Abstract
We show, with miR171, that plant miRNA genes are modular independent transcription units in which the fold-back pre-miRNA is sufficient for miRNA processing, and that the upstream region contains highly specific promoter elements. Processing depends on flanking sequences within the miRNA stem-loop precursor rather than the miRNA sequence itself, and mutations affecting target pairing at the center and 5′ but not 3′ region of the miRNA compromise its function in vivo. Inactivation of the SDE1 RNA-dependent-RNA-polymerase was mandatory for accurate representation of miRNA activity by sensor constructs in Arabidopsis. Work in sde1 background revealed a near-perfect spatial overlap between the patterns of miR171 transcription and activity, supporting the idea that plant miRNAs enable cell differentiation.
Keywords: RNA silencing, microRNA, transitivity, Arabidopsis
In eukaryotes, 21-25-nt RNAs are effectors of RNA silencing, a conserved pathway leading to suppression of gene expression. These small RNAs are processed by homologs of the RNase-III Dicer (Bernstein et al. 2003) and are in two categories. Small interfering (si)RNAs (Hamilton and Baulcombe 1999; Elbashir et al. 2001a) are processed from long double-stranded RNAs produced by inverted repeats, virus replication, or RNA-dependent-RNA-polymerases (RdRps). They guide an RNA-induced silencing complex (RISC) promoting cleavage of RNAs sharing sequence homology (Hammond et al. 2000; Hutvagner and Zamore 2002; Tang et al. 2003). In plants and Caenorhabditis elegans, siRNAs initiate synthesis of secondary siRNAs through transitive RNA silencing, which requires activity of cellular RdRps (Dalmay et al. 2000; Sijen et al. 2001). Production of secondary siRNAs by the SDE1 RdRp is linked to silencing cell-to-cell movement in Arabidopsis (Himber et al. 2003).
Micro (mi)RNAs are transcribed from noncoding nuclear genes (Bartel 2004). In animals they originate from primary transcripts, the (pri)miRNAs, that are processed by the Drosha RNase-III into 60-70-nt stem-loop intermediates known as miRNA precursors or premiRNAs (Lee et al. 2003). Upon cytoplasmic export, the pre-miRNA is processed by Dicer into a mature miRNA. Most animal miRNAs are believed to inhibit translation of putative mRNA targets through imperfect complementarity to their 3′ UTR, in a process similar to that elicited by the lin-4 miRNA in C. elegans (Olsen and Ambros 1999; Bartel 2004). Recently, however, an animal miRNA was identified that specifies endonucleolytic cleavage of its biological targets (Yekta et al. 2004). From the latter and other studies (Hutvagner and Zamore 2002; Doench et al. 2003; Zeng et al. 2003), it appears that the extent of complementarity at the central region of the miRNA/target duplex essentially determines whether animal miRNAs will direct cleavage or translation inhibition.
Plant miRNAs originate from multiple genomic loci that are mainly located between protein-coding genes (Bartel and Bartel 2003). Extensive rather than partial base-pairing between plant miRNAs and target sequences has enabled identification of cellular mRNAs that are likely regulated by those molecules (Rhoades et al. 2002). Strikingly, most encode transcription factors implicated in developmental fates, leading to the proposal that plant miRNAs clear regulatory transcripts from specific daughter cell lineages and thereby enable cell differentiation. Accordingly, mutations in genes influencing miRNA biogenesis in plants, most notably in the DCL-1 Dicer homolog, cause developmental anomalies (Jacobsen et al. 1999). Experimental validation of plant miRNA targets shows that suppression of mRNA expression occurs mainly through cleavage rather than translation inhibition (Llave et al. 2002; Aukerman and Sakai 2003; Kasschau et al. 2003; Palatnik et al. 2003).
Plant miRNAs differ from animal miRNAs in many aspects. For instance, their predicted fold-back structures are much more variable in size than those of animals (Reinhart et al. 2002), and pre-miRNAs have not been compellingly detected in plants. Rules governing the activity of some plant miRNAs also appear to be less clear than those drawn from studies in metazoans. For instance, miR172, which regulates APETALA2 (AP-2) via translational repression (Aukerman and Sakai 2003; Chen 2004), has near perfect complementarity with its target sequence in the AP-2 ORF, which would paradoxically favor endonucleolytic cleavage in animals (Hutvagner and Zamore 2002). This is made harder to understand by the fact that the pairing requirements for the activity of “conventional” plant miRNAs have not been investigated thus far, reflecting our poor knowledge of plant miRNA biology, which has been largely inferred from computational predictions or analogies to animal systems rather than experimental facts. Here, we present the results of an in vivo analysis of the transcription, processing, pairing requirements, and functional relevance of the spatial distribution of miR171 in Arabidopsis.
