Abstract
The Rad1-Rad10 nuclease of yeast and its human counterpart ERCC1-XPF are indispensable for nucleotide excision repair, where they act by cleaving the damaged DNA strand on the 5′-side of the lesion. Intriguingly, the ERCC1- and XPF-deficient mice show a severe postnatal growth defect and they die at ∼3 wk after birth. Here we present genetic and biochemical evidence for the requirement of Rad1-Rad10 nuclease in the removal of 3′-blocked termini from DNA strand breaks induced on treatment of yeast cells with the oxidative DNA damaging agent H2O2. Our genetic studies indicate that 3′-blocked termini are removed in yeast by the three competing pathways that involve the Apn1, Apn2, and Rad1-Rad10 nucleases, and we show that the Rad1-Rad10 nuclease proficiently cleaves DNA modified with a 3′-phosphoglycolate terminus. From these observations, we infer that deficient removal of 3′-blocking groups formed from the action of oxygen free radicals generated during normal cellular metabolism is the primary underlying cause of the inviability of apn1Δ apn2Δ rad1Δ and apn1Δapn2Δ rad10Δ mutants and that such a deficiency accounts also for the severe growth defects of ERCC1- and XPF-deficient mice.
Keywords: Rad1-Rad10 nuclease, 3′-blocked termini, 3′-phosphoglycolate, ERCC1-XPF, inviability of apn1 apn2 rad1/rad10 mutants
Cellular DNA is damaged continuously by a variety of endogenous and exogenous factors. Abasic (AP) sites arise in DNA frequently as a result of spontaneous hydrolysis of the N-glycosylic bond and as intermediates following the removal of damaged bases by DNA glycosylases (Wallace 1997). Oxygen free radicals, generated from normal cellular metabolism or from oxidative stress resulting from the exposure of cells to DNA damaging agents such as H2O2 and ionizing radiation, can attack DNA at either a sugar or a base (Imlay and Linn 1988; Henle and Linn 1997; Wallace 1997). Attack at a sugar can cause sugar fragmentation, yielding a strand break with a 3′-terminal fragmented deoxyribose moiety. Strand breaks formed in this manner usually have a 3′-phosphate or a 3′-phosphoglycolate (3′-PG) at the terminus, which present a block to repair synthesis by DNA polymerases. Free radical attack on purines and pyrimidines in DNA produces a variety of damaged bases, as for example, 8-oxoguanine and thymine glycol, which are removed by the action of specific DNA glycosylases such as Ogg1 and Ntg1 and Ntg2, respectively. In addition to a DNA glycosylase activity, these enzymes also harbor an AP lyase activity that can incise DNA on the 3′ side of the AP site (Shinmura et al. 1997; You et al. 1998), yielding a 3′ terminus with an α,β-unsaturated aldehyde, 4 R-4-hydroxy-trans-2-pentenal (3′-dRP) moiety, which, too, is a block to synthesis by DNA polymerases.
In Saccharomyces cerevisiae, Apn1 and Apn2, which possess a class II AP endonuclease activity and also a 3′-phosphodiesterase activity, function in the removal of AP sites as well as in the removal of 3′-PG, 3′-dRP, and other such 3′-blocking groups. Apn1, which shares extensive homology with Escherichia coli endonuclease IV, is the major AP endonuclease in yeast, accounting for >90% of this activity in yeast cells, and it also has a 3′-phosphodiesterase activity (Johnson and Demple 1988). Apn2, which shares extensive homology with E. coli exonuclease III and with human Ape1 and Ape2 proteins (Johnson et al. 1998; Unk et al. 2000), accounts for <10% of the AP endonuclease activity in yeast. In addition to an AP endonuclease activity (Unk et al. 2000), Apn2 displays a robust 5′-phosphodiesterase activity (Unk et al. 2001), and this activity, but not the AP endonuclease activity, is further stimulated by proliferating cell nuclear antigen (PCNA; Unk et al. 2002).
Apn1, Apn2, and nucleotide excision repair (NER) constitute alternate pathways for the removal of AP sites in yeast (Torres-Ramos et al. 2000). The NER system is highly conserved among eukaryotes. In S. cerevisiae, a combination of Rad14, Rad4-Rad23, RPA, TFIIH, Rad1-Rad10, and Rad2 and in humans, a combination of their respective counterparts XPA, XPC-HR23B, RPA, TFIIH, XPF-ERCC1, and XPG mediates the dual incision of the damaged DNA strand, resulting in the release of an ∼30-nt lesion-containing DNA fragment (Guzder et al. 1995; Mu et al. 1995, 1996). The yeast Rad14 and Rad4-Rad23 proteins act at the damage recognition step, and following the unwinding of the duplex DNA by the Rad3 and Rad25 DNA helicase subunits of TFIIH, the Rad1-Rad10 and Rad2 nucleases incise the damaged strand on the 5′- and 3′-side of the lesion, respectively (Prakash and Prakash 2000). As in yeast, in humans, the damage is recognized by the XPA and XPC-HR23B proteins, and following the unwinding by the XPD and XPB helicases in TFIIH, the XPF-ERCC1 and XPG nucleases incise the damaged strand on the 5′- and 3′-side of the lesion, respectively (Reardon and Sancar 2002, 2003).
Many of the proteins involved in NER function also in other biological processes. Although the Rad14 and Rad4 proteins of yeast and their human counterparts XPA and XPC function only in NER, TFIIH has an essential role in transcription, and the Rad1-Rad10 nuclease and its human counterpart, XPF-ERCC1, contribute to homology-based genetic recombination (Prakash and Prakash 2000).
