Abstract
Pseudomonas aeruginosa (PA) can thrive in anaerobic biofilms in the lungs of cystic fibrosis (CF) patients. Here, we show that CrcZ is the most abundant PA14 RNA bound to the global regulator Hfq in anoxic biofilms grown in cystic fibrosis sputum medium. Hfq was crucial for anoxic biofilm formation. This observation complied with an RNAseq based transcriptome analysis and follow up studies that implicated Hfq in regulation of a central step preceding denitrification. CrcZ is known to act as a decoy that sequesters Hfq during relief of carbon catabolite repression, which in turn alleviates Hfq-mediated translational repression of catabolic genes. We therefore inferred that CrcZ indirectly impacts on biofilm formation by competing for Hfq. This hypothesis was supported by the findings that over-production of CrcZ mirrored the biofilm phenotype of the hfq deletion mutant, and that deletion of the crcZ gene augmented biofilm formation. To our knowledge, this is the first example where competition for Hfq by CrcZ cross-regulates an Hfq-dependent physiological process unrelated to carbon metabolism.
PA can form persistent biofilms in the lungs of CF patients1. Polymorphonuclear leukocytes are known to surround the biofilms in the CF lung and to consume the majority of O2 to produce reactive oxygen species, which suggested that PA biofilms may partially grow anaerobically in this environment2,3. Based on several studies, Yoon et al.4 provided first hints for anaerobic respiration in PAO1 biofilms. This has been recently supported through the quantification of compounds of the denitrification pathway in sputum of CF patients5.
In Enterobacteriaceae, the RNA chaperone Hfq is known to play a key role in riboregulation by facilitating the interaction between small regulatory RNAs (sRNAs) and their target mRNAs6. As a number of sRNAs have been shown to regulate virulence traits including biofilm formation in different bacterial pathogens7, the requirement for Hfq in RNA-mediated regulation most likely contributes to the observed attenuated virulence phenotype of respective hfq mutants6. In PAO1, a deletion of the hfq gene resulted in pleiotropic effects on growth and virulence8.
Although many putative regulatory RNAs have been identified in different PA strains9, the function of only a few has been revealed, and only three regulatory RNAs have been implicated in biofilm formation. The sRNA PhrS represents the founding member of anaerobically controlled PA sRNAs10. PhrS has been suggested to be involved in biofilm formation as it was shown to stimulate the synthesis of the Pseudomonas quinolone signal (PQS)10, which can induce the release of DNA that serves as a biofilm matrix component11. A stimulatory effect of PhrS on biofilm formation has been recently observed12. The PA protein binding RNAs RsmY and RsmZ antagonize the function of the translational regulator RsmA13. The RsmA protein is known to act as a translational repressor of psl mRNA, which prevents exopolysaccharide synthesis, and thus to control biofilm formation in a negative manner14. On the other hand, up-regulation of the RsmY/Z RNAs results in titration of RsmA, and therefore in increased biofilm formation7,13.
The aim of this study was to identify PA14 regulatory RNAs involved in anoxic biofilm formation, which is a poorly studied aspect of chronic PA infections. We show that the Hfq-binding RNA CrcZ is highly abundant under these conditions, and that it impacts on anoxic biofilm formation. Thus, in addition to its established role in carbon catabolite repression, where CrcZ acts as a decoy to abrogate Hfq-mediated translational repression of catabolic genes15, this study reveals a novel aspect of Hfq sequestration by CrcZ, that is cross-regulation of other Hfq-dependent physiological processes.
