Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease associated with aggregation of TAR DNA-binding protein-43 (TDP-43) in neuronal cells and manifests as motor neuron dysfunction & muscle atrophy. The carboxyl-terminal prion-like domain of TDP-43 can aggregate in vitro into toxic β-sheet rich amyloid-like structures. So far, treatment options for ALS are very limited and Riluzole, which targets glutamate receptors, is the only but highly ineffective drug. Therefore, great interest exists in developing molecules for ALS treatment. Here, we have examined certain derivatives of acridine containing same side chains at position 4 & 5, for inhibitory potential against TDP-43 aggregation. Among several acridine derivatives examined, AIM4, which contains polar carboxyl groups in the side arms, significantly reduces TDP-43-YFP aggregation in the powerful yeast model cell and also abolishes in vitro amyloid-like aggregation of carboxyl terminal domain of TDP-43, as observed by AFM imaging. Thus, AIM4 can be a lead molecule potentiating further therapeutic research for ALS.
Several devastating diseases such as Alzheimer’s disease, Parkinson’s disease and Amyotrophic Lateral Sclerosis (ALS) are associated with conversion and deposition of otherwise normally soluble protein into insoluble, highly ordered, fibrillar aggregates termed amyloid1. Amyloids exhibit “cross β-sheet” structural organization and can bind planar dyes like Thioflavin-T and Congo Red thereby changing the spectroscopic properties of these dyes2,3,4. Multi-faceted ongoing efforts are directed at finding agents to prevent amyloid formation and deposition in order to find therapeutic options for amyloid diseases5,6.
Amyotrophic lateral sclerosis (ALS) is a fatal disease associated with motor neuron degeneration and thus far, there is no cure or effective treatment for ALS7. Although most ALS cases are sporadic (90%), mutations in several genes have been identified that can lead to ALS pathogenesis (10% cases) among which SOD18,9,10,11, TDP-4312,13,14,15, FUS16 and hexanucleotide repeat expansions in C9orf727,17,18,19 are the most prominent. Notably, presence of TDP-43 aggregates is found in ALS cases of diverse etiology7. Thus TDP-43 is an important protein in pathology of ALS and an important target for screening of drugs for anti-ALS properties. TDP-43 is a versatile RNA/DNA-binding protein involved in cellular process such as RNA-metabolism (transcription, translation, miRNA processing, mRNA transport across nucleus)20,21,22, apoptosis23, cell division24, embryo development25, and stress response26.
In ALS cases, there is increasing evidence supporting that the TDP-43 protein converts into prion-like aggregates12,14. In particular, the carboxyl-terminal glycine-rich domain is highly aggregation-prone and can form amyloid-like aggregates in vitro as well12,27. Extensive investigations using TDP-43 aggregation models such as Drosophila and yeast cell have shown correlation between TDP-43’s aggregation and toxicity7,28,29. In fact, genetic screen using yeast model has helped uncover several candidate genes that can affect aggregation and toxicity of TDP-43 among which ataxin-2, the human homolog of yeast PBP1 protein, is now widely accepted as an ALS risk factor30. Towards finding therapeutic molecules for ALS, efforts at identifying inhibitors of TDP-43 aggregation have involved examining small molecules like methylene blue and dimebon, however, results from clinical trials have not been very encouraging31. Although, there are no consensus structures that can act as general inhibitors of amyloid protein’s aggregation, using candidate approach, certain compounds with hydrogen bonding groups attached to aromatic rings have been found to show inhibitions of several amyloid proteins, possibly due to steric interference by their aromatic rings while beta sheet structure formation by the proteins5,32,33. Recently, acridine-based compounds varying in side chains at the C-9 position of acridine skeleton, have been investigated for inhibitory potential against amyloid formation by several proteins such as insulin, HEWL and the prion protein PrPSc 34,35,36. Here we have investigated if certain imidazolium-tagged acridine derivatives containing two identical functional groups in the side arms at position 4 & 5 of acridine ring, can show inhibitory potential against TDP-43 protein. These acridine derivatives contained two imidazolium groups attached to carbon 4 & 5 of acridine to improve their hydrophilicity (Fig. 1). The nitrogen donor sites at acridine and two imidazoles are expected to enhance the hydrogen bonding interactions. Furthermore, imidazolium groups were linked with side chains such as isopropyl, ester or carboxylic acid, which display varying hydrophilicity to impart differential interaction capability with biomolecules. For comparison, another derivative of acridine, 4,5-bis(hydroxymethyl)acridine, which lacks imidazole in the side arms but contains polar hydroxymethyl groups at carbon 4 & 5 of acridine, was also examined for anti-aggregation ability against TDP-43. The effect of these compounds on in vitro aggregation of TDP-43’s C-terminal amyloidogenic fragment (TDP-432C), using tools like ThT fluorescence, circular dichroism, and AFM37. Furthermore, we have also examined if there is anti-aggregation effect on full-length TDP-43 tagged with YFP using the eukaryotic single cell yeast model by fluorescence microscopy.
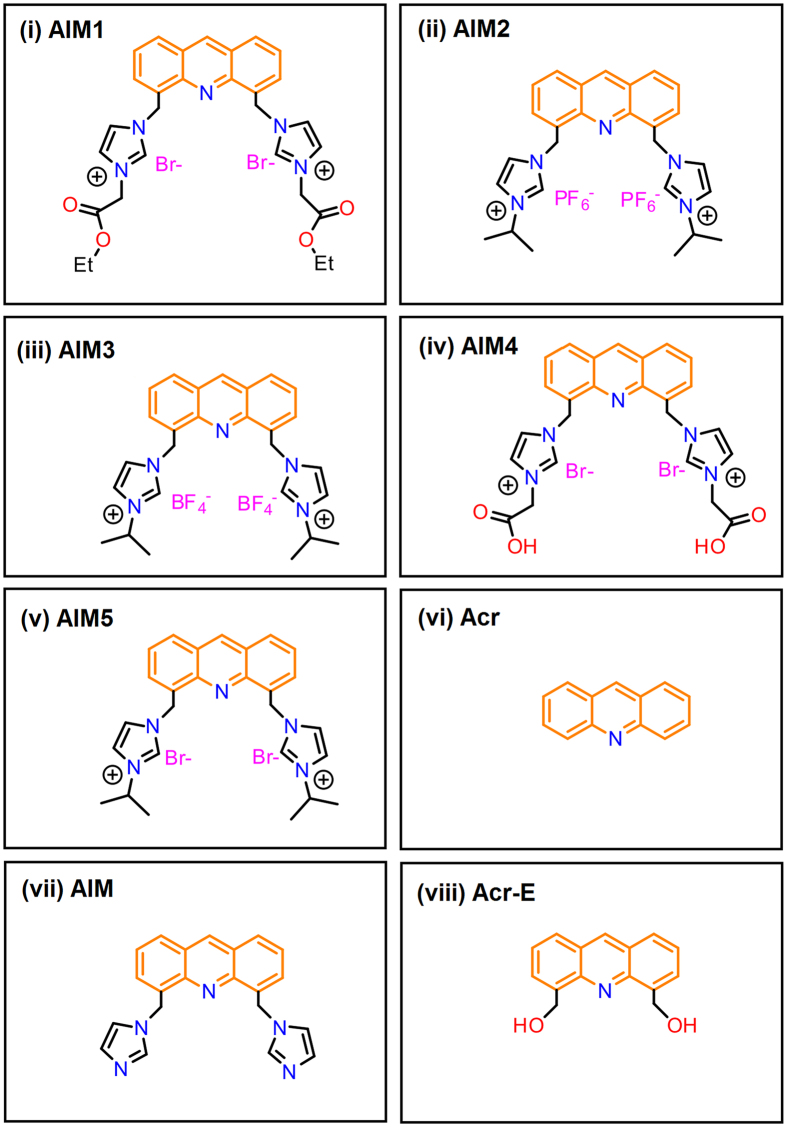
Figure 1. Structures of acridine derivatives examined for effect on TDP-432C aggregation.
AIM1: [4,5-bis{(N-ethoxycarbonyl methyl imidazolium)methyl}acridine] dibromide; AIM2: [4,5-bis{(N-isopropylimidazolium) methyl} acridine] hexafluorophosphate; AIM3: [4,5-bis{(N-isopropylimidazolium)methyl}acridine] tetrafluoroborate; AIM4: [4,5-bis{(N-carboxy methyl imidazolium)methyl}acridine] dibromide; AIM5: [4,5-bis{(N-isopropylimidazolium) methyl} acridine] dibromide; Acr: Acridine; AIM: (4,5-bis(imidazole methyl)acridine) and Acr-E: (4,5-bis(hydroxymethyl)acridine).
