Abstract
The cadicivirus IRES diverges structurally from canonical Type 1 IRESs (e.g. poliovirus) but nevertheless also contains an essential GNRA tetraloop in a subdomain (d10c) that is homologous to poliovirus dIVc. In addition to canonical initiation factors, the canonical Type 1 and divergent cadicivirus IRESs require the same IRES trans-acting factor, poly(C)-binding protein 2 (PCBP2). PCBP2 has three KH domains and binds poliovirus IRES domain dIV in the vicinity of the tetraloop. How PCBP2 binds the cadicivirus IRES, and the roles of PCBP2 and the tetraloop in Type 1 IRES function are unknown. Here, directed hydroxyl radical probing showed that KH1 also binds near the cadicivirus tetraloop. KH2 and KH3 bind adjacently to an IRES subdomain (d10b) that is unrelated to dIV, with KH3 in an inverted orientation. KH3 is critical for PCBP2's binding to this IRES whereas KH1 is essential for PCBP2's function in promoting initiation. PCBP2 enforced the wild-type structure of d10c when it contained minor destabilizing substitutions, exposing the tetraloop. Strikingly, PCBP2 enhanced initiation on mutant IRESs that retained consensus GNRA tetraloops, whereas mutants with divergent sequences did not respond to PCBP2. These studies show that PCBP2 enables the IRES to exploit the GNRA tetraloop to enhance initiation.
INTRODUCTION
Initiation of translation on most eukaryotic cellular mRNAs occurs by the 5′-end-dependent scanning mechanism, but some viral mRNAs utilize 5′end-independent internal ribosomal entry instead (1). Viral internal ribosome entry sites (IRESs) are long, structured RNA elements that mediate ribosomal recruitment to an internal location on the mRNA, are commonly located in the 5′UTR, and are classified into a few major groups on the basis of shared sequence motifs and a common structural core. The most extensively characterized picornavirus IRESs are the Type 1 IRESs, which occur in members of the genus Enterovirus such as poliovirus (PV), Coxsackievirus B3 (CVB3), enterovirus 71 (EV71) and human rhinovirus (HRV), and the Type 2 IRESs, epitomized by encephalomyocarditis virus (EMCV) and foot-and-mouth disease virus (FMDV) (2).
Type 1 IRESs (Supplementary Figure S1A) are ∼450nt long, comprise domains dII–dVI, and their 3′-border is ∼30nt (HRV) to ∼160nt (PV) upstream of the initiation codon. The 3′-border of the IRES is marked by a Yn-Xm-AUG motif (in which a Yn pyrimidine tract (n = 8–10nt) is separated by a spacer (m = 18–20nt) from a (silent) AUG triplet). Domains dIII and dVI are non-essential and vary in size and sequence, whereas dIV and dV are highly conserved. Domain dIV is ∼200nt long, has a cruciform structure and includes the apical subdomain dIVc, which radiates from the four-way junction in dIV and contains two essential motifs, an internal C-rich loop (3,4) and an apical GNRA tetraloop of unknown function (5). dV is ∼110nt long and forms an irregular hairpin with a large internal loop. The Type 2 IRESs (Supplementary Figure S1B) are also ∼450nt long, but their structure is unrelated to that of Type 1 IRESs, with two exceptions: they also have a Yn-Xm-AUG motif at their 3′-border and an essential GNRA tetraloop at the apex of the largest domain (6,7). In the FMDV IRES, the tetraloop is involved in a tertiary interaction that stabilizes the structure of the IRES (8,9), but the step in initiation that depends on this has not been established.
In vitro reconstitution and factor binding experiments have identified the outline of the mechanism of initiation on canonical Type 1 IRESs. Eukaryotic initiation factor (eIF) 4G, a constituent of the eIF4F cap-binding complex, is cleaved during enterovirus infections into an N-terminal fragment that binds eIF4E and a C-terminal fragment that binds the eIF4A DEAD-box RNA helicase and the multimeric eIF3 (10,11). The C-terminal fragment of eIF4G binds to dV, recruits eIF4A (12) and they together promote attachment of the ribosomal 43S preinitiation complex to the IRES. The 43S complex comprises a 40S subunit, eIF1, eIF1A, eIF3 and a ternary complex consisting of eIF2-GTP and aminoacylated initiator tRNA (Met-tRNAiMet). This binding step requires the interaction of IRES-bound eIF4G with eIF3, to engage the 43S complex, and eIF4A's catalytic activity, likely to restructure elements of the IRES to allow ribosomal attachment (13). Next, the 43S complex scans downstream to the initiation codon, where eIF5 and eIF5B cooperate to mediate hydrolysis of eIF2-bound GTP and movement of initiator tRNA into the peptidyl (‘P’) site of the 40S subunit, followed by release of initiation factors from the 40S subunit and joining of a 60S ribosomal subunit to form an 80S ribosome that is competent to begin translation. Initiation on Type 1 IRESs thus differs from the canonical initiation mechanism by dispensing with the need for eIF4E and the element of eIF4G to which it binds, but it also differs by requiring one or more IRES trans-acting factors (ITAFs) for formation of the 48S complex on the IRES. The poly(C) binding protein 2 (PCBP2) is an essential ITAF for Type 1 IRES function in vivo and in cell-free extracts (14–18), although other ITAFs may have an accessory role (19). PCBP2 is sufficient to complement the activity of canonical eIFs in in vitro reconstituted initiation reactions (13).
The four PCBP isoforms, PCBP1-4, each contains three K-homology (KH) domains which bind single-stranded RNA and DNA (20,21). The two consecutive domains at the N-terminus are followed by a long spacer and a third KH domain at the C-terminus (Figure 2B). Each domain has a classical type-1 KH fold, with a β1α1α2β2β3α3 topology. Binding of RNA involves a hydrophobic cleft formed on one side by α1, α2 and the intervening conserved GXXG loop, which interacts with the RNA backbone and orients four bases to the other side, which is formed by the β-sheet and a variable loop (22,23). PCBP2 engages in intricate interactions with the apical region of dIV of Type 1 IRESs, including a major interaction of KH1 with the C-rich loop in dIVc (3,4,13,24). Various functions have been proposed for PCBP2 and other ITAFs in promoting IRES-mediated initiation, including interaction with and recruitment of initiation factors, other ITAFs or the 40S ribosomal subunit to the IRES, and stabilization of IRES structure in an active conformation (25). PCBP2 recruits SRp20 to the PV IRES (26), but since SRp20 is not required for 48S complex on the PV IRES in in vitro reconstituted reactions, in which PCBP2 is essential, PCBP2 must have at least one other critical function. The presence of three KH domains in PCBP2 and their engagement in multiple interactions with the IRES would be consistent with a function being to enforce an active conformation on the IRES, although this raises the question of which subsequent interaction or conformational transition in the IRES is dependent on prior binding of PCBP2.
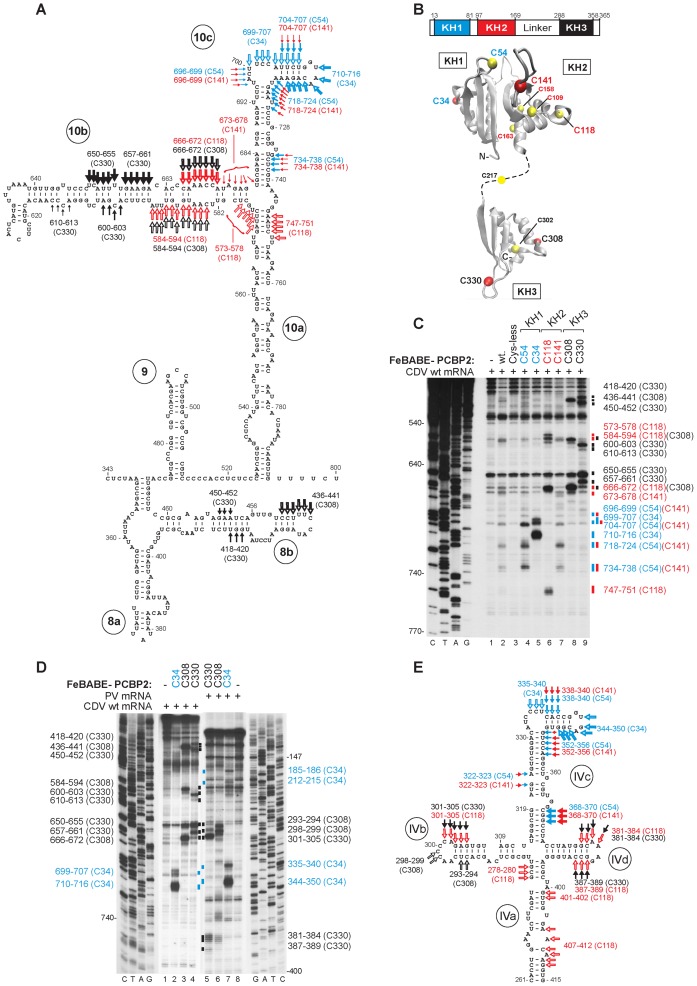
Figure 2.