Results and Discussion
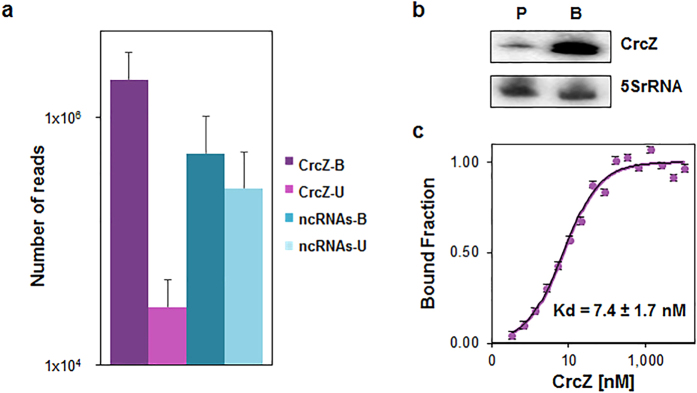
With the goal to identify regulatory RNAs that impact on anoxic biofilm formation, we concentrated on RNAs that interact with Hfq. The PA14 strain was grown in modified cystic fibrosis sputum medium (SCFM)16, which approximates to the conditions of the CF lung. Upon anaerobic biofilm growth of PA14 for 96 h in SCFM (B-96 cultures), Hfq-bound RNAs were isolated by co-immunoprecipitation with Hfq-specific antibodies. The identity of Hfq-bound and unbound putative and confirmed regulatory RNAs of PA1417 was revealed by RNAseq (Supplementary Table S1). All other reads were excluded from further analyses. Based on the total reads obtained for these RNAs (Hfq-bound and unbound), CrcZ was the most abundant PA14 regulatory RNA in B-96 cultures (Fig. 1a). The RNAseq results were mirrored by a Northern-blot analysis, showing that the levels of CrcZ RNA were ~50-fold higher in B-96 cultures than in planktonically grown PA14 cultures (OD600 = 2.0; P) (Fig. 1b). Moreover, the reads obtained for Hfq-bound CrcZ RNA outnumbered all other described or putative PA14 regulatory RNAs that interacted with Hfq by a factor of 4 (Fig. 1a). For verification, the Hfq-bound and unbound fractions were tested by RT-PCR for the presence of CrcZ RNA, ErsA RNA, which requires Hfq for function18, and RsmZ RNA, which poorly binds to Hfq19. Both, CrcZ and ErsA, were found in complex with Hfq, whereas the majority of RsmZ RNA was detected in the unbound fraction (Supplementary Fig. S1).
Figure 1. CrcZ is the major regulatory RNA bound to Hfq in anoxic biofilms.
(a) Hfq-bound RNAs were isolated from lysates of B-96 cultures after co-immunoprecipitation with Hfq-specific antibodies. The identity of Hfq-bound and unbound RNAs in the supernatant was revealed by RNAseq. The number of reads for CrcZ in the Hfq-bound fraction (CrcZ-B; dark purple bar) and unbound fraction (CrcZ-U; light purple bar) is shown in comparison to the number of reads for all other described or putative PA14 regulatory RNAs in the Hfq-bound (ncRNAs-B; blue bar) and unbound fraction (ncRNAs-U; light blue bar). Error bars, mean ± s.d. of 2 biological replicates. (b) The steady state levels of CrcZ are elevated in B-96 cultures. The PA14 strain was grown in SCFM under aerobic conditions to an OD600 of 2 (P) and for 96 hours in anoxic biofilms (B), respectively. Total RNA was isolated and 10 μg of total RNA was used for the Northern-blot analysis. (c) Kd of Hfq for CrcZ RNA revealed by microscale thermophoresis. Increasing amounts of non-labelled in vitro transcribed CrcZ were added to 20 nM fluorescently labelled Hfq protein. The dissociation constant (Kd) of CrcZ was determined as described47, and was expressed as mean EC50 ± EC50 confidence interval of 2 independent experiments.
CrcZ transcription requires the alternative sigma factor RpoN and the response regulator CbrB, which is phosphorylated by the sensor/histidine kinase CbrA20,21. A recent transcriptome analysis indicated that the cbrA gene is 2.5-fold up-regulated in B-96 cultures when compared to P cultures22. This observation may partially rationalize the up-regulation of CrcZ in B-96 cultures. On the other hand, a putative motif for the anaerobic regulator Anr was identified upstream of the RpoN-dependent crcZ promoter (Supplementary Fig. S2). As crcZ was poorly expressed in a PA14anr- strain (Supplementary Fig. S2), it remains to be shown whether the observed up-regulation of crcZ in B-96 cultures (Fig. 1b) is indeed mediated by Anr.
As the intracellular quantities of a given Hfq-bound RNA depend on its abundance as well as on its affinity for the protein, we next determined the Kd of Hfq for CrcZ using microscale thermophoresis. The Kd was determined with ~7.4 nM (Fig. 1c). The high Kd of Hfq for CrcZ can be reconciled with six A-rich stretches in CrcZ15,20 to which Hfq can bind with its distal tripartite binding motifs15,23. The majority of Hfq was co-immunoprecipitated from the lysate of B-96 cultures (Supplementary Fig. S3) and CrcZ was the most abundant Hfq-binding RNA (Fig. 1a). Therefore, we next asked whether Hfq is crucial for and whether competition for Hfq by CrcZ can interfere with anoxic growth and biofilm formation.
To test whether Hfq is crucial for anaerobiosis, the growth of PA14 and PA14Δhfq was monitored in SCFM. Under these conditions growth of the PA14Δhfq was impaired (Supplementary Fig. S4). In addition, the metabolic activity of PA14 and PA14Δhfq was assessed during anoxic growth in SCFM by employing a PrrnBP1-gfp(AGA) reporter gene24. When compared with strain PA14, the absence of Hfq resulted in a decrease of the GFP activity, indicating that PA14Δhfq is less metabolically active than the wild-type strain (Supplementary Fig. S4).