Results
AIM4 retards in vitro aggregation of TDP-432C
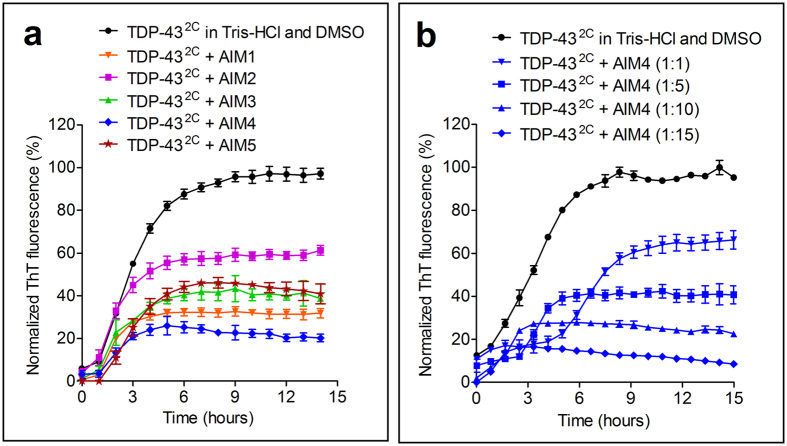
Therapeutic options for ALS are very limited and concerted research is required to find agents that can help in ALS treatment considering its highly fatal pathology. Towards this goal, here we investigated imidazolium-tagged acridine compounds for inhibitory potential against TDP-432C amyloid-like aggregation (Fig. 1). These compounds have two methylimidazolium groups at the C-4 and C-5 positions of the planar acridine skeleton and differ in the side chains attached to the imidazolium and also in the counter anions associated with the positive charge on the imidazolium groups37. By examining TDP-432C aggregation using ThT fluorescence assay, we found that among these compounds examined at 1:10 (Protein: Compound) molar ratio, the compound AIM4, which contains two carboxyl groups in the side chains, caused the maximum decrease in ThT fluorescence intensity (up to 76%) (Fig. 2a). The compound AIM1 which contains two ester groups in the side chains was the second most effective compound after AIM4 showing 65% (Fig. 2a). The compounds, AIM2, AIM3 & AIM5, which contain non-polar two isopropyl groups in the side-chains, were relatively less effective (38%, 54% & 54% inhibitions respectively) in inhibiting the ThT fluorescence kinetics of TDP-432C aggregation (Fig. 2a). Although, the backbone structure is same in the AIM2, AIM3 & AIM5 compounds, they differ in the counter ions. The PF6− counter ion in AIM2, BF4− counter ion in AIM3 & the Br− counter ion in AIM5, can play an important role in tuning the acidity of the imidazole C-H proton, which is responsible for the N(N)C-H····X, hydrogen bonding strength37,38,39. The observed difference in the TDP-432C inhibition ability of AIM2 compared to AIM3 & AIM5, may possibly be due to their differential interaction capabilities with the protein molecule due to altered hydrogen bonding abilities. Furthermore, as AIM4 & AIM5 both have bromide counter-ions, the observed comparatively higher inhibitory potential of AIM4 indicates important role of the polar carboxyl groups in the side chains. Furthermore, it concurrently also rules out the possibility that the differences in the counter-ions in AIM4 versus AIM2 & AIM3 are solely responsible for the difference in their inhibitory abilities. As these acridine compounds are also intrinsically fluorescent37, control experiments were performed to eliminate their any possible quenching effect on the ThT fluorescence (Supplementary Fig. S1). When further examined in detail, AIM4 also exerted concentration-dependent decrease in ThT fluorescence intensity and at 1:15 (Protein: Compound) molar ratio, the kinetic trend was observed to flatten to the baseline thereby suggesting complete inhibition of the TDP-432C aggregation (Fig. 2b).
Figure 2. Imidazolium-tagged acridine derivatives affect TDP-432C aggregation.
(a) Aggregation trend of TDP-432C in presence of imidazolium-tagged acridine derivatives monitored by ThT fluorescence intensity. TDP-432C (400 μM) was added with 4 mM acridine derivatives (1:10, protein: compound, molar ratio) & 450 μM ThT in aggregation buffer with 2% DMSO final and incubated at 37 °C for 15 hours with intermittent agitation (15 seconds agitation every 5 minutes). A sample lacking any compound was incubated similarly for comparison. Error bars represent standard deviation (n = 3). Percentage inhibitions in ThT fluorescence by AIM1, AIM2, AIM3, AIM4 & AIM5 were found respectively to be 65%, 38%, 54%, 76% & 54%. (b) Effect of increasing concentration of AIM4 on TDP-432C aggregation monitored by ThT fluorescence. TDP-432C (400 μM) was incubated with increasing stoichiometry of AIM4 (1:1 to 1:15; protein: AIM4) along with 450 μM ThT in aggregation buffer with 2% DMSO final and aggregation was recorded by ThT fluorescence increase with intermittent agitation at 37 °C for 15 hours. A sample lacking AIM4 was incubated and analyzed similarly for comparison. Percentage inhibition in ThT fluorescence at different protein: compound stoichiometry were found to be 37% (at 1:1), 57% (at 1:5), 72% (at 1:10) and 86% (at 1:15).
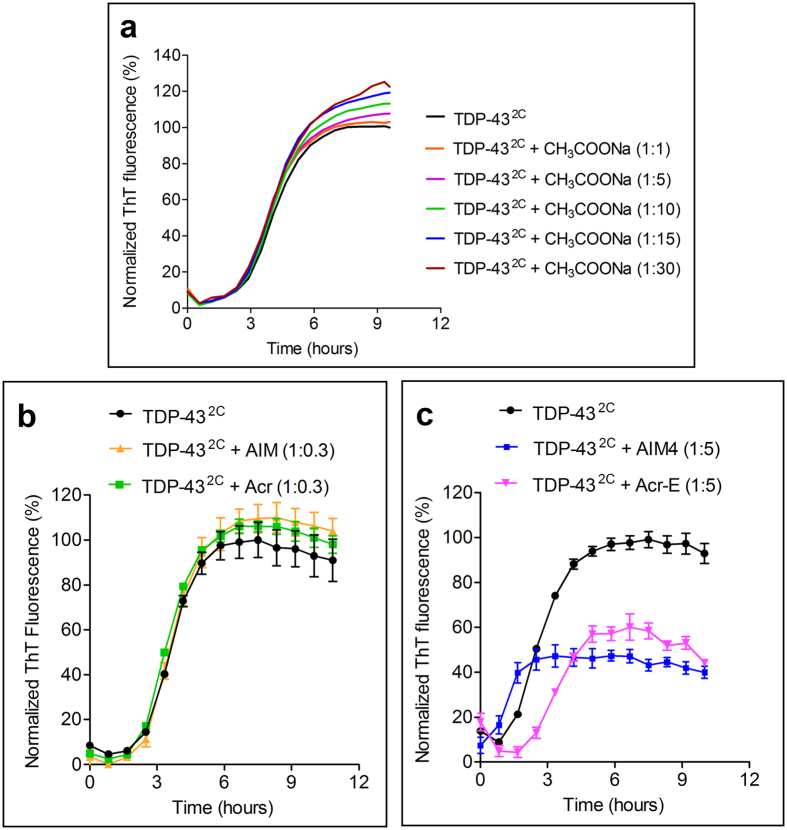
As the AIM4 compound which contains terminal carboxyl groups was more effective at the inhibition of TDP-432C aggregation, in control experiments we cross-checked whether the presence of only carboxyl group lacking the aromatic rings in AIM4 would be inhibitory. For this, when we checked effect of carboxyl group containing small compound, sodium acetate (CH3COONa) at stoichiometry comparable to as used for AIM4, no inhibition of the aggregation of TDP-432C was observed, rather & consistent with the established general kosmotropic effect of the acetate ions which enhance protein structure40, a marginal increase in the aggregation of TDP-432C was observed (Fig. 3a). Furthermore, as AIM4 also contains acridine and imidazolium groups, we attempted to examine the effect of acridine (Acr) and imidazolium tagged acridine (AIM) separately on the aggregation of TDP-432C, however, these compounds being inherently hydrophobic have very limited solubility in the aggregation buffer which originally contained the organic solvent 2% DMSO. Even after increasing the DMSO to 8%, these compounds could only be assayed at 125 μM concentrations leading to protein: compound stoichiometry of 1: 0.3, at which no inhibition of TDP-432C aggregation was observed (Fig. 3b). Additionally, another acridine derivative Acr-E (4,5-bis(hydroxymethyl)acridine) lacking the imidazolium moieties & containing two hydroxymethyl polar groups when examined, displayed higher solubility than Acr & AIM, however, lower solubility than that of AIM4. Thus, its effect on TDP-432C aggregation could be examined, even in the presence of 8% DMSO, only upto the protein: compound stoichiometry of 1:5. At this stoichiometry, Acr-E displayed inhibition of TDP-432C aggregation by 45% as compared to 55% inhibition by AIM4 (Fig. 3c). However, as Acr-E manifested limited solubility, it could not be assayed at the 1:15 (protein: compound) stoichiometry at which AIM4 caused complete inhibition of the aggregation of TDP-432C as indicated by the flattening of the ThT-fluorescence kinetics (Fig. 2b). The observed inhibition of TDP-432C aggregation by Acr-E may possibly be due to the hydrogen bonding capability of its hydroxyl groups helping in its interaction with the protein molecule and its acridine moiety causing steric hindrance to the protein’s aggregation. In the view that at similar stoichiometry, Acr-E also exhibited inhibitory effect on TDP-432C aggregation to the extent that was only marginally lower than that of AIM4, the presence of imidazolium moieties in the side arms of the acridine derivatives is apparently not essential for the inhibitory action. However, as the AIM4 displayed efficient inhibitory propensity as well as better solubility possibly owing to the presence of the imidazolium moieties, it was further analyzed as the lead inhibitory compound.