Unlike the XPA- or XPC-deficient mice, which develop normally (de Vries et al. 1995; Nakane et al. 1995; Sands et al. 1995), the ERCC1 and XPF mutant mice are runted at birth and die at ∼20 d after birth (McWhir et al. 1993; Tian et al. 2004). The reason for the severe growth defect of ERCC1/XPF mutant mice has remained unclear so far.
Here we provide genetic and biochemical evidence for a specific role of the Rad1-Rad10 nuclease in the repair of 3′-blocked termini that accumulate following treatment of yeast cells with the oxidative DNA damaging agent, H2O2. Interestingly, introduction of the rad1Δ or rad10Δ mutation into the apn2Δ strain results in a large increase in H2O2 sensitivity, whereas introduction of the other NER mutations such as rad2Δ, rad4Δ, or rad14Δ confers no significant increase in H2O2 sensitivity of the apn2Δ strain. The various genetic observations we present here support the inference that the Apn1 and Rad1-Rad10 nucleases compete with Apn2 for the repair of 3′-blocked termini, and we provide biochemical evidence that the Rad1-Rad10 nuclease proficiently removes the 3′-PG group from DNA. From these observations, we suggest that the inefficient removal of the 3′-blocked ends arising during the course of normal oxidative cellular metabolism is the primary cause of severe growth and developmental abnormalities in ERCC1/XPF-deficient mice.
Results
The rad1Δ and rad10Δ mutations enhance the H2O2 sensitivity of the apn2Δ mutant
Oxidative metabolism is the primary contributor to free radical-induced DNA damage. Superoxide anion, a product of aerobic metabolism, is converted to H2O2 by the action of superoxide dismutases. The Fenton reaction of H2O2 with a transition metal such as Fe2+ produces the DNA-damaging hydroxyl radical. If hydroxyl radical is formed in the vicinity of DNA, it leads to the abstraction of a hydrogen from the C4 atom of the deoxyribose sugar, resulting in a strand break. Strand breaks are a hallmark of DNA damage induced by oxidative agents such as H2O2, and they usually terminate with a phosphoglycolate or a phosphate group at the 3′ end (Imlay and Linn 1988; Henle and Linn 1997; Wallace 1997).
Although sensitivity to H2O2 is not increased in the apn1Δ or the apn2Δ single mutants, the apn1Δ apn2Δ double mutant displays a much higher level of sensitivity to H2O2 than the single mutants (Unk et al. 2001). This observation had suggested a redundant role for Apn1 and Apn2 in the repair of H2O2-induced DNA damage. However, because of the involvement of NER in the repair of AP sites as an alternate to Apn1 and Apn2, we have now examined whether the inactivation of various NER genes has a similar effect on the repair of H2O2-induced DNA lesions as it does on the repair of AP sites.
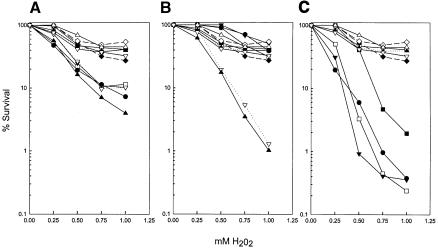
As shown in Figure 1A, H2O2 sensitivity is not significantly enhanced by the apn1Δ mutation or by a deletion of any of the NER genes RAD1, RAD2, RAD4, RAD10, and RAD14. Some increase in H2O2 sensitivity, however, occurs when the apn1Δ mutation is combined with any of these NER mutations. A strikingly different response to H2O2 is observed when the apn2Δ mutation is combined with the NER mutants. Although no significant increase in H2O2 sensitivity occurs when the apn2Δ mutation is combined with the rad2Δ, rad4Δ, or rad14Δ mutation, a large enhancement in H2O2 sensitivity is seen on deleting the RAD1 or the RAD10 gene from the apn2Δ strain (Fig. 1B). Next, we constructed triple mutant strains that harbor a deletion of any of the aforementioned five NER genes in the apn1Δ apn2Δ strain. During the course of these studies, we found that the apn1Δ apn2Δ rad1Δ and the apn1Δ apn2Δ rad10Δ mutants were inviable, and this observation has since been reported by others (Guillet and Boiteux 2002). As shown in Figure 1C, introduction of the rad2Δ, rad4Δ, or rad14Δ mutations into the apn1Δ apn2Δ strain resulted in an increase in H2O2 sensitivity.
Figure 1.
Deletion of the RAD1 and RAD10 genes, but not of other NER genes, enhances the H2O2 sensitivity of the apn2Δ strain. (A) Effect of deletions of NER genes on the H2O2 sensitivity of the apn1Δ strain. (○) Wild type; (▪) apn1Δ; (- -♦- -) rad1Δ; (- -⋄- -) rad2Δ; (----▴----) rad4Δ; (▵) rad10Δ; (▿) rad14Δ; (----▿----) apn1Δ rad1Δ; (□) apn1Δ rad2Δ; (▾) apn1Δ rad4Δ; (▴) apn1Δ rad10Δ; (•) apn1Δ rad14Δ. Because of their similar H202 sensitivities, the wild-type strain and some of the single mutant strains cannot be distinguished from one another. (B) Effect of deletion of NER genes on the H2O2 sensitivity of the apn2Δ strain. Symbols as in A except that apn2Δ instead of apn1Δ is combined with the radΔ mutations. Only the sensitivity of the apn2Δ rad1Δ (----▿----) and apn2Δ rad10Δ (▴) is greatly enhanced. (C) Effect of deletions of NER genes on the H2O2 sensitivity of the apn1Δ apn2Δ strain. (○) Wild type; (- -♦- -) rad1Δ; (- -⋄- -) rad2Δ; (----▴----) rad4Δ; (▵) rad10Δ; (▿) rad14Δ; (▪) apn1Δ apn2Δ; (□) apn1Δ apn2Δ rad2Δ; (▾) apn1Δ apn2Δ rad4Δ; (•) apn1Δ apn2Δ rad14Δ.
