Abstract
Regulation/activation of the Porphyromonas gingivalis gingipains is poorly understood. A unique 1.3-kb open reading frame downstream of the bcp-recA-vimA transcriptional unit was cloned, insertionally inactivated with the ermF-ermAM antibiotic resistance cassette, and used to create a defective mutant by allelic exchange. In contrast to the wild-type W83 strain, the growth rate of the mutant strain (designated FLL93) was reduced, and when plated on Brucella blood agar it was nonpigmented and nonhemolytic. Arginine- and lysine-specific gingipain activities were reduced by approximately 90 and 85%, respectively, relative to activities of the parent strain. These activities were unaffected by the culture's growth phase, in contrast to the vimA-defective mutant P. gingivalis FLL92, which has increased proteolytic activity in stationary phase. Expression of the rgpA, rgpB, and kgp gingipain genes was unaltered in P. gingivalis FLL93 compared to that of the wild-type strain. Further, in extracellular protein fractions a 64-kDa band was identified that was immunoreactive with the RgpB-specific proenzyme antibodies. Active-site labeling with dansyl-glutamyl-glycyl-arginyl chloromethyl ketone or immunoblot analysis showed no detectable protein band representing the gingipain catalytic domain. In vitro protease activity could be slightly induced by a urea denaturation-renaturation cycle in an extracellular protein fraction, in contrast to the vimA-defective mutant P. gingivalis FLL92. Expression of flanking genes, including recA, vimA, and Pg0792, was unaltered by the mutation. Taken together, these results suggest that the vimA downstream gene, designated vimE (for virulence-modulating gene E), is involved in the regulation of protease activity in P. gingivalis.
The expression of extracellular proteolytic activities is highly regulated in both prokaryotic and eukaryotic systems (6, 40). This regulation usually occurs at the level of expression of the protease genes, secretion, and/or processing of an inactive secreted precursor to its active form. The multiple layers of regulation are vital to ensure that expression is tightly controlled in the appropriate temporal and spatial patterns. Porphyromonas gingivalis, a black-pigmented, gram-negative anaerobe, has been implicated as an important etiological agent in adult periodontitis (19, 23, 38). While several virulence factors, including hydrolytic enzymes, fimbriae, hemagglutinin, capsule, and lipopolysaccharide, have been implicated in the pathogenicity of P. gingivalis, the strong proteolytic abilities of this organism are the focus of much attention, as they are considered to play the most significant role in virulence (25). The major proteases, called gingipains, are both extracellular and cell associated. They consist of arginine-specific protease (Arg-gingipain [Rgp]) and lysine-specific protease (Lys-gingipain [Kgp]) (25). Although glycosylation appears to be important for their activation (10, 30), there remains a gap in our knowledge regarding the regulation/activation of the P. gingivalis gingipains.
It was previously reported that the recA locus can affect the phenotypic expression and distribution of the gingipains in P. gingivalis (1, 2, 27). Using the cloned vimA gene, which is downstream of the recA gene and is part of the bcp-recA-vimA transcriptional unit, a defective mutant was constructed by allelic exchange (1). The mutant strain, designated FLL92, did not have a black pigmentation and showed increased autoaggregration in addition to a significant reduction in proteolytic, hemolytic, and hemagglutinating activities (1). For in vivo experiments using a mouse model, P. gingivalis FLL92 had dramatically reduced virulence compared to that of the wild-type W83 strain (1). While a reduction in Arg-X- and Lys-X-specific proteolytic activities was observed in P. gingivalis FLL92, transcription of the gingipain genes was unaltered in this mutant compared to that of the wild-type strain (1). Furthermore, the partially processed RgpB proenzyme was secreted in P. gingivalis FLL92 (27). Collectively these observations suggest that the vimA gene in P. gingivalis may be involved in virulence modulation via an ability to affect protease activation/maturation. In addition, appearance of the gingipain proenzyme forms and the growth-phase-dependent activation of proteolytic (27) activity have raised the possibility of multiple mechanisms for gingipain activation/maturation involving multiple bacteria-specific factors.
We have further investigated a unique 1.3-kb gene downstream of the vimA gene to determine its relationship to the bcp-recA-vimA transcriptional unit and to evaluate its role, if any, in protease activation. In this report, we have created and characterized a P. gingivalis isogenic mutant (FLL93) defective in this gene, now designated vimE. While this gene was expressed independent of vimA, its inactivation resulted in reduced Arg-X- and Lys-X-specific proteolytic activities that were not affected by the phase of growth. In addition, in vitro protease activity was slightly activated by a urea denaturation-renaturation cycle. These results suggest an important role for the vimE gene in protease activation/maturation in P. gingivalis and further confirm the requirement of multiple specific host factors in this process.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. P. gingivalis strains were grown in brain heart infusion (BHI) broth supplemented with 0.5% yeast extract (both from Difco Laboratories, Detroit, Mich.), hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%) (all from Sigma-Aldrich, St. Louis, Mo.). Escherichia coli strains were grown in Luria-Bertani broth. Unless otherwise stated, all cultures were incubated at 37°C. P. gingivalis strains were maintained in an anaerobic chamber (Coy Manufacturing, Ann Arbor, Mich.) in 10% H2, 10% CO2, 80% N2. Growth rates for P. gingivalis and E. coli strains were determined spectrophotometrically (optical density at 600 nm [OD600]). Antibiotics were used at the following concentrations: clindamycin, 0.5 μg/ml; erythromycin, 300 μg/ml; and carbenicillin, 100 μg/ml.
TABLE 1.
Plasmids and bacterial strains used in this study
| Plasmid or strain | Phenotype or description | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR-2.1 TOPO | Apr, Kmr | Invitrogen |
| pFLL80 | pCR 2.1-TOPO Pg0791:Pg0792 | This study |
| pFLL81 | pFLL80 with Pg0791 interrupted with ermF-ermAM | This study |
| pVA2198 | Spr, ermF-ermAM | 1 |
| P. gingivalis | ||
| W83 | Wild type | |
| FLL92 | vimA defective | 1 |
| FLL93 | vimE defective | This study |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ-thi-1 gyrA96 relA1 | Invitrogen |
| Top10 | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔ M15 ΔlacX74 recA1 ara139 Δ (ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
DNA isolation and analysis.
P. gingivalis chromosomal DNA was prepared by the method of Marmur (22). For plasmid DNA analysis, DNA extraction was performed by the alkaline lysis procedure of Birnboim and Doly (4). For large-scale preparation, plasmids were purified by using the QIAGEN (Santa Clarita, Calif.) plasmid maxi kit. DNA was digested with restriction enzymes as specified by the manufacturer (Roche, Indianapolis, Ind.). For DNA subcloning, the desired fragments were isolated from 0.8% agarose gels run in Tris-acetate-EDTA (TAE) buffer and then were purified by using a Gene Clean kit as recommended by the manufacturer (QBiogene, Inc., La Jolla, Calif.).
Generation of Pg0791 (vimE) mutant P. gingivalis strain.
A 2.5-kb fragment carrying the intact vimE and Pg0792 downstream genes was amplified by PCR using the P1 and P5 oligonucleotide primer (Table 2; Fig. 1). This fragment was cloned into the pCR 2.1-TOPO plasmid vector (Invitrogen, Carlsbad, Calif.) and was designated pFLL80. The ermF-ermAM cassette, which confers erythromycin/clindamycin resistance in E. coli and P. gingivalis (9), was PCR amplified from pVA2198 with Pfu turbo (Stratagene) and was ligated into the HincII restriction site of the vimE gene. The resultant recombinant plasmid, pFLL81, was used as a donor in electroporation of P. gingivalis W83.