Figure 3. Effect of acetate ions & acridine derivatives on in vitro aggregation of TDP-432C.
(a) TDP-432C (400 μM) solubilized in aggregation buffer was added with increasing concentrations of sodium acetate (1:1 to 1:30, protein: compound, molar ratio) that was also pre-dissolved in the aggregation buffer but lacking the Tris-HCl & DMSO. Aggregation trend was monitored by ThT fluorescence at 37 °C for 15 hours with intermittent agitation (15 seconds agitation every 5 minutes). Increasing stoichiometry of sodium acetate caused concomitant marginal increase in the overall ThT fluorescence. (b) Aggregation trend of TDP-432C in presence of Acr and AIM was monitored by ThT fluorescence. TDP-432C (400 μM) was added with 125 μM Acr or 125 μM AIM (protein: compound ratio of 1: 0.3) in aggregation buffer lacking Tris-HCl and containing 8% DMSO final. The protein aggregation was examined at 37 °C for 15 hours with intermittent agitation by recording ThT fluorescence intensity. A sample lacking any compound was incubated similarly for comparison. (c) TDP-432C aggregation in presence of Acr-E was monitored by recording ThT fluorescence intensity. TDP-432C (400 μM) was added with 2000 μM Acr-E or in the control experiment with 2000 μM AIM4, in aggregation buffer containing 12 mM Tris-HCl and 8% DMSO final. Samples were incubated at 37 °C for 15 hours with intermittent agitation and ThT fluorescence was recorded. A sample lacking any compound was incubated & monitored similarly for comparison. Acr-E and AIM4 were found to respectively reduce the ThT fluorescence by 45% and 55%.
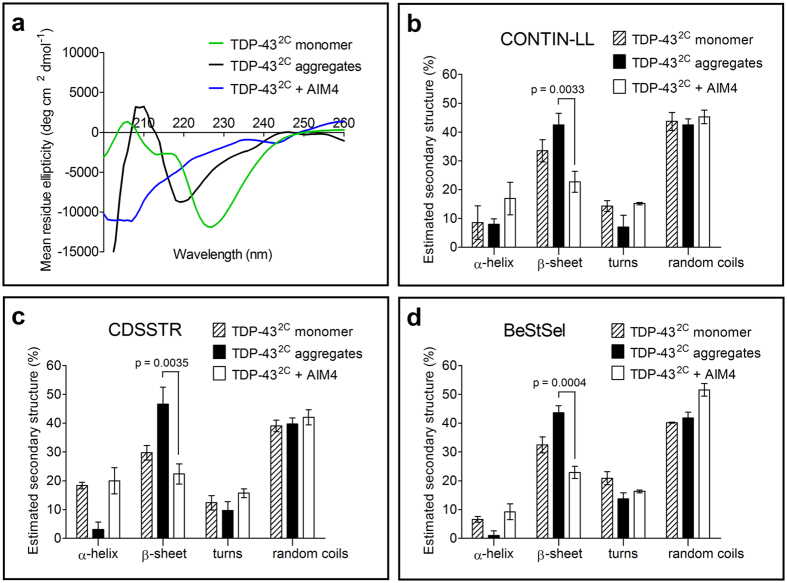
As the aggregation of TDP-43 fragments has been previously shown to be accompanied by change into amyloid-like β-sheet rich conformation12,41, we therefore examined the secondary structural features by far-UV circular dichroism (CD). As expected, TDP-432C protein aggregates displayed a negative peak at ~218 nm in the far-UV CD spectrum thus indicating a β-sheet rich structure. Strikingly, the TDP-432C samples that were pre-incubated with AIM4 and then incubated under aggregation conditions, yielded a markedly different far-UV CD spectrum suggesting change in structural features (Fig. 4a). Furthermore, estimation of secondary structural content from these spectra showed a decrease of ∼20% β-sheet content upon incubation with AIM4, thereby supporting a reduction in amyloid formation (Fig. 4b–d).
Figure 4. Effect of AIM4 on secondary structure of TDP-432C estimated by CD spectrometry.
(a) Far-UV circular dichroism (CD) spectra of TDP-432C in presence and absence of AIM4. Freshly prepared TDP-432C monomer in aggregation buffer, pre-formed TDP-432C aggregates and TDP-432C protein incubated similarly for aggregation with AIM4 were diluted in aggregation buffer and far-UV CD spectra were recorded and results have been expressed as mean residue ellipticity [ϴ]MRE. (b–d) Using the far-UV CD spectra from (a), the secondary structural contents were predicted by the online structure prediction tools: DichroWeb algorithms CONTIN & CDSSTR and server BeStSel. The relative contents of α-helix, β-sheet, turns and random coils predicted by these tools have been depicted as bar charts. The TDP-432C aggregation sample with AIM4 showed a decrease of ~20% in β-sheet content compared to those without AIM4. Error bars represent standard deviations of structural predictions obtained from the CD spectra from three samples (n = 3).
AIM4 prevents maturation of TDP-432C oligomers into amyloid aggregates
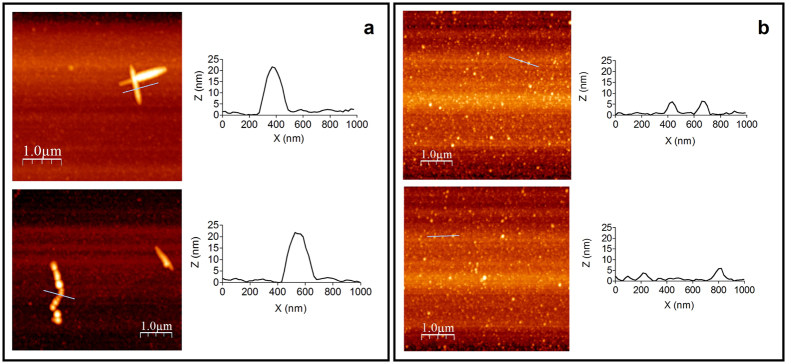
To analyze if AIM4 prevents amyloid aggregation by keeping the molecules of TDP-432C completely monomeric, we examined the morphologies of TDP-432C samples aggregated with or with incubation along with AIM4, using atomic force microscopy (AFM). Alike amyloid aggregates, TDP-432C samples lacking AIM4, displayed unbranched fibrillar structures of varying lengths (~1 to 2 μm) with an average height of ~20-25 nm (Fig. 5a). The fibers were either straight or gently curved similar to those previously observed for amyloid of certain amyloidogenic fragments of TDP-43 in AFM and TEM13,42. Suggesting prevention of the aggregation, the TDP-432C samples incubated with AIM4 lacked fibrillar structures and displayed only granular & spherical oligomeric structures with an average height of ~5 to 10 nm (Fig. 5b). This data shows absence of polymeric TDP-432C aggregates upon incubation with AIM4, and suggests that AIM4 does not prevent the formation of TDP-432C oligomers but arrests their conversion to polymeric amyloid aggregates.
Figure 5. Assessment of AIM4-mediated inhibition of TDP-432C aggregation by AFM.
