CT vs. CC, OR = 1.782, 95% CI = 1.451–2.189, P < 0.001); recessive model (TT vs. CT+CC, OR = 1.945, 95% CI = 1.545–2.447, P < 0.001); homozygous model (TT vs. CC, OR = 2.489, 95% CI = 1.876–3.301, P < 0.001); and heterozygous model (CT vs. CC, OR = 1.686, 95% CI = 1.390–2.044, P < 0.001). When the stratification analysis was conducted based on cancer type, we uncovered that the T allele was significantly associated with an increased susceptibility to colorectal cancer, nasopharyngeal carcinoma, and acute myeloid leukemia in all genetic models, and an increased risk for breast cancer only in recessive and homozygous models.
Sensitivity analysis
In order to examine the influence of individual data sets on the pooled ORs, single studies were sequentially excluded from analysis. Pooled ORs were persistent, indicating that the results were statistically stable and robust (data not shown). Sensitivity analysis of the rs13347 polymorphism in an allelic comparison and the rs11821102 polymorphism in the heterozygous model are presented in Fig. 3.
Figure 3. Sensitivity analysis of association between CD44 polymorphisms and cancer risk.
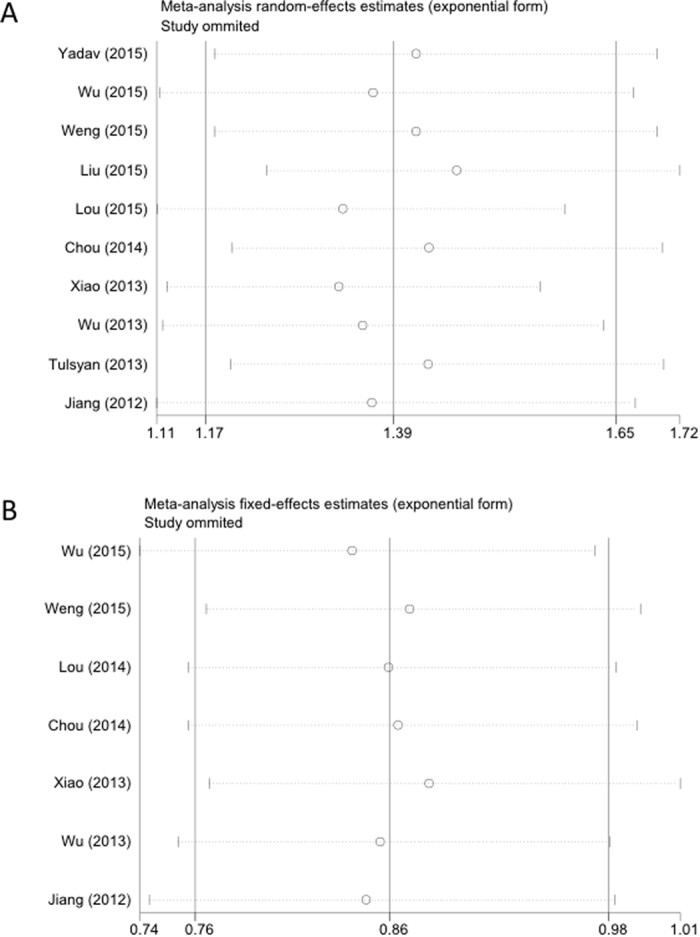
Polymorphisms represented in (A) rs13347 under the allele contrast model and (B) rs11821102 under the heterozygous model.
Publication bias
The Begg’s and the Egger’s tests were performed to quantitatively evaluate publication bias of the included studies. Asymmetry was not observed in the funnel plots of any of the polymorphisms. The funnel plots for the rs13347 polymorphism in an allelic comparison and the rs11821102 polymorphism in the heterozygous model are presented in Fig. 4, and all P-values from the two tests are listed in Table 2. These results were consistent with the absence of significant publication bias in the meta-analysis except in the case of polymorphism rs10836347. For this polymorphism, publication bias was apparent in the recessive and homozygous models. However, analysis with the trim and fill method demonstrated that the results of our study did not significantly change even after adjusting for the publication bias.