These initial studies prompted us to perform a RNAseq based comparative transcriptome analysis with B-96 cultures of PA14 and the PA14Δhfq mutant, respectively, to assess the impact of Hfq on anoxic biofilm formation in more detail. For the initial analysis a p-value (adjusted for multiple testing) of 0.05 was set as a threshold for statistical significance and the change in abundance (fold change) had to exceed ±4 for a given transcript in order to be considered differentially abundant. In addition, transcripts were also considered, if they were differentially abundant at least ±1.5-fold and part of an affected pathway or operon (Supplementary Table S2). 428 transcripts were found to be differentially expressed in the hfq mutant when compared with PA14; 296 and 132 transcripts showed at least a 4-fold increased and decreased abundance, respectively (Supplementary Table S2).
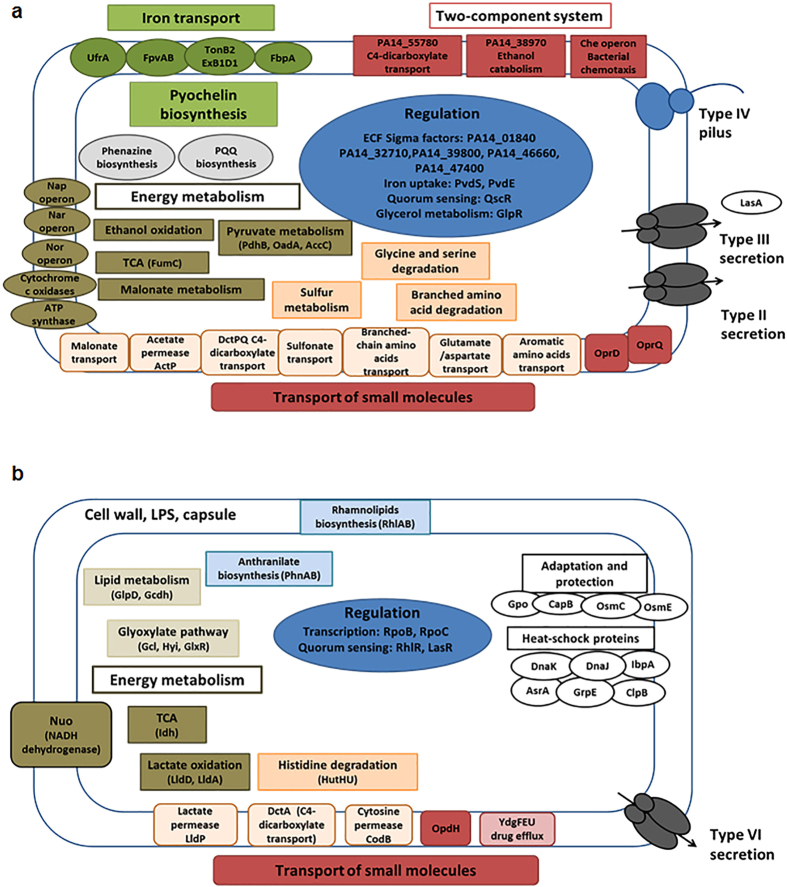
These analyses revealed functions implicated in anoxic biofilm formation, which might be under direct or indirect control of Hfq (Fig. 2, Supplementary Table S2). Among them, the transcript encoding the quorum-sensing regulator QscR, known to act as a negative regulator of LasR and RhlR25, was 9.23-fold up-regulated in the absence of Hfq (Fig. 2a, Supplementary Table S2a). Accordingly, transcripts encoding the quorum-sensing regulators RhlR and LasR, as well as downstream genes involved in rhamnolipid biosynthesis (rhlAB) were down-regulated in the absence of Hfq (Fig. 2b, Supplementary Table S2b). As an impairment of QS can result in killing of PAO1 in anaerobic biofilms4, a de-regulation of the qscR gene in PA14Δhfq could thus impact on anoxic biofilm formation.
Figure 2. Hfq regulon in B-96 cultures.
Regulatory and metabolic networks affected by Hfq. Increased (a) and decreased abundance (b) of the transcripts encoding for the corresponding proteins in PA14Δhfq versus PA14. Functional classification according to the Pseudomonas Genome database52 and KEGG (Kyoto Encyclopedia of Genes and Genome)53.
The abundance of the transcript encoding glycerol-3-phosphate dehydrogenase GlpD, which serves as a major link between carbohydrate and lipid metabolism, was 4.84-fold decreased in the absence of Hfq (Fig. 2b, Supplementary Table S2b). This observation can be reconciled with the 11.51-fold increased abundance of the glpR transcript encoding the negative regulator of glpD (Fig. 2a, Supplementary Table S2a). As glycerol metabolism plays an important role in P. aeruginosa persistence by promoting biofilm formation26, a possible de-regulation of glpD in PA14Δhfq might also interfere with anoxic biofilm formation.