AFM images of TDP-432C incubated in the absence (a) and presence of 1:10 AIM4 (b). Fibrillar aggregates of TDP-432C were observed in absence of AIM4 while oligomeric aggregates of TDP-432C were observed in the presence of AIM4. Height profiles of selected fibrillar (a) or oligomeric (b) species, marked by blue lines, were generated using the AFM image analysis software WSxM 4.0.
AIM4 reduces TDP-43-YFP aggregation in yeast cell model
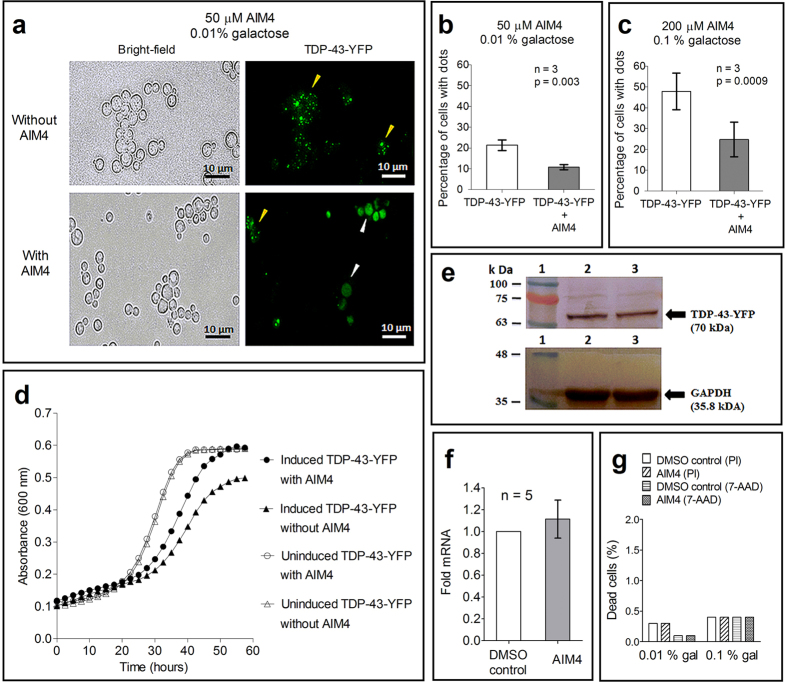
Aggregation of TDP-43 has already been shown in yeast S. cerevisiae model, where transient expression of TDP-43-YFP results in prion-like punctate fluorescent foci (dots) formation which is also accompanied by cytotoxicity28,43. Relevance and parallel of the yeast TDP-43-YFP expression model to ALS has been greatly supported by several cellular factors modulating the aggregation and toxicity in similar manner in both yeast and mammalian cell cultures30,44,45. In fact, ataxin-2, a homolog of yeast PBP1 protein, was identified as an ALS risk factor after the role of PBP1 in modulating TDP-43 toxicity was deciphered in the yeast model30,46. Therefore, owing to the relevance of the yeast TDP-43 aggregation model to ALS, we examined if the compound AIM4 also affects TDP-43 aggregation in the yeast cell. To facilitate permeation of the compound across yeast cell wall, first TDP-43-YFP aggregation was established in an erg6Δ mutant yeast which is defective in ergosterol biosynthesis that results in leakiness of the cell wall5. For this, a 64-D694 yeast strain with erg6Δ mutation was transformed with plasmid (pGAL1p-TDP-43-YFP) coding TDP-43-YFP under galactose-inducible GAL1 promoter which responds incrementally with increasing galactose concentrations in the range of 0–2%29. Formation of punctate TDP-43-YFP foci was then examined in cultures growing with or without the compound AIM4 in the growth media. We found that after 4 hours of induction with 0.01% galactose, nearly 20% of untreated cells displayed TDP-43-YFP dots (434 cells with dots, out of 1806 cells), whereas only ~10% of AIM4 treated cells formed the dots (201 cells with dots, out of 1870 cells) (Fig. 6a,b). In addition, number of TDP-43-YFP dots per cell was also markedly reduced when AIM4 was present thereby further suggesting inhibition of TDP-43 aggregation (Fig. 6a). Likewise, upon induction of TDP-43-YFP expression with 0.1% galactose, nearly 50% of yeast cells growing without AIM4 had punctate TDP-43-YFP foci (819 cells with dots out of 1723 cells) which was reduced to ~25% (369 cells with dots, out of 1729 cells) when AIM4 was present in the medium (Fig. 6c). When expression levels of TDP-43-YFP in the yeast cells incubated with or without AIM4, were estimated for protein content by western blotting and mRNA levels by quantitative-PCR, similar levels of both were found thereby eliminating the possibility that reduction in TDP-43-YFP foci formation was due to sub-critical protein levels in the presence of AIM4 (Fig. 6e,f and Supplementary Fig. S2). Furthermore, to eliminate the possibility that reduction in TDP-43-YFP foci formation is due to toxic effects of AIM4 on yeast cells, growth curves of the yeast cells that were expressing TDP-43-YFP as well as those not expressing TDP-43-YFP, were also analyzed in presence and absence of AIM4. While similar trends of growth curves were observed for the yeast cells not expressing TDP-43-YFP in presence or absence of AIM4, a relatively faster growth was observed for the yeast cells expressing TDP-43-YFP that were incubated with AIM4 versus those growing without AIM4 (Fig. 6d). However, when relative population of viable cells was assessed by flow cytometry using propidium iodide & 7-amino-actinomycin D staining of the dead yeast cells, no significant difference was observed between the yeast cells expressing TDP-43-YFP that were incubated with or without AIM4 (Fig. 6g and Supplementary Figs S3 and S4). This data supports that AIM4, at the concentrations used, is not toxic to the yeast cells, however, the relatively better growth of the TDP-43-YFP expressing cells in presence of AIM4 remains unexplained.
Figure 6. Inhibition of TDP-43-YFP aggregation by AIM4 in yeast model.
(a) Fluorescence images of wild-type human TDP-43 tagged with YFP in the absence and presence of 50 μM AIM4 in S. cerevisiae strain with erg6Δ mutation after 4 hours of induction with 0.01% galactose. Formation of fluorescent foci (yellow arrows) indicates TDP-43-YFP aggregation while diffused fluorescence (white arrows) indicates the absence of TDP-43-YFP aggregation. (b) Percentage of TDP-43-YFP containing erg6Δ yeast cells displaying fluorescent dots in the absence or presence of AIM4 after 4 hours induction with 0.01% galactose. Error bars represent standard deviations (p = 0.003, n = 3; two-tailed t-test). (c) Percentage of TDP-43-YFP containing erg6Δ yeast cells displaying fluorescent dots in absence or presence of AIM4 after 4 hours induction of expression with 0.1% galactose. Error bars represent standard deviations (p = 0.0009, n = 3; two-tailed t-test). (d) Growth curve of TDP-43-YFP plasmid containing erg6Δ yeast cells cultured in SRaf-Ura media either under induced (0.1% galactose) or un-induced state which were either treated or not treated with 200 μM AIM4. Data from triplicate cultures were averaged. (e) Effect of AIM4 (200 μM) on TDP-43-YFP protein expression levels in S. cerevisiae erg6Δ cells determined by western blotting using anti-GFP antibody. Protein bands of TDP-43 YFP and the protein loading control, endogenously expressed GAPDH (probed with anti-GAPDH antibody) have been cropped from the original image (Supplementary Fig. S2). Similar TDP-43-YFP levels were observed in absence (Lane 2) or presence (Lane 3) of AIM4. (f) Effect of AIM4 on TDP-43-YFP mRNA levels determined by Real-Time PCR. Expression of TDP-43-YFP was induced (0.01% galactose, 4 hours) either in presence of 200 μM AIM4 or DMSO buffer (control). Actin was used as the endogenous control. No significant change in TDP-43-YFP mRNA levels was observed. The error bar indicates standard deviation from five independent samples (n = 5). Cytotoxicity of AIM4 (200 μM) on erg6Δ yeast cells analyzed by propidium iodide (PI) and 7-aminoactinomycin D (7-AAD) staining using flow cytometry (total: 50,000 cells) using PE-Texas Red-A & PerCP-Cy5-5A filters respectively. Similar percentage of dead cells indicate lack of AIM4 cytotoxicity at this concentration.