Sites of interaction of PCBP2 with the CDV IRES. (A) Secondary structure model of IRES domains 8–10, showing sites of directed hydroxyl radical cleavage (DHRC) from C34 and C54 in KH1, C118 and C141 in KH2, and C308 and C330 in KH3 domains of PCBP2. Arrow size is proportional to cleavage intensity. Sites are colored to match the domains in (B). Open and filled arrows indicate DHRC from cysteines in the GXXG loop and in the invariant loop of each KH domain, respectively. (B) Schematic representation of PCBP2 showing the positions of the KH domains (upper panel). Ribbon diagrams of PCBP2 KH1-KH2 and KH3 domains (PDB: 2JZX and 2P2R) with a dashed line representing the linker between them (lower panel). Yellow spheres indicate native (C54, C109, C118, C158, C163, C217 and C302) and introduced (C34, C141, C308 and C330) cysteines. (C, D) Primer extension analysis of DHRC of (C) CDV and (D) PV IRESs from PCBP2. Sites of cleavage are indicated on both sides of the gel and (A, E) mapped onto IRES models. In (E), sites of DHRC of the PV IRES from C34 and C141 (panel D and Supplementary Figure S3D) are combined with data for DHRC from other residues (14).
Metagenomic analyses have expanded the number of known picornaviruses, including some with highly divergent IRESs and others with IRESs that appear to have been exchanged with those of unrelated viruses (27–34). Cadicivirus (CDV), a naturally occurring dicistronic picornavirus (35), has IRESs in the 5′UTR and in the intergenic region between ORF1 (encoding structural proteins) and ORF2 (encoding nonstructural proteins) that have some homology to Type 1 IRESs. The 5′UTR contains a domain (d11) that is highly homologous to domain V of canonical Type 1 IRESs, whereas d12 (18 nt long) is much shorter than e.g poliovirus dVI, and the adjacent upstream d10 (268nt long) is a Y-shaped structure with a three-way helical junction rather than the four-way junction in dIV of canonical Type 1 IRESs (36). Nevertheless, the apex of d10 has a structure that is homologous to that of dIVc and, like it, contains a C-rich internal loop and an apical GNRA tetraloop. Despite these structural differences, the same factors, including PCBP2, are required for efficient initiation on the CDV IRES as on canonical Type I IRESs (13,36). However, how PCBP2 binds to the structurally divergent CDV IRES is not known, and its function in promoting initiation on this and on canonical Type 1 IRESs has not been established.
Here, we have characterized the structure of CDV d10 and its interaction with PCBP2 in detail, using footprinting to map its binding site, directed hydroxyl radical cleavage to orient individual KH domains on the IRES and mutagenesis of the IRES and RNA-binding determinants in each KH domain to establish the functional importance of each interaction and, more generally, how factors such as IRES trans-acting factors engage functionally with structurally divergent IRESs. We have established a function for PCBP2 in enabling this IRES to exploit the function of the GNRA tetraloop in the process of internal ribosomal entry that is likely of general relevance to all Type 1 IRESs. These findings are discussed in light of current models for the mechanism of IRES-mediated initiation.
MATERIALS AND METHODS
Plasmids
Expression vectors were for His6-tagged eIF1 and eIF1A (37), eIF4A and eIF4B (38), eIF4GI736–1115 (eIF4Gm) (39), Escherichia coli methionyl tRNA synthetase (40), and wild type (wt), cysteineless and C308 and C330 single cysteine mutant forms of His6-tagged PCBP2 (13). The pET28b-PCBP2(cysteineless) vector was used (NorClone Biotech, London, Ontario) to generate four single-cysteine mutants (C34, C54, C118 and C141); the C54, C118 and C141 substitutions were combined individually with substitutions in the GXXG motif of the same KH domain (KK31-32DD, KG115-116DD and RQ305-306DD, respectively), or together, yielding mutants with C54, C118 and C141 residues and GXXG→GDDG substitutions in KH1, KH2 or KH3 domains. CDV and PV IRES transcription vectors have been described (36,41). CDV IRES domain 10 was mutated (96 Proteins, San Francisco, CA) in the Stem-DC-XL-CDV transcription vector.
Purification of initiation factors, ribosomal subunits and aminoacylation of tRNA
40S ribosomal subunits, eIF2 and eIF3 were purified from rabbit reticulocyte lysate (RRL) (42). eIF1, eIF1A, eIF4A, eIF4B, eIF4Gm, E. coli methionyl tRNA synthetase and wt and mutant forms of PCBP2 were expressed in E. coli and purified (13,42). Partially purified native tRNAiMet obtained from unfractionated calf liver tRNA (Promega) and in vitro transcribed tRNAiMet were aminoacylated using E. coli methionyl tRNA synthetase (42,43).
Assembly and analysis of ribosomal complexes
48S ribosomal complexes were assembled by incubating 1 pmol CDV IRES-containing mRNA with 1.5 pmol 40S subunits, 3.5 pmol partially purified native Met-tRNAiMet (unless stated), 2.5 pmol eIF2, 2 pmol eIF3, 7 pmol eIF1, 7 pmol eIF1A, 5 pmol eIF4A, 2 pmol eIF4B, 3 pmol eIF4Gm, 1.4 pmol wt or mutant PCBP2 as indicated, for 15 min at 37°C in 20 μl buffer A (20mM Tris pH 7.5, 100 mM KCl, 1 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine) supplemented with 1 mM ATP and 0.4 mM GTP. The assembled 48S complexes were analyzed by primer extension inhibition using avian myleoblastosis virus reverse transcriptase (AMV-RT) and a [32P]-labeled primer corresponding to CDV nt 1086–1108 (36). The resulting cDNA products were resolved by electrophoresis on 7 M urea/6% polyacrylamide sequencing gels and visualized by autoradiography.
Electrophoresis mobility shift assay
CDV IRES domain 10 (nt 510–812) was amplified using a forward primer containing a T7 promoter and transcribed in the presence of [32P]ATP (42,44). 10nM labeled RNA was incubated with PCBP2 or its single-cysteine variants at the indicated concentrations in 10 μl buffer B (20 mM Tris, 2.5 mM MgCl2, 100 mM KCl, 4% glycerol, 5 mM Heparin and 2 mM DTT) for 15 min at 37°C. After addition of 2 μl loading buffer (60% glycerol, 0.01% Bromophenol blue), samples were applied to non-denaturing 5% (40:1) acrylamide gel and resolved by electrophoresis at 200 V for 2 h at 4°C using 50 mM Tris, 50 mM boric acid, 5% glycerol and 2.5 mM MgCl2 running buffer. Bound and unbound fractions were visualized by autoradiography. The curves were fitted to the non-linear Hill equation (Frac[bound] = [PCBP2]n •Frac[bound]max/([PCBP2]n + Kn1/2,app)) using GraphPad Prism software. The dissociation constant (K1/2,app) and Hill coefficient (n) were calculated on the basis of three independent experiments.
Interaction of PCBP2 with the CDV IRES assayed by toe-printing
2 pmol monocistronic CDV mRNA was incubated with the indicated combinations of 10 pmol wt or mutant PCBP2, 8 pmol eIF4Gm and 20 pmol eIF4A for 15 min at 37°C in 40 μl buffer A supplemented with 1 mM ATP. IRES/PCBP2 complexes formed with or without eIF4Gm/eIF4A were analyzed by primer extension inhibition at 20°C for 2 h, using AMV-RT and [32P]-labeled primer corresponding to CDV nt 1016–1037 in the presence of 4 mM MgCl2 and 0.5 mM dNTPs.
Directed hydroxyl radical cleavage assay
Wt and mutant forms of recombinant PCBP2 were derivatized with Fe(II)-BABE (Dojindo Molecular Technologies, Rockville, MD) (44). 20 pmol FeBABE-derivatized protein was incubated with 4 pmol wt or mutant CDV or PV mRNA, as indicated, in 50 μl buffer A for 15 min at 37°C. Fenton chemistry was initiated by addition of 0.05% H2O2 and 5 mM ascorbic acid. Reaction mixtures were incubated on ice for 10 min, and reactions were quenched using 20 mM thiourea. Sites of cleavage were located by primer extension using AMV-RT and [32P]-labeled primer corresponding to CDV nt 834–852 (36).
Enzymatic and chemical footprinting analysis of PCBP2-IRES complexes
For enzymatic footprinting, 2 pmol wt or mutant CDV IRES was incubated alone or with 10 pmol PCBP2 in 40 μl buffer (20 mM Tris pH 7.5, 100 mM KAc, 1 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine) for 15 min at 37°C and then partially digested by incubating at 37°C for 5 min in presence of RNase T1 (2.5 × 10−3 U/ul) or RNase V1 (2.5 × 10−4 U/ul). RNA was immediately recovered by phenol/chloroform extraction followed by ethanol precipitation. Sites of cleavage were mapped by primer extension inhibition using AMV-RT and appropriate [32P]-labeled primers. For chemical footprinting, 2 pmol CDV RNA was denatured, refolded and modified alone or in the presence of 10 pmol PCBP2 in 20 μl buffer (100 mM HEPES pH 8.0, 2.5 mM MgCl2, 100 mM NaCl) at 37°C for 45 min with 1 μl DMSO or 1 μl 130 mM N-methylisatoic anhydride (NMIA) in DMSO (36). RNA was recovered by ethanol precipitation and modified sites were mapped using AMV-RT and appropriate [32P]-labeled primers.
