Abstract
Porphyromonas gingivalis is an important bacterium involved in periodontal diseases. Colonization by periodontopathogens has been associated with severe local inflammatory reactions in the connective tissue. In this study we characterized P. gingivalis-mediated infection and activation of human umbilical vein endothelial cells by using two strains of different virulence capacities, strains ATCC 53977 and DSMZ 20709. Both strains were able to adhere to and infect endothelial cells with an infection rate of 0.48% for ATCC 53977 and 0.007% for DSMZ 20709. The triggering of two signal transduction pathways in P. gingivalis-infected endothelial cells was demonstrated for both strains, with a rapid increase of p38 mitogen-activated protein kinase phosphorylation and a more delayed degradation of IκBα, followed by nuclear translocation of NF-κB. In addition, both strains induced enhanced expression of endothelial adhesion molecules E-selectin and intracellular adhesion molecule 1 (ICAM-1). Target cell activation was independent of bacterial fimbriae expression since the fimA knockout strain A7436 ΔfimA induced the same level of ICAM-1 as the corresponding wild type (A7436-WT). Thus, two P. gingivalis strains, ATCC 53799 and DSMZ 20709, infect endothelial cells and trigger signaling cascades leading to endothelial activation, which in turn may result in or promote severe local and systemic inflammation.
Porphyromonas gingivalis is an obligate anaerobic gram-negative bacterium associated with periodontal diseases. Colonization with P. gingivalis has been strongly associated with increasing pocket depth and bleeding on probing, hallmarks of periodontitis (3, 49). Immunohistochemical staining of the connective tissue of established periodontal lesions demonstrated a massive local inflammatory reaction with tissue edema, enhanced expression of endothelial adhesion molecules, local release of proinflammatory cytokines, and infiltration of polymorphonuclear granulocytes (37, 40, 44). Several recent studies have demonstrated that P. gingivalis is able to invade and activate different cell types in the surrounding tissue of the teeth (endothelial and gingival epithelial cells as well as periodontal ligament cells) (13, 33, 46, 47). Moreover, recent studies have demonstrated a transient bacteremia with potential subsequent systemic infection after a variety of dental treatment procedures (1, 20, 21, 48). Endothelial cells, therefore, can act as primary target cells during infection with P. gingivalis. Little is known about mechanisms of infection and activation of endothelial cells by P. gingivalis. Endothelial cells are key players during inflammatory reactions and are able to produce an array of proinflammatory mediators including cytokines such as CXCL8 and interleukin-6, lipid mediators like prostaglandins or platelet-activated factor, complement factor, or adhesion molecules E-selectin, intracellular adhesion molecule 1 (ICAM-1), and vascular cell adhesion molecule 1 (18). Several studies have demonstrated different virulence capacities of selected P. gingivalis strains (15, 17, 38). Using a mouse abscess model analyzing lesion size and lethality, Ebersole et al. identified several more or less virulent strains (14). Fimbriae, ceramides, different proteases, outer membrane proteins, and lipopolysaccharides (LPS) have been discussed as potential virulence factors (25, 39, 42, 45). Until now, the different strains have not been characterized in detail with regard to the expression of these potential virulence factors. Additionally, the importance of these virulence factors for different steps during infection and the activation of target cells is still controversial (19, 43).
Therefore, the first goal of our study was to assess the capability of two different P. gingivalis strains to infect or invade human umbilical vein endothelial cells (HUVEC). Expression of the endothelial adhesion molecules E-selectin and ICAM-1 was used to demonstrate subsequent target cell activation. Due to the different virulence capacities of the known strains, we used the most virulent strain (ATCC 53977), according to the study of Ebersole et al. (14), and a less virulent strain, DSMZ 20709 (equivalent to strain ATCC 33277, used by Ebersole et al.).
Mitogen-activated protein kinase (MAPK)-related signal transduction pathways are among the most widespread mechanisms of eukaryotic cell regulation (activation, stress response, differentiation, and growth). MAPK-dependent activation of transcription factors is considered to be a prerequisite for altered gene expression in stimulated target cells (for a review, see references 4 and 32). Specifically, p38 MAPK is strongly activated during inflammatory reactions and appears to be of specific importance during LPS-mediated signal transduction (22, 48).
The expression of inflammatory mediators such as cytokines or adhesion molecules relies on the activation of cytosolic transcription factors. Among the primary transcription factors, NF-κB plays a central role in the regulation of these proinflammatory molecules (5, 50) Nuclear translocation of NF-κB induced by proinflammatory stimuli (cytokines, bacteria, or bacterial toxins) is regulated by phosphorylation and degradation of its naturally occurring inhibitor IκB (16, 28, 35). Little is known about signal transduction pathways activated in target cells upon infection with P. gingivalis. Therefore, the second objective of the present study was to investigate P. gingivalis-dependent activation of host cell signal transduction pathways in human endothelial cells. In our study we demonstrated that both P. gingivalis strains induced a phosphorylation of p38 MAPK, degradation of IκBα, and translocation and activation of endothelial cell NF-κB, with subsequently increased transcription and translation of E-selectin and ICAM-1.
(Part of this research was conducted by Clemens Walter in partial fulfillment of the requirements for an MD degree from Charité, Universität Medizin Berlin.
MATERIALS AND METHODS
Materials.
Tissue culture plasticware was obtained from Becton-Dickinson (Heidelberg, Germany). MCDB-131 medium, fetal calf serum, phosphate-buffered saline (PBS), trypsin-EDTA solution, and antibiotics were from Life Technologies (Karlsruhe, Germany). Brain heart infusion broth was from Difco Laboratories (Detroit, Mich.). Monoclonal antibodies directed against E-selectin (clone 1.2B6) and ICAM-1 (clone 15.2) were obtained from Dianova (Hamburg, Germany). All other reagents were from Sigma (München, Germany) and were of analytical grade.
Preparation of HUVEC.
