Abstract
Two-component systems (TCS) and small regulatory RNAs (sRNAs) are both widespread regulators of gene expression in bacteria. TCS are in most cases transcriptional regulators. A large class of sRNAs act as post-transcriptional regulators of gene expression that modulate the translation and/or stability of target-mRNAs. Many connections have been recently unraveled between these two types of regulators, resulting in mixed regulatory circuits with poorly characterized properties. This study focuses on the negative feedback circuit that exists between the EnvZ-OmpR TCS and the OmrA/B sRNAs. We have shown that OmpR directly activates transcription from the omrA and omrB promoters, allowing production of OmrA/B sRNAs that target multiple mRNAs, including the ompR-envZ mRNA. This control of ompR-envZ by the Omr sRNAs does not affect the amount of phosphorylated OmpR, i.e. the presumably active form of the regulator. Accordingly, expression of robust OmpR targets, such as the ompC or ompF porin genes, is not affected by OmrA/B. However, we find that several OmpR targets, including OmrA/B themselves, are sensitive to changing total OmpR levels. As a result, OmrA/B limit their own synthesis. These findings unravel an additional layer of control in the expression of some OmpR targets and suggest the existence of differential regulation within the OmpR regulon.
INTRODUCTION
Regulation of gene expression plays a crucial role in the rapid adaptation of bacteria to their environment, a process that allows them to colonize and survive in various media. This control can take place at all stages of gene expression and via a great diversity of precise molecular mechanisms.
For instance, examples of transcriptional control have been reported at the level of transcription initiation, elongation or termination, and the regulators can be DNA- or RNA-binding proteins, but also RNA molecules. Among the most widely used regulators of transcription in bacteria are the two-component systems (TCS), which are typically composed of a sensor protein and its cognate response regulator (1). In response to specific stimuli, the sensor controls the phosphorylation status of the regulator by acting as a kinase and/or as a phosphatase. In the phosphorylated state, the regulator typically controls transcription of multiple genes. More than 30 TCS have been identified in the Escherichia coli genome and allow both sensing and adaptation to the environment, in response to extremely diverse input signals, such as phosphate limitation, low magnesium, pH or osmolarity.
EnvZ-OmpR is one of the most extensively studied TCS (2). EnvZ autophosphorylates upon sensing cognate stimuli and controls the level of phosphorylated OmpR (OmpR-P). Even though the precise signal detected by EnvZ is still unclear, it is known that signals such as increasing osmolarity, temperature or acid pH (among others) induce the activity of the EnvZ-OmpR TCS (3). OmpR directly controls the transcription of several genes, in particular those encoding the major porins in E. coli, OmpC and OmpF. While ompC expression is activated in conditions of high osmolarity (high OmpR-P), OmpR represses or activates ompF expression in high and low osmolarity, respectively (4 and references therein). In addition to these two genes, OmpR also directly controls transcription of other genes such as dtpA (a.k.a. tppB) (5), flhDC (6), csgD (7) and bolA (8), that respectively encode an oligopeptide transporter, the regulator for flagella biogenesis, the activator of curli production and a transcriptional regulator also involved in controlling the switch from a planktonic to a sessile lifestyle (9). Note that regulation of dtpA by EnvZ-OmpR was investigated in detail; interestingly, it is different from regulation of both ompC and ompF in that dtpA transcription does not seem to respond to changes in osmolarity (5). Furthermore, OmpR was found to bind to several dozen sites in both E. coli and Salmonella in a recent ChIP-on-chip analysis, indicating the existence of more direct targets (10). Although the OmpR regulons were found to differ in the two species, with only a few common targets, OmpR controls genes involved in pH homeostasis and acid stress response in both E. coli and Salmonella. In agreement with this, OmpR appears to play an important role in acid resistance (11). At least 4 small RNAs (sRNAs) are also known to be controlled by EnvZ-OmpR: MicC, MicF, OmrA and OmrB (12–14) (see below).
Regulatory RNAs have emerged as major regulators of gene expression in recent years, in virtually all organisms. In bacteria, they are typically rather short (a few hundred nts or less) and are therefore referred to as small RNAs (sRNAs). So far, the best characterized class corresponds to sRNAs that can base-pair with target-mRNA(s) encoded by another locus of the genome and, as a result, regulate either positively or negatively, their translation and/or stability (15). This base-pairing is in most cases mediated by short and imperfect sRNA-mRNA duplexes and in several bacterial species, such as E. coli for instance, requires the action of an hexameric RNA chaperone called Hfq (16,17).
Base-pairing sRNAs are involved in the regulation of diverse cellular functions as crucial as iron homeostasis or quorum-sensing. Interestingly, many sRNAs target genes that encode outer membrane proteins (OMP) or are involved in synthesis of membrane appendages such as curli or flagella (14,18–22); this is true in particular for the various OmpR-dependent sRNAs. MicC and MicF were, for instance, shown to repress expression of ompC and ompF porin genes, respectively (13,23). In addition, MicC also represses the synthesis of OmpD, another abundant porin in Salmonella (24). OmrA/B are involved in membrane remodeling as well, since they negatively regulate the synthesis of several OMPs, such as the OmpT protease and the CirA, FecA and FepA receptors for iron-siderophore complexes (25). They also repress the synthesis of several transcriptional regulators such as CsgD (18) and FlhD2C2 (21), the key regulators for curli formation and flagella biogenesis respectively, as well as the EnvZ-OmpR TCS, their own transcriptional activator (26).
sRNAs play major roles in shaping gene expression in bacteria. They are involved in different regulatory circuits, and they most often participate in mixed regulatory networks that combine transcriptional and post-transcriptional controls, mediated by proteins and sRNAs respectively (27,28). While the occurrence of sRNAs in these circuits is clearly recognized, the properties they can provide are still poorly understood in most cases. sRNAs were in particular reported in feedforward motifs (FFM), where a regulator controls expression of a target gene both directly and indirectly via control of another regulator. The Spot 42 sRNA participates, for instance, in a multioutput coherent FFM with the global regulator CRP to repress expression of multiple genes involved in the use of diverse non-preferred carbon sources. Interestingly, this mixed FFM allowed an accelerated response following circuit deactivation, a dynamic property that had not been observed before with purely transcriptional FFM, suggesting a unique role for sRNAs in these motifs (29). FFM can also involve positively acting sRNAs: RprA sRNA is an activator of the synthesis of RicI, a membrane protein that inhibits conjugation, both by direct base-pairing with the ricI mRNA and via positive control of RpoS. Because the resulting mixed coherent FFM operates with an AND-logic (i.e. both RprA and RpoS are required for ricI activation), it ensures that synthesis of RicI, and hereby conjugation inhibition, occurs under sustained activation of RprA only, with a delay in comparison to the synthesis of RpoS (30).
Another recurrent motif involving sRNAs and regulatory proteins is the feedback circuit where one regulator controls and is controlled by a second regulator. This study focuses on one example of such a feedback: the direct repression of ompR-envZ expression by OmrA/B, whose transcription is activated by this TCS. Our findings reveal an exquisite elaboration in the way in which the well-studied EnvZ-OmpR TCS works, which allows for autocontrol of OmrA/B.
MATERIALS AND METHODS
Strains and plasmids, general microbiology techniques
Strains used in this study are listed in Supplementary Table S1 and their construction is described in supplementary material. Briefly, mutant alleles were either obtained from existing sources or engineered by lambda Red recombination, and moved by transduction when required. Strains were grown in LB, and when necessary, antibiotics were used at the following concentrations: 150 μg/ml ampicillin, 25 μg/ml kanamycin, 10 μg/ml chloramphenicol and 10 μg/ml tetracycline. When required, expression of various sRNAs from pBRplac derivatives was induced using 100 μM IPTG. The Phusion DNA polymerase was used for cloning and amplification of DNA fragments for recombineering.
OmpR-His purification and in vitro phosphorylation
Protein OmpR was expressed and purified from strain BL21(DE3) transformed with a pET15b plasmid carrying the ompR gene with a 6His tag at its N-terminus under control of a T7 promoter (31). Purification was mostly as described previously. Briefly, cells were diluted 250-fold from an overnight culture into fresh LB-Ampicillin and grown to exponential phase. When the optical density at 600 nm reached 0.6, synthesis of OmpR-His was induced with addition of 1 mM IPTG for 3 h. Cells were then centrifuged and, after freezing, pellets were resuspended in A buffer (20 mM sodium phosphate buffer pH 7.44, 0.5 M NaCl, 10 mM imidazole), and protease inhibitors (Complete mix, Roche) and 1mg/ml lysozyme were added. After sonication and clarification by centrifugation, the supernatant was loaded onto a column of Ni-NTA agarose resin (Macherey-Nagel) equilibrated in A buffer. After washing, OmpR-His was eluted with increasing concentrations of imidazole. Fractions containing sufficiently pure OmpR-His were then pooled and dialyzed against TEGD buffer (20 mM Tris–HCl pH 7.6, 0, EDTA 0.1 mM, glycerol 5%, NaCl 0.4M, DTT 0.1mM).