Figure 4. Begg’s funnel plot assessing evidence of publication bias in eligible studies used in the study.
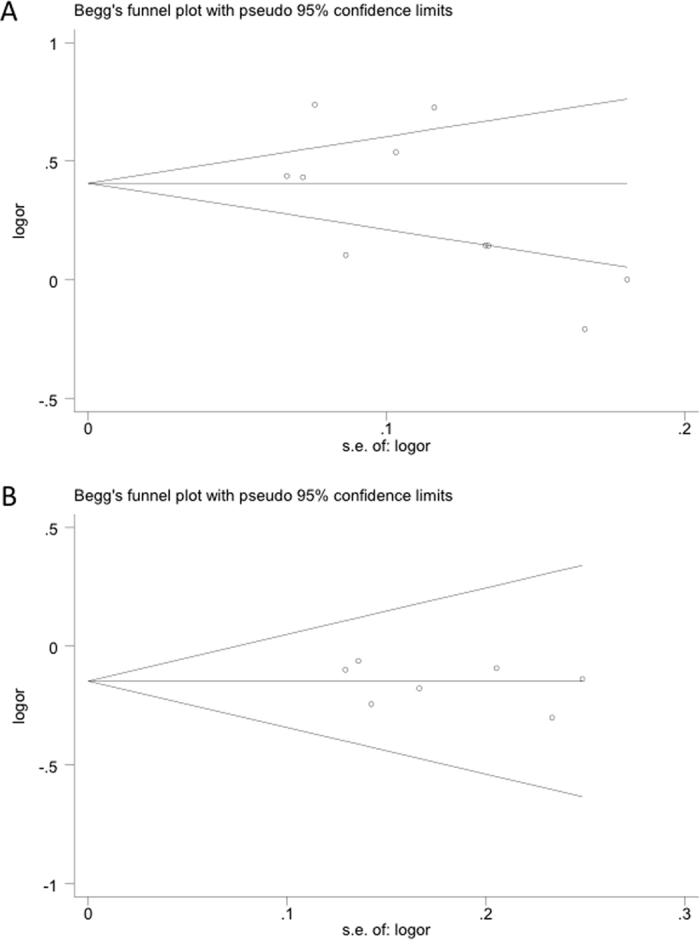
Polymorphisms represented in (A) rs13347 under the allele contrast model and (B) rs11821102 under the heterozygous model. The horizontal line in the figure represents the overall estimated log (OR). The two diagonal lines indicate the pseudo 95% confidence limits of the effect estimate. Log (OR): log-transformed OR; OR: odds ratio.
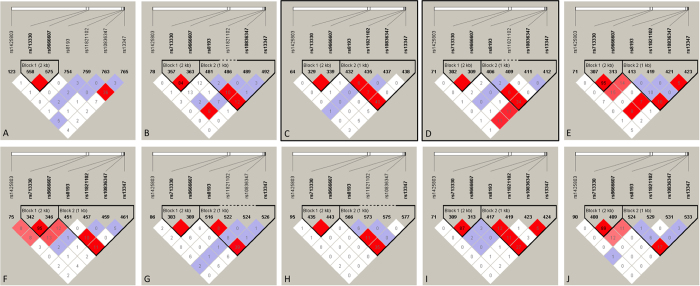
LD analysis across populations
In order to investigate the relationship between polymorphisms in the different populations, LD analysis was performed to test for the existence of bins in the region containing the seven CD44 polymorphisms evaluated, including rs10836347, rs11821102, rs13347, rs1425803, rs713330, rs8193, and rs9666607 (Fig. 5). Relatively low LD values between rs13347 and rs9666607 polymorphisms (D’ = 0.709, r2 = 0.07) were calculated for ASW populations, while complete LD (D’ > 0.90) was observed for the other ethnic populations examined. The rs713330 polymorphism was in complete LD with rs9666607 (D’ = 1.0, r2 > 0.80) for all populations. LD plots for the ASW population yielded a high LD value between rs13347 and rs11821102 polymorphisms, moderate LD values for CEU, FIN, GIH, ITU, and TSI populations, and lower LD values for CHB, CHS, GBR, and JPT populations.