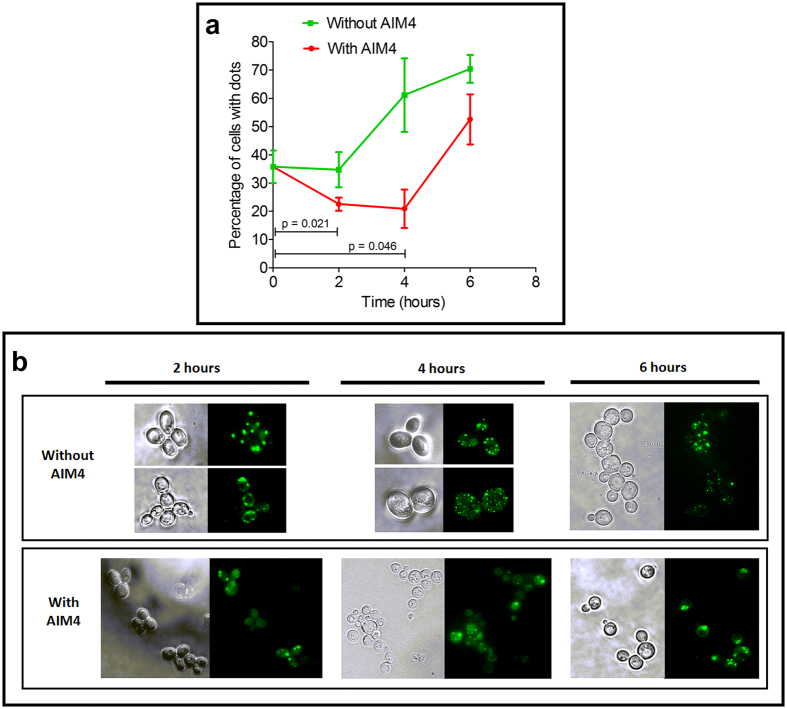
Next, to examine whether AIM4 has any effect on preformed TDP-43-YFP foci, AIM4 was added to yeast cells expressing TDP-43-YFP after the formation of the TDP-43-YFP punctate foci. When we examined the cells after 4 hours of AIM4 treatment, compared to the control untreated samples, the AIM4 treated sample manifested markedly lesser number of cells containing the TDP-43-YFP punctate foci thereby suggesting of dissolution of the aggregates (Fig. 7a). Additionally, the AIM4 treated cells displayed relatively fewer number of TDP-43-YFP punctate foci per cell and with bigger sizes compared to the untreated cells which displayed multiple TDP-43-YFP punctate foci per cell and of smaller sizes (Fig. 7b). When examined after 6 hours of AIM4 treatment, an increase in the number of TDP-43-YFP punctate foci containing cells was observed, possibly due to the sustained expression of the TDP-43-YFP (Fig. 7a,b). However, the cells continued to display fewer number of TDP-43-YFP punctate foci per cell with relatively bigger sizes (Fig. 7a,b). Taken together, the data support the ability of AIM4 to influence the aggregation of TDP-43-YFP in yeast cells.
Figure 7. Effect of AIM4 on pre-formed TDP-43-YFP aggregates.
(a) Disaggregation of pre-formed aggregates by TDP-43-YFP S. cerevisiae erg6Δ cells were induced for 4 hours at 30 °C using 0.01% galactose to express TDP-43-YFP which resulted in formation TDP-43-YFP punctate foci and the percentage of foci were counted. Subsequently, aliquots from these cells containing the TDP-43-YFP punctate foci, were transferred into fresh media also containing 0.01% galactose but one set added with 200 μM AIM4 and the other lacking AIM4. After two hours the cells were examined for the percentage of TDP-43-YFP punctate focAliquots were passaged into their respective original media with our without AIM4 and at the intervals of 2, 4 & 6 hours the percentage of TDP-43-YFP punctate foci were estimated by fluorescence microscopy. Data from triplicate samples were used to calculate standard deviation and represented as error bars. Notably, from the initial 35.5 ± 3.3% of cells with TDP-43-YFP punctate foci, after two hours of AIM4 treatment, the number of foci containing cells decreased to 22.56 ± 1.3%, which was a significant decrease (p = 0.0215, n = 3, two-tailed t-test) thereby suggesting dissolution of the aggregates. (b) Representative fluorescence microscopy images of TDP-43-YFP expressing yeast cells from (a) have been shown for comparison. Yeast cells incubated with AIM4 displayed relatively fewer cells with TDP-43-YFP punctate foci. Also, the sizes of the TDP-43-YFP punctate foci were relatively larger in the AIM4 treated cells compared with the untreated cells. Additionally, the AIM4 treated cells manifested fewer TDP-43-YFP punctate foci per cell whereas the untreated cells had multiple TDP-43-YFP punctate foci per cell.
Discussion
Several small molecules targeting amyloid aggregation, have been examined for therapeutics of amyloidosis disorders47,48. For examples, an anti-amyloid drug tafamidis, which acts by arresting the aggregating protein transthyretin into normally folded tetrameric form, has been shown to slow disease progression which has led to its FDA-approval for treatment of transthyretin amyloidosis49,50. So far, there is no success in finding anti-amyloid drugs for treatment of ALS. Towards this goal, we examined here if imidazolium derivatives of acridine differing in functional groups, have any anti-amyloid properties.
We found that the acridine’s imidazolium derivatives that have non-polar isopropyl groups in the arms (AIM2, AIM3 & AIM5) were ineffective as anti-TDP-43 aggregation inhibitors. Presence of ester groups in the arms (AIM1) displayed very mild inhibitory effect on TDP-43 aggregation, whereas presence of carboxyl groups (AIM4) was a more potent inhibitor. In contrast, carboxyl group from sodium acetate not only failed to inhibit TDP-43 aggregation, it rather stimulated the aggregation as expected due to its inherent kosmotropic effect of increasing protein structure40,51. Another acridine derivative, Acr-E, which lack the imidazolium groups but has two hydroxymethyl polar side chains also showed partial inhibition of the TDP-43 aggregation however, due to its limited solubility in the aggregation buffer, it could not be assayed at the required higher stoichiometry necessary for the complete inhibition of in vitro TDP-43 aggregation. Thus, among the various acridine derivatives investigated here AIM4 was most effective for inhibition of TDP-43 aggregation. Possible binding via hydrogen bonding or charge-charge interactions between negatively charged carboxyl groups of AIM4 & positive charges on protein side chains such as from lysine & arginine residues or an additive effect of both of these, may be responsible for the observed inhibitory effect by AIM4. Notably, the TDP-432C fragment contains four lysine & eight arginine residues in its sequence and at pH 7.5 that is used for the aggregation and inhibition studies, they are expected to bear positive charges on their side chains12. It is known that effective amyloid aggregation requires alignment of β-strands into β-sheets which are held by steric zipper inter-locking of the side chains and formation of the cross-β arrangement2. Interaction with any extrinsic aromatic agents, or even point mutations into proline which has bulky side-chain, can perturb the cross-β arrangement and inhibit the amyloid aggregation52. It appears here that interaction via carboxyl groups helps AIM4 to establish proximity to TDP-43 and the presence of acridine imidazolium bulky rings impart the steric hindrance thereby preventing the cross-β amyloid structure formation by TDP-432C.
The observed formation of oligomers displaying relatively less β-sheet secondary structure in presence of AIM4, suggests that these oligomers might be off-pathway oligomers and would be distinct from oligomers leading to TDP-43 amyloid fibrils. Similar in vitro off-pathway oligomer formation has also been previously documented in presence of small molecule inhibitors of Alzheimer’s Aβ peptide’s amyloid aggregation53.
The observed capabilities of AIM4 to inhibit in vitro amyloid aggregation of the carboxyl terminal TDP-432C fragment and to also reduce full-length TDP-43-YFP’s aggregation in the yeast cell, show its propensity as an anti-TDP-43 aggregation agent. The lack of complete abolition of TDP-43-YFP aggregation in the yeast cell and only an overall reduction in the level of aggregation, & the decreased number of aggregates per cell, may be due to its partial efficacy in the cellular model. Alternatively, it may also be a reflection of the continuous de novo aggregation of the newly formed TDP-43-YFP protein that is being over-expressed by induction with sustained presence of galactose in the medium or due to modest cellular uptake of AIM4, therefore its cellular concentrations not reaching levels that may be required for complete abolition of TDP-43-YFP aggregation.
It is known that TDP-43 has multiple interactions with other proteins, RNA & DNA molecules in a cell as part of its biological function7. In fact, sequestration of TDP-43 in stress granules in a cell which comprises of other proteins such as TIA-1 & RNA molecules has been proposed as a priming mechanism for aggregation of TDP-437. In the view that AIM4 also has binding ability to nucleic acids37, the observed reduction in TDP-43-YFP aggregation could also be due to a combinatorial effect of interaction of AIM4 with TDP-43 as well as any other cellular components that may be involved in aiding the aggregation of TDP-43.