HUVEC were isolated from human umbilical cord veins and identified according to a previously described method (26). Briefly, cells obtained from collagenase digestion were washed, resuspended in MCDB-131 medium-10% fetal calf serum, and seeded into gelatin-coated 6-, 24-, or 96-well plates. Only confluent monolayers of primary cultures were used for all experiments.
Bacterial strains.
P. gingivalis ATCC 53977 was from the laboratory of H. K. Kuramitsu (State University at Buffalo, Buffalo, N.Y.), P. gingivalis DSMZ 20709 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany). Strains were characterized and tested for purity by HAIN life science (Nehren, Germany) and by means of a microbiological test kit ID 32A (BioMerieux, Marcy l'Etoile, France). Wild-type A7436 (A7436-WT) and the corresponding fimA knockout mutant A7436 ΔfimA were provided by J. Hayashi and H. K. Kuramitsu (unpublished data). All stocks were grown in brain heart infusion broth containing cysteine (0.1%), yeast (0.5%), vitamin K (0.1%), and hemin (0.25%) in an anaerobic chamber (Thermo Life Science) in an atmosphere of 85% N2, 5% H2, and 10% CO2 for 3 to 5 days, and erythromycin (10 μg/ml) was added to the A7436 ΔfimA culture. Aliquots were stored at −70°C. An aliquot of the initial stock solution was used for each experiment without subculturing.
P. gingivalis infection assay.
Thawed aliquots of both strains were grown to mid-logarithmic phase, centrifuged (1,500 × g for 5 min at 37°C) and resuspended in HUVEC media. A multiplicity of infection (MOI; as determined spectrophotometrically by species-specific curves) of 0.01 to 100 was used to infect HUVEC. Bacteria were centrifuged onto HUVEC monolayers in order to enhance the rate of infection (350 × g for 5 min). After incubation at 37°C for 1.5 h, the supernatant was replaced with fresh medium containing gentamicin (0.5 mg/ml) and metronidazole (0.1 mg/ml) in order to kill remaining extracellular (EC) bacteria. Plates were processed for experiments at the times indicated.
Scanning electron microscopy.
HUVEC were grown on glass slides in 24-well plates, infected at an MOI of 100 as described above, and fixed with 8% Karnowsky's solution (2% glutaraldehyde and 4% paraformaldehyde [PFA] in PBS) for 24 h at 4°C. After washing with HEPES buffer, samples were dehydrated with graded ethanol, critical-point dried with CO2, sputter coated with 10 nm of gold, and examined with a scanning electron microscope with 5 kV in the secondary mode (LEO 1530 Field emission scanning electron microscope; LEO Elektronenmikroskopie GmbH, Oberkochen, Germany).
Antibiotic protection and invasion assays.
Invasion of endothelial cell monolayers by P. gingivalis strains was quantified by determining the number of CFU recovered following antibiotic treatment by using an antibiotic protection assay as recently described by Deshpande et al. and Madianos et al. (10, 34). Briefly, confluent HUVEC monolayers in 6-well plates were infected at an MOI of 50 (final concentration, 5 × 107 bacteria) and incubated for 1.5 or 4 h (at 37°C in 5% CO2). After 1.5 h, unattached bacteria were removed by washing the monolayer twice with sterile PBS. Half of the monolayers were lysed with sterile H2O (30 min). Lysates were diluted and plated on Columbia agar plates containing vitamin K (0.1%) and hemin (0.25%) and incubated for 7 days under strictly anaerobic conditions. On the other half of the monolayers, external adherent cells were killed by incubating the infected monolayers with fresh medium containing gentamicin (0.5 mg/ml) and metronidazole (0.1 mg/ml). This concentration of antibiotics was sufficient to completely kill 108 bacteria per ml in 1 h (data not shown). The antibiotic did not affect the morphology or viability of the endothelial cells as detected by lactate dehydrogenase release. Controls for antibiotic killing of P. gingivalis were included in all experiments. After 4 h the remaining HUVEC were also lysed, and lysates were diluted and plated on agar plates (see above). CFU of invasive organisms were then enumerated. Invasion was expressed as the percentage of the initial inoculum recovered after antibiotic treatment and endothelial cell lysis. Following this protocol, we recovered different fractions of bacteria; the first fraction, isolated after 1.5 h, represented all bacteria of the initial inoculum that adhered to and/or invaded the endothelial cells (total, or EC bacteria plus intracellular [IC] bacteria). The second fraction (after 4 h), recovered from cells in parallel wells to which antibiotic was added, represent only those bacteria that were internalized (IC bacteria). According to the method published by Dorn et al. (11) EC P. gingivalis organisms that only adhered but did not invade endothelial cells were calculated by subtracting IC bacteria (isolated after 4 h of incubation) from the amount of bacteria we were able to isolate after 1.5 h of incubation (total, or first fraction). Control experiments confirmed the viability of P. gingivalis after exposure to water for 30 min or to HUVEC media for 1.5 h or after incubation in air for up to 5 h (data not shown). The numbers of CFU of at least five independent experiments were counted.
Cell surface ELISA.
Expression of different adhesion molecules on monolayers of endothelial cells preincubated with P. gingivalis was determined by cell surface enzyme-linked immunosorbent assay (ELISA) as described previously (31). Briefly, confluent pretreated HUVEC monolayers in 96-well flat-bottom microtiter plates were washed and fixed with 2% PFA for 10 min. Human immunoglobulin (Ig) was used to reduce nonspecific binding and primary mouse antibodies were added for 25 min. Thereafter, cells were washed three times and exposed to a horseradish peroxidase-conjugated rabbit anti-mouse Ig antibody for 20 min. After the cells were washed, ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] (Sigma, München, Germany) was added for 10 min. Data are indicated as optical densities at 405 nm.
Western blotting.