Figure 5. LD analyses for CD44 polymorphisms in populations from the 1000 genomes Phase 3.
The number in each cell represents the r2 value, with red, blue, and white cells representing complete LD or high LD values, moderate LD values, and low LD values or no LD between polymorphisms, respectively. Population descriptors are represented in (A) ASW: African ancestry in Southwest USA; (B) CEU: Utah residents with Northern and Western European ancestry; (C) CHB: Han Chinese in Beijing, China; (D) CHS: Southern Han Chinese, China; (E) FIN: Finnish in Finland; (F) GBR: British in England and Scotland; (G) GIH: Gujarati Indians in Houston, Texas; (H) ITU: Indian Telugu in the UK; (I) JPT: Japanese in Tokyo, Japan; (J) TSI: Toscani in Italy. The rs numbers are SNP IDs taken from National Center for Biotechnology Information (NCBI). LD: linkage disequilibrium; SNP: single nucleotide polymorphism.
Discussion
The genetic basis for the development of cancer involves inherited as well as somatic components. While familial predispositions to cancer have been more easily identified, those genetic factors more generally affecting populations in conjunction with environmental factors have been more difficult to identify. Thus, the focus has become polymorphisms in well-known cancer associated genes, such as CD44. The meta-analysis performed here with data pooled from 5,788 cases and 5,852 controls supports previous findings that a significant association between CD44 polymorphism rs13347 and cancer risk exists under all genetic models. Thus, the T allele emerged as a risk marker for cancers, especially in Chinese, which is a finding consistent with the results of most of the included studies.
The full-length CD44 gene is located on chromosome 11 and consists of 20 exons and 19 introns1,5. Transcripts for the CD44 gene undergo complicated alternative splicing, which results in many functionally distinct protein isoforms, such as CD44 standard (CD44s) and CD44 variant isoforms (CD44v), which account for the heterogeneity in this protein family1,4,5. It is widely accepted that CD44 is expressed as multiple transcriptional variants, but the 3′UTR, which is ~3000 nucleotides long, is in fact relatively conserved2,14. The CD44 polymorphism rs13347 has been the subject of intense investigation in cancer susceptibility studies primarily due to its location in the 3′UTR of the CD44 gene/transcripts10,13,14,16,17. The polymorphism rs13347 may lead to alterations in mRNA and/or protein levels of CD44 due to the change of the C and the T allele. In transient transfections using a construct in which the reporter gene is modulated by the CD44 3′ UTR, the presence of the rs13347 T allele led to increased transcriptional activity relative to the C allele13,15,16,17. Similarly, by immunohistochemistry and Western blotting, carriers of the rs13347 variant genotypes (CT and TT) were shown to exhibit dramatically increased CD44 than CC carriers in breast cancer, acute myeloid leukemia, nasopharyngeal carcinoma, and colorectal cancer13,15,16,17. Additionally, functional analyses demonstrated that hsa-mir-509–3p binds and negatively regulates the transcription of CD44 more strongly in C allele carriers than in T allele carriers13,15,16,17. These results support previous studies that have implicated hsa-mir-509–3p as a tumor suppressor gene in the development of human cancer29,30,31.
In addition, when the stratification analyses were conducted based on ethnicity and source of control for the rs13347 polymorphism, we identified a significant association in all the genetic models for Chinese but not Indians, only in population-based studies. Moreover, we performed LD analyses to determine the LD between the related polymorphisms. The rs713330 polymorphism was in complete LD with rs9666607 for all populations. Similarly, the high LD values between rs13347 and rs9666607 polymorphisms were observed in all populations except the ASW population. LD plots for the ASW population yielded a high LD value between rs13347 and rs11821102 polymorphisms, for CEU, FIN, GIH, ITU, and TSI populations, moderate LD values, and for CHB, CHS, GBR, and JPT populations, lower LD values. The limited number of studies and sample size may account for this discrepancy; therefore, future studies are necessary to verify these results.