In vitro translation
Wild type and mutant CDV mRNAs (0.4 pmol) were translated in the Flexi RRL system (Promega) (20 μl reaction volume) supplemented with 0.5 mCi/ml [35S]methionine (43.5 TBq/mmol) for 60 min at 37°C. Translation of the truncated viral capsid polyprotein (ΔP1) was analyzed by gel electrophoresis using NuPAGE 4–12% Bis‐Tris‐Gel (Invitrogen), followed by autoradiography and quantification on a phosphoimager.
Analysis of 40S/IRES and 40S/eIF3/IRES complexes by sucrose density gradient centrifugation
2 pmol [32P]UTP-labeled wt or mutant CDV IRES (nt 341–1108) were incubated with 6 pmol 40S subunit alone or with 6 pmol eIF3. The IRES/40S and IRES/40S/eIF3 complexes were analyzed by centrifugation through 10–30% sucrose density gradients prepared in buffer A. The optical density of fractionated gradients was measured at 260 nm and the presence of [32P]-labeled mRNA was monitored by Cherenkov counting.
RESULTS
The interaction of PCBP2 with d10 of the CDV IRES
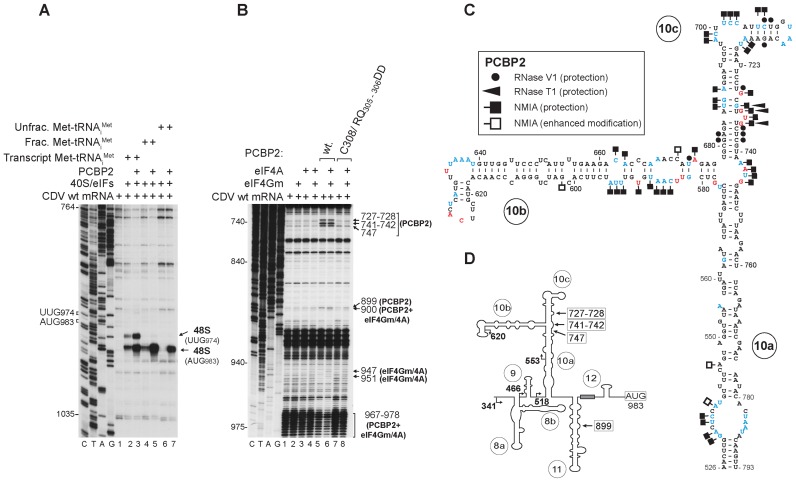
PCBP2 is required for efficient 48S complex formation at the CDV initiation codon AUG983 in in vitro reconstitution experiments done using ‘unfractionated’ Met-tRNAiMet (prepared by aminoacylation of total native mammalian tRNA) (Figure 1A, lanes 6 and 7; (36)). The PCBP2-dependence of IRES-mediated initiation was strongly reduced in reactions reconstituted using partially purified native mammalian total tRNA (‘fractionated’ Met-tRNAiMet) (from which many of the heterogeneous low molecular weight contaminants present in ‘unfractionated’ Met-tRNAiMet had been removed (36)) or in vitro transcribed ‘transcript’ Met-tRNAiMet (Figure 1A, lanes 2–5). In the latter instance, 48S complex formed at the near-cognate codon UUG974 as well as at AUG983.
Figure 1.
Specific interaction of PCBP2 with the CDV IRES. (A) Toe-print analysis of 48S complex formation in vitro on the wt IRES in the presence of 40S subunits, eIFs, PCBP2 and ‘transcript’, ‘fractionated’ native or ‘unfractionated’ native Met-tRNAiMet, as indicated. Toe-prints caused by 48S complexes assembled on AUG983 and on the near-cognate codon UUG974 are indicated on the right. (B) Toe-prints (indicated on the right) caused by interaction of the indicated combinations of wt or [C308/RQ305-306DD]-PCBP2, eIF4Gm and eIF4A with the IRES. (C) Secondary structure model of IRES domain 10 showing residues at which modification by NMIA or cleavage by RNase T1 or V1 was altered by inclusion of PCBP2 (symbols as described in the inset panel; data from Figure 6C, Supplementary Figures S2B, S4A and S4B). (D) Model of the IRES, with arrows showing the borders of 5′-terminal truncations of the 5′UTR and (boxed numbers) the PCBP2 toe-prints identified in panel (B).
Toe-prints due to binding of PCBP2 to the CDV IRES were not apparent at 37°C, but at 20°C they appeared near the three-way helical junction in d10, as well as weakly in d11 (Figure 1B, lane 5; Figure 1D). At 20°C, eIF4A/eIF4Gm induced weak reverse transcriptase (RT) stops at nt 947 and 951, downstream of their binding site in d11 (36) and enhanced changes in the pattern of RT stops induced by PCBP2 at nt 899–900 and nt 967–978 (Figure 1B, lanes 4 and 6). These observations indicate that PCBP2 binds primarily to the apex of d10, and suggest that it cooperates, directly or indirectly, with eIF4A/4Gm in inducing conformational changes upstream of the initiation codon. PCBP2's affinity for CDV nt 510–812 (Kd = 36 nM) (Supplementary Figure S2A) is comparable to that for PV dIV (Kd ∼ 15 nM) and CVB3 dIV (Kd ∼ 35 nM) (3,4); the Hill coefficient (n = 1.9) suggests positive cooperativity in binding of PCBP2 to CDV d10, consistent with PCBP2 having multiple RNA-binding KH domains.
The structures of CDV d10 and dIV in canonical Type 1 IRESs differ significantly, raising the question of how PCBP2 binds the CDV IRES. RNAse V1 and T1 footprinting showed that PCBP2 protected the IRES from cleavage near the base of d10b and in d10c (Figures 1C and 6C, Supplementary Figure S2B). The site protected from RNAse V1 cleavage at nt 727 coincides with a PCBP2 toe-print. Footprinting done using N-methylisatoic anhydride (NMIA) also revealed protection in this area, at the apex of d10c, near the base of d10b, at the base of d10a and at the d10a/d10b/d10c junction (Figures 1C, 6A(i) and B, Supplementary Figure S4A and S4B). Interaction of PCBP2 with the IRES increased NMIA modification of a few nucleotides in d10a and d10b, likely due to induced conformational changes in these regions.
Figure 6.
Role of PCBP2 in stabilizing IRES domain 10 in relation to the GNRA tetraloop. (A) Sequence and structure of the apex of domain 10 in (i) wt and (ii, iii) GNRA tetraloop mutants of the CDV IRES color-coded to show reactivity to NMIA and marked to show residues exposed to RNase T1 (see panels B and C). Sites at which PCBP2 altered NMIA modification and RNase T1 cleavage are indicated by symbols (lower inset panel). (B, C) Primer extension analysis of (B) chemical (NMIA) modification and (C) enzymatic (RNase T1) cleavage of the wt CDV IRES and its variants (panel A) in the presence/absence of PCBP2.
Interactions of PCBP2's KH domains with the IRES mapped by directed hydroxyl radical cleavage
PCBP2's three KH domains all bind to the apical region of dIV in Type 1 IRESs, with the strongest interactions involving dIVb and dIVc (3,4,13,24). With the exception of d10c, the structure of the element of the CDV IRES to which PCBP2 binds differs significantly from dIV. To investigate how the distinct sequence and structure of the CDV IRES affect binding of PCBP2, we employed directed hydroxyl radical cleavage (DHRC). In this approach, Fe(II) is site-specifically tethered by the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE) to a (unique) cysteine residue on the surface of an RNA-bound protein and used to generate hydroxyl radicals by ascorbic acid/H2O2 treatment, cleaving RNA at nearby sites that can be identified by primer extension inhibition. Hydroxyl radicals have a small radius of action, yielding strong cleavage from 0 to 22 Å from the tethering position, medium-strength cleavage from 12 to 36 Å, and weak cleavage from 20 to 44 Å (45). Thus if the structure of the protein to which Fe(II) is tethered is known, then cleavage at multiple sites allows localization and orientation of this protein relative to its RNA ligand.
The seven naturally-occurring cysteine residues in PCBP2 can be replaced to yield an active, cysteine-less form of PCBP2 (13) that was used to generate ‘single cysteine’ variants. Each KH domain has an RNA-binding cleft that accommodates four RNA bases and is formed by α-helix 1, α2 and an invariant GXXG connecting motif on one side, and β-strand 2 and a variable loop on the other (20,21). Cysteine residues replaced a residue either immediately adjacent to the GXXG motif (C34 in KH1, C118 in KH2 or C308 in KH3) or in the variable loop (C54 in KH1, C141 in KH2 or C330 in KH3) in each domain, yielding six single-cysteine variants (Figure 2B). They bound with a slightly reduced affinity and cooperativity to CDV d10 (e.g. Supplementary Figure S3A). This impairment is consistent with their slightly reduced activity in promoting initiation on the IRES (Supplementary Figure S3C, lanes 3–5).