TABLE 2.
Primers used in this study
| Name | Description | Sequence |
|---|---|---|
| P1 | Pg0791 (vimE) forward | 5′ATGAATACTGAAGTAATCCAC 3′ |
| P2 | Pg0791 (vimE) reverse | 5′ TTGGTCGATGGCCGTTTCGTA 3′ |
| P3 | Pg0791 (vimE) reverse (intragenic) | 5′ TCAGCACTTCCTTGCACAAC 3′ |
| P4 | Pg0792 forward (intragenic) | 5′ CAGTCGCACGGCGATGTGTTT 3′ |
| P5 | Pg0792 reverse (intragenic) | 5′ GCAGGATGGAGCGGGAGTATTC 3′ |
| P6 | Pg0792 reverse | 5′ TTCACGCGAAAAAGTGGAGAT 3′ |
| P7 | vimA forward | 5′ ATGCCCATCCCTCTATACCTG 3′ |
| P8 | vimA reverse | 5′ TACCTGTTTTTGCTGACCGG 3′ |
| P9 | recA forward | 5′ ATGGCAGAAGAAAAGATACCC 3′ |
| P10 | recA reverse | 5′ TGAATGTTTGTTGCGAATGG 3′ |
| P11 | pro-kgp forward | 5′ ATGAGGAAATTATTATTGCTGATCG 3′ |
| P12 | pro-kgp reverse | 5′ TTGAAGAGCTGTTTATAAGCTGTTT 3′ |
| P13 | pro-rgpA forward | 5′ ATGAAAAACTTGAACAAGTTTGTTTC 3′ |
| P14 | pro-rgpA reverse | 5′ CCGGTGTGTAACGCCCTG 3′ |
| P15 | pro-rgpB forward | 5′ CGACCGATGAAGACTTCG 3′ |
| P16 | pro-rgpB reverse | 5′ TGAAGACGCTCTTATAGATATCTT 3′ |
| P17 | erythromycin forward | 5′ TATTAGGCCTATAGCTTCCGCTATT 3′ |
| P18 | erythromycin reverse | 5′ AATAGGCCTTAGTAACGTGTAACTTT 3′ |
FIG. 1.
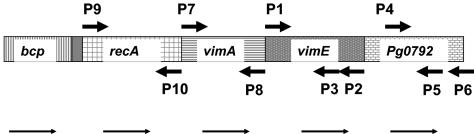
Diagram of the recA locus and the two downstream genes. The diagram shows the open reading frame (from www.oralgen.lanl.gov) of the recA locus, and the two downstream genes are shown with the indicated primer locations.
Electroporation.
Electroporation of cells was performed as previously reported (9). Briefly, 1 ml of an actively growing culture of P. gingivalis was used to inoculate 10 ml of BHI broth supplemented with yeast extract, hemin, and vitamin K, which was then incubated overnight at 37°C. Ninety milliliters of the same prewarmed (37°C) medium was then inoculated with 3 ml of the overnight culture and was incubated for an additional 4 h until an OD600 of 0.7 was obtained. The cells were then harvested by centrifugation at 3,000 × g for 15 min at 4°C and were washed twice in 50 ml of electroporation buffer (10% glycerol, 1 mM MgCl2; filter sterilized and stored at 4°C). The final cell pellet was resuspended in 0.5 ml of electroporation buffer. A 100-μl sample of resuspended cells and 5 μg of DNA was placed in a sterile electrode cuvette (0.2-cm gap). The cells were then pulsed with a Bio-Rad (Hercules, Calif.) gene pulser at 2,500 V for 9.5 ms and then were incubated on ice for 5 min. The cell suspension was then added to 0.5 ml of BHI broth supplemented with yeast extract, hemin, and vitamin K and was incubated for approximately 16 h. A 100-μl sample was plated on solid medium containing erythromycin and was incubated anaerobically at 37°C for 5 to 10 days.
Growth curve analysis.
Actively growing P. gingivalis W83, FLL92, and FLL93 cultures were inoculated at an initial OD600 of 0.2 in BHI broth supplemented with 0.5% yeast extract (both from Difco Laboratories), hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%) (all from Sigma-Aldrich). Aliquots (500 μl) were taken every 4 h, and growth rate was determined by spectrometer.
Hemolytic activity assay.
Hemolytic activity was determined as previously reported (11). Briefly, bacterial cells from overnight cultures were harvested by centrifugation (10,000 × g for 30 min) using a Sorvall RC5C centrifuge, washed three times with phosphate-buffered saline (PBS), and then resuspended to a final OD600 of 1.5. Sheep erythrocytes (Hemostat Laboratories, Dixon, Calif.) were harvested by centrifugation (4,400 × g for 25 min) and washed with 1× PBS until the supernatant was visually free of hemoglobin pigment. The washed erythrocytes were suspended to a final concentration of 1% in 1× PBS. Hemolytic activity was determined by mixing an equal volume of bacterial cells with 1% erythrocytes in PBS. This mixture was then incubated at 37°C. Samples (500 μl) were withdrawn every 2 h and then were centrifuged (1,300 × g for 5 min) in an Eppendorf 5403 centrifuge. The OD405 was determined by spectrophotometry. As a negative control, erythrocytes were used alone.
Hemagglutination studies.
Hemagglutinin activity was determined as previously reported (11). Twenty-four-hour cultures of P. gingivalis W83, FLL92, and FLL93 cells were harvested by centrifugation (10,000 × g, 15 min). Cells were washed twice in 1× PBS and were resuspended to a final OD600 of 1.5. Sheep erythrocytes were washed twice with 1× PBS and were resuspended to a final concentration of 1% in 1× PBS. An aliquot (100 μl) of the bacterial suspension was serially diluted twofold with 1× PBS in round-bottom 96-well microtiter plates. An equal volume (100 μl) of 1% sheep erythrocytes was mixed with each dilution and was incubated at 4°C for 3 h. Hemagglutination was visually assessed, and the hemagglutination titer was determined as the last dilution that showed complete hemagglutination.
Cell fraction preparation.
One-liter cultures of P. gingivalis strains FLL92 and W83 were grown from actively growing cells. Preparations of whole-cell culture, cell-free medium, cell suspension, vesicles, and vesicle-free medium were made as previously reported (27). The whole-cell culture fraction is a sample of the culture after the bacterium has been grown to a specific growth phase. This sample has the bacterial cells suspended in the growth medium, and the enzyme activity includes the gingipains that are attached to the bacterial cell surface plus those that are secreted in the culture medium. After centrifugation the cell pellet was resuspended, and the enzyme activity in this sample (the cell suspension fraction) represents the gingipains that are attached to the bacterial cell surface, while the enzyme activity in the supernatant (the cell-free medium fraction) includes the gingipains that are secreted in the culture medium. Secreted gingipains can either be associated with vesicles or be soluble in the culture medium; thus, ultracentrifugation of the cell-free fractions will yield a vesicle pellet and a supernatant of soluble gingipains.
Preparation of P. gingivalis extracellular fractions.
One-liter cultures of P. gingivalis strains FLL92, FLL93, and W83 were grown from actively growing cells. Cells were harvested by centrifugation at 10,000 × g for 30 min. The cell-free culture fluid was precipitated with 37.5 or 60% acetone (−20°C), and the protein pellet was resuspended in 7 ml of 100 mM Tris-HCl buffer (pH 7.4), dialyzed for 24 h against the same buffer, and then stored on ice or at 0°C. The presence of Arg-X- and Lys-X-specific cysteine protease activities was determined by microplate reader (Bio-Rad).