PA can thrive under anaerobic conditions by acquiring ATP from glycolysis, pyruvate fermentation27, the arginine deiminase pathway28 as well as from denitrification29. When compared with PA14, the absence of Hfq resulted in increased abundance of transcripts encoded by the nar, nap and nor operons, encoding enzymes required for denitrification (Fig. 2a, Supplementary Table S2a). In contrast, the transcripts encoding the nitrite reductase (nir-operon) and the nitrous dioxide reductase (nos operon) did not show a differential abundance. However, a number of nuo transcripts, encoding subunits of the NADH dehydrogenase, were down-regulated in the absence of Hfq (Fig. 2b, Supplementary Table S2b). The NADH dehydrogenase is required for anaerobic growth in the presence of nitrate30. It contributes to the intracellular redox balance, i.e. the NADH/NAD+ ratio31, and its activity is coupled to the energizing processes of the membrane and ATP synthesis32, which in turn is required for sustained anoxic biofilm formation.
A redox imbalance can impact on the fitness of bacteria33. As the transcriptome analysis indicated a lower abundance of some nuo transcripts as well as of other transcripts encoding dehydrogenases (glpD; lldA; lldD; Supplementary Table S2b) in the absence of Hfq, we next asked whether Hfq affects the intracellular NADH/NAD+ ratio. The NADH/NAD+ ratios were determined in B-96 cultures of strains PA14, PA14ΔcrcZ, PA14(pcrcZ) and PA14Δhfq, respectively. When compared with strains PA14 and PA14ΔcrcZ, an increase in NADH/NAD+ ratio was observed in PA14Δhfq (Fig. 3). As the NADH/NAD+ ratio is crucial for the function of metabolic pathways, it seems reasonable to speculate that the reduced metabolic activity of the PA14Δhfq strain (Supplementary Fig. S4) results at least in part from a redox imbalance.
Figure 3. The absence of Hfq causes a redox imbalance.
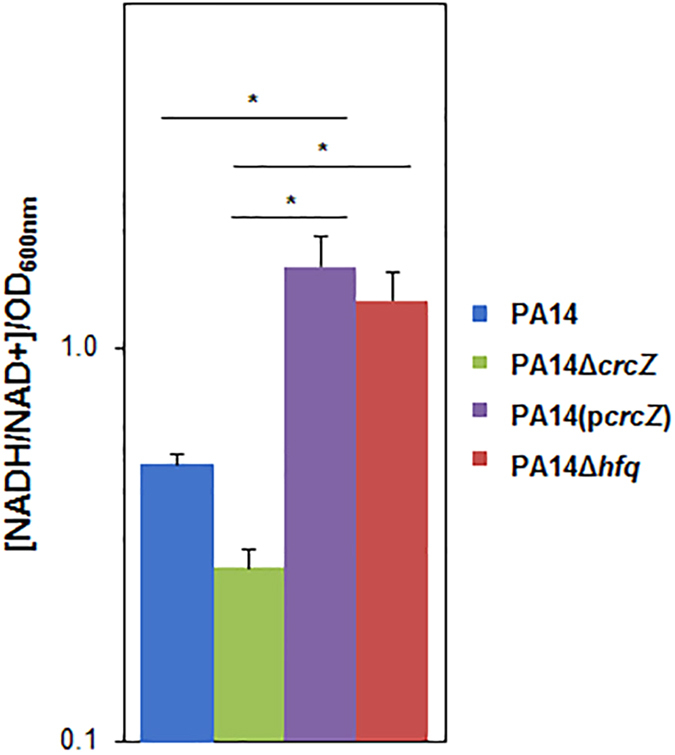
The NADH/NAD+ levels were assessed in B-96 cultures of PA14 (blue bar), PA14ΔcrcZ (green bar), PA14(pcrcZ) (violet bar) and PA14Δhfq (red bar) grown in SCFM48. Error bars, mean ± s.d. N = 3 biological replicates with 3 technical replicates. *P < 0.05, analysed by one-way ANOVA with the Tukey’s HSD post hoc test.
Since the deletion of the crcZ gene augmented the metabolic activity of strain PA14ΔcrcZ (Supplementary Fig. S4) and over-expression of a plasmid borne copy of crcZ mirrored both the anoxic growth phenotype and the increased NADH/NAD+ ratio of the PA14Δhfq strain (Supplementary Fig. S4, Fig. 3), we next examined whether differential CrcZ levels affect anoxic biofilm formation. First, anoxic biofilm formation was assessed by a static crystal violet assay34 in B-96 cultures of the strains PA14, PA14ΔcrcZ, PA14(pcrcZ) and PA14Δhfq, respectively. When compared with PA14, anoxic biofilm formation was increased in PA14ΔcrcZ, whereas ectopic over-production of CrcZ (Supplementary Fig. S5) reduced anoxic biofilm formation to a level comparable with the PA14Δhfq strain (Supplementary Fig. S5).