For determination of p38 MAPK phosphorylation in HUVEC, cells were starved for 12 h in serum-free medium, stimulated as indicated, washed twice in ice-cold HEPES buffer, pH 7.4, containing 100 mM sodium fluoride, 2 mM sodium vanadate, and 15 mM sodium pyrophosphate. Cells were then harvested on ice by scraping with lysis buffer containing 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and leupeptin, pepstatin, and antipain at a concentration of 2 μg/ml each. After removal of the cell debris by centrifugation, cell lysates were subjected to sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis and blotted on Hybond-ECL membranes (Amersham Biosciences, Freiburg, Germany). Each lane contains 80 μg of protein (ascertained by Bradford assay). Immunodetection of phosphorylated p38 MAPK was carried out with phospho-specific p38 MAPK antibodies (Cell Signaling, Frankfurt, Germany). Degradation of IκBα was analyzed in HUVEC lysates by using a rabbit polyclonal antibody (Santa Cruz Biotechnologies, Santa Cruz, Calif.) as described previously (22). In all experiments, ERK2 kinase (Santa Cruz Biotechnologies) was detected simultaneously to confirm equal protein loads. Proteins were visualized by incubation with secondary IRDye 800- or Cy5.5-labeled antibodies (23) by using an Odyssey infrared imaging system (LICOR Inc., Bad Homburg, Germany).
Phospho-p38 MAPK ELISA.
Cell culture, stimulation, and protein extraction were performed as described above, and commercial immunoassay kits (P-p38 MAPK; BioSource Europe SA, Nivelles, Belgium) were used. Assays were carried out according to the manufacturer's instructions. Assay standards or cell lysates were incubated for 2 h at room temperature in anti-P-p38-coated 8-well flat-bottom microtiter strips and then aspirated and washed four times. Detection antibody was added to each well and incubated for 1 h. After washing, wells were exposed to horseradish peroxidase-conjugated anti-rabbit antibody for 30 min. Afterwards, stabilized chromogen was added for 30 min. Absorption was measured at 450 nm after the addition of stop solution. The results were normalized for protein concentrations.
Confocal laser scanning microscopy analysis of NF-κB nuclear translocation.
After stimulation of HUVEC grown on Thermanox slides (Falcon Culture Slide; Becton Dickinson, Rutherford, N.J.) with 107 bacteria/ml (MOI, 100) for 15, 45, 90, and 120 min, cells were fixed with 3% PFA for 20 min and permeabilized with 1% Triton for 15 min. To reduce nonspecific background staining, cells were treated with 5% goat serum for 30 min in PBS before the addition of antibodies. For NF-κB p65 staining, the primary antibody (polyclonal rabbit anti-human NF-κB p65; Santa Cruz Biotechnology) was incubated overnight at 4°C. Bound antibodies were detected with Alexa Fluor 488-conjugated goat anti-rabbit monoclonal antibody (4°C, overnight) (Molecular Probes, Eugene, Oreg.). After each step cells were washed three times with sterile PBS. Coverslips were sealed, and cells were analyzed with the use of a Pascal 5 confocal laser scanning microscope (Zeiss, Jena, Germany).
Statistical methods.
A one-way analysis of variance was used for data shown in the figures (see Fig. 2 to 5). A P value of <0.05 was considered to be significant.
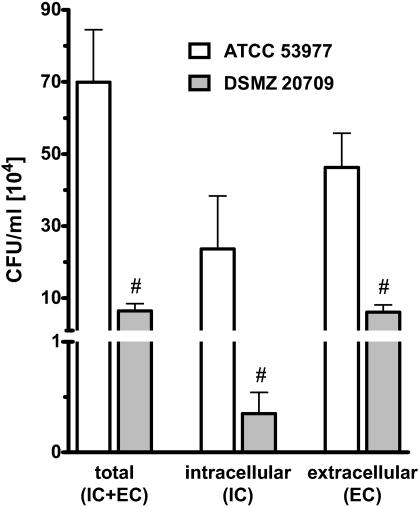
FIG. 2.
Invasion of P. gingivalis was analyzed by an antibiotic protection assay. ATCC 53977 or DSMZ 20709 at an MOI of 50 (5 × 107 bacteria to 106 endothelial cells) was added to HUVEC and further incubated as described in Materials and Methods. The total represents bacteria adhered or already invaded after 1.5 h of exposure to target cells; 1.4% of strain ATCC 53977 (7 × 105 CFU/ml) bacteria and 0.12% of DSMZ 20709 (6.4 × 104 CFU/ml) bacteria could be detected in the endothelial cell lysates. IC indicates bacteria found in endothelial cell lysates after 1.5 h of incubation followed by antibiotic treatment for 2.5 h. EC indicates bacteria that only adhered to HUVEC. Approximately 2.3 × 105 CFU/ml and 3.5 × 103 CFU/ml (34% and 5.4%, respectively) of adhered P. gingivalis ATCC 53977 and DSMZ 20709, respectively, invaded endothelial cells. Data presented are the means ± SEM of four separate experiments. #, P < 0.05 comparing ATCC 53977 versus DSMZ 20709.
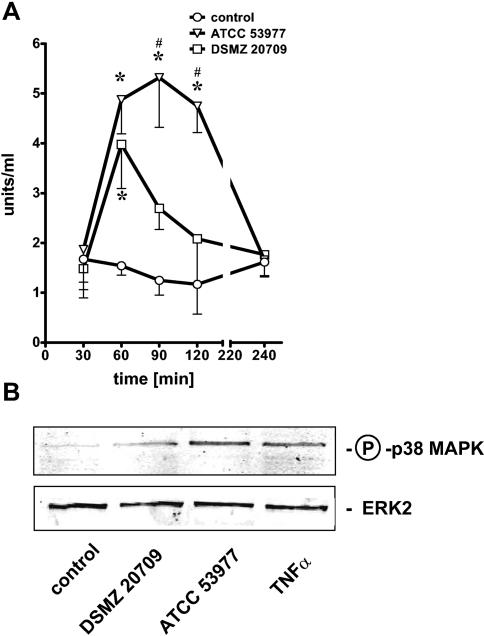
FIG. 5.