None of the eligible studies for CD44 polymorphism rs11821102 indicate a positive correlation with statistical power associated with cancer; yet pooled analysis demonstrated that the minor allele of rs11821102 polymorphism might be related to a decreased cancer risk in Chinese. The polymorphism rs11821102 is also located in 3′UTR of the CD44 gene13,17; however, no functional assay has been performed to explore how this polymorphism influences CD44 expression or function to date. The limited number of studies and sample size may account for this discrepancy; therefore, larger, preferably population-based case-control studies, as well as mechanistic studies involving CD44 polymorphism rs11821102, are warranted to validate our findings and to further investigate the role of CD44 in the development of cancer.
Although we have conducted a comprehensive retrieval for all eligible studies and polymorphisms, which markedly expanded the sample size and revealed some associations not observed in previous work, some limitations in this study exist. First, statistically significant heterogeneity was confirmed in rs13347 but not in other polymorphisms within all genotype models, and stratified analysis was used to explore the source of heterogeneity. Subgroup analysis by source of controls demonstrated that the between-study heterogeneity was still significant. When subgroup analysis was performed by ethnicity, the heterogeneity was dramatically reduced in Indians but not in Chinese. However, stratified analysis based on cancer type demonstrated that the heterogeneity was not significant in studies performed on nasopharyngeal carcinoma under all genetic models or on breast cancer under recessive and homozygous models. Therefore, the between-study heterogeneity might be mainly attributed to various cancer types. Second, our analysis was limited to individuals of Asian descent, mainly Chinese. LD between different polymorphisms varied across diverse ethnic populations. It, therefore, remains uncertain as to whether the result can be generalized to other ethnic populations.
To the best of our knowledge, this is the first meta-analysis to date to investigate the associations of CD44 polymorphisms with cancer risk. In summary, our study suggests CD44 polymorphism rs13347 can serve as a risk factor for cancer, particularly in Chinese. Our findings also indicate that the minor allele of rs11821102 polymorphism may be associated with a decreased susceptibility to cancer in Chinese.
Additional Information
How to cite this article: Qi, Q. et al. Associations of five polymorphisms in the CD44 gene with cancer susceptibility in Asians. Sci. Rep. 6, 39485; doi: 10.1038/srep39485 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by the Natural Science Foundation of China (grants 81402060 and 81572487), the Shandong Provincial Natural Science Foundation (grants BS2014YY033 and BS2012YY016), the Special foundation for Taishan Scholars (grants ts20110814 and tshw201502056), the Fundamental Research Funds of Shandong University, the Department of Science & Technology of Shandong Province (grants 2015GGE27101 and 2015ZDXX0801A01), the University of Bergen, Helse Bergen, Norway, and the Norwegian Centre for International Cooperation in Education (SIU)(UTF-2014/10047).
Footnotes
Author Contributions X.G.L. and J.W. designed the study. Q.C.Q. and J.W.W. contributed to the collection, analysis and interpretation of data as well as drafting the manuscript. A.J.C., B.H., and G.L. also analyzed data and contributed to the writing of the manuscript. Q.C.Q. and J.W.W. prepared all figures and tables. All authors reviewed the manuscript.