In vitro phosphorylation was performed prior to each transcription reaction. For this purpose, 10 μM OmpR-His was incubated in phosphorylation buffer (50 mM Tris–HCl pH 7.6, 20 mM MgCl2, 50 mM KCl) in presence of 25 mM acetylphosphate (Sigma reference 01409) at 37°C for 2 h with moderate shaking. Phosphorylation was systematically checked by Phos-Tag electrophoresis and found to be reproducibly around 50%.
In vitro transcription
In vitro transcription reactions were performed according to (32) and (33) using plasmid DNA as templates. 50 ng template DNA was incubated at 37°C for 10 min in the reaction buffer (40 mM HEPES pH 7.4, 60 mM potassium glutamate, 2 mM MgCl2, 0.05% Nonidet P-40) in presence of NTPs (final concentration of ATP, CTP and GTP is 0.2 mM, 10 nM UTP, 2.5 μCi [α-32P] UTP) and 10 mg/ml bovine serum albumin (BSA). A constant volume of phosphorylated or unphosphorylated OmpR-His (or buffer for the control) was added at the required concentration and incubation was continued for 10 min. 1 μl of E. coli RNA polymerase (Epicentre) at 0.5 units/μl was then added and transcription was allowed to proceed for 20 min at 37°C; it was then stopped by addition of 1 volume of stop solution (95% formamide, 25 mM EDTA, 0.05% xylene cyanol and bromophenol blue). Transcription products were analyzed on a 6% sequencing gel next to 5′ labeled pBR322/MspI ladder. Quantification was performed with ImageQuant software and used RNAI transcript as a reference to normalize the different reactions.
Phos-Tag electrophoresis
Protein extraction and gel electrophoresis for separation of phosphorylated and unphosphorylated OmpR was mostly as described previously (34) with the following modifications. Briefly, cells from 5 ml of culture were collected by centrifugation and resuspended in Bugbuster detergent mix (Merck) at a concentration equivalent to 40 OD600/ml. 1/4 volume of SDS loading buffer 5× (250 mM Tris–HCl pH 6.8, 8% sodium dodecyl sulfate (SDS), 25% glycerol, 572 mM β-mercaptoethanol, 0.10% bromophenol blue) was then added. After a new centrifugation, supernatant was collected and 20 μl were loaded onto a Phos-Tag containing gel. Gel composition and electrophoresis was as in (34). After migration, gel was equilibrated in 1 mM ethylenediaminetetraacetic acid (EDTA) to chelate Mn2+ ions, and transfer was then as for standard protein gels (see below).
Western blot
After a 500-fold dilution from an overnight culture into fresh medium, cells were grown to exponential phase, collected by centrifugation and pellets were resuspended in SDS-loading buffer containing DTT (Biolabs). A volume equivalent to 0.15 A600 was then loaded on a 10% SDS-PAGE gel and after separation, proteins were transferred onto an Hybond C-super membrane (GE Healthcare). Membrane was blocked for 1 h prior to overnight incubation with the anti-OmpR antibody diluted 1500-fold in phosphate-buffered saline (PBS)–0.05% Tween 20. After washing and incubation with the secondary antibody, detection was performed using the Covalight chemiluminescent reagent kit (Covalab).
β-Galactosidase activity
Cells were diluted 500-fold into fresh medium from an overnight culture and grown to an optical density of 0.4 at 600 nm. The β-galactosidase activity was then measured following Miller's protocol (35) and results presented here correspond to the average of at least two independent experiments (except for Figure 4C where a representative experiment is shown, with triplicates presented in Figure S6). For the results that are shown as ratios, activities are given in Supplementary Table S3.
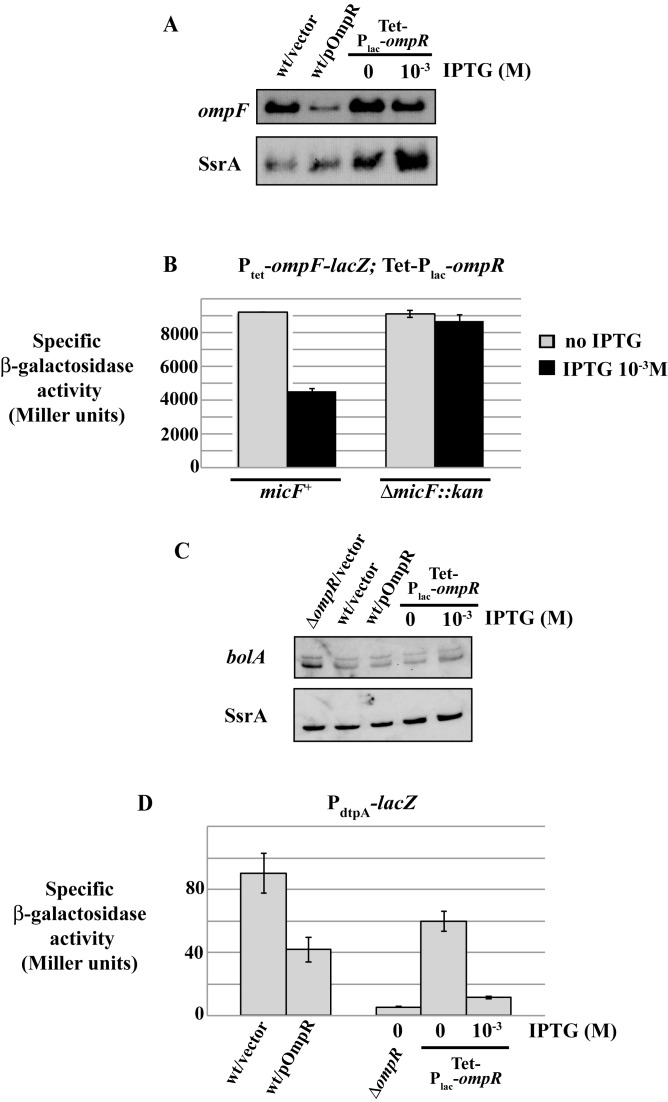
Figure 4.
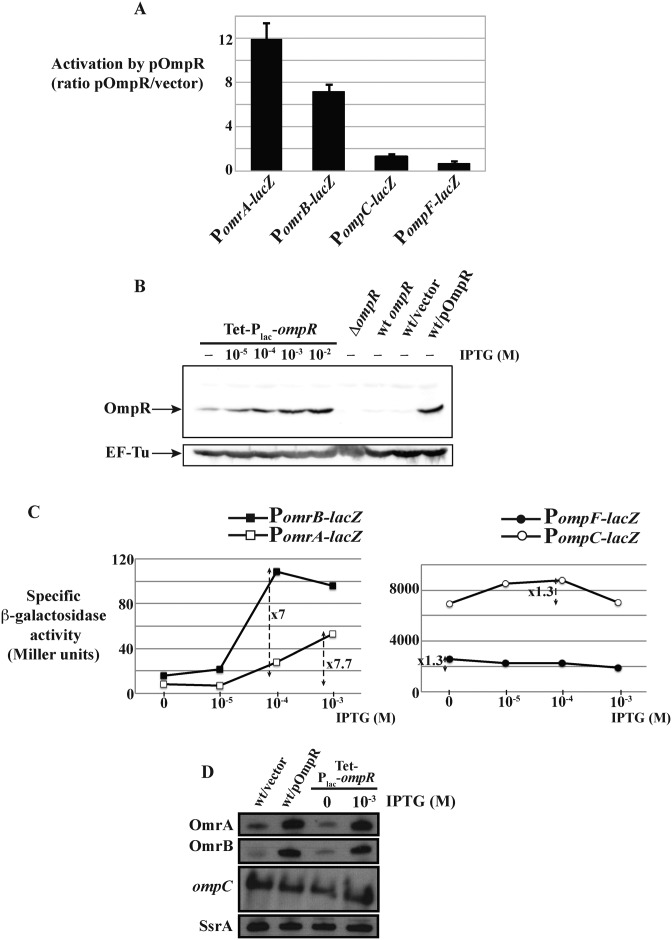
Different patterns of robustness in the OmpR regulon. (A) The activation of OmrA, OmrB, ompC or ompF promoter fusions (strains MG1004, MG1005, MG1892 and MG1690 respectively) was calculated as the ratio between the β-galactosidase activities of strains transformed by pOmpR and the pHDB3 empty vector respectively. Activities in presence of pHDB3 were, in Miller units, 33 (PomrA), 87 (PomrB), 7313 (PompC) and 3510 (PompF). (B) Western blot analysis of OmpR levels from cells carrying ompR-envZ operon expressed from its own promoter (wt ompR, strain MG1988) or from the Tet-Plac-ompR construct (strain MG2000). Cells were grown to exponential phase in LB supplemented or not with IPTG at the indicated concentrations. As controls, the level of OmpR was also analyzed from strain MG1004 transformed with pHDB3 or pOmpR, and from the ΔompR strain MG1811. Immunoblot detection of EF-Tu is shown as a loading control. (C) The β-galactosidase activity of the same promoter fusions as in panel (A) was followed when ompR-envZ expression was modulated from the Tet-Plac-ompR allele by increasing IPTG concentrations. Results of a representative experiment are shown here and two additional independent repeats are shown in Supplementary Figure S6. Strains used in this experiment are MG2000, MG2002, MG2004 and MG2006. (D) Levels of OmrA, OmrB sRNAs and of ompC mRNA were followed by northern blot when changing OmpR levels. SsrA is used as a loading control. Strains are MG1004 (wt) and MG2000 (Tet-Plac-ompR).