Production of rabbit polyclonal antibodies against the RgpB proenzyme.
Extracellular proteins were prepared from the culture supernatant of P. gingivalis FLL92 (vimA defective) grown to exponential phase (OD600 of 0.8). The cells were harvested by centrifugation (10,000 × g, 30 min), and the supernatant was further clarified by filtration through a 0.45-μm-pore-size membrane (Millipore Corporation, Billerica, Mass.). The cell-free culture fluid was precipitated with 37.5% acetone (−20°C), and the protein pellet was resuspended in 7 ml of 100 mM Tris-HCl buffer (pH 7.4), dialyzed for 24 h against the same buffer, and then stored on ice or at 0°C. The protein concentration was determined as described below. The extracellular proteins (25 μg/lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using NuPAGE 4 to 12% Bis-Tris gels. A 64-kDa band representing the partially processed RgpB proenzyme (27) was excised with a gel cutter. A total of approximately 12 mg of the RgpB proenzyme protein was excised from gels, placed in 1× PBS, and stored at −80°C. The frozen gel slices were sent to Zymed Laboratories Inc. (South San Francisco, Calif.) for the production of polyclonal rabbit RgpB proenzyme antibodies by using the manufacturer's standard protocol. Dilutions and efficiency of the antibodies were tested in the laboratory with the purified RgpB proenzyme.
Protein concentration determination.
Protein concentration was calculated spectrophotometrically by using the Warburg formula within the protein function of the Eppendorf Biophotometer (Brinkman, Westbury, N.Y.).
SDS-PAGE and immunoblot analysis.
SDS-PAGE was performed with a 4 to 12% Bis-Tris separating gel in morpholinepropanesulfonic acid-SDS running buffer (NuPAGE Novex gels; Invitrogen) according to the manufacturer's instructions. Samples were prepared (65% sample, 25% 4× NuPAGE lithium dodecyl sulfate sample buffer, 10% NuPAGE reducing agent), heated at 72°C for 10 min, and then electrophoresed at 200 V for 65 min with a XCell SureLock Mini-Cell (Invitrogen). The separated proteins were then transferred to BioTrace nitrocellulose membranes (Pall Corporation, Ann Arbor, Mich.) and processed at 15 V for 25 min with a Semi-Dry Trans-blot apparatus (Bio-Rad). The blots were probed with antibodies against specific protease domains and species-specific secondary antibodies conjugated to horseradish peroxidase (Zymed Laboratories). Immunoreactive proteins were detected by the procedure described in the Western Lightning Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Boston, Mass.). The secondary antibody was immunoglobulin G (heavy plus light chains)-horseradish peroxidase conjugate (Zymed Laboratories, Inc.).
Protease assays.
The presence of Arg-X- and Lys-X-specific cysteine protease activities was determined with a microplate reader (Bio-Rad) by the method of Potempa et al. (30).
Analysis of P. gingivalis Pg0791 mutant genes by RT-PCR.
Total RNA was extracted from P. gingivalis strains grown to early stationary phase (OD600 of 1.2 to 1.3) using a RiboPure kit (Ambion, Austin, Tex.). The primers used for reverse transcription-PCR (RT-PCR) analysis (Table 2) were specific for the recA, rgpA, rgpB, kgp, Pg0791, or Pg0792 genes. The RT-PCR (50 μl) contained 1 μg of template RNA in the Superscript One-Step RT-PCR mix (Invitrogen). Negative controls were reactions in the absence of reverse transcriptase.
Denaturation-renaturation procedure for in vitro protease activation.
Extracellular protein fractions from P. gingivalis were mixed with 8 M urea and incubated at 4°C for 1 h as described previously (11). The urea was slowly removed from the mixture by centrifugation (10,000 × g) in a Millipore filtration unit (Biomax 10K NMWL membrane, 0.5-ml volume) with the addition of increasing volumes of 100 mM Tris-HCl buffer (pH 7.4).
Fluorescence labeling of proteases by using DNS-EGR-CK.
Fluorescence labeling of proteases in the extracellular protein fraction from P. gingivalis was done as previously reported (3). Briefly, solutions of extracellular protein fractions from P. gingivalis were treated at 4°C for 10 min with equal volumes of 0.2 M Tris-HCl (pH 8.4) containing 20 mM CaCl2 and 20 mM 2-mercaptoethanol. Dansyl-glutamyl-glycyl-arginyl chloromethyl ketone (DNS-EGR-CK) (0.25 mg; Calbiochem, San Diego, Calif.) was dissolved in 600 μl of 95% (vol/vol) aqueous ethanol just before use. Fifty microliters was added to the reduced protease solution, and the reaction was allowed to proceed at 4°C until the enzyme activity (monitored by N-α-benzoyl-dl-arginine-p-nitroanilide [BApNA] hydrolysis) was abolished. Samples of the protein solution were then dried in a SpeedVac concentrator (Thermo Savant, Holbrook, N.Y.). To identify the protease band unambiguously after SDS-PAGE, labeling was also performed in the presence of 50 μM leupeptin. The dried fluorescence-labeled proteases were then treated with NuPAGE SDS-PAGE sample buffer (Invitrogen) and were subjected to electrophoresis.
RESULTS
Inactivation of the vimE gene in P. gingivalis W83 by allelic exchange mutagenesis.
Isogenic mutants of P. gingivalis W83 defective in the Pg0791 (www.oralgen.lanl.gov) or open reading frame Pg0883 (www.tigr.org) gene (designated vimE) were constructed by allelic exchange mutagenesis. The circular recombinant plasmid pFLL81, which carries the ermF-ermAM cassette in the unique HincII restriction site (base pair 909 of the open reading frame) of the vimE gene, was used as a donor in electroporation of P. gingivalis W83 (Fig. 2A). Because the plasmid was unable to replicate in P. gingivalis, we predicted that two double-crossover events between the regions flanking the erythromycin marker and the wild-type gene on the chromosome would result in replacement of a segment of the wild-type gene with a fragment conferring erythromycin resistance. Following electroporation and plating on selective medium (BHI containing 10 μg of erythromycin/ml), we detected approximately 61 erythromycin-resistant colonies after a 5-day incubation period. To compare their phenotypic properties to those of wild-type strain W83, all mutants were plated on Brucella blood agar plates (Anaerobic Systems Inc., San Jose, Calif.). In contrast to the wild-type strain, all mutants displayed a non-black-pigmented, non-beta-hemolytic phenotype.
FIG. 2.