C34 and C54 in KH1 induced distinct but overlapping patterns of cleavage in d10c (Figure 2A and C, lanes 4 and 5) that coincided with sites protected by PCBP2 from RNase V1 cleavage and NMIA modification (Figure 1C). The pattern of cleavage from C118 and C141 in KH2 suggested that it binds close to the base of d10b, at its junction with d10a and d10c (Figure 2A). C118 induced a staggered pattern of cleavages in the basal region of d10b, consistent with it forming a helix (Figure 2C, lanes 6 and 7). Many of the cleavages induced in d10c by C141 overlapped cleavages from C54 in KH1 (Figure 2A): KH1 and KH2 are thus likely close to each other when bound to the IRES, possibly in the back-to-back conformation seen in the KH1-KH2 crystal structure (21) (Figure 2B). The overlapping of cleavages induced by C308 (KH3) and C118 (KH2), and the identification of cleavages from C330 in their immediate vicinity (Figure 2C, lanes 6, 8 and 9) indicated that the flexibly linked KH3 binds to the IRES in the immediate vicinity of KH2. In addition to allowing PCBP2's KH domains to be mapped on the IRES, the pattern of cleavages from the different Fe-BABE-modified Cys residues also provided support for the helical nature of d10b and for distinguishing structural characteristics of d10, including the three-way junction of d10a, d10b and d10c.
The patterns of cleavage induced by C34 (Figure 2D, lane 7) and C54 (13) in dIVc of the PV IRES (Figure 2E) were almost identical to those seen here in CDV d10c, indicating that KH1 bound to these related elements in a similar manner. C34 also induced weak cleavage in the peripheral dIII of the PV IRES (Figure 2D, lane 7), which has no equivalent in the CDV IRES (36). Interestingly, cleavages at PV nts 301–305 and 381–384 from C118 in KH2 overlapped those from C330 in KH3 (Figure 2D, lanes 5 and 6; Figure 2E) whereas cleavages of the CDV IRES from C118 coincided with those from C308. Thus, although KH2 and KH3 bound in proximity to each other on PV (13) (Figure 2E) and CDV IRESs (Figure 2A), the orientation of KH3 on the CDV IRES appears to be flipped relative to that on the PV IRES.
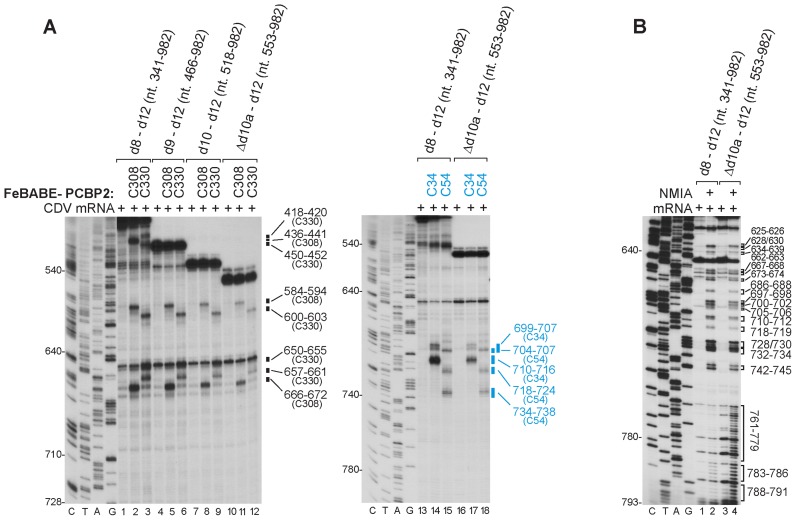
Interaction of PCBP2 with d8 of the CDV 5′UTR is not required for its binding to d10
The observation that FeBABE-linked C308 and C330 variants of PCBP2 induced cleavage in d8b (Figure 2A) could be due to the proximity of d8b and d10b in the IRES/PCBP2 complex or to binding of KH3 to independent sites. Deletion of domains 1–7 (nt 1–340) (Figure 1D) did not affect the intensity of cleavage induced by C308 and C330 in d8b or d10b (Figure 3A, lanes 1–3) but deletion of d8 (nt 1–465), d9 (nt 1–517) and part of d10a (nt 1–552) progressively reduced the intensity but did not abrogate cleavage in d10b (Figure 1D; Figure 3A, lane 4–12). The intensity of cleavage is a surrogate for binding of PCBP2 KH3 to the IRES, and its progressive reduction parallels the effect of these truncations on IRES activity in RRL (36). Deletion of nt 1–619, which additionally removed part of d10b, led to the complete disappearance of these cleavages (Supplementary Figure S3E, lanes 4–6). PCBP2 KH3 therefore binds to d10b independently of d8b.
Figure 3.
Interaction of PCBP2 with IRES domain 10 is independent of upstream domains. (A, B) Primer extension analysis of (A) DHRC from Fe(II)-derivatized C34, C54, C308 and C330 single-cysteine variants of PCBP2 and (B) chemical (NMIA) modification of 5′-terminally truncated derivatives of the CDV 5′UTR (Figure 1D). Sites of cleavage/modification are indicated on the right side of the gels.
Deletion of nt 1–552 increased the susceptibility of nucleotides on the 3′-side of d10a to NMIA modification (Figure 3B, lanes 2 and 4), but did not alter the pattern of modification of d10b and d10c. Truncation of d10b by deleting nt 1–619 did not affect d10c but enhanced modification elsewhere in the residual 5′UTR (Supplementary Figure S3F, lane 2). d10b and d10c therefore fold independently of d10a, but truncation of d10a nevertheless impaired binding of PCBP2, as reflected by the weakened cleavage of d10b and d10c by C308 and C330 noted above, and by the weakened cleavage by C34 and C54 in KH1 (Figure 3A, lanes 13–18). This effect of partially deleting d10a correlates with near-abrogation of IRES function (36), likely due to alteration of the mutual orientation of domains or of some other aspect of their tertiary structure.
The RNA-binding activities of KH1 and KH3 are key determinants of PCBP2 function in promoting initiation on the IRES
Cooperative binding of KH domains is critical for the high affinity and specificity of binding to their RNA targets of several proteins with multiple KH domains, including hnRNP-K (46) and CRD-BP (47). In all KH domain/RNA complexes for which structures have been determined, interaction of the nucleic acid backbone with the conserved GxxG loop that links α-helices 1 and 2 (Figure 4A) orients the bases into a hydrophobic groove in a manner that leads to interactions that underlie base-specific binding (22). Besides the GXXG loop, other basic residues forming the nucleic acid binding cleft, such as the arginine residue near the C-terminal end of the α2 helix, which occurs in all three domains (Figure 4A), also determine KH domain interactions with RNA (21,23). To characterize the importance of the RNA-binding activity of individual KH domains for PCBP2's function in promoting initiation on the CDV IRES, we used a conservative approach in which substitutions are introduced into the GxxG motif of individual KH domains that eliminate RNA-binding activity without compromising stability (48). To assess the influence of such substitutions in individual domains on binding of the same and of other KH domains to the IRES, we combined GxxG→GDDG substitutions with C54, C118 and C308 substitutions, as appropriate, in the same or in all three KH domains (Figure 4B) so that directed hydroxyl radical cleavage could be used to assess the specificity and (qualitatively) the affinity of binding.
Figure 4.
Contribution of individual KH domains to PCBP2 function in IRES-mediated initiation. (A) Sequence alignment of the KH domains of human PCBP2 and their secondary structure elements. (B) Ribbon diagram of KH1–KH2 and KH3 domains (PDB: 2JZX and 2P2R) showing variable (XX) residues (violet beads) of the GXXG motif (violet loop) and retained cysteines in each domain (C54 in KH1, C118 in KH2 and C308 in KH3). (C, D) Primer extension analysis of DHRC of the CDV IRES from Fe(II)-derivatized wt, C54, C118 and C308 single-cysteine and KK31-32DD, KG115-116DD and RQ305-306DD mutant forms of PCBP2 to investigate the influence of GXXG elements in one KH domain on binding of (C) the same and (D) other KH domains of PCBP2 to the IRES.
The GKKG→GDDG30-33 substitutions reduced the intensity of cleavage from C54 in KH1 particularly at nt 718–724 and 734–738 in d10c (Figure 4C, lanes 3 and 6). Similar substitutions in KH2 (GKGG→GDDG114-117) led to an even greater decrease from C118 in cleavage near the d10a/d10b/d10c junction, and to lesser reduction in cleavage from this residue in d10b (Figure 4C, lanes 4 and 7). Neither set of substitutions changed the pattern of cleavage, indicating that the specificity of binding was unaltered. Moreover, mutations in GKKG30–33 and GKGG114-117 loops of KH1 and KH2 did not alter the pattern or intensity of cleavage from cysteines in the two other domains (Figure 4D, lanes 2 and 3). On the other hand, GDDG304–307 substitutions led to the complete disappearance of cleavages from C308 in KH3 in d10b, strongly reduced cleavage intensity in d8b (Figure 4C, lanes 5 and 8), and affected binding by the other two KH domains (Figure 4D, lane 1). Binding of KH1 was abrogated, seen by disappearance of cleavage at 734–738, whereas reduced cleavage at 747–751 indicated weakened binding of KH2. Weakening or abrogation of the IRES-binding activity of KH1 and KH2 in these experiments, seen by reduced cleavage at nt 734–738 and 747–751, respectively, and caused either directly (by mutations in GXXG loop) or indirectly (due to mutation in other domains and consequent likely loss of cooperativity in binding), is correlated with the failure of PCBP2-induced toe-prints to appear in d10 (Supplementary Figure S3B, lanes 6–8). Similarly, PCBP2 with GDDG304–307 substitutions in KH3 did not form these toe-prints, indicating that its binding was also weakened, and it did not enhance structural changes in the region upstream of the initiation codon AUG983 in the presence of eIF4G/eIF4A (Figure 1B, lanes 6–8).