Results and Discussion
Modular structure of the miR171 gene
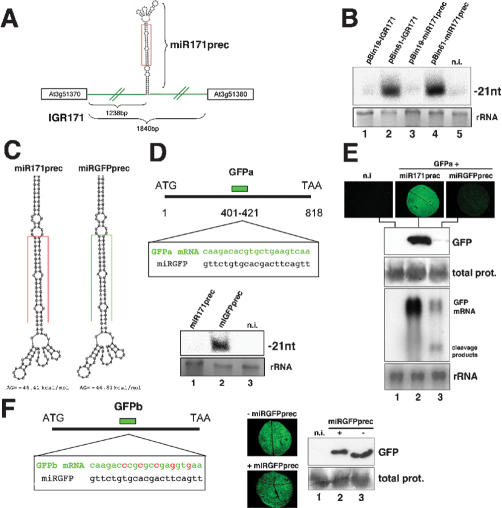
The miR171 stem-loop precursor (miR171prec) is predicted as a 123 nt-long structure located 1238 bp from the 5′ end of an 1840 bp-long IGR on chromosome 3 (IGR171, Fig. 1A; Reinhart et al. 2002). To assess whether miR171prec is indeed sufficient for miR171 synthesis in vivo, we exploited an Agrobacterium-mediated transient expression system used previously to overexpress functional miR171 in Nicotiana benthamiana leaves (Llave et al. 2002). Four days post-Agrobacterium infiltration (dpi) of pBin61-IGR171, miR171 was detected (Fig. 1B, lane 2). Its levels were much higher than those from pBin19-IGR171, which is devoid of a 35S promoter (Fig. 1B, lane 1), but they were similar to those from pBin61-miR171prec, which contained only the miR171prec sequence (Fig. 1B, lanes 2,4). Probes for the non-miR171 sequences of IGR171/miR171prec did not reveal any other small RNA (data not shown), confirming bona fide miR171 production. Therefore, the miR171prec stem-loop is sufficient for miR171 synthesis. This provides a rationale for designing generic miRNA overexpressing constructs in order to simultaneously inactivate highly related plant miRNA targets whose analysis by individual T-DNA or transposon knockouts may be prevented by functional redundancy. Further experiments showed that the 1238 bp upstream of miR171prec could drive expression of a green fluorescent protein (GFP) reporter gene (pmiR171-GFP; Supplemental Material 1), indicating a modular organization for the miR171 gene.
Figure 1.
Processing depends on structure rather than sequence of miR171 precursor. (A) Schematic of the miR171 locus (IGR171) and miR171 precursor (miR171prec). Sizes of the IGR and putative promoter-containing fragments are indicated. miR171 is boxed in red. (B) Cultures of Agrobacterium strains containing IGR171 and miR171prec constructs were injected into N. benthamiana leaves. Total RNA was extracted at 4 dpi and miR171 levels analyzed by Northern blot using 15 μg of RNA. (rRNA) Sample loading control by ethidium bromide staining; (n.i.) noninfiltrated tissue. (C) Predicted secondary structure of miR171prec (left) and miRGFPprec (right). miRNAs are boxed in red and green, respectively. (D, top) Predicted pairing between miRGFP and its target sequence in transcript GFPa. (Bottom) Gel blot analysis of 10 μg total RNA confirms synthesis of miRGFP in N. benthamiana leaves at 4 dpi. (E) Visual inspection (top), protein blot analysis (middle), and Northern analysis (10 μg total RNA, bottom) of samples cotreated with GFPa and miRGFPprec (lane 3) or miR171prec (lane 2), at 4 dpi. (F) Construct GFPb contains five successive synonymous mutations (indicated in red) at the site of miRGFP-target pairing. The experiments in E were repeated with GFPb. Total prot, sample loading controls by Coomassie staining.