HUVEC were stimulated at an MOI of 10 for each strain. ELISA for phospho-p38 MAPK (for details, see Materials and Methods) revealed a time-dependent phosphorylation of p38 MAPK in ATCC 53977- and DSMZ 20709-treated endothelial cells (A). Strain ATCC 53977 induced a prolonged phosphorylation of p38 MAPK, while the signal in DSMZ 20709-stimulated cells decreased again after 60 min. Western blot analysis confirmed p38 MAPK phosphorylation 60 min postinfection in stimulated HUVEC (MOI of 10) (B). A 12.5% polyacrylamide gel was used; ERK2 served to confirm equal loading with proteins. TNF-α (10 ng/ml; 60 min) was used as a positive control. Data presented in panel A are the means ± SEM of three separate experiments. *, P < 0.05 compared to control; #, P < 0.05 comparing DSMZ 20709 versus ATCC 53977. In panel B one representative blot of three is shown.
RESULTS
P. gingivalis strain DSMZ 20709 infects human endothelial cells.
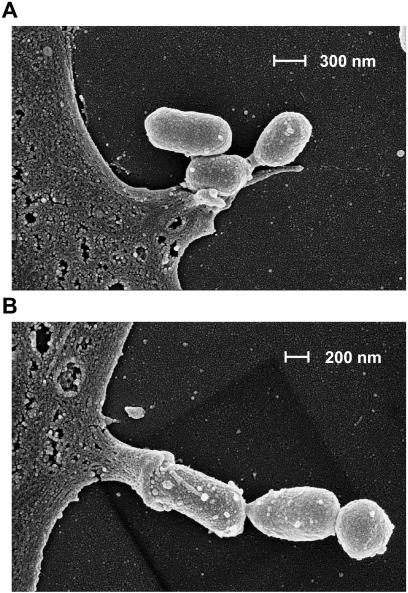
Several strains of P. gingivalis (381 and A7436) have been previously demonstrated to infect and thereby activate human endothelial cells (10, 12). We therefore infected HUVEC with strain DSMZ 20709 by using an MOI of 100, as indicated in Materials and Methods. Adhesion and infection were visualized by scanning electron microscopy (Fig. 1). After fixation at 1.5 h postinfection, visualization by scanning electron microscopy demonstrated adhesion (Fig. 1A) of P. gingivalis to the surface of endothelial cell monolayers. Adhesion is followed by subsequent internalization via a microvillus-like structure of the endothelial cell (Fig. 1B). This process may represent the initial formation of a cytoplasmic vacuole, as described by Deshpande et al. for P. gingivalis strain A7436 (10).
FIG. 1.
Adherence and interactions of P. gingivalis DSMZ 20709 to HUVEC demonstrated by scanning electron microscopy. HUVEC were infected for 1.5 h at an MOI of 100 as described in Material and Methods. Note that P. gingivalis attaches to the surface of an endothelial cell (A). Subsequent invasion appeared to be mediated by a microvillus-like structure protruding from the endothelial cell, starting to engulf the bacterium (B). A magnification of ×100,000 was used for both pictures.
Invasion efficiencies of strain ATCC 53977 and DSMZ 20709.
To quantify invasion and infection of endothelial cells, we used a modified antibiotic protection and invasion assay according to the method of Deshpande et al. and Madianos et al. (10, 34). At 1.5 h postinfection with an MOI of 50, about 1.4% (7 × 105 CFU/ml) of ATCC 53977 and 0.12% of DSMZ 20709 (6.4 × 104 CFU/ml) were in close contact to endothelial cells (adhered or already invaded) (Fig. 2, total). In the next step we evaluated the number of invasive bacteria. Thus, after 1.5 h of P. gingivalis incubation with HUVEC, the remaining EC, nonadherent bacteria were removed by washing, and adherent bacteria were killed by subsequent treatment with antibiotics for 2.5 h (see Materials and Methods). About 2.36 × 105 (ATCC 53977) or 3.5 × 103 (DSMZ 20709) CFU/ml could be recovered in the endothelial cell lysates 4 h postinfection (IC bacteria), representing an invasion rate of 0.48% for ATCC 53977 and 0.007% for DSMZ 20709 relative to the total amount of bacteria added (5 × 107 bacteria/ml).
EC bacteria are bacteria that only adhered to HUVEC and were calculated by subtraction of the amount of IC bacteria from the total amount of P. gingivalis. About 34% of adhering bacteria of strain ATCC 53977 invaded endothelial cells. For strain DSMZ 20709, we demonstrated an invasion rate of 5.4% of adhering bacteria.
P. gingivalis-induced expression of E-selectin and ICAM-1 in human endothelial cells.
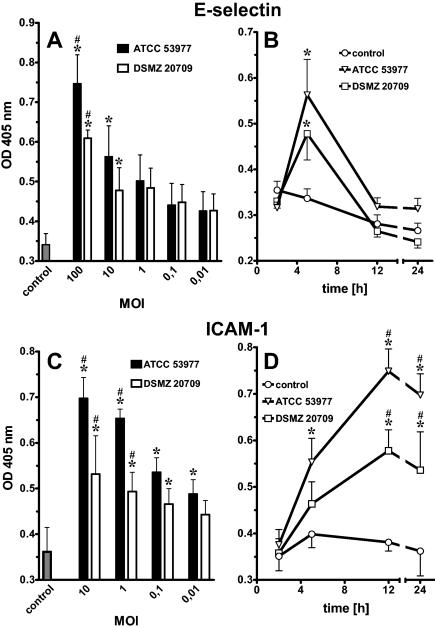
Endothelial adhesion molecule expression was determined by cell surface ELISA. Both strains induced a time- and dose-dependent increase in the endothelial cell adhesion molecules E-selectin and ICAM-1. E-selectin expression in P. gingivalis-stimulated endothelial cells peaked at 5 h and declined to almost baseline after 12 to 24 h (Fig. 3A and B). Even concentrations as low as 103 bacteria/ml were able to induce E-selectin responses (Fig. 3A). ICAM-1 surface expression in P. gingivalis-stimulated HUVEC cells increased at 4 to 8 h postinfection, peaked at 12 h, and remained elevated up to 24 h (Fig. 3C and D) and longer (data not shown). At an MOI of 10 or 1, ICAM-1 expression was more pronounced in ATCC 53977-stimulated HUVEC than in DSMZ 20709-treated endothelial cells.