Overall, this study finds AIM4 as a useful small molecule candidate with anti-TDP-43 aggregation property. Further studies by chemical modifications to increase its efficacy in cellular models and examining its efficacy in mammalian models of TDP-43 aggregation, could further its applicability. Expectedly, for a disease as debilitating & dreaded as ALS, any result towards finding a therapeutic molecule, would be a great step forward.
Methods
Materials
Ni-NTA agarose was purchased from Qiagen (USA). Thioflavin-T (ThT), ampicillin, chloramphenicol, sodium dodecyl sulfate (SDS), dithiothreitol (DTT), sodium sulfate (Na2SO4), sodium acetate (CH3COONa), phenylmethanesulfonyl fluoride (PMSF), isopropyl β-D-1-thiogalactopyranoside (IPTG) and imidazole were procured from Sigma-Aldrich (USA). Guanidine hydrochloride (GdnHCl) and urea were purchased from SRL (India) and Affymetrix (USA) respectively. Bradford’s protein concentration estimation reagent was from Bio-Rad (USA). EDTA-free protease inhibitor cocktail was purchased from Roche Diagnostics (Switzerland). Mica sheets (Grade V) were purchased from SPI Supplies (USA). Yeast nitrogen base and dextrose were procured from HiMedia (India) while raffinose and galactose were purchased from Sigma-Aldrich (USA). Potassium hexafluorophosphate was from Sigma-Aldrich (USA) and Acridine & bromomethyl methyl ether were procured from Alfa-Aesar (USA). Imidazolium derivatives of acridine ([4,5-bis{(N-ethoxycarbonyl methyl imidazolium)methyl}, acridine] dibromide (AIM1); [4,5-bis{(N-isopropylimidazolium) methyl} acridine] hexafluorophosphate (AIM2); [4,5-bis{(N-isopropylimidazolium)methyl}acridine] tetrafluoroborate (AIM3); [4,5-bis{(N-carboxy methyl imidazolium) methyl}acridine] dibromide (AIM4)) and [4,5-bis{(N-isopropylimidazolium)methyl}acridine] dibromide (AIM5) were synthesized as reported37. Acr-E [4,5-bis(hydroxymethyl)acridine] was synthesized as previous described54 AIM [4,5-bis(imidazole methyl)acridine] was synthesized as previously reported39. Organic solvents were purified according to standard procedures and freshly distilled under argon atmosphere prior to use55.
Plasmids and yeast strain
Escherichia coli Rosetta 2 (DE3) cells were obtained from Novagen, USA. E. coli expression plasmid pET15b-His-TDP-432C which codes for carboxyl terminal aa: 193-414 of TDP-4312 and yeast expression plasmid pRS416-pGAL1-TDP-43-YFP(URA3) (Addgene: 27447), were kind gifts of Prof. Susan Liebman, University of Nevada, Reno, USA. Saccharomyces cerevisiae strain, L-3341 MATα ade1-14 leu2-3,112 trp1-289 ura3-52 lys9-A21 erg6::TRP1 [PIN+], a derivative of 64-D strain was also a kind gift of Prof. Susan Liebman, University of Nevada, Reno, USA.
Recombinant protein expression and purification
Recombinant over-expression and purification of soluble TDP-432C under denaturing conditions was performed as described previously with minor modifications12. Briefly, Rosetta 2 (DE3) cells were transformed with pET15b-His-TDP-43-2C and induced to express TDP-432C for 4 hours by adding 1 mM IPTG. Cells were then harvested and lysed by ultra-sonication in lysis buffer containing 6 M GdnHCl in phosphate buffered saline (PBS), pH 7.5 along with protease inhibitor cocktail and 1 mM PMSF. The cell extract was pre-cleared by centrifugation at 13,000xg for 10 minutes at 4 °C and the supernatant was passed through Ni-NTA agarose column pre-equilibrated with 6 M GdnHCl at pH 7.5. The Ni-NTA column was then washed (wash buffer: PBS, pH 7.5 with 6 M GdnHCl and 10 mM imidazole) and protein was eluted with buffer containing 250 mM imidazole (elution buffer: PBS, pH 7.5 with 6 M GdnHCl and 250 mM imidazole). For assessing homogeneity, aliquots from the purified fractions were first diluted 10 fold in PBS pH 7.5, which resulted in precipitation of the protein and allowed for removal of GdnHCl. The precipitated protein was pelleted (14000× g for 15 min at 4 °C) and re-suspended in PBS, pH 7.5 and then checked on SDS-PAGE for homogeneity. Protein concentration was determined by Bradford’s method & from absorbance at 280 nm. The molar extinction coefficient of TDP-432C was 19605 M−1 cm−1, as determined by the von Hippel method56.
Amyloid aggregation and detection
TDP-432C purified in 6 M GdnHCl was first diluted 10-fold in PBS which resulted in its precipitation. The precipitate was separated and re-suspended in PBS containing 4 M urea to obtain the stock protein solution for amyloid aggregation. For allowing amyloid aggregation, the protein was diluted to 400 μM in aggregation buffer (PBS, pH 7.5, 500 μM DTT; 2.5 M urea final). Samples were agitated overnight in a shaker pre-maintained at 37 °C and then analyzed for formation of amyloid aggregates. To obtain kinetic trend of amyloid aggregation, the TDP-432C protein present in aggregation buffer, as above, was also added with ThT at 1:1 (protein: ThT) ratio and then incubated in multimode microplate reader with intermittent agitation and amyloid aggregation was detected by increase in ThT emission fluorescence assayed as below.
ThT binding assay
Upon binding with amyloid aggregates, the fluorescence emission intensity of ThT tremendously increases at 485 nm when excited at 442 nm3. Kinetics of TDP-432C aggregation was monitored by measuring ThT fluorescence intensity at 485 nm every five minutes using SpectraMax M5e (Molecular Devices) microplate multi-mode reader upon excitation at 442 nm. Samples were agitated for 15 seconds between two reads.
Circular dichroism (CD)
Amyloid aggregation is accompanied by conformational change in the aggregating protein to β-sheet-rich structure57. To analyze secondary structure, far-UV (190–260 nm) CD spectra were recorded using Jasco 1500 spectropolarimeter and 1 mm path length quartz cuvette. Pre-formed TDP-432C aggregates & samples incubated with AIM4 were diluted to 0.06 mg/ml using two-fold dilute PBS and spectra were recorded at a scan speed of 50 nm/minute. Likewise, CD spectrum of the protein sample (0.8 mg/ml) was also recorded before aggregation to examine the monomeric protein’s structural features. All spectra were baseline corrected and results were expressed as Mean Residue Ellipticity, [ϴ]MRE, using the formula: [ϴ]MRE = (ϴ *MRW)/10*c*l, where ϴ represents ellipticity in millidegrees, c is concentration of protein in g/ml, l is path length in cm, and the mean residue weight (MRW) was taken as 112. Relative secondary structural features were estimated from the CD spectra at online servers DichroWeb using the algorithms CONTIN and CDSSTR and reference set SP175 optimized for 190–240 nm range58,59,60,61 and BeStSel refined for precise β-sheet content estimation62.
Atomic force microscopy (AFM)
Morphology of amyloid aggregates can be analyzed by AFM13,63. Morphologies of TDP-432C protein aggregates were visualized by Nanoscope V scanning probe microscope (Bruker Instruments). Aliquot (5 μl) of pre-formed aggregates of TDP-432C obtained using the method described above were diluted five-fold in the same buffer and deposited on a freshly cleaved mica sheet and incubated for 15–30 minutes for adsorption. The samples were washed several times with deionized water and excess water removed from sides by Whatman filter paper. The mica sheets were left to air-dry at RT for 30 minutes and were imaged in tapping mode using OTESPA silicon cantilever with an aluminium reflective coating on the backside (Bruker AFM probes) with a spring constant of 42 N/m and tip radius ∼7 nm. Imaging was carried out in height mode with a scan rate of 1 Hz. Images were analysed for TDP-432C aggregate’s height and length using the software WSxM64.