Processing depends on structure rather than sequence of miR171prec
Next, the miR171 sequence within miR171prec was replaced by a 21 nt-long stretch of sequence complementary to nucleotides 401-421 of the 818-nt GFP mRNA (miRGFPprec) such that authentic mismatches and free energy were preserved (Fig. 1C,D). If structure rather than sequence of miR171prec was required for miRNA maturation, miRGFPprec should produce an artificial miRNA with antisense sequence of GFP (miRGFP). At 4 dpi, a 21-nt RNA was indeed detected with a probe specific for miRGFP in pBin61-miRGFPprec-treated, but not pBin61-miR171prec-treated leaves (Fig. 1D, lanes 1,2). The detected miRGFP mediated endonucleolytic cleavage of the GFP mRNA when coexpressed with construct pBinGFPa (Fig. 1D) from which miRGFP sequence originated (Fig. 1E, lane 3). This cleavage was miRGFP-specific, as it was not observed in pBin61-miR17-1prec-treated leaves (Fig. 1E, lane 2). Cleavage was also not observed upon coexpression with a GFP construct (pBinGFPb) in which the region complementary to miRGFP carried five synonymous mutations (Fig. 1F). Thus, miRGFP had all the attributes of a genuine miRNA, and we conclude that structure rather than sequence of the excised portion of miR171prec is required for processing, although the precise structural contribution of the non-miR171 portion of miR171prec remains to be addressed. These results also indicate that miR171prec can be readily exploited to produce high levels of any functional, artificial miRNAs to be used in plant reverse genetics. Stable transformation with pBin61-miRGFPprec indeed allowed specific and dose-dependent suppression of a GFP transgene in Arabidopsis (Supplemental Material 2).
Target pairing requirements for miR171 function in vivo
We then investigated the pairing parameters influencing miR171 function in vivo. Inspired by previous work in Drosophila (Brennecke et al. 2003), we used a GFP sensor system in which the 3′ part of the GFP ORF was transcriptionally fused to the miR171 target sequence, as found in the SCL6-IV mRNA (GFP-171.1, Fig. 2A). GFP-171.1 was delivered into N. benthamiana leaves, either on its own or with pBin61-miR171prec. At 4 dpi, the GFP and GFP mRNA from GFP-171.1 were barely detectable, as expected, whereas they were high in samples infiltrated with a GFP construct devoid of the miR171 target sequence (Fig. 2A,B, lanes 1,2,9).
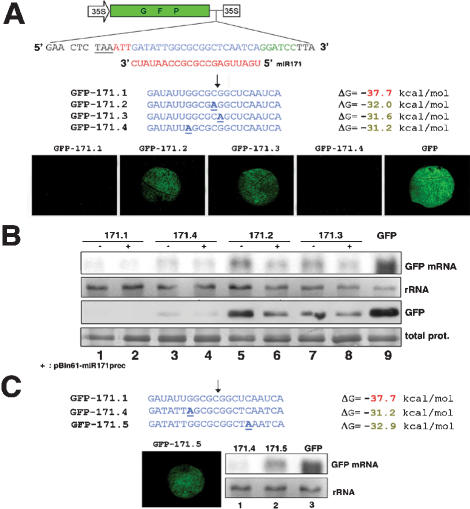
Figure 2.
Target pairing requirements for miR171 function in vivo. (A) Schematic of the GFP-miR171 target sequence fusions. The GFP stop codon is underlined. The three nucleotides in red were used as linkers. Nucleotides in green located 3′ of the miR171 target sequence (blue) define a BamHI restriction site for generic cloning. Nucleotide substitutions in miR171 targets are in bold and underlined. The predicted ΔG values for the miR171-target duplex are on the right. Arrow indicates prominent cleavage site for the SCL6-IV mRNA, as mapped by Llave et al. (2002). The pictures of N. benthamiana leaves were taken at 4 dpi. (B) Gel blot analysis of 5 μg total RNA (top) and total protein (bottom) extracted from the tissues in A. Plus sign (+) indicates cotreatment with miR171prec. The antibody was GFP-specific. (C) ΔG values of the pairing of miR171 to target sequences in GFP-171.4 and GFP-171.5. The experiment was as described in A and B. Total prot and rRNA, sample loading controls, as in Figure 1.