FIG. 3.
Endothelial cell adhesion molecules quantified by cell surface ELISA were up-regulated in P. gingivalis-stimulated HUVEC in a dose- and time-dependent manner. E-selectin expression peaked at 5 h and declined to baseline after 24 h. Dose dependency was demonstrated 5 h postinfection (A). Expression of ICAM-1 peaked at 12 to 24 h. Dose dependency was demonstrated 24 h postinfection (C). An MOI of 10 (106 bacteria/ml) was used for both time courses (B and D). Even the lowest concentrations were able to stimulate the expression of both adhesion molecules. Data presented are the means ± SEM of four separate experiments. *, P < 0.05 compared to control; #, P < 0.05 comparing ATCC 53977 versus DSMZ 20709; OD, optical density.
P. gingivalis-induced expression of ICAM-1 is independent of FimA.
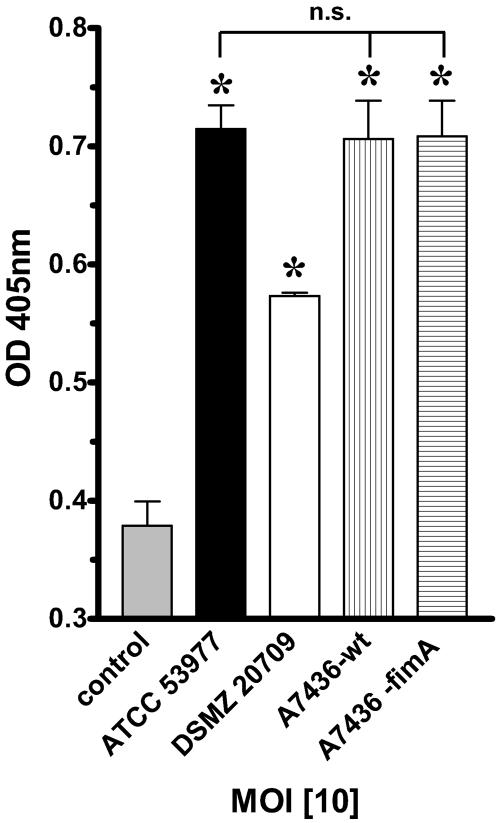
Since there is controversy about the role of fimbriae in the adhesion to and subsequent activation of target cells, we next investigated the role of the major fimbria FimA in P. gingivalis-mediated ICAM-1 expression on HUVEC. ICAM-1 expression was determined by cell surface ELISA after 24 h (MOI of 10 for all strains). We used the fimA knockout mutant A7436 ΔfimA and compared it to its corresponding wild-type A7436-WT strain as well as to strains ATCC 53977 and DSMZ 20709. Cell surface analysis for ICAM-1 expression demonstrated that P. gingivalis strain A7436-WT was equipotent to strain ATCC 53977 (Fig. 4). Interestingly, strain A7436 ΔfimA induced the same level of ICAM-1 expression as the corresponding wild-type strain (A7436-WT).
FIG. 4.
P. gingivalis-induced expression of ICAM-1 on HUVEC is independent of fimA. ICAM-1 expression was determined by cell surface ELISA after 24 h. An MOI of 10 (106 bacteria/ml) was used for all strains. A7436-WT was equipotent to ATCC 53977. The fimA knockout strain A7436 ΔfimA induced the same level of ICAM-1 expression compared to the level in the corresponding wild-type strain. Data presented are the means ± SEM of three separate experiments. *, P < 0.05 compared to control. OD, optical density; n.s., not significant.
P. gingivalis-induced phosphorylation of p38 MAPK in HUVEC.
Recent evidence supports the essential role of p38 MAPK in the inflammatory response of target cells. Using a phospho-p38-MAPK ELISA, we demonstrated a time-dependent enhanced phosphorylation of p38 MAPK in the presence of P. gingivalis strains. Effects peaked at 60 min after stimulation with strain DSMZ 20709 and decreased to almost baseline after 120 min (Fig. 5A). Strain ATCC 53977 induced a more prolonged effect. Phosphorylation remained elevated up to 120 min and thereafter decreased to baseline (240 min). Western blot analysis of P. gingivalis-stimulated HUVEC confirmed enhanced p38 MAPK phosphorylation after 60 min of stimulation with either strain (Fig. 5B); tumor necrosis factor alpha (TNF-α) was used as a positive control for inducing p38 MAPK expression.
P. gingivalis-induced IκBα degradation and NF-κB translocation.
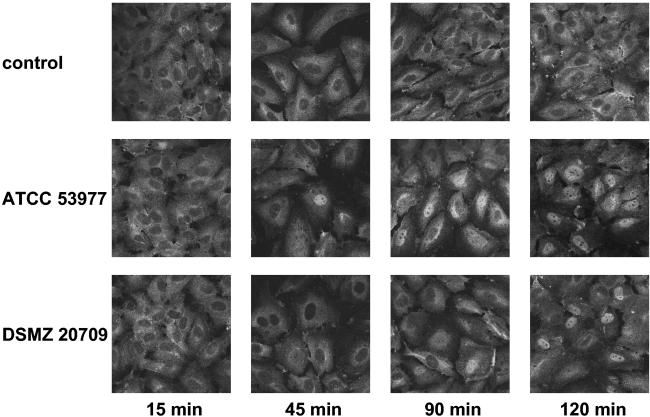
Recent publications have demonstrated the importance of NF-κB for the regulation of the transcriptional activities of the E-selectin and ICAM-1 genes (5, 7, 50). Western blot analysis with strain ATCC 53977 demonstrated a time-dependent degradation of IκBα, with maximal effects 2 h postinfection (Fig. 6). NF-κB translocation in P. gingivalis-infected endothelial cells was also demonstrated by immunofluorescence microscopy. Both strains induced a nuclear translocation of NF-κB starting at 45 (ATCC 53977) to 90 min (DSMZ 20709) postinfection. The signal remained elevated up to 120 min with both strains (Fig. 7) and decreased to almost baseline levels after 180 min (data not shown).
