Abstract
The Dishevelled/EGL-10/Pleckstrin (DEP) domain-containing (DEPDC) proteins have seven members. However, whether this superfamily can be distinguished from other proteins based only on the amino acid sequences, remains unknown. Here, we describe a computational method to segregate DEPDCs and non-DEPDCs. First, we examined the Pfam numbers of the known DEPDCs and used the longest sequences for each Pfam to construct a phylogenetic tree. Subsequently, we extracted 188-dimensional (188D) and 20D features of DEPDCs and non-DEPDCs and classified them with random forest classifier. We also mined the motifs of human DEPDCs to find the related domains. Finally, we designed experimental verification methods of human DEPDC expression at the mRNA level in hepatocellular carcinoma (HCC) and adjacent normal tissues. The phylogenetic analysis showed that the DEPDCs superfamily can be divided into three clusters. Moreover, the 188D and 20D features can both be used to effectively distinguish the two protein types. Motif analysis revealed that the DEP and RhoGAP domain was common in human DEPDCs, human HCC and the adjacent tissues that widely expressed DEPDCs. However, their regulation was not identical. In conclusion, we successfully constructed a binary classifier for DEPDCs and experimentally verified their expression in human HCC tissues.
The Dishevelled (first recognized in Drosophila)1, EGL-10 (first confirmed in Caenorhabditis elegans)2 and Pleckstrin (first identified in mammals)3 (DEP) domain-containing (DEPDC) proteins were discovered to have seven members, namely DEPDC1-DEPDC7. Most of which were found involving in signal transduction4,5,6. The domain is defined as a specific combination of secondary structures organized into a characteristic three-dimensional structure or fold, and generally as a transmembrane region or functional region. Meanwhile, different combinations of domains can generate the diverse range of proteins found in nature. Therefore, the identification of domains that occur within proteins can provide insights into their functions. DEP domains (approximately 80 amino acids) are usually globular protein domains, which may facilitate translocation of the homologous protein to the plasma membrane. The structure of the mouse Dishevelled 1 DEP domain is characterized by a three-helix bundle (H1-H3), a β-hairpin “arm” with two β-strands (B1 and B2) lying between H1 and H2, and two short β-strands (B3 and B4) at the C-terminal region as revealed by nuclear magnetic resonance (Fig. 1)7. Three α-helices composed of highly conserved hydrophobic amino acids stabilize the core structure of this domain. The DEP domain is usually located in the residue range of 402-495. Among these areas, Lys434, Asp445 and Asp448 create a strong electric dipole interaction with regulators upstream of Dvl. This interaction is an important mechanism in the transduction of the Wnt signaling pathway.
Figure 1. Molecular model of mouse Dishevelled 1 DEP domain structure with the lowest target function.
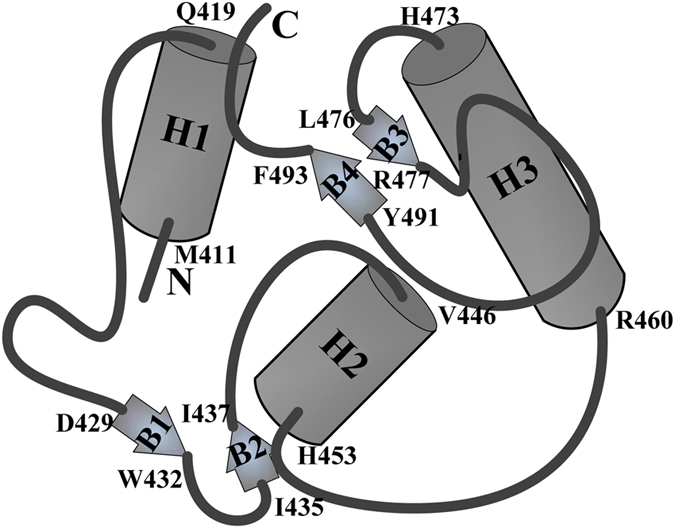
The diagram shows the residue range from the N-terminal 402aa to C-terminal 495aa, including three α helices(H1, H2, and H3) and four β sheets (B1–B4). The numbers in the figure indicate the beginning and end of the corresponding secondary structures. Many hydrophobic residues were situated in H1, H2, H3, B3, B4 to construct the hydrophobic core of DEP domain. Human Dishevelled 1 DEP domain possesses the same structure except for the amino acid position.
DEPDC1 is a highly conserved protein that was first found to over-express in bladder cancer cells8 in 2007; hence, this protein has been identified as a therapy target9. Furthermore, DEPDC1 is related to several types of cancer and contributes to carcinogenesis. For example, DEPDC1 is over-expressed in colorectal cancer10 and up-regulated in lung adenocarcinomas11, and as one of the 16 genes with concomitant genomic alterations that participate in breast cancer12. DEPDC1 could also be involved in the activation of NF-κB cell survival as a transcriptional repressor13. One of its members, DEPDC1B coordinated the de-adhesion events along the DEPDC1B/RhoA/PTPRF axis on mitotic dynamics during zebrafish development14. Moreover, the expression of DEPDC1B, which is a potential Rho GTPase-activating protein correlated to oral cancer6, might be repressed by Pitx2 when contributing to signaling pathways15. Yuan16 discovered the high DEPDC1 expression in hepatocellular carcinoma (HCC) tissues at the mRNA and protein levels. Given the higher DEPDC1 expression in HCC patients, the overall survival and disease free survival rate become poorer. In response to the anti-tubulins in C. elegans, MCL1 (a member of Bcl-2 family) occurrs via an evolutionarily conserved signaling pathway that involves the DEPDC protein LET-99 (DEPDC1 homolog in mammals). This concept suggests that DEPDC1 participates in the anti-tubulin drug-induced apoptotic cell death pathway17,18. Therefore, DEPDC1 is identified as a novel tumor-related gene19.
DEPDC2 is broadly expressed in human and zebrafish20, and it is a candidate molecular marker for human embryonic stem cells21. Three transcription factors (OCT4, SOX2, and NANOG) are required for the transcriptional regulation of DEPDC222, which may be a candidate gene linked to febrile seizures of epilepsy23. DEPDC2 is also called the phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 (PREX2), which is a Rac guanine nucleotide exchange factor (Rac-GEF). Previous reports have stated that PREX2 is over-expressed in human HCCs24,25 and melanomas; PREX2 also acts as a PTEN binding and inhibiting protein26,27. Although some studies have revealed limited information of DEPDC2, its exact function remains largely unknown.