In animals, activated RISC preferentially cleaves target RNAs between nucleotides 10 and 11 relative to the 5′ end of the complementary guide RNA (Elbashir et al. 2001b; Haley and Zamore 2004). Similarly, cleavage by most plant miRNAs usually maps near or at the center of the miRNA:target pair (Llave et al. 2002; Kasschau et al. 2003). For instance, cleavage of the SCL6-IV transcript by miR171 is prevalent between the 11th and 12th nucleotides of the miRNA complement (Llave et al. 2002). Mutations at both positions were introduced into GFP-171.1, leading to GFP-171.2 and GFP-171.3, respectively (Fig. 2A). Both significantly reduced GFP mRNA cleavage without affecting translation, whereas a mutation at the seventh nucleotide from the 5′ end of the miR171 target (construct GFP-171.4) did not (Fig. 2A,B, lanes 3-8), despite the fact that miR171-pairing energy was similar to that of GFP-171.2 and GFP-171.3 (Fig. 2A). Therefore, intact pairing within the central region of the miRNA complement is key for cleavage and this, reciprocally, applies to the miRNA itself, as a single mutation at the 11th nucleotide of miRGFP abolished its function (Supplemental Material 3). These results are similar to those obtained with siRNAs in animals (Ding et al. 2003). Notably, excess miR171 levels from pBin61-miR171prec overcame the effects of the GFP-171.2 and GFP-171.3 mutations (Fig. 2B, lanes 5-8), showing that artificially elevated miRNA levels may bias the results of experimental target validation in plants, as shown with animal miRNAs (Doench and Sharp 2004).
Mutations in the 5′ portion of animal miRNAs have far more severe consequences on their function than mutations in the 3′ portion (Doench and Sharp 2004). In fact, the 5′ end of siRNAs disproportionatly contributes to target RNA-binding energy in vitro (Haley and Zamore 2004). We tested whether a similar rule applied to miR171 in vivo. Because the GFP-171.4 mutation had no effect on cleavage (Fig. 2A,B), we engineered a symmetrical lesion with regard to the prevalent cleavage site, causing a mismatch with the sixth-most 5′ nucleotide of miR171 (GFP-171.5, Fig. 2C). This mismatch reduced cleavage efficacy (Fig. 2C, lanes 1,2) to the same extent as mismatches in GFP-171.2 and GFP-171.3 (data not shown), indicating that miR171 activity displays unequal tolerance to a 5′ as opposed to a 3′ symmetrical mismatch. Analysis of additional mismatches in other plant small RNAs will be required to determine whether these initial observations with miR171 define general rules.
miRNAs trigger transitive RNA silencing of sensor constructs in transgenic Arabidopsis
Next, we sought to exploit construct GFP-171.1 to monitor noninvasively miR171 activity in Arabidopsis. Activity of miR171 within a particular tissue would be diagnosed as an absence of GFP due to cleavage of the GFP-171.1 sensor, whereas lack of miR171 would allow GFP expression. This system should be independent of silencing-unrelated fluctuations of the natural miR171 targets and should thus provide an unbiased picture.
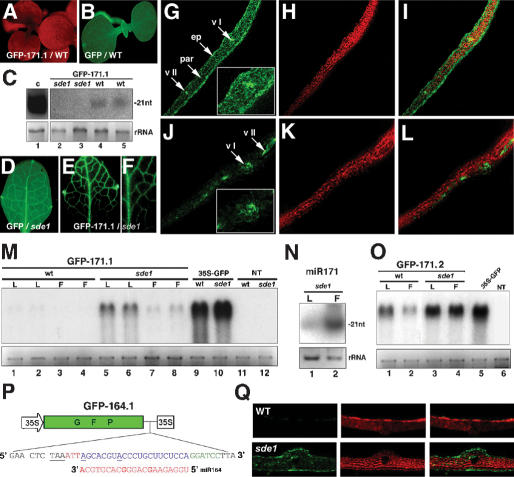
Although GFP was uniformely expressed in most pBin61-GFP transformants (Fig. 3B), it was absent from all of the 50 pBin61-GFP-171.1 tranformants analyzed (Fig. 3A). It was surprising that the GFP mRNA was below the detection limit in both leaves and flowers (Fig. 3M, lanes 1-4), because target cleavage by miR171 occurs exclusively in flowers, where this miRNA accumulates (Fig. 3N; Llave et al. 2002). Based on our previous work with tissue-specific siRNAs (Himber et al. 2003), this extensive GFP silencing likely resulted from amplification and movement of secondary siRNAs