The effects of the binding defects caused by the GXXG substitutions were correlated with changes in activity in supporting initiation on the IRES in the in vitro reconstitution assay. In control experiments, the C54, C118 and C308 single-cysteine substitutions led to only a minor reduction in 48S complex formation relative to wt PCBP2 (Supplementary Figure S3C, lanes 3–5). This observation is consistent with the slightly reduced IRES-binding activity of these mutants (Supplementary Figure S3A). Substitutions in the GKKG30-33 motif of KH1, which strongly reduced this domain's IRES-binding activity, and in the GRQG304-307 motif, which reduced binding of all three domains to the IRES, both led to complete loss of function in promoting initiation (Supplementary Figure S3C, lanes 6, 8, 9 and 11), whereas the analogous substitution in KH2 led to a less pronounced defect in binding to the IRES and 48S complex formation (Supplementary Figure S3C, lanes 7 and 10). The level of initiation in the presence of GKKG→GDDG30-33 and GRQG→GDDG304-307 mutant forms of PCBP2 was equivalent to that observed in the absence of PCBP2 in reactions containing ‘fractionated’ Met-tRNAiMet (Figure 1A, lane 4). These results are consistent with the finding that PCBP2 with similar substitutions in KH2 retained partial activity, but PCBP2 harboring similar substitutions in KH1 or KH3 was unable to rescue PV IRES function in PCBP2-depleted HeLa cell extracts (18).
PCBP2 enables the apical GNRA tetraloop in d10 to stimulate 48S complex formation
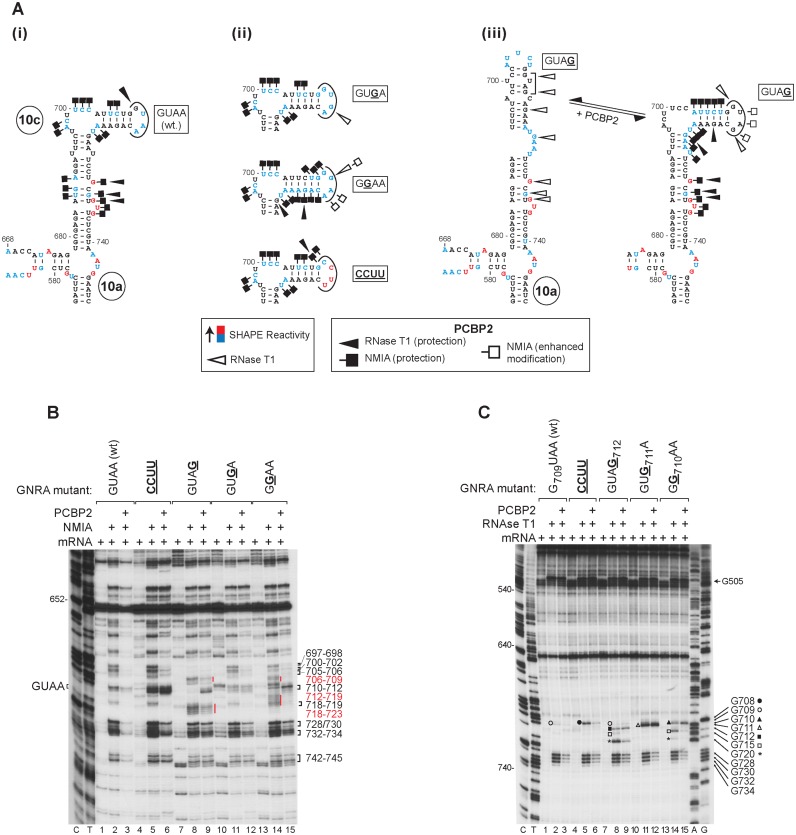
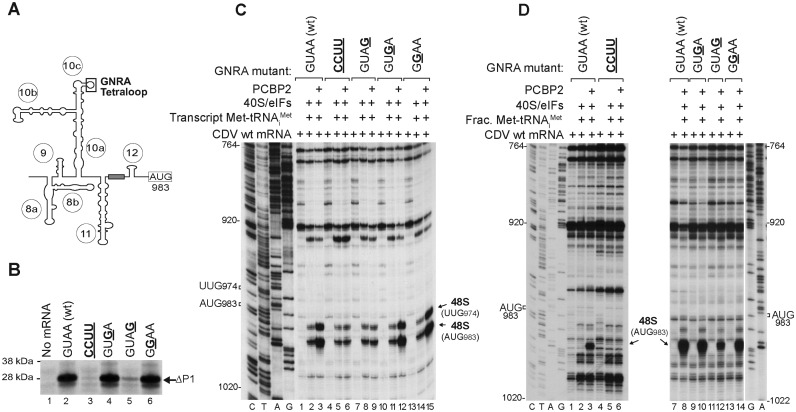
Like the apical subdomain IVc in canonical Type I IRESs, the homologous d10c in the CDV IRES contains an apical GNRA tetraloop (in which N is any nucleotide; R is A or G, and the loop is stabilized by interactions that include an unusual G-A base-pair between the first and fourth residues in the loop (49) (Figure 5A). This motif has been reported to be an important cis-acting element in Type 1 PV IRES function (5), and intriguingly, an essential GNRA tetraloop is located at a similar position (i.e. at the apex of the large central domain) in the structurally unrelated Type 2 IRESs, including EMCV and FMDV (6,7). Analysis of the tetraloop in the CDV IRES by translation of mutant mRNAs in RRL showed that GUAA→CCUU and GUAA→GUAG substitutions strongly affected IRES function, whereas substitutions of purine residues in the second and third positions in the tetraloop in the GUAA→GGAA and GUAA→GUGA mutant mRNAs had no effect (Figure 5B). This pattern of altered activity was recapitulated in in vitro reconstituted initiation reactions that included partially purified total native mammalian tRNA (Figure 5D). When ‘fractionated’ Met-tRNAiMet was replaced by ‘transcript’ Met-tRNAiMet, the level of 48S complex formation was the same on wt and tetraloop mutant mRNAs in the absence of PCBP2 (Figure 5C, lanes 2, 5, 8, 11 and 14). However, inclusion of PCBP2 enhanced 48S complex on wt, GUAA→GUGA and GUAA→GGAA mutant mRNAs, but not on GUAA→CCUU and GUAA→GUAG mutant mRNAs (Figure 5C, lanes 3, 6, 9, 12 and 15). Although ‘fractionated’ Met-tRNAiMet differs from transcript Met-tRNAiMet in containing modified nucleotides, this difference can therefore not account for the requirement for PCBP2 for the IRES to stimulate initiation on IRESs with an intact GNRA tetraloop. A difference between ‘fractionated’ and transcript Met-tRNAiMet that is likely more relevant is the presence of low molecular weight RNA contaminants in the former. Their presence results in assembly reactions that more closely resemble the cellular environment in which the IRES must compete with other RNAs for access to initiation factors. In conclusion, PCBP2 enables the IRES to exploit the function of the tetraloop to enhance 48S complex formation.
Figure 5.
Importance of the GNRA tetraloop for CDV IRES function. (A) Model of the IRES showing the GNRA tetraloop motif. (B) Translational activity of IRES-containing mRNAs with the indicated GNRA tetraloop substitutions. The ΔP1 translation product indicated on the right is a truncated form of the viral capsid polyprotein. (C, D) Toe-print analysis of 48S complex formation on these mRNAs in vitro in the presence of 40S subunits, eIFs, ‘fractionated’ native or transcript Met-tRNAiMet and/or PCBP2 as indicated.