RNA extraction and northern-blot analysis
Total RNA was extracted from cells grown to exponential phase after a 500-fold dilution from overnight culture into fresh medium, using hot phenol as previously described (14). A constant amount of total RNA was then separated on an 8% acrylamide gel in TBE 1× (for northerns for OmrA/B, bolA and SsrA) or on a 1% agarose gel in MOPS 1× (for ompC, ompF and SsrA) prior to electric transfer onto an Hybond-N+ membrane (GE Healthcare). Detection was then performed using 5′ end biotinylated oligonucleotides (sequence in Supplementary Table S2) and the Brightstar biodetect kit (Ambion) following manufacturer's instructions.
RESULTS
The OmpR response regulator directly activates transcription of omrA and omrB genes
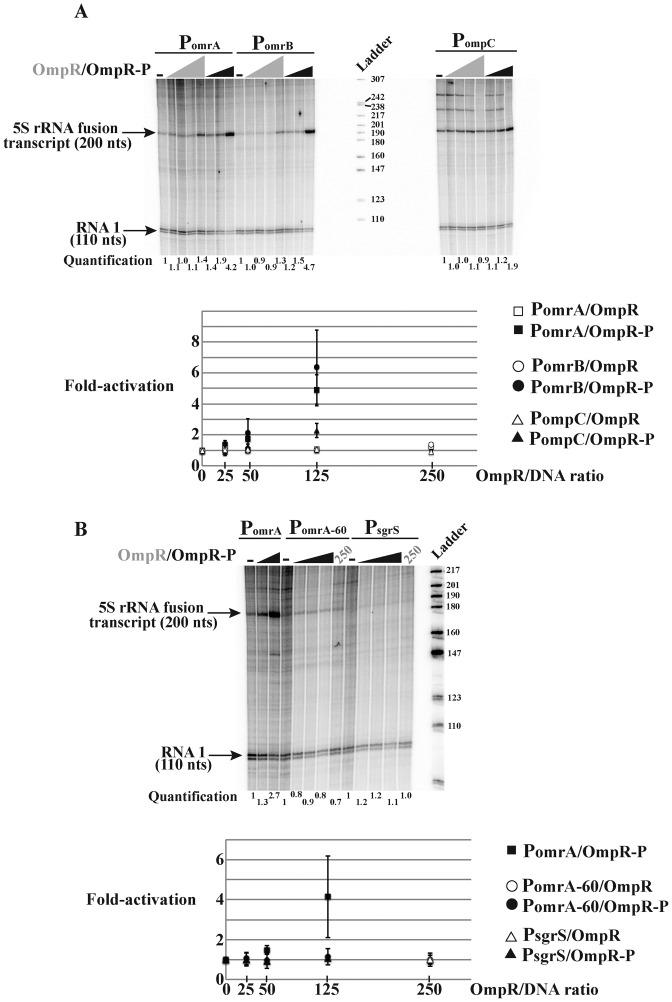
Our previous results identified OmpR as an activator of omrA/B transcription; both OmrA and OmrB levels increased with high osmolarity or upon procaine addition, in an EnvZ- and OmpR-dependent process (25). Furthermore, recent OmpR ChIP experiments suggest direct binding of OmpR to these promoters, indicating that activation by OmpR is direct regulation (10). To further investigate this, in vitro transcription reactions were performed using templates driven from the omrA or omrB promoter in presence of increasing amounts of OmpR, phosphorylated or not; the ompC promoter was included as a positive control for activation by OmpR (Figure 1). All three promoters, (starting 114 nts upstream of the TSS for omrA/B or 437 nts upstream for ompC), were cloned in the pRLG770 vector (36), allowing the production of ∼200 nt-transcripts carrying the first 50 nts of each RNA fused to 5S rRNA. An N-terminal His6-tagged version of OmpR (31) was purified and, when indicated, phosphorylated in vitro in the presence of acetylphosphate. The efficiency of the phosphorylation reaction was systemically assessed for each experiment and was reproducibly found to be around 50% (Supplementary Figure S1C). In these experiments, OmpR or OmpR-P was used at 25-, 50- and 125-fold excess over the template DNA (in molar ratio), which corresponds to a concentration of respectively 15, 30, 75 nM; an additional concentration of 250-fold (or 150 nM) was also included for unphosphorylated OmpR. As a comparison, total OmpR cellular concentration was estimated at 3500 OmpR molecules per cell (ca. 3.5 μM) (37) with ∼25% being phosphorylated in the presence of procaine or sucrose (34). The transcription products were separated on a sequencing gel and normalized to the RNA1 transcript (Figure 1A; a full representative gel is shown in Supplementary Figure S1). Transcription of ompC was, as expected, dependent on OmpR-P, but only at the highest concentration; in this condition, ompC transcription was activated by 1.9-fold relative to the control without OmpR. OmpR-P had a stronger effect on the omr promoters: a 50-fold excess of OmpR-P allowed activation of transcription from both the omrA and omrB promoters (by 1.9- and 1.5-fold, respectively, relative to the controls without OmpR) while a 125-fold excess further increased omr transcription (by 4.2- and 4.7-fold respectively). In contrast, the unphosphorylated form of OmpR did not significantly activate transcription from these three promoters. As an additional control, transcription driven by two non-OmpR-regulated promoters was studied as well. The first control promoter is a truncated version of PomrA, starting only 60 nts upstream of the TSS, which is not sufficient to allow activation by OmpR in vivo (14). The second control is the promoter of SgrS, another Hfq-dependent sRNA, whose transcription responds to phosphosugar stress via the SgrR activator and is independent of OmpR (38 and see below). Addition of OmpR, phosphorylated or not, had no effect on transcription originating from either control promoter, showing that the in vitro activation of omrA, omrB or ompC transcription is specific (Figure 1B). Altogether, these results strongly suggest that the previously reported control of OmrA or OmrB by EnvZ-OmpR is direct.
Figure 1.
OmpR-P activates omrA and omrB transcription in vitro. (A) In vitro transcription reactions were performed using plasmid templates carrying regions −114 to +50 of either the OmrA or OmrB promoter, or −437 to +50 of ompC promoter (relative to P1 transcription start site), fused to 5S to give a 200 nt transcript. When indicated, OmpR was added at a molar ratio of 25, 50, 125 and 250 with the template DNA, and OmpR-P at a molar ratio of 25, 50 and 125. Products were separated on a sequencing gel and a portion of a representative gel is shown where transcripts driven from PomrA, PomrB or PompC promoter and the internal control RNA1 are visible. The ladder corresponds to radiolabeled pBR322/MspI digestion products and indicated sizes refer thus to double-stranded DNA fragments. Quantification of 5S rRNA fusion transcripts was performed after normalization to RNA1 and the level is arbitrarily set at 1 for control without OmpR for each promoter. The graph below represents the average activation of transcription from PomrA, PomrB or PompC in presence of increasing OmpR or OmpR-P concentrations, as calculated from two independent experiments. Error bars indicate standard deviations and, when not visible, are smaller than the symbols. (B) Similar in vitro transcription reactions were performed using templates carrying a short version of omrA promoter (PomrA-60, carries nts −60 to +50 relative to TSS) or the sgrS promoter (from nts −70 to +50). OmpR-P was used at a molar ratio of 25-, 50- and 125-fold with template DNA, and OmpR at 250-fold. PomrA was used here as a positive control for activation by 50- or 125-fold OmpR-P. Full gels are shown in Supplementary Figure S1.