DEPDC3 has no related literature in PubMed, but it is also known as the G-protein-coupled receptor 155 (Gpr155), which is conserved among mammals and may be a candidate gene for type 2 diabetes in mouse models28. As a 17-transmembrane (TM) protein, Gpr155 is widely expressed in adult mouse tissues, and it has a carrier domain, a G-protein-interacting region and a DNA-binding domain29. Gpr155 might also play an important role in the GABAergic neurotransmission involved in sensory information processing and memory30. However, specific information on Gpr155 protein function has not been elucidated.
DEPDC4 is also a signaling molecule involved in G-protein-coupled receptor (GPCR) signaling pathways. The gene sequences of this protein were disrupted by rearrangement in the gibbon genome during mapping around the breakpoint regions, as compared with the human genome31. Hawthorne32 reported that one single nucleotide polymorphism (SNP) in its 3′-untranslated region was associated with the high-grade myopia MYP3 gene. However, the function of DEPDC4 remains largely unknown.
DEPDC5 is a protein that is strongly associated with various of familial epilepsies33, such as familial focal epilepsies34, familial temporal lobe epilepsy35, and autosomal dominant nocturnal frontal lobe epilepsy36. Pippucci37 discovered that two different truncating mutations of the DEPDC5 gene are involved in uncommon presented focal epilepsy with auditory features based on whole-exome sequencing. Several DEPDC5-related hereditary mutations are correlated with focal epileptic spasms in a cohort of patients and controls. In addition, DEPDC5 variation might be the most frequent gene in epileptic spasms; its variants are associated with focal cortical dysplasia (FCD) type IIA38. As an important anti-rapamycin regulatory gene, DEPDC5 mutations play an inhibitory role in the malformations of cortical development (MCD) of epilepsy. Moreover, DEPDC5 is also a signaling molecule involved in the phosphoinositide 3-kinase (PI3K)-AKT-mTOR pathway and an important component of the GAP activity on RAGs complex1 (GATOR1), which is a negative regulator of mTOR. Research on SNPs suggested that DEPDC5 polymorphisms affect the progression of HBV-related liver disease39. Miki40 identified an intronic SNP variant of DEPDC5 in a Japanese patient with chronic HCV-related HCC. However, when HCV-positive HCC recurrence patients who have undergone hepatectomy, no correlations were found between the DEPDC5 genetic polymorphism and the recurring patients41.
DEPDC6 is also known as the DEPDC mTOR-interacting (DEPTOR) protein, which can only be found in vertebrates. mTOR is a conserved serine/threonine-protein kinase forming mTOR complex [mTORC] 1 and mTORC242. As an important member of mTOR complex, human DEPTOR contains two N-terminal DEP domains and one C-terminal PDZ domain. In addition, DEPTOR is an endogenous inhibitor of both mTORCs via its PDZ domain43. Therefore, DEPTOR is involved in the mTOR-dependent signaling pathway; specifically, DEPTOR inhibits the mTORC1/PI3K pathway and activats Akt, whereas mTOR is a strongly negative regulator of autophagy, which is correlated with diabetes mellitus and energy metabolism44. The DEPTOR-mTOR signaling pathway was regulated by glutamine administration in colitis-associated colorectal cancer mice45. Moreover, DEPTOR binding weakened for both mTOR complexes when mutations in the FRAP-ATM-TTRAP (FAT) domain in clear cell renal cell carcinoma. Consequently, point mutations in the FAT domain promoted mTORC1 and mTORC2 activity, as well as increased cancer cell proliferation, thereby decreasing DEPTOR binding and indicating poor patient prognosis46. Abnormally high DEPTOR expression activated the PI3K-AKT pathway and was regarded as a poor prognostic biomarker, which has been found in various types of solid neoplasms such as esophageal squamous cell carcinoma47, breast cancer48,49, and HCC50. However, DEPTOR was also reported playing dichotomous functions of the proliferation and metastasis in breast cancer51.
Three years ago, DEPDC7 was called LOC91614 and relatively unknown; however, our group has given close attention to this gene and studied it for several years. Our previous studies showed the high level of DEPDC7 differential expression in HCC tissues and hepatoma cell lines; the gene is closely correlated with the proliferation, migration, and invasion capacity as verified by RNA interference researches52. Furthermore, DEPDC7 can interact with CARMA2 and CARMA3 proteins as a positive regulator as well as active NF-κB signal transduction pathway5. A recent report indicated that DEPDC7 DNA intron hypomethylation may be correlated with depression53, DEPDC7 deletion may also be one factor of azoospermia in cryptorchidism patients, thereby implying its influence on reproduction54. Consequently, further studies on DEPDC7 are needed.
Machine learning, developed from computational learning and pattern recognition theory, makes computers able to learn without being explicitly programmed, and it has been widely used to devise prediction models55. Classification machine learning models can be validated by many accuracy estimation methods and evaluated by tools for classification model assessment. Random Forest(RF) is an ensemble classifier that has been proven to be robust in classification issue with high dimensional data, which is often employed in handling bioinformatics problems56. Although the class predictions are averaged multiple deep decision RF trees, the final model prediction is based on the majority vote57.
Based on the sequence and other physicochemical properties of each protein, we discriminated and predicted the DEPDC proteins from non-DEPDC proteins with a machine learning algorithm by extracting the 188D and 20D feature vectors and constructing a binary classifier for this purpose in this study. Subsequently, we searched for the main motifs of human DEPDC proteins, which are related to their functional domains. Finally, we performed experimental verification of human DEPDC gene expression with qRT-PCR in HCC and adjacent normal tissues.