PCBP2 stabilizes IRES d10 in a conformation in which the GNRA tetraloop is exposed
Tetraloops commonly participate in tertiary interactions in RNAs by binding with distal receptors such as helical minor grooves and asymmetric internal loops (50), but can also function as recognition sites for proteins (51). The GNRA tetraloop in the FMDV IRES engages in intramolecular RNA–RNA interactions that contribute to the structural organization and stability of the IRES (8,9). We therefore compared the structures of wt and tetraloop mutant variants of the CDV IRES. Free energy minimization (52) suggested that the apical region of d10 had the same structure in each instance, with the exception of d10c in the GUAA→GUAG mutant, which could exist in two equally stable conformations (Figure 6A(iii)). The results of NMIA modification of the wt and mutant IRESs were consistent with the predicted structures, showing that the GUAG mutant was predominantly in the variant rather than the canonical conformation (Figure 6B, lanes 2 and 8; Figure 6A(iii)), and that the loop itself in the case of GUAA→CCUU or the adjacent stem in GUAA→GGAA mutant RNAs is significantly more exposed (Figure 6B, lanes 2, 5 and 14; Figure 6A(ii)). The enhanced exposure of loop residues to chemical modification in the GUAA→CCUU mutant is consistent with the loss of interactions that stabilize GRNA tetraloops, such as the G-A base-pair between the first and fourth residues. Inclusion of PCBP2 with wt and tetraloop mutant forms of the IRES led to strong protection from NMIA modification in the pyrimidine-rich ACUUCC697–702 loop in d10c, and to nucleotides in the helix between it and the tetraloop (Figure 6B). The apical tetraloop and mutant variants thereof were thus not shielded by PCBP2, and remained solvent-accessible (Figure 6B, lanes 3 and 12) or even became more accessible to NMIA modification (Figure 6B, lanes 6, 9 and 15). Changes in the pattern of NMIA modification of the GUAG mutant indicated that binding of PCBP2 enforced its folding into the canonical conformation (Figure 6A(iii)), seen by the reduction in modification at nts. 706–708 and 718–723 and the appearance of modification at nt 710–712 (Figure 6B, lanes 8 and 9). These observations were supported by enzymatic footprinting, which showed that the predicted unpaired G residues G710, G711 and G712 in GGAA, GUGA and GUAG mutant RNAs were accessible to cleavage by RNAse T1 whether or not PCBP2 was present (Figure 6C). Other residues e.g. G708 in the CCUU mutant and G715/G720 in GUAG and GGAA mutants that were accessible to RNAse T1 in PCBP2's absence were strongly shielded in its presence. The tetraloop thus remains exposed when PCBP2 binds to the apical region of d10c, and substitutions in this motif do not impair binding of PCBP2 to the IRES. This conclusion is supported by the appearance of DHRC in the pyrimidine-rich loop (nts. 697–707) and near the GNRA tetraloop (nts. 710–716) in d10c from FeBABE-modified PCBP2-[C34], which monitors binding of KH1 to this region (Supplementary Figure S4C).
The accessibility of the tetraloop to NMIA modification in the IRES suggests that it does not function as part of a tetraloop/receptor unit that might determine the structural organization of the IRES, as has been suggested for the Type 2 FMDV IRES (9,10). Consistently, none of the tetraloop mutations led to changes in NMIA modification elsewhere in the IRES (Supplementary Figure S4A and S4B; data not shown). The alternative possibility is that the essential function of the tetraloop in the activity of the CDV IRES involves an interaction with a component of the translation apparatus, which must be limited to the 40S subunit, initiator tRNA and those initiation factors (eIFs 1, 1A, 2, 3, 4A, 4B and 4Gm) used for 48S complex formation in reactions in which the defect caused by tetraloop mutations is apparent (Figure 5D). The level of binding of 40S subunits and eIF3 to wt and the inactive GUAA→CCUU mutant form of the IRES did not differ (Figure 4D).
Mutational analysis of structural and cis-acting elements in d10 of the IRES
We analyzed aspects of the function of a panel of mutants (Figure 7A) to test the predicted structural model of the IRES, to validate PCBP2 binding sites, and to determine the importance of these and other sequence motifs for IRES function.
Figure 7.
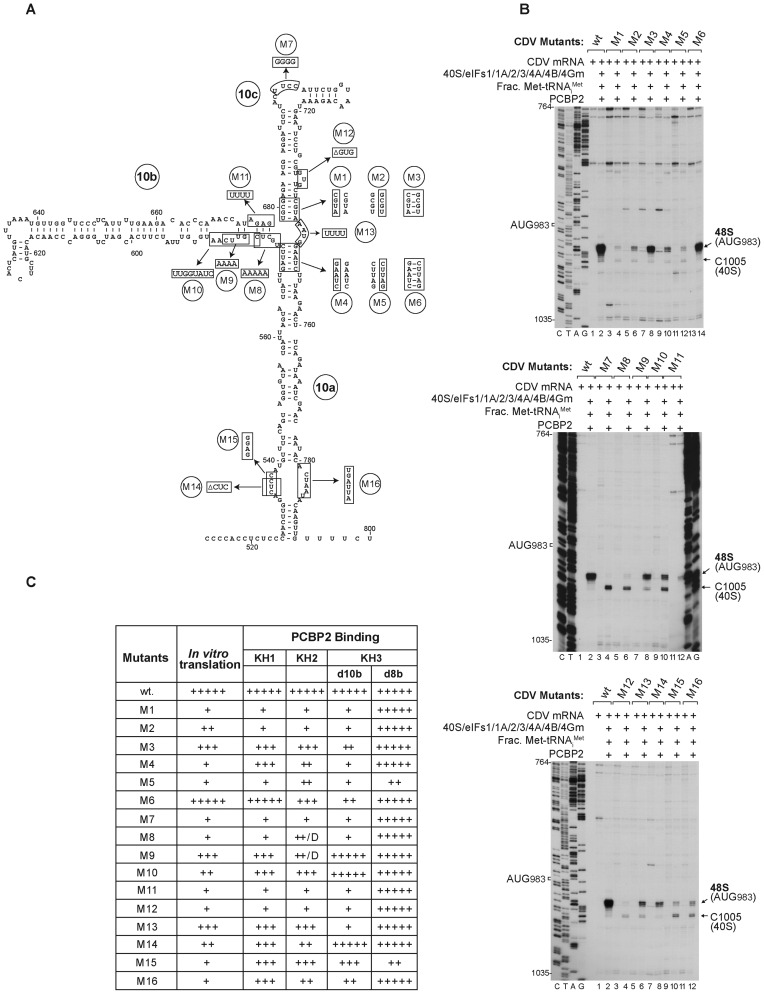
Mutational analysis of sequence and structural elements in IRES Domain 10. (A) Secondary structure model of IRES domain 10 showing mutations (i) in apical and basal stems of domains 10a and 10c (M1-M6 respectively), (ii) in the subapical pyrimidine-rich bulge of domain 10c (M7), (iii) at the base of domain 10b (M8–M11) and (iv) in the internal loops and bulges of domain 10a and 10c (M12–M16). (B) Toe-print analysis of 48S complex formation on wt and mutant CDV mRNAs (as indicated) in vitro in the presence of 40S subunits, eIFs, ‘fractionated’ native Met-tRNAiMet and PCBP2. Toe-prints corresponding to 48S complexes assembled at AUG983 and caused by binding of 40S subunits are indicated on the right. (C) Summary of the efficiency of translation of wt and M1–M16 mutant CDV mRNAs and of the interaction of mutant IRESs with individual KH domains of bound PCBP2 (D = defective/aberrant binding).
Substitutions that destabilized helices flanking the junction of d10a and d10c strongly reduced (mutant M2) or abrogated IRES-mediated translation in RRL (mutants M1, M4 and M5), but IRES function was partially or wholly restored by the compensatory substitutions in mutants M3 and M6 that re-established base-pairing (Supplementary Figure S5A, upper panel). Consistently, 48S complex formation in in vitro reconstituted assembly reactions was strongly impaired by the destabilizing substitutions in mutants M1, M2, M4 and M5, but was partially or wholly restored by combining these substitutions to re-establish base-pairing in M3 and M6 (Figure 7B, top panel). The incomplete restoration of activity of the M3 mutant may be because substitutions in it overlap a conserved sequence element in Type 1 IRES (25), but may simply reflect folding of a proportion of IRESs with substitutions at two locations into an alternative, inactive conformation. Monitoring of DHRC by FeBABE-modified PCBP2-C54 showed that destabilization of the basal helix of d10c (mutants M1 and M2) and the apical helix of d10a (mutant M5) abrogated binding of PCBP2's KH1 domain, and that re-establishment of base-pairing (M3 and M6) restored binding (Supplementary Figure S5D). Interestingly, destabilization of the apex of d10a in M4 did not affect binding of KH1. Cleavage from C118 in KH2 and C308 in KH3 were completely abrogated (M1 and M2) or reduced (M4 and M5) by destabilizing mutations; compensatory substitutions (M3 and M6) lead to its partial restoration, particularly at nts. 666–672 and 747–751 from PCBP2-C118 and at nt 666–672 from PCBP2-C308 (Supplementary Figure S5G). Taken together, these results strongly support the existence of the proposed three-way helical junction in d10.
Substitutions in regions of the IRES that footprinting (Figure 1C) and DHRC analysis (Figure 2A) had implicated as likely binding sites for PCBP2's KH1 and KH2 domains led to strong or almost complete loss of function (mutants M9 and M10, and M7, M8 and M11, respectively) (Supplementary Figure S5A, center panel). Analogous defects were apparent in in vitro reconstituted 48S complex formation reactions (Figure 7B, center panel). Consistently, binding of all three KH domains was almost completely abolished by M7, M8 and M11 substitutions (Supplementary Figure S5E) whereas binding of KH1 and KH2 was moderately reduced by M9 and M10 substitutions and binding of KH3 was unaffected. Mutations in the base of d10b led to the appearance of aberrant cleavage at nts 743–746 in mutant M8, and at nts. 580–584 and nt 602 in mutant M9 from C118 (Supplementary Figure S5H). The correlation between defects in binding of PCBP2 to d10 in mutant IRESs and in their ability to support internal initiation is wholly consistent with the importance of PCBP2 for IRES function. Notably, cleavage was observed in d8 even in the case of inactive IRES mutants, indicating that PCBP2's interaction with d10 is of primary importance for the function of the IRES.