Inhibition of TDP-432C in vitro aggregation
Imidazolium-tagged acridine derivatives were synthesized as reported earlier37. Stock solutions (50 mM) of the acridine derivatives (AIM1-5) were prepared by first dissolving in 100% dimethyl sulfoxide (DMSO) and then adjusting the volume with 400 mM Tris-HCl buffer to obtain final pH of 7.5 and final DMSO at 25%. All solutions were filtered through 0.2 μm PVDF membrane filter. Inhibitory potential of acridine compounds (AIM1-5) against TDP-432C aggregation was investigated using ThT fluorescence assay. For this, 1:10 protein to compound molar ratio was examined using DMSO at 2% final in the aggregation buffer. The ThT fluorescence observed for TDP-432C aggregation in the absence of any compound was taken as 100%. Stock solutions of acridine (Acr), Acr-E (4,5-bis(hydroxymethyl)acridine) and AIM (4,5-bis(imidazole methyl)acridine) were prepared in 100% DMSO and diluted to 8% DMSO final in the aggregation buffer to assess their effect on the aggregation of TDP-432c. Additional biophysical analysis tools like circular dichroism & AFM were also employed subsequently to confirm the inhibition of TDP-432C aggregation.
Inhibition of TDP-43-YFP aggregation in yeast
Yeast culture and transformation
S. cerevisiae cells with erg6 deletion, were transformed with a plasmid encoding full-length TDP-43 tagged with YFP using a modified transformation protocol previously described for erg6 deletion yeast cells65. ERG6, which encodes a protein involved in the ergosterol biosynthetic pathway when deleted enhances membrane permeability for small compounds in yeast66. Yeast media and growth conditions were followed as per standard protocols67. Briefly, the erg6Δ yeast cells were grown overnight at 30 °C in plasmid selective synthetic complete media lacking uracil and containing 1% raffinose as the sugar (SRaf-Ura) until they reached mid-log phase. Different concentrations of galactose (ranging from 2% to 0.001%) were added to induce de novo aggregation of TDP-43-YFP from the GAL1 promoter. In order to investigate the effect of acridine compounds on the aggregation pattern of TDP-43-YFP, different concentrations of the compounds (from 50 μM to 200 μM) were also added along with galactose.
Fluorescence microscopy
Aliquots of yeast cells expressing TDP-43-YFP with or without an inhibitor, were withdrawn at 2, 4 and 6 hours and YFP fluorescence pattern was monitored using Leica DM2500 fluorescence microscope using 100X, oil immersion objective lens. Images were acquired in bright-field mode as well as through GFP-filter. Formation of punctate YFP foci indicated aggregation of TDP-43-YFP.
Growth Curve
To analyze if presence of AIM4 has any effect on the growth of the yeast cells, growth kinetics was monitored. For this, erg6Δ yeast cells expressing TDP-43-YFP using 0.1% galactose as well as those not expressing TDP-43-YFP, were grown respectively in SRaf-Ura with 0.1% galactose or without galactose and the growth was recorded by absorbance at 600 nm every 5 minutes for a period of over 70 hours using SpectraMax M5e multi-mode reader (Molecular Devices).
Western blotting
To examine whether the expression levels of TDP-43-YFP protein was altered when the yeast cells were induced with 0.01% galactose & grown in the presence or absence of AIM4 (200 μM) and western blot analysis was performed. Yeast cells were lysed by vortexing with glass beads at 4 °C in CelLytic™ Y (Cat. C4482S, Sigma-Aldrich) yeast cell lysis reagent as per the manufacturer’s instructions. The cell lysate was then pre-cleared of the cellular debris at 4 °C by centrifugation at 2000 rpm for 3 minutes using a refrigerated table-top micro-centrifuge (Micro CL21R, ThermoFisher Scientific). The supernatant was carefully decanted and total protein concentration was estimated using Pierce™ BCA Protein Assay Kit (Cat. 23225, ThermoFisher Scientific). Normalized amounts of equal total protein were electrophoresed on 10% SDS-PAGE and the proteins were electro-blotted to polyvinylidene difluoride (PVDF) membrane which was then incubated with western blot blocking buffer (Cat. ML044, Hi-Media Labs) for 90 minutes and washed with PBS containing 0.1% Tween-20 (PBS-T). The membrane was probed for the TDP-43-YFP using mouse monoclonal anti-GFP primary antibody (Cat. G6795, Sigma-Aldrich) at 1:500 dilutions followed by incubation with anti-mouse secondary antibody tagged with alkaline phosphatase (APP) (Cat. A3562, Sigma-Aldrich) at 1:2000 dilution. Protein levels of GAPDH were examined for protein loading controls. The membrane was probed using rabbit monoclonal anti-GAPDH primary antibody (custom-made, Genescript) at 1:1000 dilutions followed by incubation with anti-rabbit secondary antibody tagged with alkaline phosphatase (Cat. A3562, Sigma-Aldrich) at 1:2000 dilution. Then, BCIP/NBT liquid substrate (Cat. B6404, Sigma-Aldrich) which leads to development of color band due to action of APP was used to visualize the levels of the TDP-43-YFP protein. A molecular weight standard (Cat. SM0671, Fermentas), electrophoresed and electro-blotted simultaneously was used for assignments of molecular weights to the TDP-43-YFP and GAPDH proteins.
Real-Time PCR
To estimate the mRNA levels of TDP-43-YFP, the erg6Δ mutant yeast expressing TDP-43-YFP in presence or absence of AIM4 were grown for 4 hours at 30 °C in presence of 0.01% galactose respectively after treatment with 200 μM of AIM4 or DMSO buffer as the control. Cells were harvested & lysed by glass beads and RNA was isolated by the standard acidic phenol-chloroform extraction method. The extract was treated with DNase I enzyme to eliminate genomic DNA contamination and was further purified using RNeasy MinElute Cleanup Kit (Qiagen: 74204). cDNA was prepared using High Capacity cDNA kit (Applied Biosystems) and real-time PCR was carried out using SYBR Green (Fast SYBR from Applied Biosystems) on Mx3005 P qPCR System (Agilent). ACT1 (Actin) was used as the endogenous control and amplified using 0.5 μM of forward (5′TGGATTCCGGTGATGGTGTT3′) & reverse (5′CAGCAGTGGTGGAGAAAGAGTA3′) primers were used. Likewise, for TDP-43, 0.5 μM of forward (5′GGACGATGGTGTGACTGCAAAC3′) & reverse (5′CAAAGGCAAAGGCCCTGAATGG3′) primers were used. Data was represented as fold-change in mRNA level in presence of AIM4 by comparison with no inhibitor sample.
Flow Cytometry
To examine any cytotoxic effects of AIM4 on yeast cells, flow cytometry was employed using propidium iodide and 7-AAD (7-amino-actinomycin D) staining of dead yeast cells. First, the yeast cells bearing the TDP-43-YFP plasmid were grown overnight at 30 °C in SRaf-Ura broth medium where the expression of TDP-43-YFP is turned off. Next morning, aliquots of cells from the overnight culture were transferred to fresh SRaf-Ura broth medium and allowed to grow to mid-log phase (OD600nm: 0.5–0.6). Subsequently, TDP-43-YFP expression from its GAL1 promoter was induced by addition of galactose to either 0.01% or 0.1% final and the cells were allowed to grow either without and with AIM4 (200 μM final) for 4 hours. Then, more than one million cells were harvested, washed and stained with 7-AAD (Cat. 555815, BD Biosciences) & propidium iodide (Cat. P4864, Sigma-Aldrich) and incubated for 15 minutes in dark. Finally, 50000 Cells were analyzed in flow cytometer (FACS AriaIII, BD Biosciences) in the filters PerCP-Cy5-5A for 7-AAD and PE-Texas Red-A for the propidium iodide.
Additional Information
How to cite this article: Prasad, A. et al. An acridine derivative, [4,5-bis{(N-carboxy methyl imidazolium)methyl}acridine] dibromide, shows anti-TDP-43 aggregation effect in ALS disease models. Sci. Rep. 6, 39490; doi: 10.1038/srep39490 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. C.S. Sharma & Mr. S. Mattaparthi from Dept. of Chemical Engineering, IIT-Hyderabad and Mr. J. Joseph from Dept. of Electrical Engineering IIT-Hyderabad, for useful help with AFM data acquisition. We also thank Prof. Susan W Liebman, University of Nevada Reno, USA for kindly gifting the yeast strain and plasmids. AP, GR & AG are thankful to MHRD, Govt. of India, for research fellowship. VS thanks UGC, Govt of India, for Senior Research Fellowship. GP thanks DST, Govt. of India, for research grant (No. SB/S1/IC-07/2014) that partly supported this work. We also thank IIT-Hyderabad funded by MHRD, Govt. of India, for research infrastructure & support.
Footnotes
Author Contributions A.P., G.R., V.S., A.G., M.V. and A.V. performed the experiments. V.T., G.P. and B.K.P. directed the studies. G.P. and B.K.P. conceived the studies and analyzed the data. A.P., V.S. and B.K.P. wrote the manuscript.