Results
Phylogenetic analysis of positive Pfam corresponding longest protein sequences
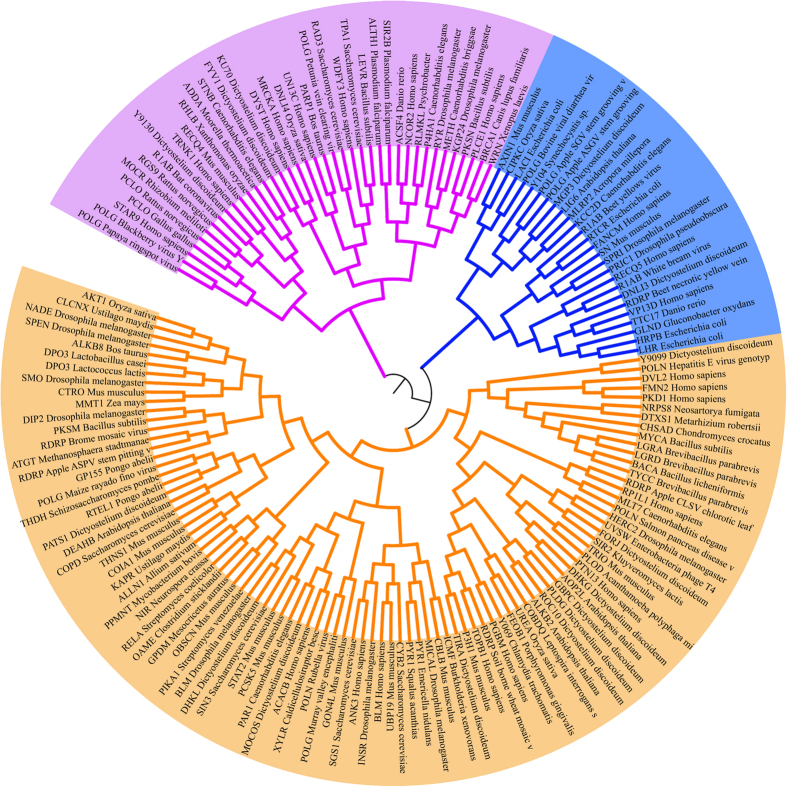
Based on the neighbor-joining algorithm and bootstrap method for phylogeny test, the number of bootstrap replicates was set to 500, and the tree was out-grouped with the “Root On Midpoint” option58. Subsequently, we built a robust circular polar phylogenetic tree of the whole positive 160 Pfam-containing sequence members (Fig. 2). The general presentation of this tree can obviously distinguish all members into three main classes, Cluster I–III, which included a total of 71 species. From the figure, the Cluster Icontained 45 species (right), Cluster II kept 24 species (bottom left), and Cluster III had 19 species (upper left). Among these clusters, the 21 human proteins were distributed between Clusters I and II.
Figure 2. Rooted neighbor-joining tree based on p-distance in a circular polar form as reconstructed from 160 full-length sequences of the positive Pfam family.
Reclassification of positive and negative proteins
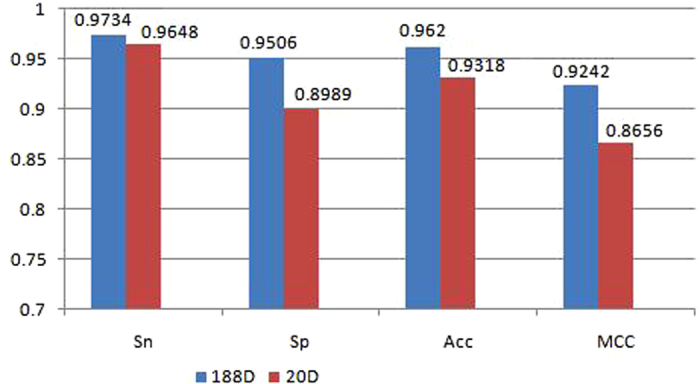
We obtained data on the 188D and 20D feature vectors from the positive and negative groups for import into the Weka explorer. The results showed that the correctly classified rates were 96.2% and 93.2%, whereas the ROC area reached 0.995 and 0.983. The confusion matrix is shown as Table 1. The four common measured features are illustrated in Fig. 3.
Table 1. Confusion matrix from RF classifier with 188D and 20D methods.
| Cases | DEP domain containing proteins(188D) | Non DEP domain containing proteins(188D) | DEP domain containing proteins(20D) | Non DEP domain containing proteins(20D) |
|---|---|---|---|---|
| Positive cases | 11027 | 523 | 10929 | 1070 |
| Negative cases | 301 | 10062 | 399 | 9515 |
Figure 3. Sn, Sp, Acc, and MCC values listed for 188D and 20D methods.
Analysis of human DEPDC proteins for phylogenetics and conserved motif composition
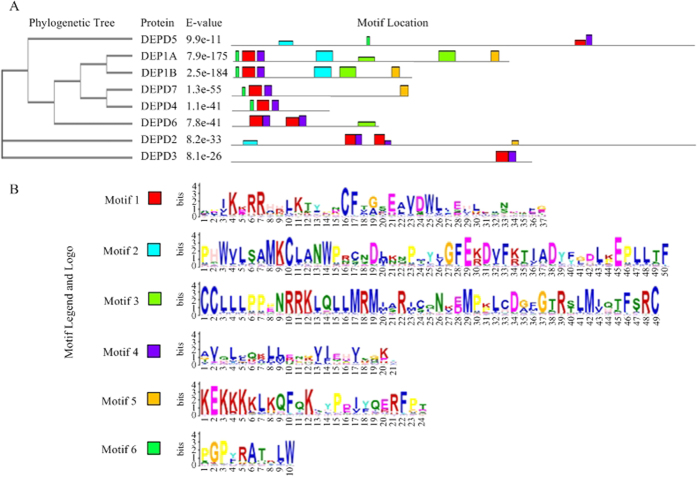
Our phylogenetic analyses with MAFFT revealed that the eight human DEPDC members can be divided into three main groups (Fig. 4 and Table 2). Group I includes DEPDC5, DEPDC1A, DEPDC1B, DEPDC7, DEPDC4, and DEPDC6. Group II includes DEPDC2 only. Group III includes DEPDC3 only. Thus, the six members of group I possess the more sequence similarities than the other two groups. To a certain extent, this indicated that Group I proteins shared evolutionary origins. Among the six searched motifs, Motif 1 and 4 comprise the DEP domain in all members, whereas Motif 2 and 3 are part of the GTPase-activator protein for Rho-like small GTPases (RhoGAP) domain located near the C-terminus. Small GTPases act as molecular switches to control the active or inactive effectors.
Figure 4. Motifs of human DEPDC proteins as found by the MEME/MAST system.
(A) Eight human DEPDC protein sequences were initially used for MSA to construct a phylogenetic tree with the MAFFT program before searching for the motifs with the MEME/MAST software. (B) The corresponding six-motif legends and logos as visualized for human DEPDC proteins (details in Table 2).