due to miR171-triggered transitive RNA silencing of the sensor transgene. Indeed, secondary siRNAs upstream of the 21-nt stretch of complementarity between the GFP-171.1 mRNA and miR171 were detected in those plants (Fig. 3C, lanes 4,5). Because transitive RNA silencing is strictly dependent upon the SDE1 RdRp, those experiments were repeated in an sde1 mutant background. Most GFP-171.1 transformants had a highly vein-specific GFP phenotype in leaves (Fig. 3E,F). GFP was absent in the epidermis, present at only a low level in the underlying parenchyma, and high in class I, II, and III veins (Fig. 3J-L). The GFP mRNA levels were higher in leaves than in flowers (Fig. 3M, lanes 5-8), a pattern inversely correlated with the accumulation of miR171 in those respective organs, as anticipated from accurate sensor function. As expected, the sde1 pBin61-GFP plants were uniformly green (Fig. 3D,G-I) and had high GFP mRNA levels in leaves (Fig. 3M, lane 10). This was also the case for sde1 plants transformed with the cleavage-resistant pBin61-GFP-171.2 construct (Figs. 2A,B, 4A-C). However, the GFP mRNA levels were lower than in pBin61-GFP plants (Fig. 3O, cf. lanes 3,4 and 5). Thus, some cleavage still occured in the GFP-171.2 transformants, in agreement with the transient expression data (Fig. 2B). Notably, the GFP mRNA levels were significantly lower in GFP-171.2 plants with an SDE1 background, indicating that the GFP-171.2 transcript was also subject to transitivity (Fig. 3O, cf. lanes 1,2 and 3,4).
Figure 3.
miR171-triggered transitive RNA silencing of GFP-target fusion constructs. (A) GFP-171.1 plants with an SDE1 genetic background are uniformly silenced for GFP. Pictures were taken under UV illumination without a band-pass filter so that tissues were visible due to chlorophyll red fluorescence. (B) GFP transformants with the SDE1 background are uniformly green. (C) Gel blot analysis of 8 μg of total RNA for detection of secondary GFP siRNAs. RNA was extracted from leaves of the progeny of two independent lines as depicted in A (lanes 4,5) or E and F (lanes 2,3). (C, lane 1) RNA from GFP silenced Arabidopsis provided a positive control. (D) Closer view of a leaf from a GFP transformant with the sde1 background. (E,F) Vein-specific GFP phenotype of the GFP-171.1 plants with the sde1 background. (G-I) Tranverse section of the leaf in D observed under CLSM. (vI) Primary vein; (vII) secondary vein; (ep) epidermis; (par) parenchyma. (Inset) Closer view of the tissue near the primary vein. The green channel was cut off in H to monitor chlorophyll fluorescence only. The panel in I is an overlay of G and H revealing the epidermal layer. (J-L) Same as G-I but with a leaf from a GFP-171.1 plant, as depicted in E,F. (M) Gel blot analysis of two independent GFP-171.1 lines in the SDE1 (lanes 1-4) or sde1 (lanes 5-8) background. Total RNA was extracted from leaves (L) or inflorescences (F), and 5 μg was subjected to gel blot analysis. Leaf RNA from a GFP line with SDE1 (lane 9) or sde1 (lane 10) background provided positive controls, and leaf RNA from nontransformed plants (SDE1 or sde1 background) provided negative controls (lanes 11,12). (N) miR171 levels in leaves (L) and inflorescences (F) of nontransgenic sde1 plants, as assessed by gel blot analysis of 10 μg total RNA. (O) Same as in M but with tissues from representative GFP-171.2 lines in SDE1 or sde1 background. (P) Sensor construct for miR164. The target sequence is as found in the NAC1 transcript (At1g56010). (Q) Transversal leaf sections of GFP-164.1 transformants with a wild-type (WT, top) or sde1 background (bottom).
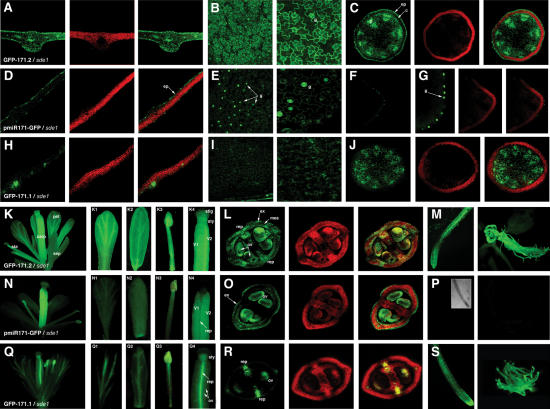
Figure 4.