FIG. 6.
Degradation of IκBα was demonstrated to confirm P. gingivalis-mediated NF-κB translocation. Cells were stimulated with strain ATCC 53977 (MOI of 10). Total cell protein was subjected to Western blotting, and IκBα and ERK2 were detected by specific antibodies as indicated in Materials and Methods. A 12.5% polyacrylamide gel was used. TNF-α (10 ng/ml; 60 min) was used as a positive control. One representative gel (of three) is shown.
FIG. 7.
NF-κB activation in P. gingivalis-stimulated HUVEC was analyzed by using immunofluorescence. HUVEC grown on Thermanox slides were stimulated with both strains for the times indicated. Cells were fixed and permeabilized as indicated in Materials and Methods, incubated with a polyclonal rabbit anti-human NF-κB p65 antibody, followed by the addition of an Alexa Fluor 488-conjugated goat anti-rabbit Ig antibody. The top row shows mock-treated control cells. Note a time-dependent increasing fluorescence intensity in the nuclei of P. gingivalis-stimulated HUVEC, demonstrating the nuclear translocation of NF-κB starting after 45 (ATCC 53977) to 90 min (DSMZ 20709) postinfection. Representative pictures (of three independent experiments; magnification, ×640) are shown.
DISCUSSION
The present study demonstrates that two P. gingivalis strains, ATCC 53977 and DSMZ 20709, can infect and activate human endothelial cells. Activation was followed by enhanced expression of the endothelial adhesion molecules E-selectin and ICAM-1, as demonstrated previously with another strain of P. gingivalis (29). The major fimbriae (FimA) were not essential for P. gingivalis-mediated ICAM-1 expression. In addition, we identified two key signal transduction pathways—phosphorylation of p38 MAPK and nuclear translocation of NF-κB—that were activated after stimulation of endothelial cells with both strains. These observations add important new properties to this bacteria regarding its capacity to initiate a cascade of events leading to endothelial cell activation.
The potential of different P. gingivalis strains to induce adhesion molecule expression on different target cells is somewhat controversial and has been discussed previously (9, 24). Results may vary due to the use of different strains, the growth conditions of these strains, and the target cells used in different in vitro systems (29). In our model of cell activation we used (i) primary liquid broth-cultured bacteria in the mid-log phase of growth made from frozen bacterial stocks and (ii) freshly isolated human endothelial cells. The approach with primary isolated endothelial cells seemed especially important since different studies have demonstrated that these cells lose several of their properties during subculture (2, 27).
Several groups have recently described different virulence capacities of selected P. gingivalis strains (36, 43). Since these strains have not been characterized in detail, however, it still remains unclear which mechanisms are responsible for the variability in infection. Diverse rates of bacterial adherence and target cell infection, expression of different virulence factors, and various levels of host cell sensitivity may influence the severity of an infection with P. gingivalis (15, 29).
It has been established that the invasion efficiencies of different P. gingivalis strains usually are very low, ranging from 0.04 to 0.3% (10). In our model both strains were able to invade endothelial cells but with striking differences relative to infectivity. While strain ATCC 53977 infected HUVEC at a rate recently described for many other P. gingivalis strains (10), we could demonstrate only a very low invasion efficiency of 0.007% for strain DSMZ 20709. However, these data were consistent with studies published by Eick et al. and Deshpande et al. (10, 15). Interestingly, both strains were able to activate endothelial signal transduction pathways (phosphorylation of p38 MAPK and translocation of NF-κB) and to induce the expression of adhesion molecules in a similar manner. Although there were some differences in the intensity of the cellular responses (early decrease of p38 MAPK phosphorylation, delayed translocation of NF-κB, and reduced expression of E-selectin and ICAM-1 with strain DSMZ 20709), the overall effects were the same with both strains, indicating that infectivity is an important virulence factor but not the only one. In our in vitro model with isolated endothelial cells, both strains (even the less invasive strain DSMZ 20709), therefore, turned out to be equally virulent, inducing similar levels of target cell activation. These results are in contrast to the in vivo study of Ebersole et al., which used a mouse abscess model and analyzed lesion size and lethality to identify several more or less virulent strains. In their model, strain ATCC 53977 appeared to be the most virulent (14). Little is known about the expression of different virulence factors in strains DSMZ 20709 and ATCC 53977, especially in regard to factors mediating adhesion or the invasion of target cells. Several studies have suggested that fimbriae, proteases, or ceramides are key factors during the process of target cell infection and activation (29, 39, 51). In our model of target cell activation, expression of fimbriae (FimA) appeared not to be required for P. gingivalis-mediated ICAM-1 expression; the fimA knockout strain A7436 ΔfimA induced the same level of ICAM-1 expression as the corresponding wild type (A7436-WT). These results appear to be in contrast to the study by Khglatian et al. (29), but this might be due to the different mutants applied in both studies. The mutant used by Khglatian (DPG3) is apparently pleiotropic, since it also is affected in expression of the proteases arginine-specific gingipain, Rgp, and lysine-specific-gingipain Kgp as well as some adhesins (6). In contrast, the A7436 ΔfimA mutant expresses normal levels of the proteases (J. Hayashi and H. K. Kuramitsu, unpublished results).
Different groups have identified the importance of p38 MAPK in the signal transduction of P. gingivalis-stimulated monocytes (8). Ogawa et al. demonstrated that fimbriae extracted from P. gingivalis were able to stimulate p38 MAPK phosphorylation in human monocytes (41). These data were supported by Darveau et al., who used P. gingivalis LPS with human monocytes. However, in that study, LPS failed to induce phosphorylation of p38 MAPK in endothelial cells (8). By using viable bacteria in our model, we were able to demonstrate by Western blotting and ELISA that ATCC 53977 as well as DSMZ 20709 were able to stimulate the phosphorylation of p38 MAPK.