Although DNA strand breaks with 3′-blocked termini are an important feature of oxidative damage, AP sites are also formed following the removal of oxidatively damaged bases by DNA glycosylases. However, because all of the major DNA glycosylases that remove damaged bases resulting from oxidative reactions possess, in addition, a 3′ lyase activity that cleaves DNA on the 3′ side of the AP site, their action also generates a blocked 3′ terminus with a dRP moiety. Nevertheless, to ensure that the specific enhancement of H2O2 sensitivity in the apn2Δ rad1Δ and the apn2Δ rad10Δ strains was due to a role of the Rad1-Rad10 nuclease in the repair of oxidative DNA lesions such as the removal of the 3′-PG group, and not due to its role in the removal of AP sites, we compared the methyl methane sulfonate (MMS) sensitivity of the rad1Δ and rad10Δ mutations in combination with the apn1Δ and apn2Δ mutations. MMS alkylates adenine at the N3 position and guanine at the N7 position, forming 3-methyl adenine and 7-methyl guanine, respectively. An N-methyl purine DNA glycosylase removes these and a variety of other alkylated bases (Roy et al. 1994; Bjoras et al. 1995). Because this glycosylase lacks a 3′-lyase activity, removal of alkylated bases in strains lacking the Apn1 and Apn2 endonucleases or the NER system would result in the accumulation of stable AP sites without the further processing to 3′ dRP moieties.
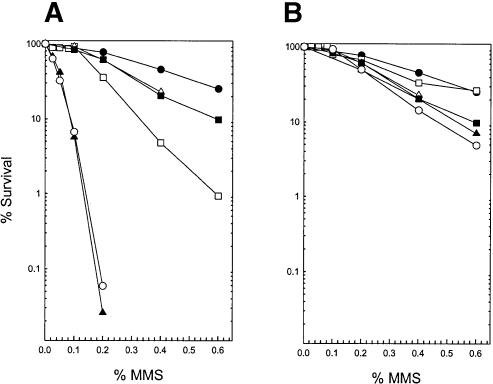
Previously, we have shown that deletion of the APN1 gene engenders an increase in MMS sensitivity, whereas deletion of the NER genes RAD2, RAD4, or RAD14, which had been examined in the previous study, conferred a level of MMS sensitivity that was intermediate between that of the wild-type strain and the apn1Δ strain (see Fig. 1A in Torres-Ramos et al. 2000). Deletion of APN2, however, had no effect on MMS sensitivity, and deletion of APN2 from the NER-defective mutant strains, rad2Δ, rad4Δ, or rad14Δ, did not cause any additional increase in MMS sensitivity (see Fig. 1B in Torres-Ramos et al. 2000). Simultaneous deletion of APN1 and APN2, however, conferred a large increase in MMS sensitivity, and introduction of any of the rad2Δ, rad4Δ, or rad14Δ mutations into the apn1Δ apn2Δ strain led to a further synergistic increase in MMS sensitivity (see Fig. 1C in Torres-Ramos et al. 2000). The defect in the removal of AP sites, as analyzed by alkaline sucrose gradient sedimentation, paralleled the MMS sensitivity profile of these various mutant strains (Torres-Ramos et al. 2000). These observations had led to the inference that Apn1 makes a major contribution to the repair of AP sites, whereas Apn2 plays a relatively minor role, and only in the absence of Apn1 does the need for Apn2 and NER become paramount. In accord with these previously reported observations, a synergistic enhancement in MMS sensitivity is observed when the apn1Δ mutation is combined with the rad1Δ or the rad10Δ mutation (Fig. 2A), whereas introduction of the apn2Δ mutation in either the rad1Δ or the rad10Δ mutant has no effect of MMS sensitivity (Fig. 2B). Thus, although the pattern of MMS sensitivity conferred on the apn1Δ and apn2Δ strains by the rad1Δ and rad10Δ mutations resembles that for the other NER mutants described earlier (Torres-Ramos et al. 2000), the pattern of H2O2 sensitivity that results on combining the rad1Δ or rad10Δ mutation with the apn2Δ mutation is strikingly different from that of the other NER mutants (Fig. 1B). From these observations, we infer that for the repair of AP sites, the Rad1-Rad10 nuclease functions in collaboration with the other protein components of NER, but for the repair of H2O2-induced DNA lesions, the Rad1-Rad10 nuclease plays a specific role that is unrelated to NER.
Figure 2.
Effect of rad1Δ and rad10Δ mutations on the MMS sensitivity of apn1Δ and apn2Δ strains. (A) MMS sensitivity of apn1Δ when combined with rad1Δ or rad10Δ mutations. (•) Wild type; (▵) rad1Δ; (▪) rad10Δ; (□) apn1Δ; (▴) apn1Δrad1Δ; (○) apn1Δrad10Δ. (B) MMS sensitivity of apn2Δ when combined with the rad1Δ or rad10Δ mutations. (•) Wild type; (▵) rad1Δ; (▪) rad10Δ; (□) apn2Δ; (▴) apn2Δ rad1Δ; (○) apn2Δ rad10Δ.