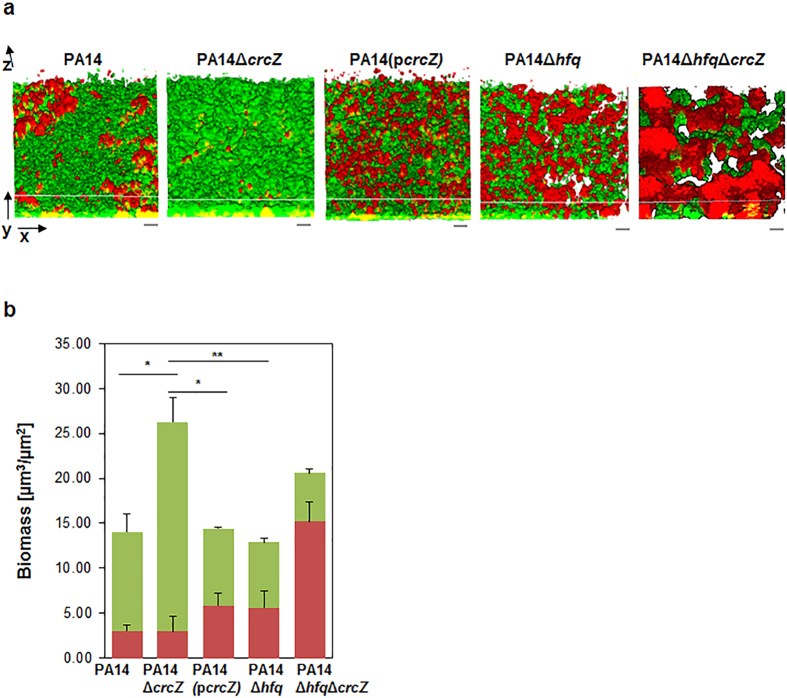
Second, the viability and biomass of B-96 cultures of PA14, PA14ΔcrcZ, PA14(pcrcZ) and PA14ΔhfqΔcrcZ was assessed by live/death staining and visualized by confocal laser scanning microscopy (Fig. 4). When compared with PA14, a marked increase in both, the total and live cell biomass was observed for PA14ΔcrcZ (Fig. 4). In contrast, when compared to PA14, an increase of dead cells was observed in anoxic biofilms of the CrcZ over-producing strain PA14(pcrcZ) and of PA14Δhfq (Fig. 4). Moreover, the PA14ΔhfqΔcrcZ displayed a marked increase in dead cell biomass and decrease of viable cells (Fig. 4). Taken together, these results corroborate the idea the competition for Hfq by CrcZ perturbs metabolic pathways, which results in diminished anoxic growth and biofilm formation in P. aeruginosa.
Figure 4. Biofilm formation of B-96 cultures of PA14, PA14ΔcrcZ, PA14(pcrcZ), PA14Δhfq and PA14ΔhfqΔcrcZ.
(a)The distribution of live and death cells in B-96 cultures of PA14, PA14ΔcrcZ, PA14(pcrcZ), PA14Δhfq and PA14ΔhfqΔcrcZ were visualized by CLSM. Live cells (green) were stained with Syto 9 fluorescent dye, whereas dead cells (red) were visualized with propidium iodide. Three-dimensional images of biofilms (x, y, z-axes, scale bars represent 10 μm). The CrcZ levels in the different strains are shown in Supplementary Fig. S5a. (b) The biomass of live (green) and death (red) cells of B-96 cultures of PA14, PA14ΔcrcZ, PA14(pcrcZ), PA14Δhfq and PA14ΔhfqΔcrcZ were quantified from CLSM vertical image stacks using Comstat2 software51. Error bars, mean ± s.d. from three independent experiments. *P < 0.05, **P < 0.01, analysed by one-way ANOVA with the Tukey’s HSD post hoc test.
The CrcZ levels are known to increase in the presence of poor carbon sources or at a low carbon to nitrogen ratio20. Under these conditions, Hfq titration by CrcZ permits the uptake and utilization of non-preferred carbon sources15. When compared to planktonically grown cultures, the CrcZ levels were strongly increased in anoxic biofilms, which provides a further ambience where CrcZ can impact on Hfq function. Given the large Hfq regulon19 (Fig. 2) and the pivotal role of Hfq in PA physiology and virulence8, it appears worthwhile to explore crcZ regulation under a variety of conditions. A better understanding of the signals leading to CrcZ over-production could be valuable in view of developing strategies to attenuate the role of Hfq in PA pathogenicity.