The expression of inflammatory mediators like cytokines or adhesion molecules depends on the activation of transcription factors. Among the primary transcription factors, NF-κB plays a central role in the regulation of these proinflammatory molecules (5, 50). While Watanabe et al. failed to measure a NF-κB activation in gingival epithelial cells with strain ATCC 33277 (52), we could clearly demonstrate that both strains DSMZ 20709 and ATCC 53977 were able to activate degradation of IκBα and subsequent nuclear translocation of NF-κB in endothelial cells.
The interpretation of our study is limited to cultured human large vessel endothelial cells. For an exact analysis of P. gingivalis-related alterations of endothelial function in clinical disorders, it would be desirable to also study human small vessel endothelial cells especially of the periodontium. However, the isolation and culture of these cells in sufficient quantities are difficult, and therefore the applicability of the data presented to human disease must be verified in further studies.
In conclusion, we present evidence that two different strains of P. gingivalis, ATCC 53977 and DSMZ 20709, can infect HUVEC and activate different signal transduction pathways; p38 MAPK phosphorylation, degradation of IκBα, and activation and translocation of NF-κB occurred within minutes. Within hours, increased surface expression of E-selectin and ICAM-1 was observed. This effect was apparently independent of fimA expression in at least strain A7436. Taken together, the data presented suggest that P. gingivalis infection triggers a cascade of events that could lead to endothelial damage as well as local and systemic inflammation.
Acknowledgments
This work was in part supported by the Deutsche Forschungsgemeinschaft to C.W., J.Z., and J.-P.B. (GRK 325), to M.K. and N.S. (Kr 2197/1-1), to N.S. (BMBF-NBL3 and BMBF-CAPNetz), and to S.H. (BMBF-NBL3) and by a research grant from Universität Medizin Charité to N.P. (89846093).
We gratefully acknowledge the excellent technical assistance of Kerstin Möhr and Verena Kanitz.
Editor: V. J. DiRita
REFERENCES
- 1.Amar, S., N. Gokce, S. Morgan, M. Loukideli, T. E. Van Dyke, and J. A. Vita. 2003. Periodontal disease is associated with brachial artery endothelial dysfunction and systemic inflammation. Arterioscler. Thromb. Vasc. Biol. 23:1245-1249. [DOI] [PubMed] [Google Scholar]
- 2.Bonfanti, R., B. C. Furie, B. Furie, and D. D. Wagner. 1989. PADGEM (GMP140) is a component of Weibel-Palade bodies of human endothelial cells. Blood 73:1109-1112. [PubMed] [Google Scholar]
- 3.Burdon, K. L. 1928. Bacterium melaninogenicum from normal and pathogenic tissues. J. Infect. Dis. 42:161-171. [Google Scholar]
- 4.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. C., and A. M. Manning. 1995. Transcriptional regulation of endothelial cell adhesion molecules: a dominant role for NF-kappa B. Agents Actions Suppl. 47:135-141. [DOI] [PubMed] [Google Scholar]
- 6.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, T., A. Williams, G. I. Johnston, J. Kim, R. Eddy, T. Shows, M. A. Gimbrone, Jr., and M. P. Bevilacqua. 1991. Structure and chromosomal location of the gene for endothelial-leukocyte adhesion molecule 1. J. Biol. Chem. 266:2466-2473. [PubMed] [Google Scholar]
- 8.Darveau, R. P., S. Arbabi, I. Garcia, B. Bainbridge, and R. V. Maier. 2002. Porphyromonas gingivalis lipopolysaccharide is both agonist and antagonist for p38 mitogen-activated protein kinase activation. Infect. Immun. 70:1867-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darveau, R. P., M. D. Cunningham, T. Bailey, C. Seachord, K. Ratcliffe, B. Bainbridge, M. Dietsch, R. C. Page, and A. Aruffo. 1995. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect. Immun. 63:1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshpande, R. G., M. B. Khan, and C. A. Genco. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn, B. R., J. N. Burks, K. N. Seifert, and A. Progulske-Fox. 2000. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol. Lett. 187:139-144. [DOI] [PubMed] [Google Scholar]
- 12.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67:5792-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 2001. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 69:5698-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebersole, J. L., L. Kesavalu, S. L. Schneider, R. L. Machen, and S. C. Holt. 1995. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1:115-128. [DOI] [PubMed] [Google Scholar]
- 15.Eick, S., J. Rodel, J. W. Einax, and W. Pfister. 2002. Interaction of Porphyromonas gingivalis with KB cells: comparison of different clinical isolates. Oral Microbiol. Immunol. 17:201-208. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 17.Grenier, D., and D. Mayrand. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hack, C. E., and S. Zeerleder. 2001. The endothelium in sepsis: source of and a target for inflammation. Crit. Care Med. 29:S21-S27. [DOI] [PubMed] [Google Scholar]
- 19.Hajishengallis, G., M. Martin, R. E. Schifferle, and R. J. Genco. 2002. Counteracting interactions between lipopolysaccharide molecules with differential activation of Toll-like receptors. Infect. Immun. 70:6658-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heimdahl, A., G. Hall, M. Hedberg, H. Sandberg, P. O. Soder, K. Tuner, and C. E. Nord. 1990. Detection and quantitation by lysis-filtration of bacteremia after different oral surgical procedures. J. Clin. Microbiol. 28:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzberg, M. C., and M. W. Weyer. 1998. Dental plaque, platelets, and cardiovascular diseases. Ann. Periodontol. 3:151-160. [DOI] [PubMed] [Google Scholar]
- 22.Hippenstiel, S., B. Schmeck, J. Seybold, M. Krüll, C. Eichel-Streiber, and N. Suttorp. 2002. Reduction of tumor necrosis factor-alpha (TNF-α) related nuclear factor-kappaB (NF-κB) translocation but not inhibitor kappa-B (Iκ-B)-degradation by Rho protein inhibition in human endothelial cells. Biochem. Pharmacol. 64:971-977. [DOI] [PubMed] [Google Scholar]