Table 2. Conserved motifs of human DEPDC proteins identified by the MEME system.
| Motif | Width | Best possible match |
|---|---|---|
| 1 | 37 | MPIKKRRHHLKTYPNCFTGSEAVDWLYEHLMANDNFG |
| 2 | 50 | PHWVLSAMKCLANWPRCNDMNNPMYVGFEKDVFKTIADYFGDLPEPLLTF |
| 3 | 49 | CCLLLPPPNRRKLQLLMRMMARMCQNKDMPPLCDGFGTRTLMIQTFSRC |
| 4 | 21 | AVQLCQKLMEHHVIEHVTGKW |
| 5 | 24 | KEKKKKLKQFQKCYPDIYQERFPT |
| 6 | 10 | PGPYRATCLW |
Human DEPDC proteins were expressed in HCC and their paired adjacent tissues
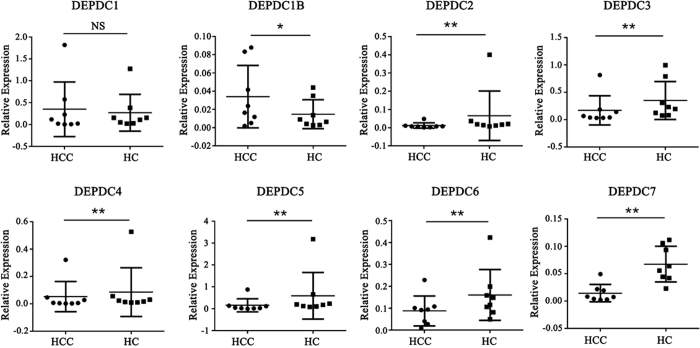
Human DEPDCs were broadly expressed in HCC. Most genes showed significant differential expression (Fig. 5). The expression level of DEPDC genes in HCC samples and paired adjacent normal tissues (HC) were analyzed with quantitative real-time PCR (qPCR). As shown in Fig. 5, DEPDC1B was up-regulated as compared with HC. The other six genes were down-regulated, with the exception of DEPDC1 whose difference was not significant. The combined data suggest the significant variations among DEPDC family genes.
Figure 5. Expression of human DEPDC genes in HCC samples compared with paired adjacent normal tissues (HC) presented as mean ± standard error.
*P < 0.05, **P < 0.01 (nonparametric Mann–Whitney U-test). NS, not significant.
Discussion
The DEPDC protein family includes seven members, which contain the DEP domain as their common characteristic; these proteins can mediate protein-to-protein interaction and membrane targeting of signal molecules5,18,59,60. Consequently, most DEPDCs are involved in signal transduction and closely related to several other signal molecules. For instance, these molecules participate in the Wnt61, mTOR, NF-κB, PI3K-AKT4, and G-protein signaling pathways62. Besides the DEP domain, some DEPDCs contain other domains such as PDZ and GAP, which play various roles in protein-to-protein interaction.
In this study, we performed binary-class classification of the DEPDC and non-DEPDC family. First, a phylogenetic tree was built from all the longest positive Pfam-containing sequences and discovered that the DEPDC family can be divided into three clusters. Based on protein sequence and isual-chemical characteristics, we extracted the 188D and 20D feature vectors on the positive and negative datasets by machine learning, as predicted by RF classifier. We obtained relatively high effects on the correctly classified rates and the AUC value. Similarly, the Sn, Sp, Acc, and MCC values reached relatively superior results. To the best of our knowledge, we are the first to categorize the DEPDC family; thus, no corresponding pre-existing data is available for comparison. Third, the MEME Suite is suitable for screening common sequence motifs from a set of amino acid sequences63. Thus, we applied this technique to analyze the main motifs of human DEPDCs and found that these proteins can be divided into three groups by phylogenetic analyses. Group I contains six of the eight DEPDC members, thereby suggesting their closer relationship and evolutionary origin. On the other hand, some motifs comprise the DEP and RhoGAP domains.
Finally, we also used experimental methods to verify the gene expression of the human DEPDC family in HCC and adjacent normal tissues. From the qRT-PCR results, we observed that human DEPDCs were widely expressed in the cancer tissue. However, direction of the expression was not identical. Our experiment showed that DEPDC1 expression was not affected between the two tissues. DEPDC1B was up-regulated in HCC, whereas the other six genes (DEPDC2- DEPDC7) were down-regulated in HCC. This trend may indicate their different functional correlation and great variation. Further study on the regulation and function of these genes with HCC or other related cancers is needed.
In conclusion, we have successfully constructed a binary-class model algorithm to split the DEPDCs with non-DEPDCs, and the 160 positive Pfam-containing members can be differentiated into three main clusters. The eight human DEPDCs can be divided into three groups from molecular phylogenetic analyses and the conserved DEP and RhoGAP domains were discovered by MEME tools. Finally, we verified the eight human DEPDCs mRNA expression with experiment in human HCC tissues.
Methods
Data retrieval and treatment
The primary sequences of DEPDC proteins and the control Pfam proteins (as FASTA files) were retrieved from the Universal Protein Resource (UniProt) database (www.uniprot.org)64. To reduce the sequence redundancy and improve analytical performance, the raw data were preprocessed by the CD-HIT program (http://cd-hit.org) for merging the sequence similarities65, which has been widely used in bioinformatics66. In the present study, the sequence identity cut-off was set at 0.90, whereas the other default parameters were used to avoid bias during categorization. We obtained 1416 DEPDC sequences for the positive dataset; the negative samples were identified from the control proteins after removing the positive samples. Finally, we acquired 10585 entries as negative dataset.
Multiple sequence alignment (MSA) and phylogenetic tree construction based on positive Pfam
All positive DEPDCs were applied to extract their corresponding protein families (returned Pfam number) from the Uniprot “Family & Domains” section. After excluding the identical and redundant entries, we acquired 229 unique Pfam numbers (the names begin with PF). To construct a phylogenetic tree67,68,69, we initially extracted the longest sequence from each Pfam containing member of the positive dataset (in.fasta format) and identified 160 sequences by combining the same records. Second, these sequences were subjected to perform MSA with default parameters of CLUSTAL X2.170. We performed the “Do Complete Alignment” and provided the resulting alignments in an.aln CLUSTAL file as output. Finally, the.aln file was transformed into a.meg format for the MEGA 6 program71 to construct a neighbor-joining tree with p-distance model. Other default parameters were kept, whereas phylogenetic analysis was isualized in a circular polar tree.
Prediction analyses of DEPDCs
To fully uncover the important information hidden in protein sequences, we constructed two feature extraction algorithms: the 188-dimensional(188D) and the simplified 20D feature vectors. First, we extracted the feature vectors from the positive and negative protein sequence dataset by a previously developed novel machine learning method72,73,74. We transformed all the positive and negative datasets into the corresponding protein family information (Pfam number files). Meanwhile, the sequence evolutional information, the k-skip-n-gram model, physicochemical properties, local PsePSSM, and other features were obtained75. Subsequently, we extracted the longest sequence from both datasets of each Pfam protein family(as.fasta files). Finally, the.fasta files were assembled into 188D and 20D feature vectors. A detailed description of the methods are shown in refs 72 and 76.
After the above mentioned process, the resulting feature vectors are imported into weka (http://www.cs.waikato.ac.nz/ml/weka/), which is a machine learning workbench. In weka, we filtered the vector data with the synthetic minority over-sampling technique(SMOTE)77,78,79 and changed the positive instances from the 100% into 700% to overcome the highly imbalanced data. the vector data were automatically classified by visualization and cross-validation analysis80,81,82,83,84,85. Based on the optimal features in some preliminary trials on the same dataset, we finally selected RF module to distinguish the two classes and utilize the ten-fold cross-validation model.