OmrA and OmrB are highly sensitive to various mutants of EnvZ-OmpR TCS
While OmpR regulates many genes, the details of regulation are different for different promoters, allowing differential expression of members of this regulon. For instance, ompC transcription is induced at high osmolarity, whereas ompF transcription is repressed at high osmolarity, but activated at low osmolarity (2). Furthermore, the analysis of dtpA expression in different mutants of envZ and ompR showed that it behaved differently from both ompC and ompF (5). We were interested in comparing omrA and omrB regulation by EnvZ and OmpR to that for ompC and ompF, the best-studied OmpR-regulated genes, with the expectation that this should provide a better sense of when expression of these sRNAs is physiologically important. For this purpose, we chose to analyze expression of lacZ fusions to the promoters of omrA/B in strains carrying different mutant alleles of envZ or ompR. As controls, expression of ompC- or ompF-lacZ promoter fusions was measured in the same mutants.
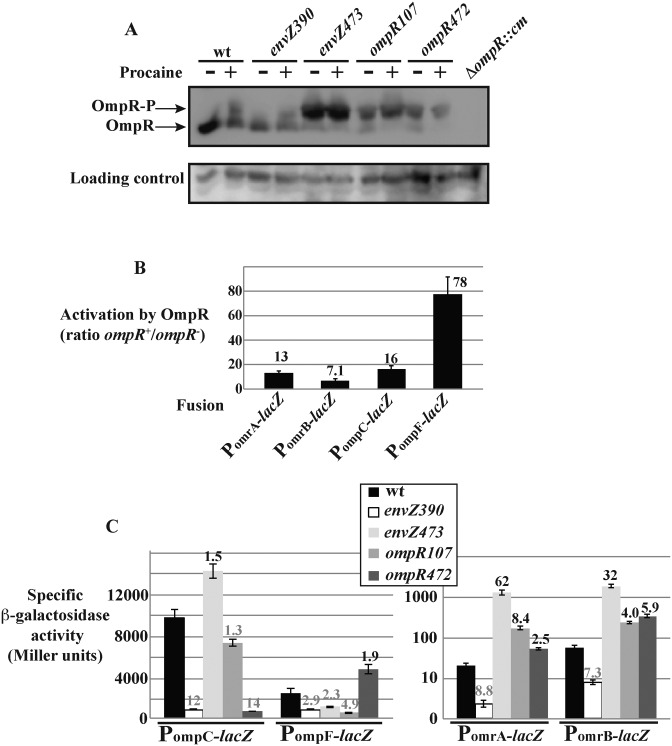
The envZ mutant alleles used were envZ390 and envZ473. EnvZ390 was isolated in a screen for EnvZ mutants that were deficient in kinase activity but that retained phosphatase activity (39). With such a mutant, the level of phosphorylated OmpR is therefore expected to be low. In contrast, the EnvZ473 mutant retained kinase activity, but lost its phosphatase activity; the level of OmpR-P should therefore be high (40,41). This was confirmed in vivo through protein electrophoresis in acrylamide gels containing Phos-Tag, a phosphate chelator that allows separation of phosphorylated proteins (42), followed by immunoblotting with an OmpR specific antibody (a kind gift from Dr Ann Stock). In the wt strain, OmpR-P was detected after treatment with procaine, an anesthetic known to activate the EnvZ-OmpR TCS. In the envZ390 and envZ473 strains, our results confirmed the respectively low and high levels of OmpR-P, with or without procaine (Figure 2A; an independent experiment is shown in Supplementary Figure S2A). In the envZ473 mutant strain, note that the vast majority of total OmpR was phosphorylated, which differs from what was observed by Barbieri et al. (34). These differences were found to be due to the different strains used in the two studies, an MC4100 derivative in (34) and an MG1655 derivative in this study (Supplementary Figure S2B). Another striking result from Figure 2A is that the level of total OmpR was significantly increased in the envZ473 allele. This result was confirmed when the same extracts were separated on a classical sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (Supplementary Figure S2C). This increase is most likely due to a transcriptional up-regulation as expression of a promoter fusion between PompR and lacZ was increased by more than 10-fold in the same mutant (Supplementary Figure S2D).
Figure 2.
Transcription of OmrA and OmrB sRNAs is strongly dependent on EnvZ-OmpR TCS. (A) The effect of various mutants in envZ or ompR on OmpR phosphorylation in vivo was analyzed by western blot following protein separation in presence of Phos-Tag. Samples were taken from strains DJ480 (wt), MG1670 (envZ390), MG1671 (envZ473), MG1672 (ompR107), MG1673 (ompR472) and MG1035 (ΔompR::cm) grown in LB to exponential phase, before or after addition of 10 mM procaine for 10 min. A non-specific band obtained from cross-hybridization with an anti-EF-Tu antibody is shown as a loading control. (B) The activation of promoter fusions to omrA, omrB, ompC or ompF was calculated as the ratio between the activity of those fusions in ompR+ and ompR− strains. The average and the standard deviations from at least two independent experiments are shown. Strains used here are, for ompR+ and ompR− respectively, MG1004 and MG1811 (PomrA fusion), MG1005 and MG1812 (PomrB fusion), MG1863 and MG1891 (PompC fusion) and MG1690 and MG1810 (PompF fusion). In this experiment, the β-galactosidase activities in the ompR+ strain were, in Miller units, 19.1 (PomrA fusion), 49.4 (PomrB), 6010 (PompC) and 3270 (PompF). (C) The β-galactosidase activity of the same promoter fusions in wt strains or in the same set of envZ-ompR mutants used in (A) was measured in LB medium in exponential phase. wt, envZ390, envZ473, ompR107 and ompR472 strains are respectively MG1892, MG1893, MG1894, MG1895 and MG1896 (PompC fusion), MG1690, MG1692, MG1694, MG1696, MG1698 (PompF fusion), MG1004, MG1299, MG1300, MG1301, MG1302 (PomrA fusion) and MG1005, MG1303, MG1304, MG1305 and MG1306 (PomrB fusion). Numbers above the bars indicate repression- or activation-fold (in gray or black respectively) in expression relative to the wt strain; note that right-hand panel uses log scale.
Two mutants of the OmpR response regulator were also used. These were isolated in a screen for OmpR mutants yielding either an OmpC−, OmpF+ phenotype (ompR2 class) or, in contrast, an OmpC+, OmpF− phenotype (ompR3 class) (43). The mutant representatives of these two classes used in this study are, for ompR2, ompR472, where Val203 is changed to Met and, for ompR3, ompR107, where Arg15 is changed to Cys. These two mutants were reported to be defective in dephosphorylation (44,45). In previous in vitro studies, OmpR107 bound to ompC and ompF promoters similarly to the wt protein, whereas OmpR472 displayed a higher affinity for the ompF promoter region and a weaker affinity for ompC (44,46). Again, Phos-Tag electrophoresis confirmed the increase in the OmpR-P/OmpR ratio in these two mutant strains in vivo (Figure 2A and Supplementary Figure S2A).
We then compared the effect of the various mutants on omrA, omrB, ompC and ompF transcription. For this purpose, we constructed lacZ fusions to the promoters of each of these genes, carrying all upstream sequences known to be required for regulation by OmpR (see Supplementary Figure S3 for a schematic representation of the different fusions used in this work, and experimental procedures for details). As expected, expression of the four resulting fusions was strongly dependent on ompR: the β-galactosidase activities were decreased from 7- to 78- fold in an ompR− strain in the absence of procaine (Figure 2B). A similar pattern was observed in the presence of procaine (data not shown).