Repair of H2O2-induced DNA strand breaks is impaired in apn1Δ apn2Δ, apn2Δ rad1Δ, and apn2Δ rad10Δ strains
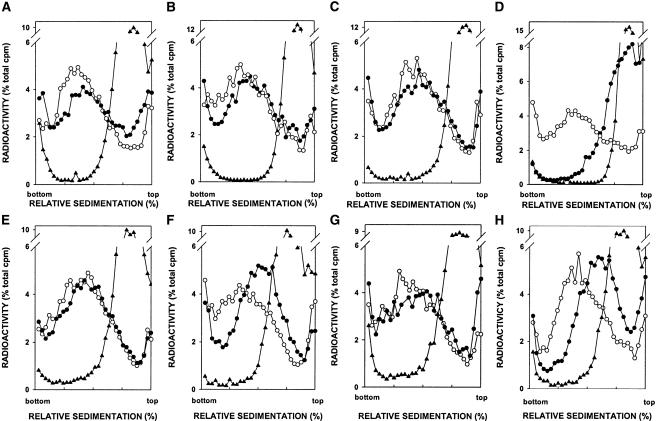
Next, we examined the repair of H2O2-induced DNA strand breaks by alkaline sucrose gradient sedimentation in wild type; in apn1Δ, apn2Δ, rad1Δ, and rad10Δ single mutants; and in apn1Δ apn2Δ, apn2Δ rad1Δ, and apn2Δ rad10Δ double mutants (Fig. 3). Sedimentation in alkaline sucrose gradients of chromosomal DNA immediately after a 1-h treatment of yeast cells with H2O2 revealed low molecular weight DNA, indicative of strand breaks, in all of the strains examined. Incubation of cells in H2O2-free medium for 4 h led to reformation of nativesized DNA in wild-type as well as in apn1Δ, apn2Δ, rad1Δ, and rad10Δ single mutants, but not in apn1Δ apn2Δ, apn2Δ rad1Δ, and apn2Δ rad10Δ strains. The apn2Δ rad1Δ and apn2Δ rad10Δ mutants, however, are not as defective in strand rejoining as is the apn1Δ apn2Δ strain. That the defect in the repair of H2O2-induced DNA breaks in the apn2Δ rad1Δ and apn2Δ rad10Δ mutants was specifically due to the inactivation of the RAD1 and RAD10 genes and not due to a defect in NER was confirmed from the observation that deletion of RAD14 in the apn2Δ strain had no adverse effect on repair (data not shown).
Figure 3.
Alkaline sucrose gradient analysis of DNA from wild-type and mutant yeast strains treated with H2O2. (A) Wild type. (B) apn1Δ. (C) apn2Δ. (D) apn1Δ apn2Δ. (E) rad1Δ. (F) apn2Δ rad1Δ. (G) rad10Δ. (H) apn2Δ rad10Δ. (○) Untreated cells; (▴) cells treated with H2O2 for 1 h; (•) cells treated with H2O2 for 1 h and then given a 4-h repair period.
Rad1-Rad10 catalyzes the removal of the 3′-PG group
The Rad1-Rad10 nuclease acts in a structure-specific manner, cleaving 3′-ended single-stranded DNA at its junction with duplex DNA (Bardwell et al. 1994). To examine the ability of Rad1-Rad10 to remove 3′-PG, we constructed two different DNA substrates, PG1 and PG2 (Fig. 4). In PG1, a 70-nt template oligomer is hybridized to a 5′-end-labeled 35-nt oligomer that contains either a PG at the 3′ terminus or no modification; in addition, this template is hybridized to a 34-nt oligomer, and that creates a 1-nt gap between the 3′-PG and the downstream 34-nt oligomer. In PG2, we hybridized a 40-nt oligomer that has a PG at the 3′ end or no modification, to the 70-nt template oligomer. This 40-nt oligomer differs from the 35-nt oligomer in having five additional nucleotides at the 3′ end that form a single-stranded flap structure. Downstream, the template is hybridized to the same 34-nt oligomer as in PG1.
Figure 4.
DNA substrates used in this study. See text for details. The asterisk indicates the radioactively labeled terminus.
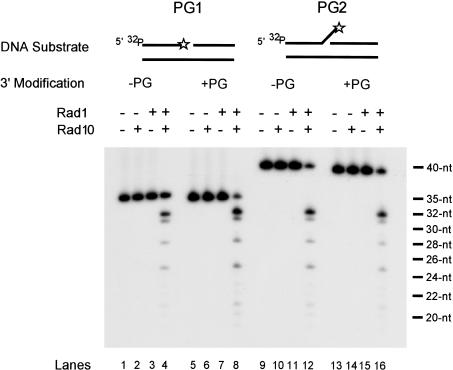
As shown in Figure 5, the Rad1-Rad10 nuclease displays similar activities on the 3′-PG and unmodified DNAs, with the prominent incision site being 3 nt into the duplex region, with additional incisions occurring farther into the duplex region. Importantly, the level and the pattern of incision activity on the 3′-PG-containing and unmodified DNAs is almost identical, regardless of whether the enzyme acts on the 1-nt gapped substrate or the flap substrate. As expected, the nuclease activity requires both of the proteins, because Rad1 or Rad10 alone display no activity.
Figure 5.
Removal of 3′-PG group by the Rad1-Rad10 nuclease. 5′-End labeled DNA substrate PG1 or PG2 (50 fmole) with a 3′-PG modification (lanes 5-8,13-16) or unmodified (lanes 1-4,9-12) was incubated with equimolar amounts (50 fmole) of Rad1, Rad10, or Rad1-Rad10 protein complex for 30 min at 30°C. Lanes 1, 5, 9, and 13 contained DNA substrate alone in reaction buffer. Reaction products were analyzed by electrophoresis on a 10% sequencing gel with oligonucleotide size markers (indicated on the far right) and autoradiography. The oligomer carrying the 3′-PG modification or unmodified and labeled with 32P at the 5′ end is indicated with an asterisk at the 3′ end.