To measure the performance quality of the statistical classification more intuitively in machine learning, we calculated four common parameters for evaluating the classifier: sensitivity (Sn), specificity (Sp), accuracy (Acc) and Matthew’s correlation coefficient (MCC). The formulas of these parameters are86,87,88,89:
 |
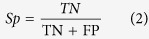 |
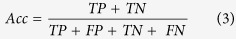 |
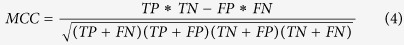 |
where TP(true positive) indicates the number of true DEPDCs that are correctly predicted, TN(true negative) indicates the number of true non-DEPDCs that are correctly predicted, FP(false positive) is the number of true non-DEPDCs that are predicted to be DEPDCs, and FN(false negative) is the number of true DEPDCs that are predicted to be non-DEPDCs.
Conserved motif composition analysis of human DEPDCs
Before motif searching, the eight human DEPDCs (DEPDC1A, DEPDC1B, DEPDC2-7) were implemented by MSA and forwarded to the MAFFT server (http://www.ebi.ac.uk/Tools/msa/mafft/) to generate a phylogenetic tree. MAFFT has been significantly improved in accuracy and reduced CPU time as compared with CLUSTALW; the algorithm is also faster than T-COFFEE90,91. The default parameters were kept unchanged. The MEME Suite (http://meme-suite.org/, 4.11.2 version) was performed for conserved motif analysis. A motif can be assumed as a conserved sequence pattern that repeatedly occurs in a group of related sequences. MEME is a powerful motif-based sequence analysis suite, which can promptly discover novel, ungapped motifs by integrating various sequence analysis tools for proteins, DNA and RNA92. The maximum motif number was set to 6 and the remaining parameters were set as default values.
Gene expression analysis for experimental verification with quantitative real-time PCR in human tissues Patients and samples
Pathological sections were obtained from 8 patients with HCC at the Fuzhou Dongfang Hospital in 2015. The pathology slides and institutional pathology reports were reviewed by the pathologists by following the evidence-based practice guidelines in the standardized pathological diagnosis of primary liver cancer.
Gene expression analysis with quantitative real-time PCR
The primers were designed by the PrimerQuest tool of IDT, which is freely available at https://sg.idtdna.com/primerquest/Home/Index. A BLAST search of the sequences was used to assure that only the selected gene were targeted. The gene and sequence information of the primers are presented in Table 3. The sequences were synthesized by Sangon Biotech. The total RNA was isolated from formalin-fixed paraffin-embedded (FFPE) samples with the FFPE RNA Purification Kit (AmoyDx, China), according to the manufacturer’s instructions. cDNA synthesis was performed on 5 μg of RNA in a 100 μL sample volume with the PrimeScriptTM RT reagent Kit (Takara), as recommended by the manufacturer. Real-time PCR was performed on the Step-OneTM Real-Time PCR system (Applied Biosystems) with the SYBER Green qPCR Supermix (Roche) under universal thermal cycling parameters (95 °C for 30 sec, 40 cycles of 30 sec at 95 °C and 5 sec at 60 °C). The comparative Ct method was used to quantify gene expression93. The target gene expression level was normalized to the expression of the housekeeping gene β-actin within the same sample (−ΔCt), where the relative expression of each gene was calculated with 2−ΔCt.
Table 3. Human DEPDC and internal control genes and their primer sequences information.
| Gene | Primer sequences | Tm(°C) | %GC | Product(bp) |
|---|---|---|---|---|
| DEPDC1-F | GAAGCAGTGGATTGGCTTTATG | 62 | 45.5 | 136 |
| DEPDC1-R | CCCACCTCCCTTTGATATCTTC | 62 | 50.0 | |
| DEPDC1B-F | GGAAATTCTGAAAGTCCCTTTGG | 62 | 43.5 | 98 |
| DEPDC1B-R | CCATATCAGCTCCTGGGTATTT | 62 | 45.5 | |
| DEPDC2-F | GAGCACAAAGCCAAGAGAGA | 62 | 50.0 | 100 |
| DEPDC2-R | TCCTACAGCATGCACAACAG | 62 | 50.0 | |
| DEPDC3-F | GCAGAGAAATGGTGGAACTCT | 62 | 47.6 | 105 |
| DEPDC3-R | CTCCTGGTGCTACAGGAAATAC | 62 | 50.0 | |
| DEPDC4-F | GAACCGTAGAGATGGCTTCTG | 62 | 52.4 | 101 |
| DEPDC4-R | GGGCCTGAAGAGAGTGAATAAT | 62 | 45.5 | |
| DEPDC5-F | CTCCTGTGGCTTCTTGTTAGT | 62 | 50.0 | 106 |
| DEPDC5-R | TGATGTTGAGTGGGATGAAGAG | 62 | 50.0 | |
| DEPDC6-F | TTGTGGTGCGAGGAAGTAAG | 62 | 47.6 | 107 |
| DEPDC6-R | CCGTTGACAGAGACGACAAA | 62 | 45.5 | |
| DEPDC7-F | ACCTTCCACTTCTTGACTCCTTAC | 57.8 | 45.8 | 155 |
| DEPDC7-R | CGAGAGCCACTCATCTTCCTG | 57.5 | 57.1 | |
| β-actin-F | CGTGCGTGACATTAAGGAGAAG | 57.2 | 50.0 | 176 |
| β-actin-R | GGAAGGAAGGCTGGAAGAGTG | 57.5 | 57.1 |
Note: F, forward; R, reverse.
Statistical analysis
The nonparametric Mann–Whitney U-test94 was applied for the statistical comparison of normal and cancerous tissues. The significance level was P < 0.05.
Ethical Statements
The study is approved by the ethics committee of the Fuzhou Dongfang Hospital and the experimentation is conducted in accordance with the Declaration of Helsinki and Good Clinical Practice (GCP).
Additional Information
How to cite this article: Liao, Z. et al. Identification of DEP domain-containing proteins by a machine learning method and experimental analysis of their expression in human HCC tissues. Sci. Rep. 6, 39655; doi: 10.1038/srep39655 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
The work was supported by the Natural Science Foundation of Fujian Province of China (No. 2016J01152), and National Natural Science Foundation of China (No. 61370010, No. 61573235, No. 61272315, No. 61302139).