The effect of the envZ390 allele was similar to that of the ompR deletion for omrA, omrB and ompC fusions (compare fold activation in Figure 2B to fold repression in Figure 2C), suggesting that OmpR-P is strictly required for activation at these promoters. While ompF transcription was significantly decreased as well in the envZ390 context (by 3-fold), the effect of this mutant allele was in this case not nearly as strong as the ompR deletion strain, that decreased expression of the ompF fusion by 78-fold (Figure 2B and C). This is possibly due to the presence of residual OmpR-P in this mutant that may be sufficient to activate ompF. In line with this, OmpR-P is still detectable by Phos-Tag in the envZ390 background in presence of procaine (Figure 2A). The three other mutants, each of which has increased levels of OmpR-P, activated transcription from both the omrA and omrB promoters, with envZ473 being by far the most efficient, since it increased activity of the omrA and omrB promoter fusions by 62- and 32-fold respectively (Figure 2C, note logarithmic scale). These effects differed from those observed with the fusions to ompC and ompF promoters. For instance, activation of ompC or ompF fusion in the envZ473, ompR107 or ompR472 mutants was never more than 2-fold (Figure 2C), and as expected from previous work, envZ473 and ompR107 repressed the ompF fusion, while ompR472 repressed the ompC fusion (41,43). The basal level of transcription of OmrA/B is strikingly lower than ompC/F. To rule out that those envZ/ompR mutants affect OmrA/B via non-specific regulation of poorly transcribed genes, the effect of these mutants was analyzed on control promoter fusions. These controls were lacZ fusions to the short version of OmrA promoter or to the SgrS promoter, i.e. the same control promoters as used for the in vitro transcription reactions. None of these fusions was significantly affected by the different envZ/ompR mutants, even though their expression in wt cells is lower than the expression of the Omr promoter fusions (Figure 2 and Supplementary Figure S4A). This confirms that the effect of EnvZ-OmpR on OmrA/B transcription is specific and not due to a low transcription level.
These effects on the omr fusions were also different from what was reported with the dtpA gene, whose transcription was unaffected in envZ473 and repressed in ompR107 and ompR472 strains (5). Therefore, omrA and omrB are differentially regulated by EnvZ-OmpR in comparison to the other well-studied members of the regulon and they display a stronger dependence on changes in total OmpR and/or OmpR-P levels than ompC and ompF.
ompF and ompC have long non-coding leaders and are subject to post-transcriptional regulation by the MicF and MicC sRNAs, themselves OmpR-dependent. While the ompF and ompC promoter fusions used above did not include the leaders and sites of sRNA interaction, the effects of the different envZ and ompR alleles were very similar to that shown previously with ‘long’ transcriptional fusions isolated by Silhavy and coworkers and carrying the leaders together with the beginning of the ompC/F ORF (41,47) (fusions ompC-lacZ and ompF-lacZ, Supplementary Figure S4, panels B and C, compare to PompC-lacZ and PompF-lacZ fusions used in Figure 2). This suggests that the effect of the mutants used here on ompC/F is primarily at the level of transcription initiation. As a further confirmation, fusions were created in which Ptet replaced the ompF or ompC promoter, but the leader of these genes and the translation start site was retained. These fusions were much less affected by loss of OmpR (Supplementary Figure S4B, Ptet-ompC/F-lacZ). Post-transcriptional regulation of ompF expression is further examined later in this study.
OmrA/B-mediated repression of ompR does not change OmpR-P levels
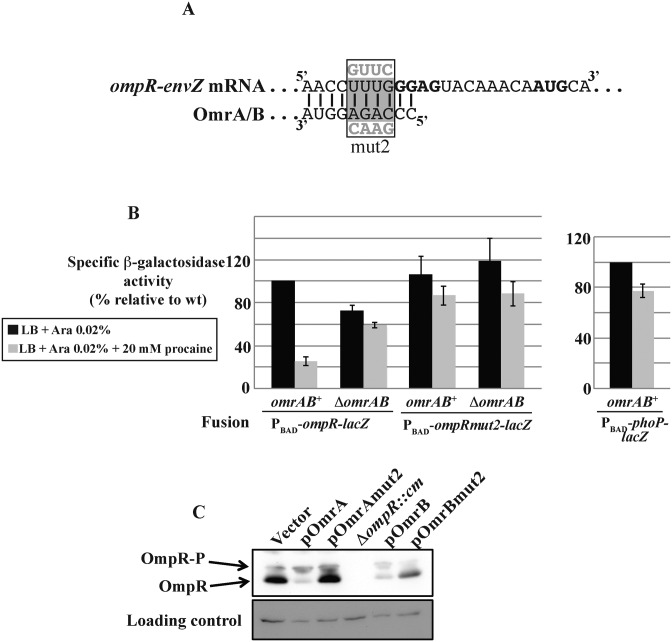
OmrA/B sRNAs regulate the expression of multiple targets through their 5′ conserved region; one target is the bicistronic operon encoding their own transcriptional regulator, the EnvZ-OmpR TCS (Figure 3A). We showed previously that, when overproduced, OmrA/B directly pair to the ompR translation initiation region and repress ompR expression as well as the levels of ompR-envZ mRNA (26). This control of ompR-envZ by OmrA/B also occurs upon induction of expression of OmrA/B from their natural locus. This was assayed by comparing expression of a PBAD-ompR-lacZ translational fusion in wt and ΔomrAB cells when the EnvZ-OmpR TCS was activated using 20 mM procaine. Expression of the wt fusion was repressed 4-fold upon addition of procaine. This repression is specific to ompR as it was not observed on another PBAD-driven translational fusion (PBAD-phoP-lacZ). Furthermore, it is clearly due to the induction of the OmrA/B sRNAs, since procaine did not affect the wt fusion in a ΔomrAB strain, and it did not affect a fusion carrying the ompRmut2 change that abolished the control by OmrA/B (Figure 3B).
Figure 3.
Control of ompR expression by OmrA/B changes OmpR, but not OmpR-P levels. (A) Seed-pairing interaction between OmrA/B 5′ end and ompR-envZ mRNA (26). Shine-Dalgarno sequence and start codon of ompR are in bold, and mut2 changes in OmrA/B or ompR mRNA are shown in gray. (B) The β-galactosidase activity of an ompR-lacZ translational fusion, wt or with the mut2 change, expressed from a PBAD inducible promoter was measured in omrAB+ or omrAB− strains in a medium supplemented or not with 20 mM procaine. Strains used in this experiment are MG2174, MG2175, MG2176 and MG2177. As a control, the activity of a PBAD-phoP-lacZ translational fusion (strain MG1425) was measured in the same media. Activities are expressed as % of the wt fusions in omrAB+ cells in the absence of procaine; these reference activities are, in Miller units, 2238 (ompR fusion) and 542 (phoP fusion). (C) In vivo phosphorylation of OmpR was analyzed upon overproduction of OmrA/B or of their mut2 derivatives in strain DJ480. An extract from a ΔompR::cm strain (MG1035) was loaded on the same gel. A non-specific band revealed with the anti-OmpR antibody was used as a loading control.
Interestingly, a mathematical model of OmpR-P levels performed by Batchelor and Goulian predicted that OmpR-P was robust (i.e. insensitive) to changes in ompR or envZ expression, as long as total OmpR levels remained much greater than EnvZ. This prediction was validated when transcription of ompC and ompF was followed over a wide range of OmpR or EnvZ levels (48,49 and personal communication). Therefore, one would predict that modulating expression of ompR-envZ by OmrA/B should not affect the level of OmpR-P and regulation of ompC and ompF. To test whether levels of OmpR phosphorylation changed with changing OmpR levels, proteins were extracted from cells overproducing either wt Omr or a derivative mutated in the 5′ end (mut2) that no longer repressed ompR (Figure 3A and 26). These extracts were then subjected to electrophoresis on a Phos-Tag containing gel followed by a western blot analysis to monitor both unphosphorylated OmpR and OmpR-P levels. The results were in complete agreement with the previously reported robustness: overproduction of wt OmrA/B, but not of the mutant versions, decreased the total amount of OmpR as expected, but the level of OmpR-P remained unchanged (Figure 3C; an independent repeat is shown in Supplementary Figure S5). This robustness, however, leaves unexplained what the physiological role of regulating ompR-envZ expression by the OmrA/B sRNAs might be. Is this truly a negative feedback circuit, if the levels of OmpR-P, the presumably active form of the protein, do not change?
Not all OmpR targets are as robust as ompC/F to changes in OmpR levels
A first hint toward resolving this question came from the fact that in earlier work we were able to isolate a plasmid overexpressing OmpR from its native promoter as a multicopy activator of OmrA/B (hereafter referred to as pOmpR (14)). This indicates that at least some OmpR targets are sensitive to changes in ompR expression. Indeed, expression of omr fusions was strongly activated by this plasmid (roughly 12- and 7-fold for OmrA and OmrB, respectively), while expression of the ompC/F promoter fusions was only modestly changed (1.3-fold activation or 1.4-fold repression respectively), in agreement with the robustness of these two latter targets (Figure 4A). This difference in behavior suggested a qualitative difference in the response of the omrA/B promoters, compared to the ompC/F promoters.