Rad1-Rad10 can digest duplex DNA in a 3′ → 5′ manner
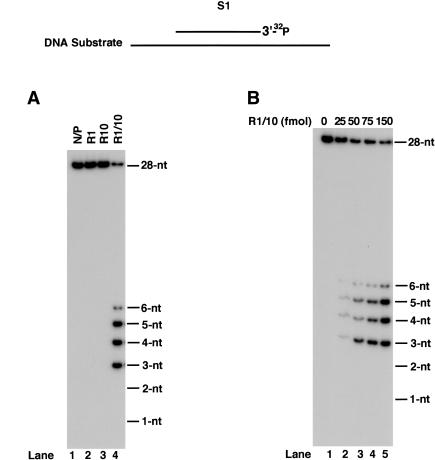
The proficient ability of Rad1-Rad10 nuclease to act on the single nucleotide gapped substrate, PG1, raised the possibility that it can remove nucleotides from the 3′ end acting in an exonucleolytic manner. To examine this, we constructed a partial duplex DNA substrate in which a 70-nt oligomer was annealed in its middle to a 27-nt oligomer (Fig. 4). Although the Rad1 or Rad10 protein alone showed no activity, the Rad1-Rad10 enzyme degraded DNA from the 3′ end, releasing products 3-6 nt in length (Fig. 6A), and even with increasing enzyme concentrations, the nearly equivalent amounts of 3-, 4-, and 5-nt products, and a much lower amount of a 6-nt fragment, remained nearly constant (Fig. 6B).
Figure 6.
3′ → 5′ exonucleolytic activity of Rad1-Rad10 complex. (A) Rad1-Rad10 catalyzes DNA degradation from the 3′ end. One-hundred femtomoles purified Rad1 (lane 2), Rad10 (lane 3), or Rad1-Rad10 complex (lane 4) were incubated with 3′-end-labeled DNA substrate S1 (200 fmole) for 10 min at 30°C in standard reaction buffer. The control reaction shown in lane 1 contained no Rad1 or Rad10. (B) Effect of Rad1-Rad10 concentration on 3′ → 5′ degradation.
Discussion
The Rad1-Rad10 nuclease is an integral component of the NER ensemble, cleaving the damaged DNA strand on the 5′ side of the lesion. Here we provide genetic and biochemical evidence for a novel role of this nuclease in removing the 3′-PG group formed in DNA from oxidative damage.
The rad1Δ and rad10Δ mutations differ markedly in their effect on H2O2 sensitivity from deletions of other NER genes such as RAD2, RAD4, and RAD14. Specifically, the rad1Δ and rad10Δ mutations greatly enhance the H2O2 sensitivity of the apn2Δ strain, whereas the rad2Δ, rad4Δ, or rad14Δ mutations have no such effect. Deletion of any of the NER genes, including that of RAD1 or RAD10, from the apn1Δ strain, however, confers only a small increase in H2O2 sensitivity over that in the respective single mutants, whereas deletion of APN1 from the apn2Δ strain results in a large increase in H2O2 sensitivity. Accordingly, the rate of repair of DNA strand breaks induced by H2O2 treatment is greatly reduced in the apn1Δ apn2Δ strain and also in the apn2Δ rad1Δ and apn2Δ rad10Δ strains. From these observations, we infer that in wild-type cells, Apn2 plays a prominent role in the repair of H2O2-induced DNA lesions, and only in the absence of Apn2 does the role of Apn1 and Rad1-Rad10 become crucial.
Although H2O2 primarily inflicts strand breaks in DNA, the incidence of AP sites could also increase in H2O2-treated yeast cells. Relative to strand breaks, however, the increase in the incidence of AP sites would be far less. Because of the involvement of Apn1, Apn2, and Rad1-Rad10 in the repair of AP sites as well as in the removal of 3′-PG, we carried out experiments to ensure that the effects of rad1Δ and rad10Δ mutations in H2O2-treated yeast cells arose predominantly from a defect in the removal of 3′-blocking groups and not from a defect in AP removal. For this, we compared the effects of the rad1Δ and rad10Δ mutations with deletions of the other NER genes in apn1Δ and apn2Δ mutant cells that had been treated with MMS or H2O2. Removal of MMS-induced base damage by a DNA glycosylase such as Mag1 would generate AP sites that are removed by the competing pathways of Apn1, Apn2, and NER. Our inference that, in contrast to the repair of AP sites wherein the Rad1-Rad10 nuclease functions in conjunction with NER proteins, the H2O2-induced 3′-blocking groups are removed by the competing action of Apn1, Apn2, and the Rad1-Rad10 nuclease but where Rad1-Rad10 acts independently of other NER protein components, is supported by the observation that although the MMS sensitivity of the rad1Δ or rad10Δ mutation when introduced into the apn1Δ or the apn2Δ strain (Fig. 2) is very similar to that of deletions in the other NER genes (Torres-Ramos et al. 2000), for H2O2, only the deletion of the RAD1 or RAD10 gene, and not of other NER genes, enhances the sensitivity of the apn2Δ strain (Fig. 1B).
The critical role of Apn2 in the removal of 3′ blocking groups, inferred here from genetic studies, is supported by biochemical studies indicating that the 3′-phosphodiesterase activity of Apn2 is 30- to 40-fold more active than its AP endonuclease activity (Unk et al. 2001). Apn1, on the other hand, constitutes the major AP endonuclease activity (∼90%) in yeast cells (Johnson and Demple 1988). Accordingly, in the absence of Apn1, the need for Apn2 and NER becomes paramount for AP repair, and this is reflected in the greatly enhanced MMS sensitivity and greatly reduced rate of repair of AP sites when the apn1Δ mutation is combined with either apn2Δ or with deletions of any of the NER genes (Torres-Ramos et al. 2000). On the other hand, the rather small increase in MMS sensitivity observed in the apn2Δ mutation when it is combined with a deletion of any of the NER genes suggests a more subsidiary role of Apn2 and NER in the repair of AP sites (Torres-Ramos et al. 2000).