- 23.Hippenstiel, S., M. Witzenrath, B. Schmeck, A. Hocke, M. Krisp, M. Krüll, J. Seybold, W. Seeger, W. Rascher, H. Schutte, and N. Suttorp. 2002. Adrenomedullin reduces endothelial hyperpermeability. Circ. Res. 91:618-625. [DOI] [PubMed] [Google Scholar]
- 24.Huang, G. T., D. Kim, J. K. Lee, H. K. Kuramitsu, and S. K. Haake. 2001. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect. Immun. 69:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 26.Jaffe, E. A., R. L. Nachman, C. G. Becker, and C. R. Minick. 1973. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Investig. 52:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston, G. I., R. G. Cook, and R. P. McEver. 1989. Cloning of GMP-140, a granule membrane protein of platelets and endothelium: sequence similarity to proteins involved in cell adhesion and inflammation. Cell 56:1033-1044. [DOI] [PubMed] [Google Scholar]
- 28.Karin, M., and Y. Ben Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 29.Khlgatian, M., H. Nassar, H. H. Chou, F. C. Gibson III, and C. A. Genco. 2002. Fimbria-dependent activation of cell adhesion molecule expression in Porphyromonas gingivalis-infected endothelial cells. Infect. Immun. 70:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krüll, M., A. C. Klucken, F. N. Wuppermann, O. Fuhrmann, C. Magerl, J. Seybold, S. Hippenstiel, J. H. Hegemann, C. A. Jantos, and N. Suttorp. 1999. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 162:4834-4841. [PubMed] [Google Scholar]
- 31.Krüll, M., R. Nost, S. Hippenstiel, E. Domann, T. Chakraborty, and N. Suttorp. 1997. Listeria monocytogenes potently induces up-regulation of endothelial adhesion molecules and neutrophil adhesion to cultured human endothelial cells. J. Immunol. 159:1970-1976. [PubMed] [Google Scholar]
- 32.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 33.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madianos, P. N., P. N. Papapanou, U. Nannmark, G. Dahlen, and J. Sandros. 1996. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect. Immun. 64:660-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May, M. J., and S. Ghosh. 1998. Signal transduction through NF-kappa B. Immunol. Today 19:80-88. [DOI] [PubMed] [Google Scholar]
- 36.McKee, A. S., A. S. McDermid, R. Wait, A. Baskerville, and P. D. Marsh. 1988. Isolation of colonial variants of Bacteroides gingivalis W50 with a reduced virulence. J. Med. Microbiol. 27:59-64. [DOI] [PubMed] [Google Scholar]
- 37.Moughal, N. A., E. Adonogianaki, M. H. Thornhill, and D. F. Kinane. 1992. Endothelial cell leukocyte adhesion molecule-1 (ELAM-1) and intercellular adhesion molecule-1 (ICAM-1) expression in gingival tissue during health and experimentally induced gingivitis. J. Periodontal Res. 27:623-630. [DOI] [PubMed] [Google Scholar]
- 38.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Shlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 39.Nichols, F. C. 1998. Novel ceramides recovered from Porphyromonas gingivalis: relationship to adult periodontitis. J. Lipid Res. 39:2360-2372. [PubMed] [Google Scholar]
- 40.Nylander, K., B. Danielsen, O. Fejerskov, and E. Dabelsteen. 1993. Expression of the endothelial leukocyte adhesion molecule-1 (ELAM-1) on endothelial cells in experimental gingivitis in humans. J. Periodontol. 64:355-357. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, T., Y. Asai, M. Hashimoto, and H. Uchida. 2002. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur. J. Immunol. 32:2543-2550. [DOI] [PubMed] [Google Scholar]
- 42.Ogawa, T., H. Uchida, and K. Amino. 1994. Immunobiological activities of chemically defined lipid A from lipopolysaccharides of Porphyromonas gingivalis. Microbiology 140:1209-1216. [DOI] [PubMed] [Google Scholar]
- 43.Ozmeric, N., N. R. Preus, and I. Olsen. 2000. Genetic diversity of Porphyromonas gingivalis and its possible importance to pathogenicity. Acta Odontol. Scand. 58:183-187. [DOI] [PubMed] [Google Scholar]
- 44.Page, R. C., and H. E. Schroeder. 1976. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab. Investig. 34:235-249. [PubMed] [Google Scholar]
- 45.Pattamapun, K., S. Tiranathanagul, T. Yongchaitrakul, J. Kuwatanasuchat, and P. Pavasant. 2003. Activation of MMP-2 by Porphyromonas gingivalis in human periodontal ligament cells. J. Periodontal Res. 38:115-121. [DOI] [PubMed] [Google Scholar]
- 46.Sandros, J., P. N. Madianos, and P. N. Papapanou. 1996. Cellular events concurrent with Porphyromonas gingivalis invasion of oral epithelium in vitro. Eur. J. Oral Sci. 104:363-371. [DOI] [PubMed] [Google Scholar]
- 47.Sandros, J., P. Papapanou, and G. Dahlen. 1993. Porphyromonas gingivalis invades oral epithelial cells in vitro. J. Periodontal Res. 28:219-226. [DOI] [PubMed] [Google Scholar]
- 48.Sconyers, J. R., J. J. Crawford, and J. D. Moriarty. 1973. Relationship of bacteremia to toothbrushing in patients with periodontitis. J. Am. Dent. Assoc. 87:616-622. [DOI] [PubMed] [Google Scholar]
- 49.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 50.Voraberger, G., R. Schafer, and C. Stratowa. 1991. Cloning of the human gene for intercellular adhesion molecule 1 and analysis of its 5′-regulatory region. Induction by cytokines and phorbol ester. J. Immunol. 147:2777-2786. [PubMed] [Google Scholar]
- 51.Wang, P. L., M. Shinohara, N. Murakawa, M. Endo, S. Sakata, M. Okamura, and K. Ohura. 1999. Effect of cysteine protease of Porphyromonas gingivalis on adhesion molecules in gingival epithelial cells. Jpn. J. Pharmacol. 80:75-79. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe, K., O. Yilmaz, S. F. Nakhjiri, C. M. Belton, and R. J. Lamont. 2001. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect. Immun. 69:6731-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]