Substitution or deletion of nucleotides in internal loops in d10c (M12), at the d10a/d10c junction (M13) and in d10a (M14, M15 and M16) all strongly impaired or even abrogated IRES function assayed by in vitro translation (Supplementary Figure S5A, bottom panel) and consistently, in in vitro reconstituted initiation reactions (Figure 7B, bottom panel). The substitutions in M12 strongly impaired binding of PCBP2, whereas those in M13, M14, M15 and M16 had a much weaker effect (Figure 7C, Supplementary Figure S5F, S5I). These mutations were therefore responsible for impairing 48S complex formation at a stage other than binding of PCBP2 to the IRES. Notably, mutations at the apex of d10c (M7 and M12) and at the base of d10b (M8) led to the appearance of a strong toe-print at nt 1005 in in vitro reconstitution assays (Figure 7B) that systematic factor omission experiments identified as being caused by direct binding of a 40S subunit to the IRES (Supplementary Figure S5B). This toe-print was as prominent as that of the 48S complex assembled on mutants M10, M15 and M16. A weak toe-print at this position caused by binding of the 40S subunit that had not previously been noted (36) was also apparent on the wt IRES (Supplementary Figure S5C) and the other mutants (Figure 7B).
DISCUSSION
Specificity of binding of PCBP2 to the CDV IRES
PCBP2 is required for initiation on canonical Type 1 IRESs and on the divergent Type 1 IRES located in the CDV 5′UTR. Here, we determined that PCBP2 binds specifically to the CDV IRES and mapped the principal binding site to the apical region of d10. The affinity of PCBP2 for a fragment of the CDV IRES encompassing d10 (Kd = 36 nM) is comparable to that for PV dIV (Kd ∼ 15 nM) and CVB3 dIV (Kd ∼ 35 nM) (3,4). The concentration of PCBP2 in the cytoplasm of HeLa cells (∼100 nM) (17) is such that PCBP2 would saturate its binding site on these IRESs.
The proposed structure of CDV d10, initially based on free energy minimization and the pattern of sequence covariation in CDV isolates (36), has been validated here by the results of chemical and enzymatic probing, directed hydroxyl radical probing and mutational analyses. With the exception of the apical d10c region, the proposed structure of d10 is very different from that of e.g. PV dIV, a consequence of the multiple recombination events that have shaped the CDV genome (35). The question of how PCBP2 engages functionally with structurally divergent IRESs is of interest because of the emerging appreciation of the importance of recombination in the acquisition of IRESs by viral genomes and in the evolution of their structural diversity (e.g. 27–34,53). PCBP2 binds specifically via its KH1 domain to an exposed internal C-rich loop in the apical subdomain IVc of Type 1 IRESs (3,4,13,24) and to the CDV IRES by interaction with the closely related d10c, protecting an analogous pyrimidine-rich loop from NMIA modification (Figure 1C). This site of interaction is critical for the function of the CDV IRES (this report) and for canonical Type 1 IRESs (3,4). Correlation of the PCBP2 footprint, the sites of DHRC from C141 and particularly from C118, and the effects of substitutions in the CDV IRES indicates that the KH2 domain binds to d10b near the three-way junction with d10a and d10c, a location that is comparable to the binding of KH2 at the four-way junction in canonical Type 1 IRESs (13). Although KH3 has a high degree of mobility in solution (21), it is in close proximity to KH2 in CDV IRES-bound PCBP2, as also noted for PCBP2 bound to PV and EV71 IRESs (13). However, whereas the KH3 binding site on these and other canonical Type 1 IRESs is centered on the pyrimidine-rich loop motif at the apex of domain IVb (3,4,13,24), the KH3 binding site on the CDV IRES is located centrally in d10b. Moreover, the orientation of KH3 relative to KH2 is flipped, so that when PCBP2 is bound to e.g. the PV IRES, C330 in KH3 is closer than C308 to C118 in KH2, whereas the positions of C308 and C330 relative to C118 are reversed in PCBP2 bound to the CDV IRES. It remains to be determined whether these adaptations to the distinct structure of the region of the CDV IRES to which KH2 and KH3 bind have specific functional consequences, or whether binding of e.g. KH3 to unrelated sites on canonical and divergent Type 1 IRESs serves simply to anchor PCBP2 on the IRES and to promote cooperative binding of the other domains. This function is suggested by the observation that substitutions in the GXXG motif of KH3 that abrogates its binding to the IRES led to severe defects in binding of KH1 and KH2 domains.
Interestingly, similar substitutions in KH1 and KH2 domains weakened but did not abrogate binding of the domain in question and had little or no effect on binding of the other two domains. Whereas KH3 may bind independently to the IRES, KH1 and KH2 could bind as a preformed unit that is stabilized by a large hydrophobic interface between domains (21), and this could lead to masking of a binding defect in either KH1 or KH2. Binding of a preformed KH1/KH2 unit with a defective GXXG motif in one constituent KH domain to the IRES would deliver that domain to the correct location, where its binding might be stabilized by interactions with the KH domain outside the hydrophobic cleft bordered by GXXG and variable loops. Notably, substitutions outside the GXXG motif impair binding of PCBP2 to PV dIV and reduce PCBP2's ability to promote initiation on the PV IRES (18). Moreover, foot-printing data for the CDV IRES/PCBP2 complex (this report) and for complexes formed by binding of PCBP2 to PV and CVB3 IRESs (3,4) show that PCBP2 protects extensive regions of these IRESs outside the tetranucleotide pyrimidine-rich motif that binds to the hydrophobic cleft.
PCBP2's role in enabling the IRES to utilize the function of an apical GNRA tetraloop to enhance initiation
In light of the major structural differences between CDV and canonical Type 1 IRESs, retention of the conserved d10c (equivalent to dIVc) suggests that it is functionally important. A key element in it is the internal pyrimidine-rich loop that is bound by KH1 of PCBP2, but a second motif of interest is the apical GNRA tetraloop. Although experimental data have not been presented, this element has been described as being critical for PV IRES function (5). Although present in a different structural context, a GNRA tetraloop is located at a similar position in Type 2 IRESs, such as those of EMCV and FMDV, and is essential for their function (6,7). Substitutions of nucleotides at the first and fourth positions strongly impaired CDV IRES function whereas purine substitutions at the second and third positions had no effect, consistent with sequence requirements for the function of this motif in Type 2 IRESs. Nevertheless, the function of the tetraloop in the CDV IRES, and possibly other Type 1 IRESs, may differ from its role in Type 2 IRESs: observations that inactivating tetraloop mutations are responsible for structural disruption of the FMDV IRES have led to proposals that its tetraloop is responsible for enforcing a specific tertiary structure (8,9). By contrast, no analogous structural disruption was detected as a consequence of introducing such mutations into the CDV IRES. Although GNRA tetraloops are primarily implicated in the establishment of tertiary interactions in complex RNAs (50), they can also function as recognition sites for proteins (51), and this possibility must be entertained as a possibility in the present case.
Significantly, mutant forms of the CDV IRES, even with substitutions in the tetraloop that abrogated IRES function in in vitro translation, were nevertheless as active as the wt IRES in in vitro reconstitution experiments in which ‘fractionated’ Met-tRNAiMet had been replaced by in vitro transcribed Met-tRNAiMet and PCBP2 had been omitted. Inclusion of PCBP2 enhanced initiation on the wt IRES and on variants with GGAA and GUGA tetraloops that conformed to the GNRA consensus sequence but not on mutants with sequences that diverged from this consensus. Various potential non-exclusive functions for PCBP2 and other ITAFs in promoting IRES function have been suggested, including interaction with and recruitment of initiation factors or other ITAFs to the IRES, recruitment of a 40S ribosomal subunit via simultaneous interaction with it and the IRES and (stabilization of IRES structure in an active conformation (25). We found no evidence for direct recruitment or stabilization of binding of 40S subunits to the IRES by PCBP2 (36), and although PCBP2 binds SRp20 and recruits it to the PV IRES, and this has been implicated in trans-activation of its function (26), this role can be excluded in the present case, because PCBP2 was required in the absence of SRp20. Instead, we determined that PCBP2 enables the IRES to utilize a cis-acting RNA element, namely the GNRA tetraloop, to enhance IRES function, and that it appears to execute this function by enforcing a specific orientation on this element. Thus, PCBP2 can suppress structural defects in the relevant region of the CDV IRES e.g. the tendency to misfolding caused by the GUAA→GUAG tetraloop substitution, suggesting that its multiple interactions with elements of d10 that support the apical tetraloop and associated helix promote adoption of a specific conformation on this element. In this model, the extensive interactions of PCBP2's KH1 domain with d10c would clearly be critical for its function and would account for the strict requirement for this domain in initiation on Type 1 IRESs (17,18).