CrcZ represents the first protein-binding RNA that limits biofilm formation. In contrast, several base-pairing sRNAs have been implicated in the regulation of biofilm formation in bacteria7. For example, the Vibrio cholerae VqmR RNA inhibits biofilm formation by translational silencing of the vpsT mRNA, encoding a transcriptional regulator of biofilm development35, whereas the Salmonella Typhimurium ArcZ RNA positively regulates rpoS36, which promotes biofilm formation37. Interestingly, over-expression of ArcZ RNA changed the profile of Hfq-bound sRNAs and mRNAs, which suggested that ArcZ titrates Hfq and thereby might also post-transcriptionally cross-regulate other genes36.
The biological function of CrcZ in limiting biofilm formation in PA might be associated with the complex environment in the CF lung. Recent studies indicated that polymorphonuclear leukocytes restrict the growth of PA in CF lungs, presumably by a high consumption of oxygen38. This could be linked with an increased CrcZ-mediated sequestration of Hfq under anoxic conditions and cessation of further biofilm formation, which in turn could be advantageous in terms of balancing the immune response and thus for establishing a long-term chronic infection.
Methods
Bacterial strains, plasmids and growth conditions
The strains and plasmids used in this study are listed in Supplementary Table S3. The synthetic cystic fibrosis sputum medium (SCFM) was prepared as described16. The concentration of FeSO4·7H2O was increased to 100 μM and that of KNO3 to 100 mM. This was done to allow for increased anaerobic biofilm formation after 96 h, which was required for the extraction of sufficient amounts of RNA for subsequent RNAseq analysis. For anoxic growth, 25 ml of bacterial cultures in SCFM were inoculated to an initial OD600 of 0.05 and then split in 1 ml aliquots into 5 ml polypropylene tubes. The cultures were incubated statically for 96 hours at 37 °C in a 2.5-liter anaerobic jar containing a gas pack (AN25; AnaeroGen, Oxoid, United Kingdom).
RNAseq and CoIP RNAseq library construction and sequence analysis
The RNAseq analysis with total RNA prepared from two biological replicates of B-96 cultures was performed as recently described22. Hfq-bound RNAs were isolated from lysates of B-96 cultures after co-immunoprecipitation with Hfq-specific antibodies as described15. The Hfq-unbound fraction included all RNAs isolated from the supernatant after co-immunoprecipitation.
Total RNA from all samples was isolated using the TRIzol reagent (Ambion) according to the manufacturer’s instructions. The samples were DNase I treated, followed by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. The Ribo-Zero rRNA Removal Kit (Gram-Negative Bacteria) Magnetic kit (Epicentre Biotechnologies) was used to deplete rRNA from samples used for RNAseq. Libraries were constructed using the NEBNext® Ultra™ Directional RNA Library Prep Kit from Illumina. RNA sequencing has been performed at the Next Generation Sequencing Facility (VBCF, Vienna, Austria). The samples were subjected to Illumina 100 nt single end sequencing. PCR and index primers were removed from the raw reads with cutadapt39 and quality control was performed with FastQC40. Mapping of the samples against the PA UCBPP-PA14 reference genome (accession number NC_008463) was performed with segemehl41,42. Mapped sequencing data were split by strand and further processed for automatic UCSC Genome Browser43 visualization with the ViennaNGS toolbox44. Read counting for subsequent DESeq45 differential gene expression analysis was performed with BEDtools46. Only reads corresponding to putative or confirmed regulatory RNAs of PA14 were considered in the analysis.
Northern-blot analyses
Total RNA was isolated by the hot phenol method as previously described10. The steady-state levels of CrcZ and 5 S rRNA (loading control) were determined using 10 μg of total RNA. The RNA samples were denatured for 10 min at 65 °C in loading buffer containing 50% formamide, separated on a 6% polyacrylamide/8 M urea gel, and then transferred to a nylon membrane by electroblotting. The RNAs were cross-linked to the membrane by exposure to UV light. The membranes were hybridized with gene-specific 32P-end-labelled oligonucleotides: K3 (CrcZ) and I26 (5 S rRNA), respectively (Supplementary Table S4). The hybridization signals were visualized using a PhosphorImager (Molecular Dynamics).
In vitro transcription of CrcZ
For in vitro transcription of CrcZ the AmpliScribe T7-Flash Transcription Kit (Epicentre Biotechnologies) was used according to the manufacturer’s instructions. The 426 nt PCR fragment generated with the oligonucleotides E6 and C6 (Supplementary Table S4) served as template.