However, the pOmpR plasmid carries a truncated ompR-envZ operon with only two-thirds of envZ ORF and it is not clear whether the resulting EnvZ protein (if produced) is functional or not. Another construct was therefore engineered on the chromosome to modulate transcription of the whole operon: the promoter region of ompR was replaced by a PLlacO-1 (hereafter Plac) promoter inducible with IPTG, preceded by a tetracycline resistance gene so that this allele can be easily moved between strains. As visible in Figure 4B, this construct allowed us to increase total OmpR by varying concentrations of IPTG, with the level of OmpR at 10−2 M IPTG being similar to the level of OmpR produced from the pOmpR plasmid mentioned above. Furthermore, even in the absence of IPTG, this construct synthesizes significantly more OmpR than from the chromosomal wt promoter. When combined with the different OmpR target fusions, changing OmpR levels from this construct using a range of IPTG from 0 to 10−3 M, we observed that transcription of omrA/B fusions increased significantly (between 7- and 8-fold), while again, expression of ompC/F fusions varied much more modestly (1.3-fold) (Figure 4C and Supplementary Figure S6).
To rule out the possibility that these differences between OmrA/B and ompC/F expression were inherent to the fusions, the levels of the OmrA/B sRNAs and of the ompC mRNA were analyzed by northern blot in similar experiments to those above, where OmpR levels were changed using either the pOmpR plasmid or by induction of the Tet-Plac-ompR construct with IPTG. As previously, while OmrA/B increased greatly upon OmpR overproduction, ompC mRNA was not significantly affected (Figure 4D). The levels of ompF mRNA were also followed and results are discussed below (see Figure 7 and accompanying text).
Figure 7.
Robustness and non-robustness of other OmpR targets. (A) The same RNA samples used in Figure 4D were analyzed by northern blot probed for ompF and for SsrA as a loading control. (B) MicF sRNA is involved in the post-transcriptional control of ompF upon OmpR overproduction. The β-galactosidase activity of Ptet-ompF-lacZ translational fusion was measured with and without IPTG induction of ompR overexpression in micF+ and micF− cells (strains MG2127 and MG2167 respectively). (C) bolA mRNA levels were analyzed by northern blot in strains MG1004 (wt) transformed with pHDB3 or pOmpR, MG1811 (ΔompR) transformed with pHDB3 and in MG2000 (Tet-Plac-ompR) with or without IPTG. SsrA levels were probed as well and used as a loading control. (D) Expression of a dtpA-lacZ promoter fusion was followed in a set of strains with various OmpR levels (MG2138 (wt), MG2169 (ΔompR) and MG2170 (Tet-Plac-ompR)).
Altogether, our data confirm that the levels of OmpR-P are robust to changes in ompR-envZ expression and that this translates into robust transcriptional control of some targets such as ompC and ompF. However, this is not true for all OmpR targets. In particular, omrA/B transcription is not robust and expression of these two sRNAs varies greatly with changing levels of the OmpR protein.
OmrA/B limit their own synthesis through feedback control
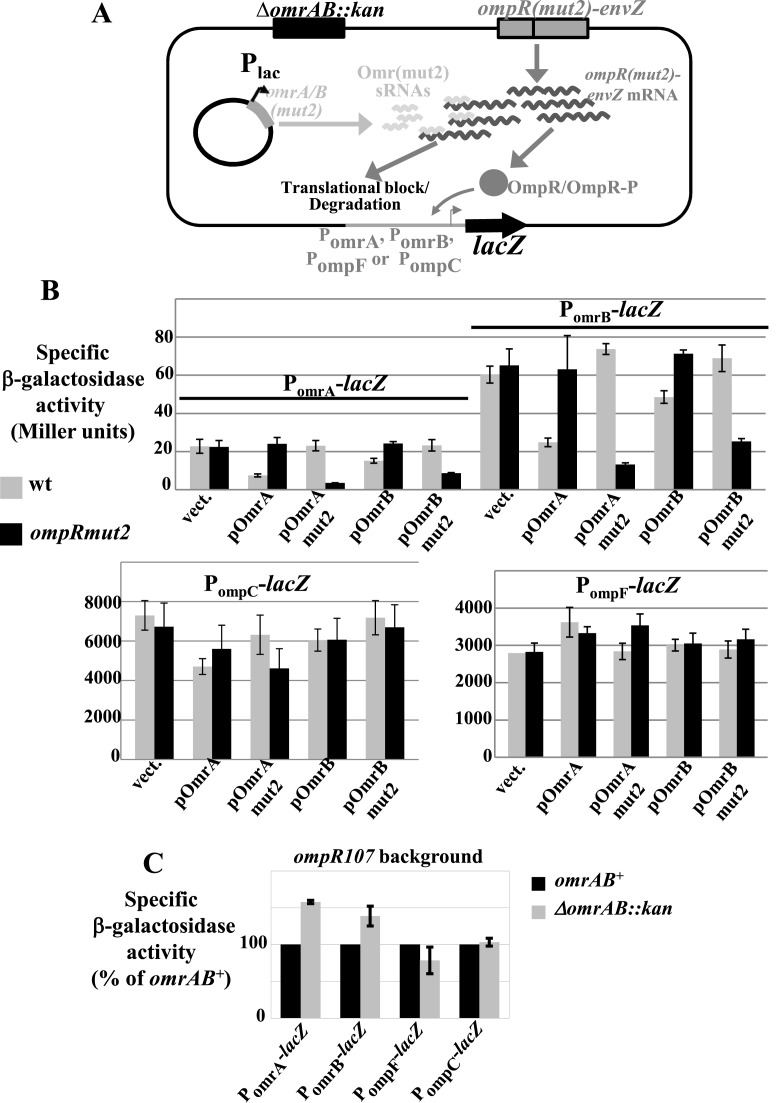
One prediction that can be made in the light of these results is that, by regulating ompR translation, OmrA/B sRNAs should not modify the expression from robust promoter targets such as ompC/F but should modulate that of non-robust targets such as the omrA and omrB promoters. To test this, the activity of the OmrA/B- and ompC/F- promoter fusions was monitored upon overexpression of either OmrA/B or their mut2 derivative that no longer represses ompR-envZ (Figure 5A). The overproduction of the different Omr variants was achieved using plasmids carrying the corresponding omr gene under control of an IPTG-inducible promoter in strains deleted for the omrAB chromosomal genes. As a control, the same experiment was carried out in an isogenic strain where the chromosomal copy of ompR-envZ carries the compensatory change to mut2, i.e. its expression is no longer controlled by wt OmrA/B but is controlled by the mutant OmrA/Bmut2. This ompRmut2 mutation consists of a change in nts −13 and −17 (relative to the ompR ATG start codon). Therefore, it is not expected to change the amino acid sequence of OmpR, and it does not significantly affect ompR expression, at least when expressed from a PBAD promoter (26).
Figure 5.
Feedback control in OmrA/B expression. (A) Scheme depicting the experiment performed in (B): the β-galactosidase activity of promoter fusions to OmrA, OmrB, ompC and ompF was measured upon overproduction of OmrA, OmrB or of their mut2 derivatives, in strains with a wt copy of ompR (gray bars) or with a mutant version carrying the ompRmut2 compensatory change to OmrA/Bmut2 (black bars). Strains used here are MG1014, MG1666, MG1017, MG1667, MG1901, MG1902, MG1686, MG1688 (panel B, see strain table for details). (C) Activity of the same set of promoter fusions was compared in omrAB+ and omrAB− strains in the ompR107 background. Values are represented as % of wt strain for each fusion; the activities of the wt strains were, in Miller units, 135 (PomrA fusion), 173 (PomrB), 547 (PompF) and 5042 (PompC). Strains used are MG1301, MG1908, MG1305, MG1909, MG1696, MG1910, MG1895 and MG1911.
The presence of plasmid pOmrA decreased expression of both PomrA- and PomrB-lacZ fusions, by 3- and 2.4-fold, respectively, in the wt strain, but had no effect in the ompRmut2 background (Figure 5B, top panel). In contrast, overexpression of OmrAmut2 did not significantly change the activity of either fusion in the wt context, but decreased it in presence of the ompRmut2 variant, clearly showing that the observed repression is due to the down-regulation of ompR-envZ by OmrA. Similar results were obtained when OmrB (or OmrBmut2) was overproduced, but with somewhat lower repression factors. Interestingly, repression of either fusion was always higher in the mut2/mut2 situation than in the wt/wt. There are at least two hypotheses that could explain this observation; these models are not mutually exclusive. First, it is possible that the sRNA-mRNA duplex formation and/or stability was increased by the compensatory changes. Alternatively, since all targets of OmrA/B known so far are regulated through the 5′ end region of these sRNAs (18,21,26), it is plausible that, in the context of the compensatory changes, ompRmut2 is the only target (or at least one among very few) of OmrA/Bmut2. The end result is that, in the absence of other targets, the level of OmrA/Bmut2 available to base-pair with ompRmut2 is increased compared to the wt/wt situation.