The key questions raised by this aspect of the present study are the identity of the binding target for the tetraloop of Type 1 IRESs, and how its interaction with the tetraloop influences the initiation process. As noted above, candidates are limited to those constituents of in vitro reconstituted reactions in which the requirement for integrity of the tetraloop is manifested (Figure 5C). A possibility is that the PCBP2-dependent function of the tetraloop is connected to the process of eIF4G/eIF4A-mediated preparation of a binding site for the 43S complex. This hypothesis is suggested by observations that PCBP2 influences eIF4G/eIF4A-mediated conformational changes immediately upstream of the initiation codon (Figure 1B) and that despite possessing a typical Type 1 dIV-like structure, the hybrid IRESs of Aichi virus and several other picornaviruses are not dependent of the integrity of the GNRA tetraloop and do not require PCBP2 for initiation (29,54). These IRESs have an eIF4G/eIF4A binding site that is typical of Type 2 rather than Type 1 IRESs, and attachment of 43S complexes to the initiation codon, which is sequestered in a stable hairpin at the 3′-border of the IRES, is additionally dependent on DHX29.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [AI51340 and AI123406 to C.H., GM59660 to T.P.]. Funding for open access charge: NIH/NIAID [AI51340].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niepmann M. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta. 2009;1789:529–541. doi: 10.1016/j.bbagrm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Gamarnik A.V., Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sean P., Nguyen J.H., Semler B.L. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology. 2009;389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaminski A., Hunt S.L., Gibbs C.L., Jackson R.J. Internal Initiation of mRNA translation in eukaryote. Genet. Eng. 1994;16:115–155. [PubMed] [Google Scholar]
- 6.López de Quinto S., Martínez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson M.E., Seamons R.A., Belsham G.J. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Miragall O., Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Miragall O., Ramos R., Ramajo J., Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamphear B.J., Kirchweger R., Skern T., Rhoads R.E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 11.Gradi A., Svitkin Y.V., Imataka H., Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Breyne S., Yu Y., Unbehaun A., Pestova T.V., Hellen C.U. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sweeney T.R., Abaeva I.S., Pestova T.V., Hellen C.U. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014;33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blyn L.B., Chen R., Semler B.L., Ehrenfeld E. Host cell proteins binding to domain IV of the 5′ noncoding region of poliovirus RNA. J. Virol. 1995;69:4381–4389. doi: 10.1128/jvi.69.7.4381-4389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blyn L.B., Towner J.S., Semler B.L., Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik A.V., Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 17.Silvera D., Gamarnik A.V., Andino R. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 1999;274:38163–38170. doi: 10.1074/jbc.274.53.38163. [DOI] [PubMed] [Google Scholar]
- 18.Walter B.L., Parsley T.B., Ehrenfeld E., Semler B.L. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002;76:12008–12022. doi: 10.1128/JVI.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology. 2015;479-480:457–474. doi: 10.1016/j.virol.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenn S., Du Z., Lee J.K., Tjhen R., Stroud R.M., James T.L. Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution. Nucleic Acids Res. 2007;35:2651–2660. doi: 10.1093/nar/gkm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Z., Fenn S., Tjhen R., James T.L. Structure of a construct of a human poly(C)- binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J. Biol. Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valverde R., Edwards L., Regan L. Structure and Function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 23.Nicastro G., Taylor I.A., Ramos A. KH-RNA interactions: back in the groove. Curr. Opin. Struct. Biol. 2015;30:63–70. doi: 10.1016/j.sbi.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Zell R, Ihle Y., Effenberger M., Seitz S., Wutzler P., Görlach M. Interaction of poly(rC)-binding protein 2 domains KH1 and KH3 with coxsackievirus RNA. Biochem. Biophys. Res. Commun. 2008;377:500–503. doi: 10.1016/j.bbrc.2008.09.156. [DOI] [PubMed] [Google Scholar]
- 25.Jackson R.J., Howell M.T., Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 26.Bedard K.M., Daijogo S., Semler B.L. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellen C.U., de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuter G., Pankovics Knowles P. N.J., Boros Á. Two closely related novel picornaviruses in cattle and sheep in Hungary from 2008 to 2009, proposed as members of a new genus in the family Picornaviridae. J. Virol. 2012;86:13295–13302. doi: 10.1128/JVI.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweeney T.R., Dhote V., Yu Y., Hellen C.U. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J. Virol. 2012;86:1468–1486. doi: 10.1128/JVI.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boros A., Pankovics P., Reuter G. Avian picornaviruses: molecular evolution, genome diversity and unusual genome features of a rapidly expanding group of viruses in birds. Infect. Genet. Evol. 2014;28:151–166. doi: 10.1016/j.meegid.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 31.Boros Á., Pankovics P., Simmonds P., Pollák E., Mátics R., Phan T.G., Delwart E., Reuter G. Genome analysis of a novel, highly divergent picornavirus from common kestrel (Falco tinnunculus): the first non-enteroviral picornavirus with type-I-like IRES. Infect. Genet. Evol. 2015;32:425–431. doi: 10.1016/j.meegid.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Asnani M., Kumar P., Hellen C.U. Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology. 2015;478:61–74. doi: 10.1016/j.virol.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng T.F., Sachsenröder J., Reuter G., Knowles N.J., Delwart E., Johne R. Rabovirus: a proposed new picornavirus genus that is phylogenetically basal to enteroviruses and sapeloviruses. Arch. Virol. 2015;160:2569–2575. doi: 10.1007/s00705-015-2523-y. [DOI] [PubMed] [Google Scholar]
- 34.Drexler J.F., Corman V.M., Lukashev A.N., van den Brand Gmyl J.M. A.P., Brünink S., Rasche A., Seggewiβ N., Feng H., Leijten L.M., Hepatovirus Ecology Consortium et al. Evolutionary origins of hepatitis A virus in small mammals. Proc. Natl. Acad. Sci. U.S.A. 2015;112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo P.C., Lau S.K., Choi G.K., Huang Y., Teng J.L., Tsoi H.W., Tse H., Yeung M.L., Chan K.H., Jin D.Y., et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 2012;86:2797–2808. doi: 10.1128/JVI.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asnani M., Pestova T.V., Hellen C.U. Initiation on the divergent Type I cadicivirus IRES: factor requirements and interactions with the translation apparatus. Nucleic Acids Res. 2016;44:3390–3407. doi: 10.1093/nar/gkw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova T.V., Borukhov S.I., Hellen C.U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 38.Pestova T.V., Hellen C.U., Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolupaeva V.G., Lomakin I.B., Pestova T.V., Hellen C.U. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomakin I.B., Shirokikh N.E., Yusupov M.M., Hellen C.U., Pestova T.V. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova T.V., Hellen C.U., Wimmer E. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology. 1994;204:729–737. doi: 10.1006/viro.1994.1588. [DOI] [PubMed] [Google Scholar]
- 42.Pisarev A.V., Unbehaun A., Hellen C.U., Pestova T.V. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 43.Pestova T.V., Hellen C.U. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolupaeva V.G., de Breyne S., Pestova T.V., Hellen C.U. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007;430:409–439. doi: 10.1016/S0076-6879(07)30016-5. [DOI] [PubMed] [Google Scholar]
- 45.Joseph S., Weiser B., Noller H.F. Mapping the inside of the ribosome with an RNA helical ruler. Science. 1997;278:1093–1098. doi: 10.1126/science.278.5340.1093. [DOI] [PubMed] [Google Scholar]
- 46.Paziewska A., Wyrwicz L.S., Bujnicki J.M., Bomsztyk K., Ostrowski J. Cooperative binding of the hnRNP K three KH domains to mRNA targets. FEBS Lett. 2004;577:134–140. doi: 10.1016/j.febslet.2004.08.086. [DOI] [PubMed] [Google Scholar]
- 47.Barnes M., van Rensburg G., Li W.M., Mehmood K., Mackedenski S., Chan C.M., King D.T., Miller A.L., Lee C.H. Molecular insights into the coding region determinant- binding protein-RNA interaction through site-directed mutagenesis in the heterogeneous nuclear ribonucleoprotein-K-homology domains. J. Biol. Chem. 2015;290:625–639. doi: 10.1074/jbc.M114.614735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hollingworth D., Candel A.M., Nicastro G., Martin S.R., Briata P., Gherzi R., Ramos A. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012;40:6873–6886. doi: 10.1093/nar/gks368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heus H.A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 50.Fiore J.L., Nesbitt D.J. An RNA folding motif: GNRA tetraloop-receptor interactions. Q. Rev. Biophys. 2013;46:223–264. doi: 10.1017/S0033583513000048. [DOI] [PubMed] [Google Scholar]
- 51.Thapar R., Denmon A.P., Nikonowicz E.P. Recognition modes of RNA tetraloops and tetraloops-like motifs by RNA binding proteins. Wiley Interdiscip. Rev. RNA. 2014;5:49–67. doi: 10.1002/wrna.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapoor A., Kumar A., Simmonds P., Bhuva N., Singh Chauhan L., Lee B., Sall A.A., Jin Z., Morse S.S., Shaz B., et al. Virome Analysis of Transfusion Recipients Reveals a Novel Human Virus That Shares Genomic Features with Hepaciviruses and Pegiviruses. Mbio. 2015;6:e01466–15. doi: 10.1128/mBio.01466-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y., Sweeney T.R., Kafasla P., Jackson R.J., Pestova T.V., Hellen C.U. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.