Comparison of the miR171 transcription and activity patterns in transgenic Arabidopsis. (A) Transversal section of a leaf from a GFP-171.2 representative T2 line with the sde1 background and the corresponding overlays, as in Figure 3. (B) Longitudinal views of the leaf in A. The second panel is an enlargement. (C) Transversal section of the central stem of the plant in A. (D-G) Same as A-C but for a pmiR171-GFP transformant (sde1 background). (H-J) Same as A-C but for a GFP-171.1 transformant (sde1 background). (K) Typical flower of a GFP-171.2 line with the sde1 genetic background. GFP expression is high in all organs (petal [K1], sepal [K2], stamen [K3], and carpel [K4]). (L) Transversal section of the carpel in K4 and the corresponding overlays, observed by CLSM. (M) Roots from a GFP-171.2 representative line (sde1 background). (N-P) Same as K-M, but with tissues from pmiR171-GFP lines (sde1 background). The inlay in P shows the root imaged under transmitted light. (Q-S) Same as K-M, but with tissues from GFP-171.1 plants (sde1 background). (g) Guard cells; (ep) epidermis; (c) cortex; (sta) stamen; (carp) carpel; (pet) petal; (sep) sepal; (stig) stigma; (sty) style; (v) valve; (rep) replum; (mes) mesocarp; (ex) exocarp; (ov) ovule.
Sensor constructs for miR164 and other miRNAs also provided a distorted image of miRNA cleavage in the wild-type as opposed to sde1 background (Fig. 3P,Q; data not shown), indicating that transitivity was not a peculiarity of miR171. Therefore, prior inactivation of SDE1 is mandatory for the accurate representation of miRNA activity by reporter gene fusions in transgenic plants. Moreover, the results obtained with GFP-171.2 (Fig. 3O) show that even stringent mutations within miRNA target sequences may not entirely prevent transitivity, and this may potentially bias the interpretation of standard dominant mutation experiments used to validate miRNA targets in wild-type plants. It is unlikely, however, that transitivity by miRNAs has important biological significance, as this would compromise the highly specific effect of those molecules and would be inconsistent with the strong evolutionary selection of their target sites within mRNA sequences. Rather, the very foreign nature of miRNA sensors may promote transitive RNA silencing, as evidenced in previous experiments in which exogenous siRNAs triggered transitivity against transgenic, but not endogenous mRNAs (Himber et al. 2003).
miR171 transcription and activity patterns coincide to near perfection
Recent in situ hybridizations have shown that the accumulation pattern of two plant miRNAs is complementary to the genetic activity of their respective targets (Chen 2004; Kidner and Martienssen 2004), supporting the idea that plant miRNAs ensure clearance of regulatory transcripts from specific daughter cell lineages and thereby enable cell differentiation (Rhoades et al. 2002). This idea also applies to animal miRNAs. For instance, the temporally regulated transcription of let-7 readily correlates with temporal transitions in cell fates in C. elegans (Johnson et al. 2003). One key aspect of the “clearance model” is that it predicts a very tight spatial overlap between the transcription and activity of miRNAs. The availability of the pmiR171-GFP construct (Supplemental Material 1) and the GFP-171.1 plants (sde1 background) provided a unique opportunity to test this hypothesis with miR171. Representative GFP-171.1 and GFP-171.2 lines were used as references for the miR171 endonucleolytic activity and ubiquitous GFP expression, respectively.