As expected based on the results of Figure 4, this ability of OmrA/B to repress their own synthesis through ompR regulation was not paralleled when we looked at the expression of ompC and ompF. Instead, the activity of both PompC- and PompF-lacZ fusions was unaffected, or only very weakly affected, by overexpression of OmrA/B wt or mut2 (Figure 5B, lower panels).
The experiments shown in Figure 5B were performed in LB medium, i.e. under experimental conditions where the activity of EnvZ/OmpR TCS is not expected to be strongly induced and where overproduction of OmrA/B is driven by an inducible promoter, whose activity is completely independent of EnvZ-OmpR. To get a better sense of the effect of OmrA/B when expressed from the chromosome, the activity of the same set of promoter fusions was compared in omrAB wt or deleted strains (Figure 5C). This experiment was performed in an ompR107 background where OmrA/B are induced (Figure 2). In line with results obtained with OmrA/B overproduction, deletion of omrAB reproducibly increased the activity of the fusions to the promoters of OmrA/B by ∼40 to 50%, but did not significantly affect ompC or ompF expression (Figure 5C).
Therefore, the difference in the responses of the omrA/B- and ompC/F- promoters to changing OmpR levels allows autocontrol of OmrA/B sRNAs through the negative feedback circuit that exists between EnvZ-OmpR TCS and OmrA/B; at the same time, ompC/F transcription remains resistant to the effect of OmrA/B.
A role for non-phosphorylated OmpR in activating omrA and omrB transcription?
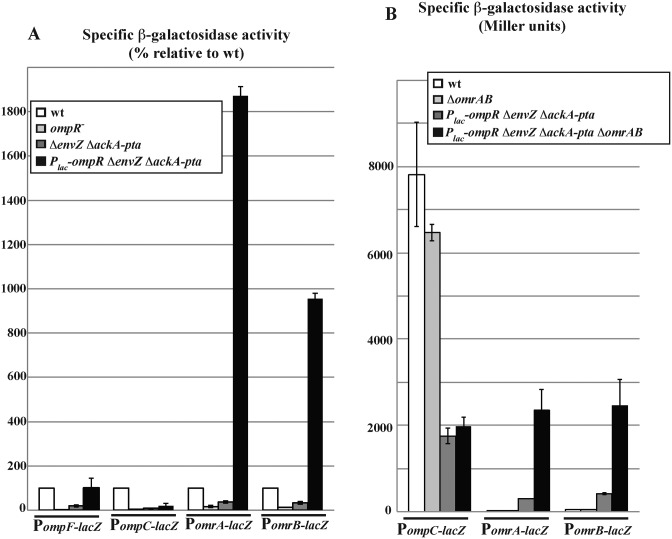
What could explain the sensitivity of non-robust promoters such as PomrA or PomrB to changing OmpR levels given that the levels of OmpR-P are themselves robust? An obvious hypothesis is that, in contrast to ompC/F, omrA/B promoters might be activated not only by OmpR-P, but also by the non-phosphorylated form of OmpR. To address this question, we first worked with mutants of the phosphorylation site of OmpR (D55A or D55N) but found that they were unable to activate omr transcription, even when overproduced (data not shown). Because it is difficult to rule out the possibility that these variant OmpR proteins might be inactive, we then decided instead to follow the activity of the different promoter fusions when using strains with a wt OmpR protein but very low level of OmpR-P. This was achieved by working in strains lacking the OmpR kinase EnvZ. However, it is known that in the absence of EnvZ, some OmpR-P is still present and is generated in large part through phosphorylation of OmpR by acetylphosphate (see e.g. 50). The envZ deletion was therefore used in conjunction with a deletion of the ackA-pta genes, required for synthesis of acetylphosphate. The resulting strains display a low expression of the four OmpR targets, almost as low as in the ompR null mutant (Figure 6A; note that each wt level is set to 100%, to allow easier comparison between the promoters, although total levels of expression are very different, see wt bars in Figure 6B). OmpR-P is thus required for expression of all these genes including omrA/B, which is consistent with the results obtained in the envZ390 mutant (Figure 2) and in the in vitro transcription reactions (Figure 1). In this ΔenvZ ΔackA-pta background, overproduction of OmpR from the Tet-Plac-ompR allele slightly increased expression of both the ompF and ompC promoter fusions, compared to the ΔenvZ ΔackA-pta strain with ompR expressed from its own promoter, suggesting that OmpR-P levels may be slightly higher (Figure 6A, compare dark gray and black bars). The stronger activation of ompF is consistent with the fact that ompF is activated at lower OmpR-P levels compared to ompC(2). The presence of OmpR-P in these strains is most likely due to cross-phosphorylation by a histidine kinase other than EnvZ or to the presence of acetylphosphate in the LB growth medium. This cross-phosphorylation is relatively inefficient, as judged by the low level of PompC-lacZ transcription, compared to a wt strain (Figure 6A, compare black and white bars). Strikingly, overexpression of OmpR in these strains leads to strong activation of both omrA and omrB transcription relative to the wt strain (by 19- and 10-fold respectively), despite the lower level of OmpR-P (Figure 6A). This shows that, even though expression of these two genes strictly requires OmpR-P, it can nonetheless be modulated by changes in the levels of unphosphorylated OmpR. While a detailed understanding of the mechanism involved is still lacking, this would set up the basis for the different robustness to OmpR levels that were observed for OmrA/B and ompC/F.
Figure 6.
Expression of omrA and omrB can be activated by non-phosphorylated OmpR. (A) The β-galactosidase activity of promoter fusions to omrA, omrB, ompC or ompF was measured in LB in exponentially growing cells with varying OmpR levels and/or phosphorylation. Activities are shown relative to that of wt cells. Strains used in this experiment are MG1690, MG1810, MG2043, MG2044 (PompF), MG1892, MG2018, MG2045, MG2046 (PompC), MG1004, MG1811, MG2039, MG2040 (PomrA), MG1005, MG1812, MG2041 and MG2042 (PomrB). The β-galactosidase activities of the different fusions in the wt strains were, in Miller units, 22 (PomrA fusion), 63 (PomrB), 11292 (PompC) and 3618 (PompF). (B) Activity of the same omrA, omrB and ompC transcriptional fusions was measured in wt cells or upon OmpR overproduction in ΔenvZ and ΔackA-pta background, both in omrAB+ or omrAB− strains. Strains are MG1892, MG2203, MG2046 and MG2206 (PompC fusion), MG1004, MG2201, MG2040 and MG2204 (PomrA), and MG1005, MG2202, MG2042 and MG2205 (PomrB).
Interestingly, activation of the OmrA and OmrB fusions upon OmpR overproduction in this low OmpR-P background was even more pronounced in ΔomrAB cells (Figure 6B). In this context, omrA or omrB transcription was increased 123- and 49-fold respectively, i.e. roughly 8- and 6-times more than in the omr+ cells. In contrast, deletion of omrAB had no effect on the decrease in ompC transcription due to the lower OmpR-P levels. This is a further demonstration that OmrA/B limit ompR overexpression and that this feedback results in lowering their own transcription.
ompF mRNA levels are not robust to OmpR
When looking at ompC mRNA by northern blot with changing OmpR levels, we found that, similarly to what was observed with the transcriptional fusion, ompC mRNA was barely affected (Figure 4D). In striking contrast however, while transcription initiation of ompF was also robust to OmpR (Figures 4 and 5), ompF mRNA levels were clearly reduced when ompR expression increased (Figure 7A). This difference should be due to an OmpR-dependent process controlling ompF expression downstream of transcription initiation. In agreement with this possibility, a translational ompF-lacZ fusion expressed from a Ptet constitutive promoter still displayed some dependency on OmpR: expression of this fusion was lower in an ompR+ than an ompR− strain (ratio of 0.64, see Supplementary Figure S4B), and was 2-fold repressed when OmpR synthesis was induced with IPTG from the Tet-Plac-ompR allele (Figure 7B). Most of this post-transcriptional regulation by OmpR appears to be due to MicF under the conditions used here, since deletion of micF abolishes the effect of overproducing OmpR (Figure 7B). This suggests that MicF is itself a non-robust OmpR target even though, given the complexity of MicF regulation (51), further experiments are required to determine whether this is due to the direct non-robust activation of micF transcription by OmpR. These experiments indicate that even though transcription of ompF is clearly robust, synthesis of the OmpF porin might nonetheless follow a non-robust behavior under some conditions because of post-transcriptional control. Furthermore, the involvement of MicF in this process does not rule out that yet other actors might play a role as well in the non-robust response of ompF mRNA levels.