Footnotes
Author Contributions Conceived and designed the experiments: Q.Z., Z.L. Performed the experiments: Z.L., X.W., Y.Z. Analyzed the data: Q.Z., Z.L., X.W. Wrote the paper: Q.Z., Z.L., X.W. All authors read and approved the final manuscript.
References
- Nagahata T. et al. Amplification, up-regulation and over-expression of DVL-1, the human counterpart of the Drosophila disheveled gene, in primary breast cancers. Cancer science 94, 515–518 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle M. R. & Horvitz H. R. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84, 115–125 (1996). [DOI] [PubMed] [Google Scholar]
- Kharrat A. et al. Conformational stability studies of the pleckstrin DEP domain: definition of the domain boundaries. Biochimica et biophysica acta 1385, 157–164 (1998). [DOI] [PubMed] [Google Scholar]
- Dibble C. C. & Cantley L. C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol 25, 545–555, doi: 10.1016/j.tcb.2015.06.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea E. L. et al. The Dishevelled, EGL-10 and pleckstrin (DEP) domain-containing protein DEPDC7 binds to CARMA2 and CARMA3 proteins, and regulates NF-kappaB activation. PloS One 9, e116062, doi: 10.1371/journal.pone.0116062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. F. et al. A putative novel protein, DEPDC1B, is overexpressed in oral cancer patients, and enhanced anchorage-independent growth in oral cancer cells that is mediated by Rac1 and ERK. J Biomed Sci 21, 67, doi: 10.1186/s12929-014-0067-1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C. et al. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nature structural biology 7, 1178–1184, doi: 10.1038/82047 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehira M. et al. Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis. Oncogene 26, 6448–6455, doi: 10.1038/sj.onc.1210466 (2007). [DOI] [PubMed] [Google Scholar]
- Obara W. et al. Cancer peptide vaccine therapy developed from oncoantigens identified through genome-wide expression profile analysis for bladder cancer. Japanese journal of clinical oncology 42, 591–600, doi: 10.1093/jjco/hys069 (2012). [DOI] [PubMed] [Google Scholar]
- Miyata Y. et al. Clinicopathological significance and prognostic value of Wilms’ tumor gene expression in colorectal cancer. Cancer biomarkers: section A of Disease markers 15, 789–797, doi: 10.3233/CBM-150521 (2015). [DOI] [PubMed] [Google Scholar]
- Okayama H. et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer research 72, 100–111, doi: 10.1158/0008-5472.CAN-11-1403 (2012). [DOI] [PubMed] [Google Scholar]
- Colak D. et al. Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women. PloS one 8, e63204, doi: 10.1371/journal.pone.0063204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y. et al. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer research 70, 5829–5839, doi: 10.1158/0008-5472.CAN-10-0255 (2010). [DOI] [PubMed] [Google Scholar]
- Marchesi S. et al. DEPDC1B coordinates de-adhesion events and cell-cycle progression at mitosis. Developmental cell 31, 420–433, doi: 10.1016/j.devcel.2014.09.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. et al. Identification of the GTPase-activating protein DEP domain containing 1B (DEPDC1B) as a transcriptional target of Pitx2. Experimental cell research 333, 80–92, doi: 10.1016/j.yexcr.2015.02.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S. G., Liao W. J., Yang J. J., Huang G. J. & Huang Z. Q. DEP domain containing 1 is a novel diagnostic marker and prognostic predictor for hepatocellular carcinoma. Asian Pacific journal of cancer prevention: APJCP 15, 10917–10922 (2014). [DOI] [PubMed] [Google Scholar]
- Denning D. P. & Hirose T. Anti-tubulins DEPendably induce apoptosis. Nature cell biology 16, 741–743, doi: 10.1038/ncb3012 (2014). [DOI] [PubMed] [Google Scholar]
- Sendoel A. et al. DEPDC1/LET-99 participates in an evolutionarily conserved pathway for anti-tubulin drug-induced apoptosis. Nature cell biology 16, 812–820, doi: 10.1038/ncb3010 (2014). [DOI] [PubMed] [Google Scholar]
- Mi Y. et al. DEPDC1 is a novel cell cycle related gene that regulates mitotic progression. BMB reports 48, 413–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons P. J., Ma L. H., Baker R. & Fricker L. D. Carboxypeptidase A6 in zebrafish development and implications for Vith cranial nerve pathfinding. PloS one 5, e12967, doi: 10.1371/journal.pone.0012967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni R., Cordero E. A., Henriques J. A. & Bonatto D. Reviewing and updating the major molecular markers for stem cells. Stem cells and development 22, 1455–1476, doi: 10.1089/scd.2012.0637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wei H. & Zhao P. X. DeGNServer: deciphering genome-scale gene networks through high performance reverse engineering analysis. BioMed research international 2013, 856325, doi: 10.1155/2013/856325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Gomez M. A. et al. Segregation of a haplotype encompassing FEB1 with genetic epilepsy with febrile seizures plus in a Colombian family. Epileptic disorders: international epilepsy journal with videotape 15, 128–131, doi: 10.1684/epd.2013.0570 (2013). [DOI] [PubMed] [Google Scholar]
- He S., Lin J., Yu S. & Sun S. Upregulation of PREX2 promotes the proliferation and migration of hepatocellular carcinoma cells via PTEN-AKT signaling. Oncology letters 11, 2223–2228, doi: 10.3892/ol.2016.4164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. et al. iDNA-Prot|dis: Identifying DNA-Binding Proteins by Incorporating Amino Acid Distance-Pairs and Reduced Alphabet Profile into the General Pseudo Amino Acid Composition. PloS ONE 9, e106691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissanu Deribe Y. et al. Truncating PREX2 mutations activate its GEF activity and alter gene expression regulation in NRAS-mutant melanoma. Proceedings of the National Academy of Sciences of the United States of America 113, E1296–1305, doi: 10.1073/pnas.1513801113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense S. M. et al. PTEN inhibits PREX2-catalyzed activation of RAC1 to restrain tumor cell invasion. Science signaling 8, ra32, doi: 10.1126/scisignal.2005840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M. et al. Searching for genomic region of high-fat diet-induced type 2 diabetes in mouse chromosome 2 by analysis of congenic strains. PloS one 9, e96271, doi: 10.1371/journal.pone.0096271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten H. J. et al. Comparison of microarray expression profiles between follicular variant of papillary thyroid carcinomas and follicular adenomas of the thyroid. BMC genomics 16 Suppl 1, S7, doi: 10.1186/1471-2164-16-S1-S7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov S. et al. GPR155: Gene organization, multiple mRNA splice variants and expression in mouse central nervous system. Biochemical and biophysical research communications 398, 19–25, doi: 10.1016/j.bbrc.2010.05.162 (2010). [DOI] [PubMed] [Google Scholar]