References
- Yan Y., Zuo X. & Wei D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl Med 4, 1033–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horta S. et al. Looking out for cancer stem cells’ properties: the value-driving role of CD44 for personalized medicines. Curr Cancer Drug Targets 14, 832–49 (2015). [DOI] [PubMed] [Google Scholar]
- Basakran N. S. CD44 as a potential diagnostic tumor marker. Saudi Med J 36, 273–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazka L., Tesarik R. & Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell Signal 26, 2234–9 (2014). [DOI] [PubMed] [Google Scholar]
- Xu H. et al. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco Targets Ther 8, 3783–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. et al. Knockdown of CD44 inhibits the invasion and metastasis of hepatocellular carcinoma both in vitro and in vivo by reversing epithelial-mesenchymal transition. Oncotarget 6, 7828–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 11, 254–67 (2011). [DOI] [PubMed] [Google Scholar]
- Schmitt M., Metzger M., Gradl D., Davidson G. & Orian-Rousseau V. CD44 functions in Wnt signaling by regulating LRP6 localization and activation. Cell Death Differ 22, 677–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L. Y., Spevak C. C., Wong G., Xia W. & Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem 284, 26533–46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. et al. CD44 regulates pancreatic cancer invasion through MT1-MMP. Mol Cancer Res 13, 9–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza I. & Miele L. Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett 341, 41–5 (2013). [DOI] [PubMed] [Google Scholar]
- Marie-Egyptienne D. T., Lohse I. & Hill R. P. Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: potential role of hypoxia. Cancer Lett 341, 63–72 (2013). [DOI] [PubMed] [Google Scholar]
- Jiang L. et al. CD44 rs13347 C>T polymorphism predicts breast cancer risk and prognosis in Chinese populations. Breast Cancer Res 14, R105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou F., Ma H. N., Xu L., Chen M. & Zhu Y. B. Two polymorphisms of CD44 3′UTR weaken the binding of miRNAs and associate with naso-pharyngeal carcinoma in a Chinese population. Eur Rev Med Pharmacol Sci 18, 2444–52 (2014). [PubMed] [Google Scholar]
- Wu H. et al. Functional polymorphisms in the CD44 gene and acute myeloid leukemia cancer risk in a Chinese population. Mol Carcinog 54, 102–10 (2015). [DOI] [PubMed] [Google Scholar]
- Wu X. M. et al. Functional Genetic Variations at the microRNA Binding-Site in the CD44 Gene Are Associated with Risk of Colorectal Cancer in Chinese Populations. PLoS One 10, e0127557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M. et al. Polymorphisms of CD44 gene and nasopharyngeal carcinoma susceptibility in a Chinese population. Mutagenesis 28, 577–82 (2013). [DOI] [PubMed] [Google Scholar]
- Chou Y. E. et al. CD44 gene polymorphisms on hepatocellular carcinoma susceptibility and clinicopathologic features. Biomed Res Int 2014, 231474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y. E. et al. CD44 gene polymorphisms and environmental factors on oral cancer susceptibility in Taiwan. PLoS One 9, e93692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng W. C. et al. Effect of CD44 gene polymorphisms on risk of transitional cell carcinoma of the urinary bladder in Taiwan. Tumour Biol (2015). [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. Association of CD44 Gene Polymorphism with Survival of NSCLC and Risk of Bone Metastasis. Med Sci Monit 21, 2694–700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K. L. et al. Association of genetic variants of cancer stem cell gene CD44 haplotypes with gallbladder cancer susceptibility in North Indian population. Tumour Biol 35, 2583–9 (2014). [DOI] [PubMed] [Google Scholar]
- Tulsyan S. et al. CD44 gene polymorphisms in breast cancer risk and prognosis: a study in North Indian population. PLoS One 8, e71073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A. et al. Association of cancer stem cell markers genetic variants with gallbladder cancer susceptibility, prognosis, and survival. Tumour Biol (2015). [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–5 (2005). [DOI] [PubMed] [Google Scholar]
- Qiu Y. et al. Genetic association of osteopontin (OPN) and its receptor CD44 genes with susceptibility to Chinese gastric cancer patients. J Cancer Res Clin Oncol 140, 2143–56 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Unique SNP in CD44 intron 1 and its role in breast cancer development. Anticancer Res 30, 1263–72 (2010). [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Nagarkatti P. S., Zhong Y., Zhang J. & Nagarkatti M. Implications of single nucleotide polymorphisms in CD44 exon 2 for risk of breast cancer. Eur J Cancer Prev 20, 396–402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. et al. miR-509-3p is clinically significant and strongly attenuates cellular migration and multi-cellular spheroids in ovarian cancer. Oncotarget (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z. et al. MicroRNA-509-3p inhibits cancer cell proliferation and migration by targeting the mitogen-activated protein kinase kinase kinase 8 oncogene in renal cell carcinoma. Mol Med Rep 12, 1535–43 (2015). [DOI] [PubMed] [Google Scholar]
- Zhai Q. et al. Identification of miR-508-3p and miR-509-3p that are associated with cell invasion and migration and involved in the apoptosis of renal cell carcinoma. Biochem Biophys Res Commun 419, 621–6 (2012). [DOI] [PubMed] [Google Scholar]