Microscale thermophoresis
Specific binding between CrcZ and Hfq was determined by microscale thermophoresis (MST)47 using the Monolith NT.115 Green/Red apparatus (Nanotemper Technologies) at the Protein Technologies Facility (ProTech, VBCF, Vienna, Austria). Labelling of 20 μM of PA14 Hfq protein with NT-642 dye was performed using the Monolith™ Antibody Labelling Kit RED-NHS (Amine Reactive; Nanotemper Technologies) according to the manufacturer’s instructions. For the measurements, the concentration of the NT-642-labelled Hfq protein was kept constant (20 nM), whereas the concentrations of non-labelled in vitro transcribed CrcZ RNA varied from 1 to 1200 nM. The binding reactions were carried out in MST-buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 10 mM MgCl2) supplemented with 0.1% Tween and 0.1% BSA. The reactants were initially incubated at 37 °C for 30 min to enable CrcZ binding with Hfq. The samples were then loaded onto NT.115 hydrophilic capillaries (Nanotemper Technologies). The data for microscale thermophoresis analysis were recorded at 25 °C using the red LED (excitation: 625 nm, emission: 680 nm); both, MST and LED Power at 40%. Data analyses were performed with NTAnalysis software.
NADH assay
The [NADH]/[NAD+] ratios were determined in B-96 cultures using an enzyme cycling-based colorimetric assay48.
Microscopy
Image acquisition was performed at the MFPL BioOptics Facility with a Zeiss LSM 700 confocal laser scanning microscope (Carl Zeiss, Germany) equipped with 405, 488, 555 and 639 nm laser diodes and two PMT channels for simultaneous monitoring of GFP (excitation, 488 nm; emission, 517 nm) and propidium iodide (excitation, 543 nm; emission, 565 nm). Images were obtained using a Plan-Apochromat 40x/1.3 Oil DIC objective. Simulated multichannel cross sections were generated using Fiji: an open-source platform for biological-image analysis and Volume Viewer Plugin49. The quantitative analyses of acquired image stacks of biofilms was performed using Comstat2 software50,51.
Statistical analysis
All experiments were performed at least in duplicate. The data shown were obtained with three biological replicates, unless indicated otherwise. Statistical significance was determined by one-way ANOVA with the Tukey’s honestly significant difference (HSD) post hoc test when more than two groups with normal distribution were compared. Levene’s test was used to test for equality (homogeneity) of variances between tested groups. *P < 0.05, **P < 0.01 and ***P < 0.001 were considered statistically significant results.
Data availability
The raw sequencing data were deposited in the NCBI sequence read archive (SRA) as a study under the accession number SRP062593 (RNAseq) and SAMN05823591 (CoIP RNAseq), respectively. All other data generated or analysed during this study are included within this article and the Supplementary information or are available from the corresponding author upon request.
Additional Information
How to cite this article: Pusic, P. et al. Cross-regulation by CrcZ RNA controls anoxic biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 6, 39621; doi: 10.1038/srep39621 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We wish to thank Dr’s S. Lory and T. Tolker-Nielson for providing materials and Dr. B. Märtens for assistance with MST. This work was supported by the Austrian Science Fund (www.fwf.ac.at/en) through the Special Research Program RNA-REG F43, subproject AF4311(UB), and the doctoral program RNA-Biology W-1207 (PP, MT).
Footnotes
Author Contributions Conceived and designed the experiments: P.P., U.B. Performed the experiments: P.P., M.T. Analysed the data: P.P., M.T., M.W., E.S., S.H., U.B. Contributed reagents/materials/analysis tools: P.P., M.T., E.S., S.H., U.B. Wrote the paper: P.P., U.B.