- Girirajan S. et al. Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome research 19, 178–190, doi: 10.1101/gr.086041.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne F. et al. Association mapping of the high-grade myopia MYP3 locus reveals novel candidates UHRF1BP1L, PTPRR, and PPFIA2. Investigative ophthalmology & visual science 54, 2076–2086, doi: 10.1167/iovs.12-11102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento F. A., Borlot F., Cossette P., Minassian B. A. & Andrade D. M. Two definite cases of sudden unexpected death in epilepsy in a family with a DEPDC5 mutation. Neurology. Genetics 1, e28, doi: 10.1212/NXG.0000000000000028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens L. M. et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nature genetics 45, 546–551, doi: 10.1038/ng.2599 (2013). [DOI] [PubMed] [Google Scholar]
- Baulac S. et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Annals of neurology 77, 675–683, doi: 10.1002/ana.24368 (2015). [DOI] [PubMed] [Google Scholar]
- Baulac S. Genetics advances in autosomal dominant focal epilepsies: focus on DEPDC5. Progress in brain research 213, 123–139, doi: 10.1016/B978-0-444-63326-2.00007-7 (2014). [DOI] [PubMed] [Google Scholar]
- Pippucci T. et al. Epilepsy with auditory features: A heterogeneous clinico-molecular disease. Neurology. Genetics 1, e5, doi: 10.1212/NXG.0000000000000005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill G. L. et al. Epileptic spasms are a feature of DEPDC5 mTORopathy. Neurology. Genetics 1, e17, doi: 10.1212/NXG.0000000000000016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N. et al. Role of IFN-ks, IFN-ks related genes and the DEPDC5 gene in Hepatitis B virus-related liver disease. Journal of viral hepatitis 21, e29–38 (2014). [DOI] [PubMed] [Google Scholar]
- Miki D. et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nature genetics 43, 797–800, doi: 10.1038/ng.876 (2011). [DOI] [PubMed] [Google Scholar]
- Motomura T. et al. Neither MICA Nor DEPDC5 Genetic Polymorphisms Correlate with Hepatocellular Carcinoma Recurrence following Hepatectomy. HPB surgery: a world journal of hepatic, pancreatic and biliary surgery 2012, 185496, doi: 10.1155/2012/185496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerill I. et al. Hypoxia Increases IGFBP-1 Phosphorylation Mediated by mTOR Inhibition. Mol Endocrinol 30, 201–216, doi: 10.1210/me.2015-1194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. An evolving role for DEPTOR in tumor development and progression. Neoplasia 14, 368–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F., Zhang M. & Chen L. 5′-Monophosphate-activated protein kinase (AMPK) improves autophagic activity in diabetes and diabetic complications. Acta pharmaceutica Sinica. B 6, 20–25, doi: 10.1016/j.apsb.2015.07.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. et al. Chemopreventive Effect of Dietary Glutamineon Colitis-Associated Colorectal Cancer Is Associated with Modulation of the DEPTOR/mTOR Signaling Pathway. Nutrients 8, doi: 10.3390/nu8050261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. P. et al. Point mutations of the mTOR-RHEB pathway in renal cell carcinoma. Oncotarget 6, 17895–17910, doi: 10.18632/oncotarget.4963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. B. et al. High DEPTOR expression correlates with poor prognosis in patients with esophageal squamous cell carcinoma. OncoTargets and therapy 8, 3449–3455, doi: 10.2147/OTT.S92862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravacci G. R. et al. Docosahexaenoic Acid Modulates a HER2-Associated Lipogenic Phenotype, Induces Apoptosis, and Increases Trastuzumab Action in HER2-Overexpressing Breast Carcinoma Cells. BioMed research international 2015, 838652, doi: 10.1155/2015/838652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas-Faja B. et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell cycle 12, 3109–3124, doi: 10.4161/cc.26173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C. H. et al. Functional characterization of glycine N-methyltransferase and its interactive protein DEPDC6/DEPTOR in hepatocellular carcinoma. Molecular medicine 18, 286–296, doi: 10.2119/molmed.2011.00331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvani J. G. et al. Deptor enhances triple-negative breast cancer metastasis and chemoresistance through coupling to survivin expression. Neoplasia 17, 317–328, doi: 10.1016/j.neo.2015.02.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z., Wang X., Lin D. & Zou Q. Construction and Identification of the RNAi Recombinant Lentiviral Vector Targeting Human DEPDC7 Gene. Interdisciplinary sciences, computational life sciences, doi: 10.1007/s12539-016-0162-y (2016). [DOI] [PubMed] [Google Scholar]
- Cordova-Palomera A. et al. Further evidence of DEPDC7 DNA hypomethylation in depression: A study in adult twins. Eur Psychiatry 30, 715–718, doi: 10.1016/j.eurpsy.2015.04.001 (2015). [DOI] [PubMed] [Google Scholar]
- Seabra C. M. et al. A novel Alu-mediated microdeletion at 11p13 removes WT1 in a patient with cryptorchidism and azoospermia. Reproductive biomedicine online 29, 388–391, doi: 10.1016/j.rbmo.2014.04.017 (2014). [DOI] [PubMed] [Google Scholar]
- Liu B. et al. Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Research 43, W65–W71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Long R. & Chou K. C. iDHS-EL: identifying Dnase I hypersensitive sites by fusing three different modes of pseudo nucleotide composition into an ensemble learning framework. Bioinformatics 32, 2411–2418, doi: 10.1093/bioinformatics/btw186 (2016). [DOI] [PubMed] [Google Scholar]
- Liu B., Wang S., Long R. & Chou K. C. iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics, doi: 10.1093/bioinformatics/btw539 (2016). [DOI] [PubMed] [Google Scholar]
- Kirchberger P. C., Sefc K. M., Sturmbauer C. & Koblmuller S. Outgroup effects on root position and tree topology in the AFLP phylogeny of a rapidly radiating lineage of cichlid fish. Molecular phylogenetics and evolution 70, 57–62, doi: 10.1016/j.ympev.2013.09.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlers K. E., Chakravarti B. & Fisher R. A. RGS6 as a Novel Therapeutic Target in CNS Diseases and Cancer. The AAPS journal 18, 560–572, doi: 10.1208/s12248-016-9899-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. et al. Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinformatics 30, 472–479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Charlat O., Zamponi R., Yang Y. & Cong F. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Molecular cell 58, 522–533, doi: 10.1016/j.molcel.2015.03.015 (2015). [DOI] [PubMed] [Google Scholar]