In contrast to Apn2, which removes 3′-PG in the form of phosphoglycolic acid (Unk et al. 2001), the Rad1-Rad10 nuclease releases this group from DNA in the form of oligomers several nucleotides in length. Thus, we expect the pattern of repair synthesis to be different pursuant to the removal of 3′-PG by these different enzymes and that would vary from the filling in of a 1-nt gap to a gap of several nucleotides.
The Rad1-Rad10 enzyme is an efficient endonuclease, able to cleave closed circular single-stranded and supercoiled double-stranded DNAs, and it cleaves 3′-ended single-stranded DNA at its junction with the duplex DNA (Sung et al. 1993; Tomkinson et al. 1993; Bardwell et al. 1994). Here we show that the Rad1-Rad10 nuclease also removes the PG group or a normal nucleotide from the 3′ end in duplex DNA. Interestingly, the pattern of incisions in the duplex region is the same regardless of whether or not the DNA substrate bears a 3′-ended single-stranded tail, and whether the 3′ end has a PG group or is undamaged. In all cases, the enzyme cleaves three or more nucleotides into the duplex region from the 3′ end.
In contrast to the inviability of the apn1Δ apn2Δ rad1Δ or the apn1Δ apn2Δ rad10Δ mutant, the apn1Δ apn2Δ strain harboring a deletion of the other NER genes, such as RAD2, RAD4, or RAD14, remains viable. Because, for the repair of AP sites, the Rad1-Rad10 nuclease acts in conjunction with all of the other proteins of the NER ensemble, the inviability of the apn1Δ apn2Δ rad1Δ or the apn1Δ apn2Δ rad10Δ mutant is unlikely to be from a defect in AP removal. On the other hand, because the Rad1-Rad10 nuclease promotes repair of H2O2-induced DNA strand breaks by removing the 3′-PG and other 3′-blocking groups without the assistance of other NER proteins, it is quite plausible that deficient removal of 3′-blocking groups formed from the action of oxygen free radicals produced during normal cellular oxidative metabolism is the prime contributor to the inviability of the apn1Δ apn2Δ rad1Δ or apn1Δ apn2Δ rad10Δ mutants. The action of DNA glycosylases such as Ntg1, Ntg2, and Ogg1, which act in the removal of oxidatively damaged bases, and which, in addition to a glycosylase activity, harbor a 3′ AP lyase activity, would further add to the generation of strand breaks with 3′-blocking groups. In support of this idea is the observation that simultaneous inactivation of NTG1, NTG2, and OGG1 in the apn1Δ apn2Δ rad1Δ yeast strain ameliorates the inviability defect to a significant degree (Guillet and Boiteux 2002).
The inviability of apn1Δ apn2Δ rad1Δ and apn1Δ apn2Δ rad10Δ strains is unlikely to be due to the involvement of Rad1-Rad10 in the single-strand annealing (SSA) pathway of genetic recombination, as this pathway represents a minor mode of genetic recombination. Moreover, even though the RAD52 group of genes plays a predominant role in homologous recombination, and RAD52 is required also for SSA (Haber 2000), deletion of neither RAD52 nor of any other gene of this epistasis group from the apn1Δ apn2Δ strain produces inviability.
The persistence of single-strand DNA breaks with 3′-blocking groups in the apn1Δ apn2Δ rad1Δ and apn1Δ apn2Δ rad10Δ strains would hinder the progression of transcription and replication, as RNA polymerases and DNA polymerases would stall at such a lesion site. In an RNA polymerase ternary complex stalled on the transcribed strand at the strand break with a 3′-blocking group (Kathe et al. 2004), the nontranscribed strand could become subject to nicking and subsequent nucleolytic degradation, as we have suggested before for the stalling of RNA polymerase at an AP site (Yu et al. 2003). However, in contrast to an AP site, where the resulting gap in the nontranscribed strand can be filled in by repair synthesis via the action of translesion synthesis DNA polymerases, no such mechanism would be available for the repair of the double-strand break ensuing from the transcriptional stalling at the strand break with a 3′-blocking group. Moreover, the presence of the 3′ group at the break site could be inhibitory to recombinational processes, leading to DNA degradation and cell death. The double-strand break formed during replication from the stalling of DNA polymerases at such a lesion site could also be similarly irreparable, causing cell death.
In humans, the XPF and ERCC1 genes encode the respective counterparts of yeast Rad1 and Rad10 proteins, and like the Rad1-Rad10 complex, the XPF-ERCC1 complex cleaves the damaged DNA strand on the 5′-side of the lesion, and it functions also in homology-based recombination (Sargent et al. 1997; Adair et al. 2000). Inactivation of either ERCC1 or XPF in mice causes severe developmental defects. In both cases, the mutant mice exhibit severe runting, with death occurring at ∼20 d after birth, and the liver of these mice contains a large number of abnormal cells with enlarged nuclei (McWhir et al. 1993; Tian et al. 2004). These developmental abnormalities of the ERCC1/XPF mutant mice cannot be attributed to a defect in NER, as mice deficient in XPA or XPC develop normally (de Vries et al. 1995; Nakane et al. 1995; Sands et al. 1995). Our inference that a deficiency in the removal of 3′-blocking groups from strand breaks induced by endogenously formed oxygen free radicals is the primary cause of the inviability of apn1Δ apn2Δ rad1Δ or apn1Δ apn2Δ rad10Δ yeast cells, leads us to propose that a similar deficiency underlies the growth defects of ERCC1/XPF mice. It is also of interest to note that mice deficient in Ape1, which is related to yeast Apn2, die during embryonic development following blastocyst formation (Xanthoudakis et al. 1996). A defect in the removal of 3′-blocking groups from endogenously generated DNA strand breaks could also be the cause of the inviability of Ape1-deficient mice.