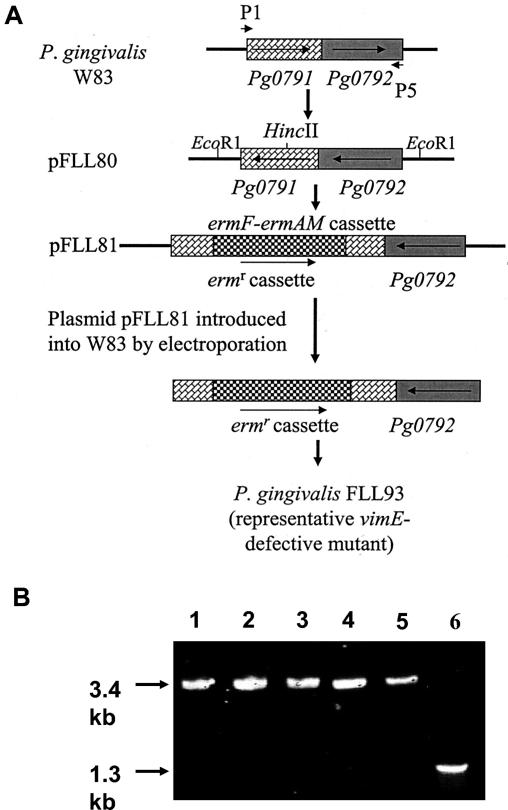
Construction of a vimE-defective mutant by allelic exchange mutagenesis and confirmation by PCR analysis. pFLL81 contains the vimE gene interrupted by an ermF-ermAM cassette (the vimE gene with flanking sequences was amplified by PCR; the ermF-ermAM cassette was purified from pVA2198). The circular recombinant plasmid pFLL81 was integrally introduced into P. gingivalis W83 by electroporation. A reciprocal recombination event between areas of homology on the chromosome and regions flanking the ermAM-ermF cassette on pFLL81 replaced the intact vimE with an inactivated copy. (B) Oligonucleotide primers specific for the vimE gene (P1 and P2; Table 2) were then used to amplify that gene from total cellular DNA from P. gingivalis. Lane 1, P. gingivalis FLL93.1 (vimE::ermF-ermAM); lane 2, P. gingivalis FLL93.2 (vimE::ermF-ermAM); lane 3, P. gingivalis FLL93.3 (vimE::ermF-ermAM); lane 4, P. gingivalis FLL93.4 (vimE::ermF-ermAM); lane 5, P. gingivalis FLL93.5 (vimE::ermF-ermAM); and lane 6, P. gingivalis W83 (wild type).
Confirmation of inactivation of vimE by PCR analysis.
Chromosomal DNA from five randomly chosen clindamycin- and erythromycin-resistant colonies and the wild-type W83 strain were analyzed by PCR to confirm inactivation in the vimE gene. If the vimE gene was interrupted by the ermF-ermAM cassette, a 3.4-kb fragment was expected to be amplified by primers P1 and P2 (Table 2). The expected 3.4- and 1.3-kb fragments were observed in the five clindamycin-resistant strains and in wild-type W83, respectively (Fig. 2B). The orientation of the ermF-ermAM cassette was also confirmed by restriction digest (data not shown). Taken together these results indicated the insertional inactivation of the chromosomal vimE gene with the 2.1-kb ermF-ermAM antibiotic cassette was successful. One mutant, designated P. gingivalis FLL93, was randomly chosen for further study. A generation time of 3 h was determined for P. gingivalis W83 and P. gingivalis FLL92, in contrast to 4.75 h for P. gingivalis FLL93.
Hemolytic activity is reduced in P. gingivalis FLL93.
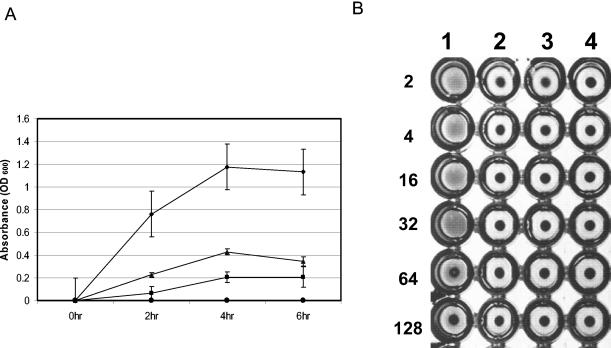
Cell- and vesicle-associated hemolysins which will liberate hemoglobin from erythrocytes are produced in P. gingivalis apparently by two distinct genes (15). Further, other hemolysin-like genes have been identified from the P. gingivalis genome project (www.oralgen.lanl.gov) (26). P. gingivalis FLL93, FLL92, and W83 were evaluated for hemolysin activity by their ability to lyse erythrocytes. As shown in Fig. 3A, there was reduced hemolytic activity in P. gingivalis FLL93 compared to that in the wild-type P. gingivalis W83 strain. Further, this level of hemolytic activity was similar to that of the vimA-defective isogenic mutant P. gingivalis FLL92 or the negative control.
FIG. 3.
Hemolytic and hemagglutinin activities of P. gingivalis FLL93 is reduced. Bacterial cells from overnight cultures were harvested by centrifugation, washed three times with 1× PBS, and then resuspended to a final OD600 of 1.5. Sheep erythrocytes were harvested by centrifugation and washed with 1× PBS until the supernatant was visually free of hemoglobin pigment. The washed erythrocytes were suspended to 1% in 1× PBS. (A) Hemolytic activity was determined by mixing equal volumes of bacterial cells with 1% erythrocytes in PBS. This mixture was then incubated at 37°C. Samples (500 μl) were withdrawn every 2 h and then were centrifuged. The OD405 was determined by spectrophotometry. As a negative control, erythrocytes were used alone. This experiment was done in triplicate, and average values are plotted (diamonds, W83; squares, FLL92; triangles, FLL93; circles, negative control). (B) Hemagglutination activity of P. gingivalis was performed on P. gingivalis W83, FLL92, and FLL93 cells that were serially diluted twofold in 1× PBS. Aliquots (100 μl) of the dilution were then mixed with an equal volume of 1% sheep erythrocytes and were incubated at 4°C for 3 h in a round-bottom microtiter plate. The hemagglutination titer was defined as the last dilution that showed full agglutination. Lane 1, W83; lane 2, FLL92; lane 3, FLL93; lane 4, negative control (sheep erythrocytes alone; Fig. 3B). Error bars indicate the standard error of the mean of three independent trials.
Hemagglutinin activity in P. gingivalis FLL93.
We assessed the hemagglutination potential of P. gingivalis FLL93 in comparison to the potentials of P. gingivalis W83 and P. gingivalis FLL92, the nonproteolytic vimA-defective mutant. In contrast to the wild-type strain, there was a reduction in hemagglutinin activity in P. gingivalis FLL93 (Fig. 3B). This level of activity was similar to those of P. gingivalis FLL92 and the negative control.
Inactivation of the vimE gene affects proteolytic activity in P. gingivalis.
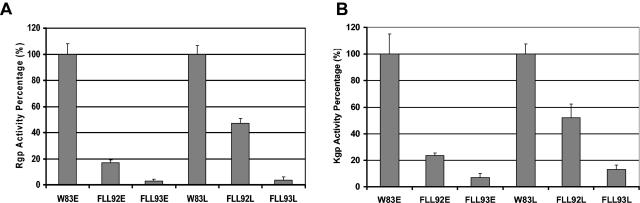
It was previously reported that the proteolytic activity of the non-black-pigmented recA-defective mutant (P. gingivalis FLL32) or vimA-defective mutant (P. gingivalis FLL92) was reduced by more that 90% compared to levels of the wild-type strain (1, 2). In addition, a late onset of proteolytic activity was also reported for the vimA mutant compared to that of the wild-type strain (27). Because hemolysis and hemagglutinin activities can be associated with gingipain activity in this organism (21, 35), P. gingivalis FLL93 was assayed for proteolytic activity by using BApNA and acetyl-lysine-p-nitroanilide (ALNA). In late-exponential-growth-phase cultures, both Arg-X and Lys-X protease activities in P. gingivalis FLL93 were very similar to that of P. gingivalis FLL92, which was reduced by approximately 90% compared to the activity of the wild-type strain (Fig. 4). In stationary-phase cultures of P. gingivalis FLL93, there was just a slight increase in Lys-X activity; however, there was no change in Arg-X activity (Fig. 4). This is in contrast to the expected late onset of proteolytic activity that was observed in P. gingivalis FLL92 (Fig. 4). The proteolytic activity of the wild type remained constant throughout this growth phase. The proteolytic activity distribution pattern was the same for both P. gingivalis FLL93 and the wild type (Fig. 5). In exponential growth phase most of the activity was cell associated; this activity was reduced in stationary phase. Taken together, these data suggest that under the same physiological conditions, the proteolytic profile for P. gingivalis FLL93 can be severely altered by mutations affecting vimE gene expression in P. gingivalis.