Analysis of tissues from the pmiR171-GFP plants (sde1 background) under a confocal laser scanning microscope (CLSM) showed a highly cell type-specific transcriptional pattern for miR171. In leaves, miR171 transcription was highest in the epidermis, particularly in stomata guard cells (Fig. 4D,E). It was very low in the mesophyll (Fig. 4D) and absent in the veins. A similar transcription pattern was found in stems (Fig. 4F,G). Remarkably, miR171 transcription coincided to near perfection with its site of endonucleolytic activity, as assessed by the specific loss of GFP expression in leaves and stems of GFP-171.1 plants (Fig. 4H-J). In inflorescences, where miR171 is most abundant (Fig. 3N), miR171 transcription was by far highest in the carpels, where GFP accumulated in the two ovary valves, but was absent in the replum, style, and stigma (Fig. 4N4). This pattern (Fig. 4N,O) also coincided with the endonucleolytic activity of miR171 in GFP-171.1 flowers (Fig. 4Q,R). For instance, in the GFP-171.1 gynoecium, GFP expression was evident only in the style and replum, and absent in the two valves, such that ovules could be detected by transparency (Fig. 4Q4). Consistent with previous studies showing high SCL6 mRNA accumulation in roots (Pysh et al. 1999), the miR171 promoter was found inactive in those organs in which, accordingly, GFP-171.1 was ubiquitously expressed (Fig. 4, cf. P and S).
Although the promoter activity of natural miR171 target genes was not examined in the present study, the exquisite spatial overlap between miR171 transcription and activity provides strong support for the idea that plant miRNAs specify cell differentiation. It also implies that the 1238-bp sequence upstream of miR171prec (Fig. 1A) contains the promoter of the miR171 gene, which is therefore an independent transcription unit. High tissue specificity and capacity to drive transcription of the GFP mRNA suggest that RNA pol II rather than RNA pol III is involved in miR171 transcription. Consistent with this idea, a recently identified 1148 bp-long EST from Arabidopsis siliques contains the whole miR171 precursor (gi 42532354), in line with the analysis of the EAT locus, which produces a 5′ capped and 3′ polyadenylated noncoding RNA representing the full-length miR172a-2 precursor (Aukerman and Sakai 2003).
This work provides new insights into plant miRNA biology. Most notably, we extrapolate from our analysis of miR171 that (1) plant miRNA genes are independent transcription units, (2) processing of miRNAs is determined by flanking sequences rather than the miRNA sequence itself; (3) 5′ and central sequences are more important than 3′ sequences in the miRNA for targeting; (4) miRNAs trigger transitive RNA silencing of foreign genes; and (5) transcription and activity patterns of miRNAs coincide to near perfection. We have also established experimental procedures that will be broadly applicable for the future characterization of many plant miRNA genes.
Materials and methods
DNA constructs
The 1840-bp intergenic region downstream of Arabidopsis locus At3g51370 was PCR-amplified from Arabidopsis genomic DNA (Col-0), cloned into pGEM-T Easy (Promega), and subsequently inserted into pBin61 and pBin19 binary vectors (Himber et al. 2003) to generate plasmids pBin61-IGR171 and pBin19-IGR171. Plasmids pBin61-miR171prec and pBin19-miR171prec containing the 123-bp miR171 precursor were generated similarly. The 35S promoter of pCK-GFP-S65C (kindly provided by D. Gilmer, Institut de Biologie Moléculaire des Plantes, Strasburg, France) was replaced by the 1238-bp upstream of miR171prec, and the expression cassette was mobilized into pBin19 to produce pmiR171-GFP. pBin61-miRGFPprec was produced as described for pBin61-miR171prec. pBinGFPa and pBinGFPb are pBin61-based constructs containing GFP sequences from plasmids mGFP5-ER and pCKGFP-S65C, respectively. pBin61-GFP, pBin61-GFP-171.1, pBin61-GFP-171.2, pBin61-GFP-171.3, pBin61-GFP-171.4, pBin61-GFP-171.5, and pBin61-GFP-164.1 were based on plasmid mGFP5-ER. The wild-type and modified miRNA target sequences were introduced downstream of the GFP coding region by PCR, using suitable reverse oligonucleotides. Amplified fusions were subcloned into pGEM-T Easy and then inserted in pBin61. ΔG values for the binding of miR171 to the tested targets were determined according to Doench and Sharp (2004).
Transgenic and mutant plants: Agrobacterium infiltration
Arabidopsis sde-1 null mutants (C24 background) were described previously (Dalmay et al. 2000). For transformation, the DNA constructs were mobilized into Agrobacterium tumefaciens strain GV3101 and introduced into plants (C24) by the floral dip method (Bechtold and Pelletier 1998). Agrobacterium-mediated transient expression was as described (Himber et al. 2003), except that all cultures were brought to a final optical density of 0.1 prior to infiltration, which prevents the onset of a generalized silencing response to the transgene constructs (Llave et al. 2002).