Materials and methods
Yeast strains
All yeast stains used in this study were derived from EMY74.7 (MATa his3-Δ1 leu2-3,-112 trp1Δ ura3-52). Deletion mutations in yeast were generated by the gene replacement method (Rothstein 1991) using the URA3 gene blaster (Alani et al. 1987).
H2O2 and MMS sensitivity
For determining sensitivity to H2O2, cells grown overnight in yeast extract-peptone-dextrose (YPD) medium were sonicated to disperse clumps, diluted in fresh YPD medium, and incubated at 30°C until they had reached midexponential phase. Stock H2O2 was diluted in water just before use and added to 1 mL cells in YPD medium. After 1 h of incubation with vigorous agitation at 30°C, 9 mL cold water was added. Appropriate dilutions of cells were plated on YPD medium for viability determinations.
For determining sensitivity to MMS, cells were grown overnight in YPD medium, sonicated to disperse clumps, washed, and resuspended in 0.05 M KPO4 buffer (pH 7.0). Appropriate dilutions of 0.5 mL MMS at twice the desired final concentration were added to 0.5-mL suspensions of cells adjusted to 3 × 108 cells per milliliter, and incubated with vigorous shaking for 20 min at 30°C. The reaction was terminated by the addition of 1 mL 10% sodium thiosulfate. Appropriate dilutions of cells were plated on YPD for viability determinations. Plates were incubated at 30°C and counted after 3 d.
Alkaline sucrose gradients
[rhoo] derivatives lacking mitochondrial DNA, obtained by ethidium bromide mutagenesis of yeast strains, were grown overnight at 30°C in synthetic complete medium (SC) containing 10 μg uracil per milliliter and 10 μCi [3H-uracil] (Moravek Biochemicals) per milliliter and collected by filtration after the cell density had reached 5 × 106 cells per milliliter. Cells were washed with water and suspended in the same volume (80 mL) of SC containing 30 μg uracil per milliliter. Cells were then incubated for 1 h at 30°C to allow for depletion of the intracellular pool of [3H-uracil] to occur. After this incubation, cells were treated with H2O2 (Fluka) at a final concentration of 0.5 mM for 1 h. Cells were collected by filtration, washed, and resuspended in 2 mL of YPD medium. One milliliter of cells was used for the 0-h sample and the other was diluted to 10 mL with YPD medium and incubated for 4 h to allow for repair to occur. Conversion of cells to spheroplasts, alkaline sucrose gradient sedimentation, and processing of samples were performed as described (Torres-Ramos et al. 1996), except that alkaline sucrose gradients were centrifuged at 21,000 rpm for 15 h and 30 min and acid precipitation of alkali-hydrolyzed samples was carried out with 1 N HCL and 0.1 M sodium pyrophosphate.
DNA substrates
A 35-nt oligomer containing a 3′-PG terminus was synthesized on CPG-glycerol support (purchased from Glen Research) at the 1-μM scale on a Perkin-Elmer Biosystems Expedite DNA synthesizer following the published method (Urata and Akagi 1993). The reaction products were separated on a preparative HPLC column and further purified on a sequencing gel to yield a homogeneous preparation of modified oligomer. A similar protocol was used to generate a 40-nt oligomer carrying a 3′-PG modification. A 70-bp double-stranded DNA substrate (PG1) with a 1-nt gap was obtained by mixing 5′-end labeled 3′-PG modified or unmodified 35-nt oligomer with an equimolar amount of a 34-nt oligomer and both annealed to a complementary 70-nt oligomer template (Fig. 4). In PG2, a 5′-end labeled 40-nt oligomer modified with a 3′-PG or unmodified was hybridized to the complementary 70-nt oligomer, and the downstream 34-nt oligomer was the same as in PG1. The resulting double-stranded DNA substrate had a 5-nt flap (GTCAGpg) at the 3′ end (Fig. 4). For examining the 3′ → 5′ exonuclease activity of Rad1-Rad10, the DNA substrate S1 was generated by annealing a 3′-end labeled 27-nt oligomer to the middle of a 70-nt oligomer template (Fig. 4). The 3′-end labeling was carried out with terminal deoxynucleotidyl transferase and [32P]ddGTP.
Nuclease assays
DNA endonuclease assays were carried out in buffer containing 20 mM Tris-HCl (pH 7.9), 5 mM MgCl2, and 100 μg/mL BSA with 50 fmole DNA substrate and an equimolar amount of the Rad1-Rad10 protein complex in a final reaction volume of 5 μL. After incubation for 30 min at 30°C, the reaction was stopped by the addition of 5 μL of formamide gel loading buffer and the reaction products analyzed on a 10% sequencing gel followed by autoradiography. For exonuclease assays, any differences in the method are noted in the figure legend.
Acknowledgments
This work was supported by grant CA41261 from the National Cancer Institute, National Institutes of Health.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1232804.