FIG. 4.
Inactivation of vimE resulted in decreased proteolytic activity of P. gingivalis. P. gingivalis was grown to late log phase (OD600 of 0.7 to 0.8) or stationary phase (OD600 of 1.4 to 1.5) in BHI broth supplemented with yeast extract, hemin, and vitamin K. Activities against Rgp (A) and Kgp (B) were tested in whole-cell culture. The results shown are representative of three independent experiments in triplicate. E, exponential phase; L, stationary phase. Error bars indicate the standard errors of the means of three independent trials.
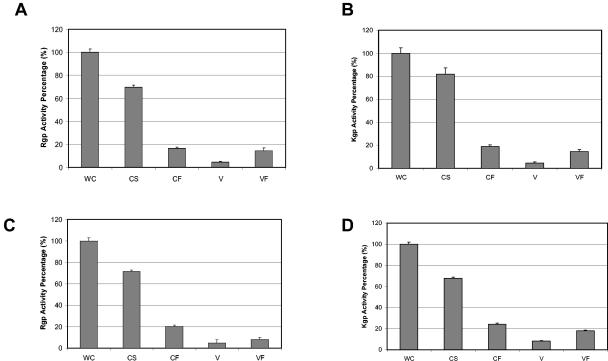
FIG. 5.
Distribution of the proteases in P. gingivalis FLL93. Activities against BApNA (A and C) and ALNA (B and D) were tested in whole-cell culture (WC), cell-free medium (CF), cell suspension (CS), vesicle medium (V), and vesicle-free medium (VF) of P. gingivalis W83 (A and B) and P. gingivalis FLL93 (C and D) grown to stationary phase (OD600 of 1.4 to 1.5). The activity of WC for each strain was assumed to equal 100%. WC = CS + CF; CF = V + VF.
Expression of the gingipain genes in the vimE-defective mutant.
The reduced proteolytic activity in P. gingivalis strain FLL93 could have been the result of alteration in transcription of the gingipain genes. There is also the possibility that the vimE gene may be involved in the posttranslational regulation of gingipain expression. In previous reports of mutations in two genes (recA and vimA) upstream of the vimE genes, a reduction in gingipain activity was also observed, although the expression of those genes was unaltered (1, 2). Furthermore, the partially processed RgpB gingipain proenzyme was identified in the vimA-defective mutant, suggesting a defect in gingipain biogenesis at the posttranslational level (27). To determine the presence of mRNA transcripts for the gingipain genes (rgpB, rgpA, and kgp), total RNA was isolated from the wild-type strain W83 and P. gingivalis FLL93 grown to stationary phase. Specific oligonucleotide primers as described in Table 2 for rgpB, rgpA, and kgp were used in RT-PCRs to amplify a predicted region of the gingipain gene transcripts. When the reverse transcriptase was present in the reaction, amplified products of the predicted size (0.8 kb for rgpB and 0.9 kb for rgpA and kgp) were observed for all three gingipain gene mRNA transcripts in both strains (Fig. 6).
FIG. 6.
Major gingipains are expressed in a vimE-defective mutant of P. gingivalis. DNase-treated total RNA extracted from P. gingivalis strains W83 and FLL93 grown to stationary phase was subjected to RT-PCR analysis.
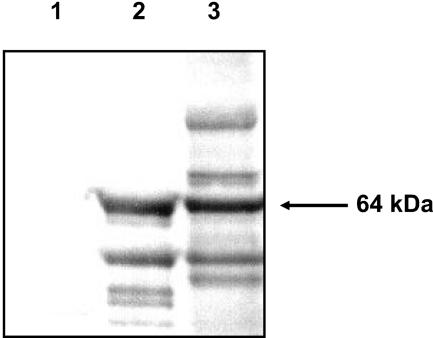
Identification of gingipain proenzyme in P. gingivalis FLL93.
The presence of mRNA transcript for the gingipain genes in P. gingivalis FLL93 may suggest a defect in gingipain biogenesis which could be similar to that of the vimA-defective (P. gingivalis FLL92) isogenic mutant previously reported (1). It was also shown previously that precipitation of extracellular proteins from P. gingivalis FLL92 with 37.5% acetone can enrich for the partially processed RgpB proenzyme (27). Extracellular fractions obtained from culture supernatants of P. gingivalis FLL93 grown to late exponential phase (OD600 of 0.8) were assessed by Western blot analysis using antibodies against the RgpB proenzyme. As shown in Fig. 7, the RgpB proenzyme-specific antibodies revealed a 64-kDa immunoreactive band in P. gingivalis FLL93, which is similar in size to the RgpB proenzyme previously reported for the vimA-defective mutant FLL92 (27). It is also noteworthy that the antibodies do not immunoreact with any proteins present in the W83 extracellular fractions.
FIG. 7.
Immunoreactivity of the RgpB proenzyme in P. gingivalis FLL93. Western blot analysis using specific RgpB proenzyme antibodies as probes was done on the extracellular fractions from P. gingivalis W83 (lane 1), FLL92 (lane 2), and FLL93 (lane 3). All lanes contained 20 μg of acetone (37.5%)-precipitated proteins from the supernatant fractions of cultures in BHI medium at exponential growth phase.
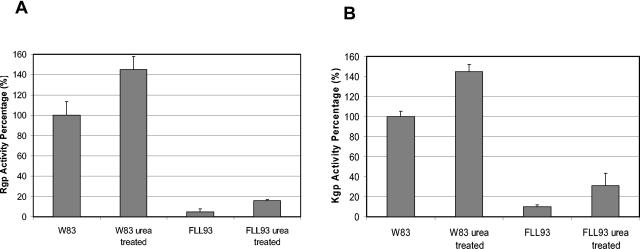
In vitro protease activation using urea.
It was previously shown that in vitro protease activity could be induced by a urea denaturation-renaturation cycle in nonproteolytic extracellular protein fractions from P. gingivalis FLL92 grown to late exponential phase (27). This was consistent with the presence of a partially processed protease in those fractions. Proteolytic cleavage is a common mechanism that can generate an active enzyme from a larger proenzyme (17). While activation can be achieved by the action of other proteolytic enzymes, we cannot rule out the possibility that an inhibitor may be present at the catalytic site of the enzyme or the possibility that the tertiary structure of the proenzyme may be incompatible with activation. Because most proteins are denatured in the presence of high concentrations of urea and some will renature if the high concentration is slowly reduced, extracellular proteins from P. gingivalis FLL93 grown to late exponential phase or stationary phase were subjected to a urea denaturation-renaturation cycle. Figure 8 shows a slight activation of Rgp and Kgp in the extracellular protein fraction from P. gingivalis FLL93 grown to late exponential or stationary phase compared to activation of the wild type.
FIG. 8.