- Tayou J. et al. Regulator of G Protein Signaling 7 (RGS7) Can Exist in a Homo-oligomeric Form That Is Regulated by Galphao and R7-binding Protein. The Journal of biological chemistry 291, 9133–9147, doi: 10.1074/jbc.M115.694075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banderas A. & Guiliani N. Bioinformatic prediction of gene functions regulated by quorum sensing in the bioleaching bacterium Acidithiobacillus ferrooxidans. International journal of molecular sciences 14, 16901–16916, doi: 10.3390/ijms140816901 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic acids research 43, D204–212, doi: 10.1093/nar/gku989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Niu B., Gao Y., Fu L. & Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682, doi: 10.1093/bioinformatics/btq003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Tang H., Ye J., Lin H. & Chou K. C. iRNA-PseU: Identifying RNA pseudouridine sites. Molecular Therapy-Nucleic Acids 5, e332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Bhagabati P., Sachan R., Kaushik A. C. & Dwivedi V. D. In Silico Analysis of Sequence-Structure-Function Relationship of the Escherichia coli Methionine Synthase. Interdisciplinary sciences, computational life sciences 7, 382–390, doi: 10.1007/s12539-015-0271-z (2015). [DOI] [PubMed] [Google Scholar]
- Zamocky M., Gasselhuber B., Furtmuller P. G. & Obinger C. Turning points in the evolution of peroxidase-catalase superfamily: molecular phylogeny of hybrid heme peroxidases. Cellular and molecular life sciences: CMLS 71, 4681–4696, doi: 10.1007/s00018-014-1643-y (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. et al. Genome wide in silico analysis of Plasmodium falciparum phosphatome. BMC genomics 15, 1024, doi: 10.1186/1471-2164-15-1024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948, doi: 10.1093/bioinformatics/btm404 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Li X., Jiang Y., Zhao Y. & G. W. BinMemPredict: a Web Server and Software for Predicting Membrane Protein Types. Current Proteomics 10, 2–9 (2013). [Google Scholar]
- Liao Z. et al. In Silico Prediction of Gamma-Aminobutyric Acid Type-A Receptors Using Novel Machine-Learning-Based SVM and GBDT Approaches. BioMed research international 2016, 2375268, doi: 10.1155/2016/2375268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z., Ju Y. & Zou Q. Prediction of G Protein-Coupled Receptors with SVM-Prot Features and Random Forest. Scientifica 2016, 8309253, doi: 10.1155/2016/8309253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Zou Q., Liao M., Lu H. & Zhao Y. A novel machine learning method for cytokine-receptor interaction prediction. Combinatorial chemistry & high throughput screening 19, 144–152 (2016). [DOI] [PubMed] [Google Scholar]
- Lin C. et al. Hierarchical classification of protein folds using a novel ensemble classifier. PloS one 8, e56499, doi: 10.1371/journal.pone.0056499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Ibata D. M., Pazos A., Orbegozo-Medina R. A., Romero-Duran F. J. & Gonzalez-Diaz H. Mapping chemical structure-activity information of HAART-drug cocktails over complex networks of AIDS epidemiology and socioeconomic data of U.S. counties. Bio Systems 132-133, 20–34, doi: 10.1016/j.biosystems.2015.04.007 (2015). [DOI] [PubMed] [Google Scholar]
- Chen K. H., Wang K. J., Adrian A. M., Wang K. M. & Teng N. C. Diagnosis of Brain Metastases from Lung Cancer Using a Modified Electromagnetism like Mechanism Algorithm. Journal of medical systems 40, 35, doi: 10.1007/s10916-015-0367-3 (2016). [DOI] [PubMed] [Google Scholar]
- Wiharto W., Kusnanto H. & Herianto H. Intelligence System for Diagnosis Level of Coronary Heart Disease with K-Star Algorithm. Healthcare informatics research 22, 30–38, doi: 10.4258/hir.2016.22.1.30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E., Hall M., Trigg L., Holmes G. & Witten I. H. Data mining in bioinformatics using Weka. Bioinformatics 20, 2479–2481, doi: 10.1093/bioinformatics/bth261 (2004). [DOI] [PubMed] [Google Scholar]
- Kumari P., Nath A. & Chaube R. Identification of human drug targets using machine-learning algorithms. Comput Biol Med 56, 175–181, doi: 10.1016/j.compbiomed.2014.11.008 (2015). [DOI] [PubMed] [Google Scholar]
- Liu B. et al. Identification of real microRNA precursors with a pseudo structure status composition approach. PloS ONE 10, e0121501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Feng P., Tang H., Ding H. & Lin H. RAMPred: identifying the N(1)-methyladenosine sites in eukaryotic transcriptomes. Scientific reports 6, 31080, doi: 10.1038/srep31080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Ding H., Feng P., Lin H. & Chou K. C. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget 7, 16895–16909, doi: 10.18632/oncotarget.7815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Deng E. Z., Ding H., Chen W. & Chou K. C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic acids research 42, 12961–12972, doi: 10.1093/nar/gku1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Feng P., Ding H., Lin H. & Chou K. C. iRNA-Methyl: Identifying N(6)-methyladenosine sites using pseudo nucleotide composition. Analytical biochemistry 490, 26–33, doi: 10.1016/j.ab.2015.08.021 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang C. J. et al. iOri-Human: identify human origin of replication by incorporating dinucleotide physicochemical properties into pseudo nucleotide composition. Oncotarget, doi: 10.18632/oncotarget.11975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. W., Lai H. Y., Tang H., Chen W. & Lin H. Prediction of phosphothreonine sites in human proteins by fusing different features. Scientific reports 6, 34817, doi: 10.1038/srep34817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Su Z. D., Wei H. H., Chen W. & Lin H. Prediction of cell-penetrating peptides with feature selection techniques. Biochemical and biophysical research communications 477, 150–154, doi: 10.1016/j.bbrc.2016.06.035 (2016). [DOI] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780, doi: 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic acids research 43, W580–584, doi: 10.1093/nar/gkv279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Johnson J., Grant C. E. & Noble W. S. The MEME Suite. Nucleic acids research 43, W39–49, doi: 10.1093/nar/gkv416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuanyuan S. et al. Reference gene selection for real-time quantitative PCR analysis on ovarian cryopreservation by vitrification in mice. Journal of assisted reproduction and genetics 32, 1277–1284, doi: 10.1007/s10815-015-0503-5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum A., Waubke K., Pehl A., Steinfeldt T. & Bartsch D. K. Suture or Stapling Are Superior in Comparison to Bipolar Sealing for Closing the Bronchi. The Thoracic and cardiovascular surgeon, doi: 10.1055/s-0036-1584951 (2016). [DOI] [PubMed] [Google Scholar]