References
- Adair G.M., Rolig, R.L., Moore-Faver, D., Zabelshansky, M., Wilson, J.H., and Nairn, R.S. 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19: 5552-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Cao, L., and Kleckner, N. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted genes. Genetics 116: 541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell A.J., Bardwell, L., Tomkinson, A.E., and Friedberg, E.C. 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265: 2082-2085. [DOI] [PubMed] [Google Scholar]
- Bjoras M., Klungland, A., Johansen, R.F., and Seeberg, E. 1995. Purification and properties of the alkylation repair DNA glycosylase encoded MAG gene from Saccharomyces cerevisiae. Biochemistry 34: 4577-4582. [DOI] [PubMed] [Google Scholar]
- de Vries A., van Oostrom, C.T.M., Hofhuis, F.M.A., Dortant, P.M., Berg, R.J.W., de Gruiji, F.R., Wester, P.W., van Keriji, C.F., Capel, P.J.A., can Steeg, H., et al. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377: 169-173. [DOI] [PubMed] [Google Scholar]
- Guillet M. and Boiteux, S. 2002. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 21: 2833-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S.N., Habraken, Y., Sung, P., Prakash, L., and Prakash, S. 1995. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270: 12973-12976. [DOI] [PubMed] [Google Scholar]
- Haber J.E. 2000. Lucky breaks: Analysis of recombination in Saccharomyces. Mutat. Res. 451: 53-69. [DOI] [PubMed] [Google Scholar]
- Henle E.S. and Linn, S. 1997. Formation, prevention, and repair of DNA damage by iorn/hydrogen peroxide. J. Biol. Chem. 272: 19095-19098. [DOI] [PubMed] [Google Scholar]
- Imlay J.A. and Linn, S. 1988. DNA damage and oxygen radical toxicity. Science 240: 1302-1309. [DOI] [PubMed] [Google Scholar]
- Johnson A.W. and Demple, B. 1988. Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidinic endonuclease: Substrate specificity and kinetics. J. Biol. Chem. 263: 18017-18022. [PubMed] [Google Scholar]
- Johnson R.E., Torres-Ramos, C.A., Izumi, T., Mitra, S., Prakash, S., and Prakash, L. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes & Dev. 12: 3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathe S.D., Shen, G.-P., and Wallace, S.S. 2004. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 279: 18511-18520. [DOI] [PubMed] [Google Scholar]
- McWhir J., Selfridge, J., Harrison, D.J., Squires, S., and Melton, D.W. 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5: 217-224. [DOI] [PubMed] [Google Scholar]
- Mu D., Park, C.-H., Matsunaga, T., Hsu, D.S., Reardon, J.T., and Sancar, A. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270: 2415-2418. [DOI] [PubMed] [Google Scholar]
- Mu D., Hsu, D.S., and Sancar, A. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271: 8285-8294. [DOI] [PubMed] [Google Scholar]
- Nakane H., Takeuchi, S., Yuba, S., Saijo, M., Nakatsu, Y., Murai, H., Nakatsuru, Y., Ishikawa, T., Hirota, S., Kitamura, Y., et al. 1995. High incidence of ultraviolet-B- or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377: 165-168. [DOI] [PubMed] [Google Scholar]
- Prakash S. and Prakash, L. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451: 13-24. [DOI] [PubMed] [Google Scholar]
- Reardon J.T. and Sancar, A. 2002. Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol. Cell. Biol. 22: 5938-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cnacers, by the human excision nuclease. Genes & Dev. 17: 2539-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. 1991. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 194: 281-301. [DOI] [PubMed] [Google Scholar]
- Roy R., Brooks, C., and Mitra, S. 1994. Purification and biochemical characterization of recombinant N-methylpurine-DNA glycosylase of the mouse. Biochemistry 33: 15131-15140. [DOI] [PubMed] [Google Scholar]
- Sands A.T., Abuin, A., Sanchez, A., Conti, C.J., and Bradley, A. 1995. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature 377: 162-165. [DOI] [PubMed] [Google Scholar]
- Sargent R.G., Rolig, R.L., Kilburn, A.E., Adair, G.M., Wilson, J.H., and Nairn, R.S. 1997. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl. Acad. Sci. 94: 13122-13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmura K., Kasai, H., Sasaki, A., Sugimura, H., and Yokota, J. 1997. 8-Hydroyguanine (7,8-dihydro-8-oxoguanine) DNA glycosylase and AP lyase activities of hOGG1 protein and their substrate specificity. Mutat. Res. 385: 75-82. [DOI] [PubMed] [Google Scholar]
- Sung P., Reynolds, P., Prakash, L., and Prakash, S. 1993. Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J. Biol. Chem. 268: 26391-26399. [PubMed] [Google Scholar]
- Tian M., Shinkura, R., Shinkura, N., and Alt, F.W. 2004. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 24: 1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson A.E., Bardwell, A.J., Bardwell, L., Tappe, N.J., and Friedberg, E.C. 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362: 860-862. [DOI] [PubMed] [Google Scholar]
- Torres-Ramos C.A., Yoder, B.L., Burgers, P.M.J., Prakash, S., and Prakash, L. 1996. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proc. Natl. Acad. Sci. 93: 9676-9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos C.A., Johnson, R.E., Prakash, L., and Prakash, S. 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20: 3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I., Haracska, L., Johnson, R.E., Prakash, S., and Prakash, L. 2000. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 275: 22427-22434. [DOI] [PubMed] [Google Scholar]
- Unk I., Haracska, L., Prakash, S., and Prakash, L. 2001. 3′-Phosphodiesterase and 3′ → 5′ exonulcease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 21: 1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I., Haracska, L., Gomes, X.V., Burgers, P.M.J., Prakash, L., and Prakash, S. 2002. Stimulation of 3′ → 5′ exonuclease and 3′-phosphdiesterase activities of yeast Apn2 by proliferating cell nuclear antigen. Mol. Cell. Biol. 22: 6480-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata, H and Akagi, M. 1993. A convenient synthesis of oligonucleotides with a 3′-phosphoglycolate and 3′-phosphoglycaldehyde terminus. Tetrahedron Lett. 34: 4015-4018. [Google Scholar]
- Wallace S. 1997. Oxidative damage to DNA and its repair. In Oxidative stress and the molecular biology of antioxidant defenses (ed. J.G. Scandalios), pp. 49-90. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Xanthoudakis S., Smeyne, R.J., Wallace, J.D., and Curran, T. 1996. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. 93: 8919-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H.J., Swanson, R.L., and Doetsch, P.W. 1998. Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry 37: 6033-6040. [DOI] [PubMed] [Google Scholar]
- Yu S.-L., Lee, S.-K., Johnson, R.E., Prakash, L., and Prakash, S. 2003. The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol. 23: 382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]