Protease activation in the extracellular protein fraction from P. gingivalis FLL93. P. gingivalis was grown to stationary phase (OD600 of 1.3) in 1 liter of BHI broth supplemented with hemin and vitamin K. Acetone (37.5%)-precipitated proteins were mixed with 8 M urea and were incubated at 4°C for 1 h. The urea was slowly removed from the mixture by centrifugation (10,000 × g) in a Millipore filtration unit with the addition of increasing volumes of 100 mM Tris-HCl buffer (pH 7.4). Activities for Rgp and Kgp were tested. Error bars indicate the standard errors of the means of three independent trials.
The Rgp active site is not labeled by DNS-EGR-CK in P. gingivalis FLL93.
DNS-EGR-CK binds irreversibly to the active site of the arginine-specific proteases of P. gingivalis and can be detected by UV illumination following electrophoresis (3). To determine if the active site or catalytic domain is exposed or accessible in Arg-gingipain from P. gingivalis FLL93, extracellular fractions that were labeled with DNS-EGR-CK was subjected to SDS-PAGE and was analyzed with a UV transilluminator. As expected, DNS-EGR-CK-treated extracellular proteins from the wild-type strain generate fluorescent bands characteristic of the arginine-specific protease catalytic domain (Fig. 9). Similarly, DNS-EGR-CK treatment of extracellular proteins from P. gingivalis FLL92, grown to stationary phase, showed a 48-kDa fluorescent band (Fig. 9). However, there was no detectable specifically labeled protein that was similar in size to the 48-kDa band in the late-exponential- or stationary-phase extracellular protein fraction from P. gingivalis FLL93 (Fig. 9). There was also no detectable specifically labeled protein that was similar in size to the 64-kDa partially processed RgpB proenzyme in the late-exponential- or stationary-phase extracellular protein fraction.
FIG. 9.
The Rgp active site is not labeled by DNS-EGR-CK in P. gingivalis FLL93 extracellular fractions. Acetone-precipitated extracellular proteins from P. gingivalis W83, P. gingivalis FLL93, and P. gingivalis FLL92 were labeled with the fluorescent irreversible inhibitor DNS-EGR-CK and were subjected to SDS-PAGE on 4 to 12% NuPAGE gels (Invitrogen). The gels were viewed under UV light to visualize labeled proteins. (A) All the lanes were labeled with DNS-EGR-CK. (B) All the lanes were labeled with DNS-EGR-CK in the presence of 50 μM leupeptin. Each lane contains 20 μg of protein. The arrow indicates the DNS-EGR-CK-labeled protein band. E, exponential phase; L, stationary phase.
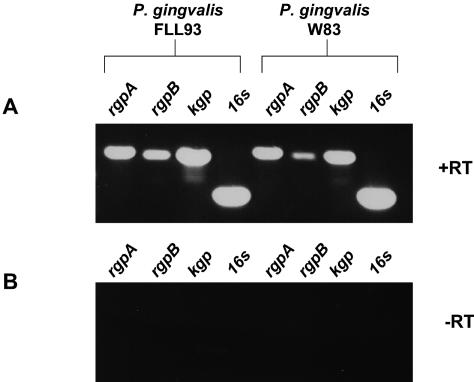
vimE can be independently expressed.
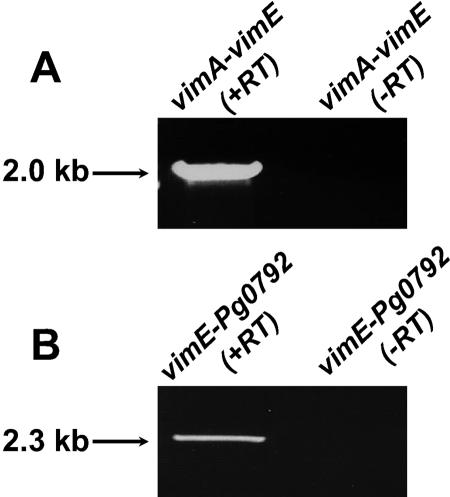
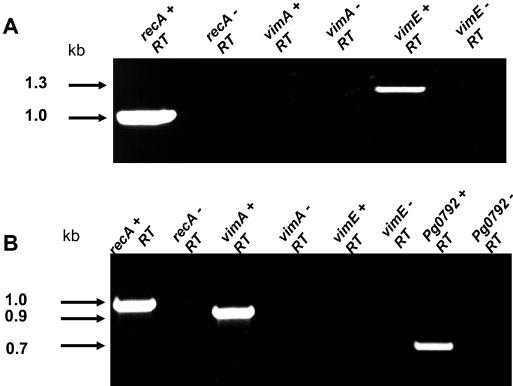
The similar phenotypic properties of the nonproteolytic recA-deficient mutant, P. gingivalis FLL32, and the vimA-defective mutant, P. gingivalis FLL92, may be explained by a polar effect (1). To determine if vimE is cotranscribed with vimA, RT-PCR using W83 RNA was performed with primers for vimA (P7) and vimE (P3). In the absence of reverse transcriptase, no amplified fragment was observed (Fig. 10). However, a 2.0-kb fragment was amplified, which is consistent with the expected size. To determine if vimE was cotranscribed with the downstream gene Pg0792, RT-PCR using W83 RNA and primers for vimE (P1) and Pg0792 (P5) were used. As shown in Fig. 10B, a 2.3-kb fragment of the size expected was amplified. Taken together, these results indicate that vimE appears to be cotranscribed with vimA or Pg0792 and may be a part of the bcp-recA-vimA transcriptional unit. In the absence of vimE there is little gingipain activation. Further, the late onset of gingipain activity in the vimA-defective mutant may suggest that the vimE gene could be independently expressed (1). RT-PCR of total RNA showed that the expected fragment for the vimE gene could be amplified from the vimA-defective mutant (Fig. 11A). In a similar experiment using RNA from the vimE-defective mutant, the downstream gene Pg0792 was amplified (Fig. 11B).
FIG. 10.
vimE can be cotranscribed with vimA or Pg0792. One microgram of total DNase-treated RNA of P. gingivalis W83 from exponential phase (OD600 of 0.6) using a RiboPure kit (Ambion) was subjected to RT-PCR. (A) Cotranscription of vimA and vimE. Lane 1, P7 and P3 plus reverse transcriptase; lane 2, P7 and P3 minus reverse transcriptase. (B) Cotranscription of vimE and Pg0792. Lane 3, P1 to P5 plus reverse transcriptase; lane 4, P1 to P5 minus reverse transcriptase.
FIG. 11.
vimE gene can be independently expressed. Total DNase-treated RNA was extracted from P. gingivalis FLL92 and P. gingivalis FLL93 grown to stationary phase (OD600 of 1.3) by using the RiboPure kit (Ambion). Samples (1 μg) were subjected to RT-PCR. (A) Expression of vimA flanking genes in P. gingivalis FLL92. (B) Expression of vimE flanking genes from P. gingivalis FLL93. +RT, with reverse transcriptase; −RT, without reverse transcriptase.
DISCUSSION
We have used a genetic approach in this study to further assess the role of specific host factors in protease regulation/activation in P. gingivalis. Several recent studies (1, 12, 36; reviewed in reference 25) have identified nongingipain genes that are involved in the modulation of gingipain activity and other virulence factors in P. gingivalis. A comparison of several of the P. gingivalis mutants from these studies has raised the possibility of multiple mechanisms for gingipain activation. Mutation in the vimA, porR, or gppX gene has shown growth-phase-dependent activation of proteolytic activity that is mostly soluble (1, 12, 36). A mechanism of activation by these genes is presently not understood. Furthermore, it is unclear if they are part of a common pathway for gingipain activation. Although these mutants had a similar proteolytic profile, inactivation of each gene did not affect the expression of the others.