References
- Chiti F. & Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- Nelson R. et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine H. 3rd. Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 2, 404–410 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk W. E., Pettegrew J. W. & Abraham D. J. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. 37, 1273–1281 (1989). [DOI] [PubMed] [Google Scholar]
- Park S. K. et al. Development and validation of a yeast high-throughput screen for inhibitors of Abeta42 oligomerization. Dis. Model. Mech. 4, 822–831 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. R. et al. Oral treatment with Cu(II)(atsm) increases mutant SOD1 in vivo but protects motor neurons and improves the phenotype of a transgenic mouse model of amyotrophic lateral sclerosis. J. Neurosci. 34, 8021–8031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S., Cozzolino M. & Teresa Carri M. Old versus new mechanisms in the pathogenesis of ALS. Brain Pathol. 26, 276–286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R. et al. Superoxide dismutase 1 and tgSOD1 mouse spinal cord seed fibrils, suggesting a propagative cell death mechanism in amyotrophic lateral sclerosis. PLoS ONE 5, e10627, 10.1371/journal.pone.0010627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y., Kaneko K., Watanabe S., Yamanaka K. & Nukina N. Intracellular seeded aggregation of mutant Cu, Zn-superoxide dismutase associated with amyotrophic lateral sclerosis. FEBS Lett. 587, 2500–2505 (2013). [DOI] [PubMed] [Google Scholar]
- Grad L. I. et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 111, 3620–3625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch C., O’Brien J. & Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. USA 108, 3548–3553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y., Kaneko K., Watanabe S., Yamanaka K. & Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J. Biol. Chem. 286, 18664–18672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T. et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 4, 124–134 (2013). [DOI] [PubMed] [Google Scholar]
- Tsuji H. et al. Molecular analysis and biochemical classification of TDP-43 proteinopathy. Brain 135, 3380–3391 (2012). [DOI] [PubMed] [Google Scholar]
- Nomura T. et al. Intranuclear aggregation of mutant FUS/TLS as a molecular pathomechanism of amyotrophic lateral sclerosis. J. Biol. Chem. 289, 1192–1202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E. et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11, 323–330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004). [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Polymenidou M. & Cleveland D. W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casafont I., Bengoechea R., Tapia O., Berciano M. T. & Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J. Struct. Biol. 167, 235–241 (2009). [DOI] [PubMed] [Google Scholar]
- Sreedharan J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala Y. M., Misteli T. & Baralle F. E. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc. Natl. Acad. Sci. USA 105, 3785–3789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton C. F. et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 285, 6826–6834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A. & Vande Velde C. Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell. Neurosci. 9, 423, 10.3389/fncel.2015.00423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesiridis G. S., Lee V. M. & Trojanowski J. Q. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum. Mol. Genet. 18, R156–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. S., McCaffery J. M., Lindquist S. & Gitler A. D. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 105, 6439–6444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. S. et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden A. C. et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M. et al. Methylene blue and dimebon inhibit aggregation of TDP-43 in cellular models. FEBS Lett. 583, 2419–2424 (2009). [DOI] [PubMed] [Google Scholar]
- Zlatic C. O. et al. Fluphenazine. HCl and Epigallocatechin Gallate Modulate the Rate of Formation and Structural Properties of Apolipoprotein C-II Amyloid Fibrils. Biochemistry 54, 3831–3838 (2015). [DOI] [PubMed] [Google Scholar]
- Ono K. et al. Potent anti-amyloidogenic and fibril-destabilizing effects of polyphenols in vitro: implications for the prevention and therapeutics of Alzheimer’s disease. J. Neurochem. 87, 172–181 (2003). [DOI] [PubMed] [Google Scholar]
- Gazova Z. et al. Acridine derivatives inhibit lysozyme aggregation. Eur. Biophys. J. 37, 1261–1270 (2008). [DOI] [PubMed] [Google Scholar]
- Korth C., May B. C., Cohen F. E. & Prusiner S. B. Acridine and phenothiazine derivatives as pharmacotherapeutics for prion disease. Proc. Natl. Acad. Sci. USA 98, 9836–9841 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B. C. H. et al. Potent inhibition of scrapie prion replication in cultured cells by bis-acridines. Proc. Natl. Acad. Sci. USA 100, 3416–3421 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju G., Vishwanath S., Prasad A., Patel B. K. & Prabusankar G. Imidazolium tagged acridines: Synthesis, characterization and applications in DNA binding and anti-microbial activities. J. Mol. Struct. 1107, 291–299 (2016). [Google Scholar]
- Xu Z., Kim S. K. & Yoon J. Revisit to imidazolium receptors for the recognition of anions: highlighted research during 2006-2009. Chem. Soc. Rev. 39, 1457–1466 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang D. et al. Acridine-based macrocyclic fluorescent sensors: self-assembly behavior characterized by crystal structures and a tunable bathochromic-shift in emission induced by H2PO4-via adjusting the ring size and rigidity. Org. Biomol. Chem. 11, 3375–3381 (2013). [DOI] [PubMed] [Google Scholar]
- Arakawa T. & Timasheff S. N. Preferential interactions of proteins with salts in concentrated solutions. Biochemistry 21, 6545–6552 (1982). [DOI] [PubMed] [Google Scholar]
- Jiang L.-L. et al. Two mutations G335D and Q343R within the amyloidogenic core region of TDP-43 influence its aggregation and inclusion formation. Sci. Rep. 6, 23928, 10.1038/srep23928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. K. et al. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J. Am. Chem. Soc. 132, 1186–1187 (2010). [DOI] [PubMed] [Google Scholar]
- Armakola M., Hart M. P. & Gitler A. D. TDP-43 toxicity in yeast. Methods 53, 238–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala Y. M. et al. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785 (2008). [DOI] [PubMed] [Google Scholar]
- Igaz L. M. et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J. Biol. Chem. 284, 8516–8524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley M. D. & Gitler A. D. Yeast genetic screen reveals novel therapeutic strategy for ALS. Rare Diseases 1, e24420, 10.4161/rdis.24420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hard T. & Lendel C. Inhibition of amyloid formation. J. Mol. Biol. 421, 441–465 (2012). [DOI] [PubMed] [Google Scholar]
- Nie Q., Du X. G. & Geng M. Y. Small molecule inhibitors of amyloid beta peptide aggregation as a potential therapeutic strategy for Alzheimer’s disease. Acta Pharmacol. Sin. 32, 545–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E. et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl. Acad. Sci. USA 109, 9629–9634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi H. et al. Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation, and mechanism of action. Angew. Chem. 42, 2758–2761 (2003). [DOI] [PubMed] [Google Scholar]
- Rubin J. et al. Ion-specific effects on prion nucleation and strain formation. J. Biol. Chem. 288, 30300–30308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R. & Lindquist S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necula M., Kayed R., Milton S. & Glabe C. G. Small molecule inhibitors of aggregation indicate that amyloid beta oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 282, 10311–10324 (2007). [DOI] [PubMed] [Google Scholar]
- Carole D. G. et al. Synthesis and antileishmanial activities of 4,5-di-substituted acridines as compared to their 4-mono-substituted homologues. Bioorg. Med. Chem. 13, 5560–5568 (2005). [DOI] [PubMed] [Google Scholar]
- Armarego W. L. F. & Chai C. L. L. Purification of Organic Chemicals In Purification of Laboratory Chemicals (Fifth Edition) 80–388 (Butterworth-Heinemann, 2003). [Google Scholar]
- Gill S. C. & von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 (1989). [DOI] [PubMed] [Google Scholar]
- Li Huiyuan, Rahimi Farid, Sinha Sharmistha, Maiti P. & Bitan G. Amyloids and Protein Aggregation-Analytical Methods In Encyclopedia of Analytical Chemistry (ed. Meyers R. A.) 1–32 (John Wiley & Sons Ltd, 2009). [Google Scholar]
- Whitmore L. & Wallace B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–673 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher S. W. & Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 (1981). [DOI] [PubMed] [Google Scholar]
- Compton L. A. & Johnson W. C. Jr. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155, 155–167 (1986). [DOI] [PubMed] [Google Scholar]
- Lees J. G., Miles A. J., Wien F. & Wallace B. A. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 22, 1955–1962 (2006). [DOI] [PubMed] [Google Scholar]
- Micsonai A. et al. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 112, E3095–3103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. L. et al. Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J. Biol. Chem. 288, 19614–19624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcas I. et al. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 78, 013705, 10.1063/1.2432410 (2007). [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Copple D. M., Kennedy B. K., Vidal M. & Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9, 3447–3456 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 350, 3–41 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.