RNA gel blot analysis analysis
RNA was extracted with Tri-Reagent (Sigma), precipitated with isopropanol, and redissolved in 50% formamide. Analysis of high- and low-molecular-weight RNA was as described (Himber et al. 2003). The probe used for GFP mRNA analysis was based on the full-length mGFP5 cDNA labeled by random priming incorporation of α-32P-dCTP. The probe used for analysis of transitivity corresponded to the 170 nt located at the 3′ end of the mGFP5 cDNA. Probes for small RNA gel blots were γ-32P-ATP end-labeled 21-mer oligonucleotides complementary to miR171, miRGFP, and miRGFP11th. Each probe was used at 20°C below the optimal dissociation temperature (Td) [Td (degrees Celsius) = 4(G + C) + 2(A + T)]; 21-nt-long and 24-nt-long end-labeled oligo-ribonucleotides were used as size markers. Blots were exposed under X-ray films and scanned.
Protein gel blot analysis
Total proteins were extracted, redissolved on 16% SDS-PAGE, and subjected to protein gel blot analysis using a rabbit polyclonal anti-GFP antibody (Invitrogen). HRP-conjugated anti-rabbit IgG was used as secondary antibody. Immunodetection was with chemiluminescent substrate (Roche Lumi-LightPLUS) and exposition to X-ray films.
GFP imaging
Pictures were taken under a Nikon SMZ15000 dissecting microscope coupled to a 100 W epifluorescence module. The LSM510 microscope (Zeiss) was used for laser scanning confocal imaging. For transversal sections, plant material was embedded in 1% low-melting-point agarose and cut transversally.
Note added in proof
While this manuscript was in press, a study of the interaction between miR165/166 and the PHABULOSA/PHAVOLUTA transcripts showed that disrupting miRNA pairing near the center of the miRNA complementary site had far milder developmental consequences than more distal mismatches. Mismatch scanning in vitro revealed more tolerance for mismatches at the center and 3′ end of the miRNA compared with mismatches to the 5′ region. These findings are in agreement with those reported here with miR171 and support a model in which the 5′ region of small RNAs nucleates pairing to their targets, both in plants and animals.
Acknowledgments
This paper is dedicated to the memory of Adilson Leite. We thank our lab colleagues for fruitful discussions. Our team is supported by an ATIP and a postdoctoral fellowship to E.A.P. from CNRS and a long-term FEBS fellowship to P.D.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.307804.
References
- Aukerman M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Bartel B. and Bartel, D.P. 2003. MicroRNAs: At the root of plant development? Plant Physiol. 132: 709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N. and Pelletier, G. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259-266. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., and Hannon, G.J. 2003. Dicer is essential for mouse development. Nat. Genet. 35: 215-217. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25-36. [DOI] [PubMed] [Google Scholar]
- Chen X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton, A.J., Rudd, S., Angell, S., and Baulcombe, D.C. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543-553. [DOI] [PubMed] [Google Scholar]
- Ding H., Schwarz, D.S., Keene, A., Affar el, B., Fenton, L., Xia, X., Shi, Y., Zamore, P.D., and Xu, Z. 2003. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell 2: 209-217. [DOI] [PubMed] [Google Scholar]
- Doench J.G. and Sharp, P.A. 2004. Specificity of microRNA target selection in translational repression. Genes & Dev. 18: 504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes & Dev. 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B. and Zamore, P.D. 2004. Kinetics analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11: 599-606. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286: 950-952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D., and Hannon, G. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature 404: 293-296. [DOI] [PubMed] [Google Scholar]
- Himber C., Dunoyer, P., Moissiard, G., Ritzenthaler, C., and Voinnet, O. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22: 4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056-2060. [DOI] [PubMed] [Google Scholar]
- Jacobsen S.E., Running, M.P., and Meyerowitz, E.M. 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231-5243. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Lin, S.Y., and Slack, F.J. 2003. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 259: 364-379. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4: 205-217. [DOI] [PubMed] [Google Scholar]
- Kidner C.A. and Martienssen, R.A. 2004. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81-84. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim J., Yim, J., Lee J., Provost, P., Radmark, O., Kim, S. et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053-2056. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. 2004. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671-680. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257-263. [DOI] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. 1999. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18: 111-119. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513-520. [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H.A., and Fire, A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465-476. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih, I.H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594-596. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]