The vimE gene, which is downstream of vimA, does not appear to show any significant similarity to any known genes (www.oralgen.lanl.gov). P. gingivalis FLL93, the isogenic mutant defective in this gene, showed reduced proteolytic activity and reduced doubling time and was not black pigmented in this study. This phenotype of P. gingivalis FLL93 could be related to the reduced gingipain activity or, in particular, reduced membrane-associated activity. This would be similar to P. gingivalis FLL32, a recA-defective mutant (2), and P. gingivalis FLL92, a vimA-defective mutant (1). These results would also be consistent with other reports (8, 21, 28, 37) of the involvement of gingipains, especially Kgp, with hemoglobin binding, absorption, and heme accumulation.
Hemolysin activity appears to be modulated by the vimE gene in P. gingivalis. In this study, P. gingivalis FLL93, the vimE-defective mutant, showed no hemolytic activity when grown on blood plates or incubated with sheep erythrocytes. Because the protease Kgp has been shown to have hemoglobinase activity (21) and plays a role in erythrocyte degradation (34), our results are not surprising because of the reduced proteolytic activity in P. gingivalis FLL93. In addition to the effect of proteolytic activity on the hemolysin potential, several hemolysin-like genes have been identified by the P. gingivalis genome project (www.oralgen.lanl.gov) (26). Furthermore, there is genetic evidence for the involvement of two distinct hemolysins in the hemolysin activity of P. gingivalis (15). Taken together, these observations may suggest regulation of those genes or gene products by the vimE gene, although we cannot rule out the effect of the gingipains on their expression. There is evidence that the gingipains are involved in the processing of other proteins (14, 39).
The vimE gene in this study appears also to affect hemagglutination in P. gingivalis. Hemagglutination of sheep erythrocytes was reduced in P. gingivalis FLL93, the vimE-defective mutant. Again, these observations are not unexpected due to the reduced proteolytic activity of P. gingivalis FLL93. The proteases RgpA and Kgp have hemagglutinating activity (35). Further, a monoclonal antibody (61BG1.3) that inhibited hemagglutination and selectively prevented the recolonization of P. gingivalis in periodontal patients was found to recognize a peptide within the adhesin domain encoded by rgpA, kgp, and hagA (5, 16). In addition to the association of the gingipains with hemagglutination in P. gingivalis, the presence of several genetically distinct genes, including hemagglutinin genes hagB and hagC, has been reported previously (20, 32). In this study, the effects of the vimE gene product on hemagglutination in P. gingivalis may also implicate the regulation of those genes or gene products by this gene.
Consistent with previous reports on two isogenic mutants (P. gingivalis FLL32, a recA-defective mutant, and P. gingivalis FLL92, a vimA-defective mutant) with reduced proteolytic activity, there was no detectable alteration of the gingipain genes in P. gingivalis FLL93. RgpB was secreted in an inactive form in the vimA-defective mutant, suggesting a role of the vimA gene in the posttranslational regulation of protease activity in P. gingivalis (27). A comparison of the extracellular proteins that used the RgpB proenzyme-specific antibodies revealed a 64-kDa immunoreactive band in P. gingivalis FLL93, which was similar in size to the partially processed RgpB proenzyme previously reported for the vimA-defective mutant P. gingivalis FLL92 (27). There was also an 80-kDa immunoreactive band which preliminary studies suggest is the full-length unprocessed RgpB that was absent in P. gingivalis W83 and P. gingivalis FLL92. Other bands may be intermediate products of the processing. In the process of maturation, the active site would become accessible. If there is a defect in this process in P. gingivalis FLL93, very little active-site labeling of the 48-kDa catalytic domain of RgpB by DNS-EGR-CK would be observed in cultures from exponential or stationary phase. As observed in this study, active-site labeling was undetected in P. gingivalis FLL93. This further confirms that there may be a defect in the activation/maturation of the gingipains. Collectively, these observations may suggest that the vimE gene product, similar to the case of VimA, is involved in the posttranslational regulation of protease activity in P. gingivalis.
An active enzyme can be generated from a larger polypeptide by autoprocessing or by the action of other proteolytic enzymes (17, 31). Several cysteine proteases are converted to their active forms by removal of the prosegment by an autocatalytic mechanism (17). There is accumulating evidence that a multicomponent maturation pathway(s), perhaps including an autolytic mechanism, may be involved in the production of Arg-X- and Lys-X-specific proteases in P. gingivalis (3, 24, 33). While it is clear that proteolytic processing of the gingipain precursors in P. gingivalis is required to produce the isoforms detected, it has been shown that denaturing and refolding of proteins can also induce the autoprocessing of RgpB (27). We have shown that there is very little activation after urea treatment in the vimA-defective mutant extracellular fraction. The presence of an 80-kDa proenzyme immunoreacting band, which was absent in the vimA mutant, may indicate that vimE is needed for the initial processing of RgpB. The processing of the 64-kDa RgpB proenzyme intermediate in vimA mutant could have been enhanced by the denaturing-renaturing experiments (27).
The distribution of the proteolytic activity in P. gingivalis FLL93 was similar to that of the wild-type strain and was in contrast to that of P. gingivalis FLL92, the vimA-defective mutant, that had mostly soluble proteolytic activity with little or no cell-associated activity (1, 27). While there was a unique late onset of Arg-X- and Lys-X-specific proteolytic activity in P. gingivalis FLL92 (27), there was little or no observed change of proteolytic activity in stationary phase in P. gingivalis FLL93, the vimE-defective mutant. Collectively, our observations could support a hypothesis that suggests that the regulation of proteolytic activity in P. gingivalis may occur by multiple mechanisms. During exponential growth phase, proteolytic activity in P. gingivalis may be regulated in a vimA-dependent manner. However, during stationary phase, an alternative pathway for protease activation may be upregulated to overcome or bypass the vimA mutation or may be activated in the absence of VimA. In addition, in the absence of the vimE gene product, as observed in this study, there is minimal activation of proteolytic activity during exponential or stationary growth phase. The specific functional role of VimE during protease activation is unclear and is being further investigated in our laboratory.
The vimE gene appears to be cotranscribed with either vimA or Pg0792 and may be a part of the bcp-recA-vimA transcriptional unit. Further, its inactivation did not have any polar effect on the expression of the downstream gene. Because both the vimE and vimA genes have a similar effect on protease biogenesis in P. gingivalis, it is likely that they may be a part of the same or different pathway(s) that is involved in protein maturation, including protease activation/maturation. Presently, there is no information on the regulation of these genes. Because gingipain activation in the vimE-defective mutant is not growth phase related and because that gene product is required for activation/maturation, both vimA and vimE genes could be regulated differently. This question is under further evaluated in our laboratory.
The effect of proteolytic activity on virulence in P. gingivalis is well documented (7, 13, 18, 29). Inactivation of the gingipain genes has been demonstrated to reduce the virulence potential of P. gingivalis. Thus, modulation of virulence in P. gingivalis may be coordinated via an ability to modulate proteolytic activity. Although not directly tested in this study, the vimE gene may be an important virulence gene. Because inactivation of the vimE gene resulted in reduction in proteolytic activity and had a pleiotropic effect on other important virulence factors, P. gingivalis FLL93 would be expected to have a reduced pathogenic potential, in contrast to the wild-type strain. This would be consistent with the vimA-defective mutant that had a phenotype similar to that of P. gingivalis FLL93, and it was dramatically less virulent than the wild-type strain in the mouse model (1).