References
- Folkesson A. et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10, 841–851 (2012). [DOI] [PubMed] [Google Scholar]
- Worlitzsch D. et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpen M. et al. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65, 57–62 (2010). [DOI] [PubMed] [Google Scholar]
- Yoon S. S. et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: Relationships to cystic fibrosis pathogenesis. Dev. Cell 3, 593–603 (2002). [DOI] [PubMed] [Google Scholar]
- Kolpen M. et al. Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. PLoS One 9, e84353 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. & Luisi B. F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G. H. & Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 (2015). [DOI] [PubMed] [Google Scholar]
- Sonnleitner E. et al. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35, 217–228 (2003). [DOI] [PubMed] [Google Scholar]
- Sonnleitner E., Romeo A. & Blasi U. Small regulatory RNAs in Pseudomonas aeruginosa. RNA Biol. 9, 364–371 (2012). [DOI] [PubMed] [Google Scholar]
- Sonnleitner E. et al. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol. Microbiol. 80, 868–885 (2011). [DOI] [PubMed] [Google Scholar]
- Yang L., Nilsson M., Gjermansen M., Givskov M. & Tolker-Nielsen T. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74, 1380–1392 (2009). [DOI] [PubMed] [Google Scholar]
- Fernandez L. et al. Interconnection of post-transcriptional regulation: The RNA-binding protein Hfq is a novel target of the Lon protease in Pseudomonas aeruginosa. Sci. Rep. 6, 26811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K., Schubert M., Allain F. H.-T. & Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253 (2008). [DOI] [PubMed] [Google Scholar]
- Irie Y. et al. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E. & Bläsi U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 10, e1004440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K. L., Aye L. M. & Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lozano M. et al. Diversity of small RNAs expressed in Pseudomonas species. Environ. Microbiol. Rep. 7, 227–236 (2015). [DOI] [PubMed] [Google Scholar]
- Ferrara S. et al. Post-transcriptional regulation of the virulence-associated enzyme AlgC by the sigma(22) -dependent small RNA ErsA of Pseudomonas aeruginosa. Environ. Microbiol. 17, 199–214 (2015). [DOI] [PubMed] [Google Scholar]
- Sonnleitner E., Schuster M., Sorger-Domenigg T., Greenberg E. P. & Blasi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59, 1542–1558 (2006). [DOI] [PubMed] [Google Scholar]
- Sonnleitner E., Abdou L. & Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 106, 21866–21871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdou L., Chou H.-T., Haas D. & Lu C.-D. Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa. J. Bacteriol. 193, 2784–2792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata M. et al. RNASeq based transcriptional profiling of Pseudomonas aeruginosa PA14 after short- and long-term anoxic cultivation in synthetic cystic fibrosis sputum medium. PLoS One 11, e0147811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link T. M., Valentin-Hansen P. & Brennan R. G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 106, 19292–19297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen L., Sternberg C. & Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6, 726–732 (2004). [DOI] [PubMed] [Google Scholar]
- Coggan K. A. & Wolfgang M. C. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14, 47–70 (2012). [PubMed] [Google Scholar]
- Scoffield J. & Silo-Suh L. Glycerol metabolism promotes biofilm formation by Pseudomonas aeruginosa. Can. J. Microbiol. 1–7, doi: 10.1139/cjm-2016-0119 (2016). [DOI] [PubMed] [Google Scholar]
- Eschbach M. et al. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186, 4596–4604 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C., Pierard A., Kley-Raymann M. & Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160, 928–934 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V. E., Bushnell D., Passador L., Brooks A. I. & Iglewski B. H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. & Paget M. S. Bacterial redox sensors. Nat Rev Micro 2, 954–966 (2004). [DOI] [PubMed] [Google Scholar]
- Williams H. D., Zlosnik J. E. A. & Ryall B. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71 (2007). [DOI] [PubMed] [Google Scholar]
- Marshall D. D., Sadykov M. R., Thomas V. C., Bayles K. W. & Powers R. Redox imbalance underlies the fitness defect associated with inactivation of the Pta-AckA pathway in Staphylococcus aureus. J. Proteome Res. 15, 1205–1212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J. H., Kadouri D. E. & O’Toole G. A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1, Unit 1B.1 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Forstner K. U., Cong J.-P., Sharma C. M. & Bassler B. L. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. USA 112, E766–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K. et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 74, 139–158 (2009). [DOI] [PubMed] [Google Scholar]
- Romling U. et al. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293, 273–285 (2003). [DOI] [PubMed] [Google Scholar]
- Kragh K. N. et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 82, 4477–4486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, pp–10 (2011). [Google Scholar]
- Andrews S. FastQC - A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- Hoffmann S. et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 5, e1000502 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S. et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 15, R34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger M. T., Fallmann J., Eggenhofer F. & Amman F. ViennaNGS: A toolbox for building efficient next- generation sequencing analysis pipelines. F1000Research 4, 50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. & Hall I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhr S. & Braun D. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. USA 103, 19678–19682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S. E., Price-Whelan A. & Newman D. K. Extraction and measurement of NAD(P)(+) and NAD(P)H. Methods Mol. Biol. 1149, 311–323 (2014). [DOI] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A. et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 1), 2395–2407 (2000). [DOI] [PubMed] [Google Scholar]
- Vorregaard M. Comstat2 - a modern 3D image analysis environment for biofilms. (Technical University of Denmark, 2008). [Google Scholar]
- Winsor G. L. et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M. & Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data were deposited in the NCBI sequence read archive (SRA) as a study under the accession number SRP062593 (RNAseq) and SAMN05823591 (CoIP RNAseq), respectively. All other data generated or analysed during this study are included within this article and the Supplementary information or are available from the corresponding author upon request.