In summary, we have constructed an isogenic mutant of P. gingivalis that is defective in the vimE gene downstream of the bcp-recA-vimA transcriptional unit. While this mutant had reduced proteolytic activity, there was no detectable reduction of the gingipain gene transcription. Further, this mutant, in contrast to the wild-type strain, showed reduced hemolytic and hemagglutinating activities. The vimE gene appears to be independently expressed, and its identification represents a potentially new mechanism for regulating proteolytic activity and virulence in P. gingivalis. It is unclear, however, if there is any interaction between the vimE and vimA gene products; this is under further investigation in the laboratory.
Acknowledgments
This work was supported by Loma Linda University, School of Dentistry, by Public Health Service grants DE13664 and DE13664-S1 from the National Institute of Dental and Craniofacial Research (to H.M.F), and by GM60507, a minority training grant from the National Institute of General Medicine.
Editor: V. J. DiRita
REFERENCES
- 1.Abaibou, H., Z. Chen, G. J. Olango, Y. Liu, J. Edwards, and H. M. Fletcher. 2001. vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect. Immun. 69:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abaibou, H., Q. Ma, G. J. Olango, J. Potempa, J. Travis, and H. M. Fletcher. 2000. Unaltered expression of the major protease genes in a non-virulent recA-defective mutant of Porphyromonas gingivalis W83. Oral Microbiol. Immunol. 15:40-47. [DOI] [PubMed] [Google Scholar]
- 3.Aduse-Opoku, J., M. Rangarajan, K. A. Young, and M. A. Curtis. 1998. Maturation of the arginine-specific proteases of Porphyromonas gingivalis W50 is dependent on a functional prR2 protease gene. Infect. Immun. 66:1594-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, V., and T. Lehner. 1997. Characterization of the Porphyromonas gingivalis antigen recognized by a monoclonal antibody which prevents colonization by the organism. J. Periodontol. Res. 32:54-60. [DOI] [PubMed] [Google Scholar]
- 6.Corcoran, M. L., D. E. Kleiner, Jr., and W. G. Stetler-Stevenson. 1995. Regulation of matrix metalloproteinases during extracellular matrix turnover. Adv. Exp. Med. Biol. 385:151-159. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, M. A., J. Aduse-Opoku, and M. Rangarajan. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192-216. [DOI] [PubMed] [Google Scholar]
- 8.Dashper, S. G., K. J. Cross, N. Slakeski, P. Lissel, P. Aulakh, C. Moore, and E. C. Reynolds. 2004. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol. Immunol. 19:50-56. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a mutant of Porphyromonas gingivalis W83 that is defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher, A., J. Aduse-Opoku, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2003. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Peptide Sci. 4:427-441. [DOI] [PubMed] [Google Scholar]
- 11.Grenier, D., S. Roy, F. Chandad, P. Plamondon, M. Yoshioka, K. Nakayama, and D. Mayrand. 2003. Effect of inactivation of the Arg- and/or Lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect. Immun. 71:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa, Y., S. Nishiyama, K. Nishikawa, T. Kadowaki, K. Yamamoto, T. Noguchi, and F. Yoshimura. 2003. A novel type of two-component regulatory system affecting gingipains in Porphyromonas gingivalis. Microbiol. Immunol. 47:849-858. [DOI] [PubMed] [Google Scholar]
- 13.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 14.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 15.Karunakaran, T., and S. C. Holt. 1993. Cloning of two distinct hemolysin genes from Porphyromonas (Bacteroides) gingivalis in Escherichia coli. Microb. Pathogen. 15:37-49. [DOI] [PubMed] [Google Scholar]
- 16.Kelly, C. G., V. Booth, H. Kendal, J. M. Slaney, M. A. Curtis, and T. Lehner. 1997. The relationship between colonization and haemagglutination inhibiting and B cell epitopes of Porphyromonas gingivalis. Clin. Exp. Immunol. 110:285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan, A. R., and M. N. James. 1998. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 7:815-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lantz, M. S. 1996. New insights into mechanisms of bacterial pathogenesis in periodontitis. Curr. Opin. Periodontol. 3:10-18. [PubMed] [Google Scholar]
- 20.Lepine, G., and A. Progulske-Fox. 1996. Duplication and differential expression of hemagglutinin genes in Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:65-78. [DOI] [PubMed] [Google Scholar]
- 21.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 23.Mayrand, D., and S. C. Holt. 1988. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 52:134-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikolajczyk, J., K. M. Boatright, H. R. Stennicke, T. Nazif, J. Potempa, M. Bogyo, and G. S. Salvesen. 2003. Sequential autolytic processing activates the zymogen of Arg-gingipain. J. Biol. Chem. 278:10458-10464. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, K. 2003. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Curr. Protein Peptide Sci. 4:389-395. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olango, G. J., F. Roy, S. M. Sheets, M. K. Young, and H. M. Fletcher. 2003. Gingipain RgpB is excreted as a proenzyme in the vimA-defective mutant Porphyromonas gingivalis FLL92. Infect. Immun. 71:3740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olczak, T., D. W. Dixon, and C. A. Genco. 2001. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol. 183:5599-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology 2000. 12:153-192. [DOI] [PubMed] [Google Scholar]
- 30.Potempa, J., J. Mikolajczyk-Pawlinska, D. Brassell, D. Nelson, I. B. Thogersen, J. J. Enghild, and J. Travis. 1998. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J. Biol. Chem. 273:21648-21657. [DOI] [PubMed] [Google Scholar]
- 31.Potempa, J., A. Sroka, T. Imamura, and J. Travis. 2003. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Peptide Sci. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 32.Progulske-Fox, A., S. Tumwasorn, G. Lepine, J. Whitlock, D. Savett, J. J. Ferretti, and J. A. Banas. 1995. The cloning, expression and sequence analysis of a second Porphyromonas gingivalis gene that codes for a protein involved in hemagglutination. Oral Microbiol. Immunol. 10:311-318. [DOI] [PubMed] [Google Scholar]
- 33.Rangarajan, M., J. Aduse-Opoku, J. M. Slaney, K. A. Young, and M. A. Curtis. 1997. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol. Microbiol. 23:955-965. [DOI] [PubMed] [Google Scholar]
- 34.Shah, H. N., and S. E. Gharbia. 1989. Lysis of erythrocytes by the secreted cysteine protease of Porphyromonas gingivalis W83. FEMS Microbiol. Lett. 61:213-218. [DOI] [PubMed] [Google Scholar]
- 35.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin finding, and hemagglutination of Porphyromonas gingivalis - construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 36.Shoji, M., D. B. Ratnayake, Y. Shi, T. Kadowaki, K. Yamamoto, F. Yoshimura, A. Akamine, M. A. Curtis, and K. Nakayama. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183-1191. [DOI] [PubMed] [Google Scholar]
- 37.Sroka, A., M. Sztukowska, J. Potempa, J. Travis, and C. A. Genco. 2001. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J. Bacteriol. 183:5609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundqvist, G. 1993. Pathogenicity and virulence of black-pigmented gram-negative anaerobes. FEMS Immunol. Med. Microbiol. 6:125-138. [DOI] [PubMed] [Google Scholar]
- 39.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wandersman, C. 1989. Secretion, processing and activation of bacterial extracellular proteases. Mol. Microbiol. 3:1825-1831. [DOI] [PubMed] [Google Scholar]