Robustness and other OmpR targets
EnvZ-OmpR directly controls transcription of other genes in addition to MicF, OmrA/B and ompC/F. One can therefore wonder whether these other OmpR targets are sensitive to changes in OmpR levels. We have tested this for two of them: bolA and dtpA. Transcription of bolA can originate from two promoters and OmpR was reported to repress transcription from the proximal promoter (8). Accordingly, two distinct bolA mRNAs were visible by northern blot and only the levels of the shorter transcript increased in an ompR deleted strain. When OmpR was overproduced, either from pOmpR or from Tet-Plac-ompR using IPTG, levels of both bolA transcripts remained unchanged, indicating that bolA transcription, like that of ompC/F, responds only to OmpR-P and is thus robust to changes in OmpR levels (Figure 7C). Regarding dtpA, transcription was followed using a transcriptional fusion carrying nts −201 to +15 of dtpA (relative to TSS) upstream of lacZ (from nt -17). Consistent with the known regulation of dtpA by OmpR (5), expression of this PdtpA-lacZ fusion was strongly reduced in the absence of OmpR. Interestingly, activity of the fusion was also decreased upon OmpR overproduction, by 2-fold in presence of pOmpR and by 5-fold when ompR expression was induced from Tet-Plac-ompR (Figure 7D). It seems therefore that dtpA transcription is activated by OmpR (or OmpR-P) at low levels but repressed at higher levels, reminiscent of the regulation of ompF by OmpR-P. More importantly for the scope of this study, this result shows that dtpA is another non-robust OmpR target whose expression is modulated in response to changes in OmpR levels. Note at last that both OmpR and OmpR-P were shown to bind the fimB promoter of an uropathogenic E. coli strain, indicating the existence of yet other targets sensitive to changes in OmpR levels (52).
DISCUSSION
The results reported here confirm and extend our understanding of the EnvZ/OmpR two-component system, how it is itself regulated and how it regulates targets. We summarize our findings in Figure 8.
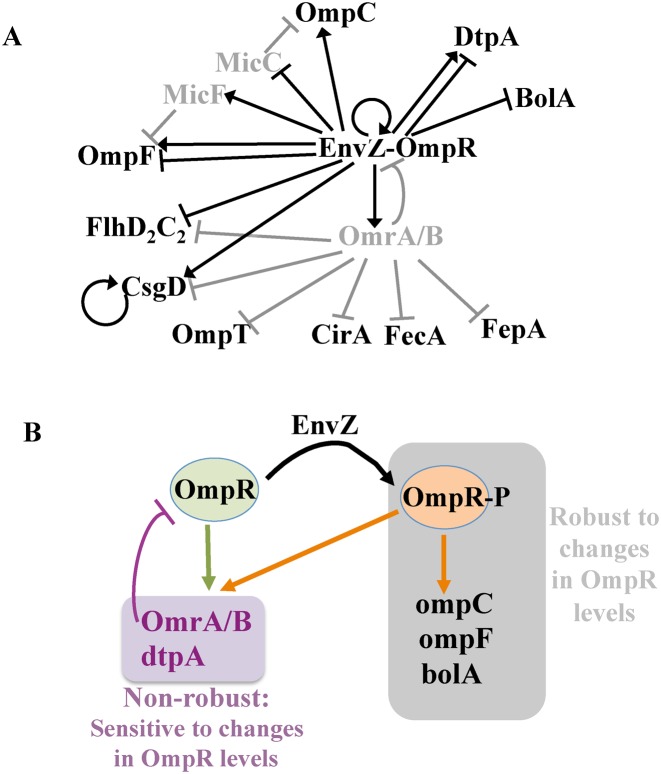
Figure 8.
Robustness and differential regulation within the OmpR regulon. (A) Regulatory circuit centered on the EnvZ-OmpR-OmrA/B feedback motif. Transcriptional and post-transcriptional regulations are in black and gray respectively, with arrows indicating activations and perpendicular bars indicating repressions. sRNAs are in gray. For simplicity, only gene products relevant to this study are indicated. Note that direct control has not been demonstrated in some aspects of this circuit (for instance, MicC control by OmpR, or OmpR autoregulation). (B) Differential regulation of robust or non-robust OmpR targets by the non phosphorylated form of OmpR. See text for more details.
A dual OmpR regulon?
OmpR-P levels were previously predicted to be robust when ompR or envZ expression varied, and accordingly ompC and ompF transcription remained constant over a wide range of EnvZ or OmpR levels, from 10-fold less to 10-fold more relative to wt levels (48). However, this robustness did not apply upon stronger variations of either EnvZ or OmpR levels (48) as well as upon overproduction of the cytoplasmic domain of EnvZ (53,54).
The use of Phos-Tag allowed us to directly confirm robustness of OmpR-P, at least upon overproduction of OmrA or OmrB sRNA. Our data furthermore show that this translates into robust transcriptional regulation of some targets such as ompC and ompF. However, this is not the case for all OmpR targets as the promoters of omrA/B and dtpA were found, for instance, to be regulated by changes in OmpR levels as well.
One prediction that can be made from these findings is that the OmpR regulon is actually composed of at least two subsets of genes. The first group, corresponding to robust targets (gray square in Figure 8B), should be sensitive only to signals affecting the level of OmpR-P, e.g. osmolarity. The second group is composed of the non-robust targets whose expression should depend on both the phosphorylation status of OmpR and its expression levels (purple square in Figure 8B). This is for instance what is observed for OmrA/B, which autoregulate their own synthesis by repressing ompR translation (Figure 5). Interestingly, control of ompR expression is not limited to the action of the Omr sRNAs, as it was reported that ompR-envZ transcription is also regulated by cAMP-CRP (via a complex pattern depending on the transcription start site), inhibited by IHF (55,56) and is possibly positively autoregulated by OmpR (10 and Supplementary Figure S2). The non-robust control of some OmpR targets is therefore expected to allow integration of multiple signals in controlling only a subset of genes of the OmpR regulon.
Robustness has been proposed to minimize output variability between cells that are subject to the same input but can present stochastic variations in the concentrations of some cellular components (57). Given the importance of the ompC/F structural genes that encode the major porins in E. coli and whose ratio will impact the entry of various beneficial or harmful compounds, one can understand why keeping their transcription independent of stochastic variations might matter. In contrast, regarding OmrA/B, it seems logical that these OmpR targets involved in feedback control of ompR expression are non-robust, which is expected to allow them to keep ompR expression levels within a given range and underlies their observed autoregulation.
Furthermore, our data indicate that bolA transcription is robust to OmpR while dtpA is not. In the future, it will be interesting to determine the nature of the other OmpR targets in order to understand a possible link between signals affecting ompR expression and the function of non-robust targets.
Finally, it is worth noting that, even if transcription of some targets is robust per se, their expression can nonetheless be modulated in response to changes in OmpR levels by a non-robust post-transcriptional step. This is what we have found for ompF for instance (Figure 7). Similarly, one can expect that OmrA/B control of flhDC and csgD will provide these targets a non-robust response to OmpR.
Mechanism of regulation of Omr sRNAs transcription by EnvZ-OmpR
An important question that is raised by the results of this study is the nature of the promoter determinants that underly the robust or non-robust regulation by OmpR, together with the molecular mechanism involved. Our data strongly suggest that unphosphorylated OmpR is involved in transcription regulation at the Omr promoters, but also possibly at the promoters of other non-robust targets. In some cases, the unphosphorylated form of a response regulator has been shown to specifically activate gene expression at some target-promoters (e.g. 58,59). However, in the case studied here, our results suggest that at least some OmpR-P is required for transcription activation at the Omr promoters. Indeed, envZ or ackA-pta mutants that lower OmpR-P reduce OmrA/B transcription as much as an ompR deletion allele (Figures 2C and 6). Furthermore, mutants of the phosphorylation site of OmpR (D55A or D55N) failed to activate Omr transcription, even when overproduced (our unpublished results), which also suggests that OmpR-P is essential for transcription activation, even at the Omr promoters.
A first hypothesis that could explain both the requirement for OmpR-P and the sensitivity to unphosphorylated OmpR is that Omr promoters could simultaneously bind both forms of OmpR. As OmpR phosphorylation has been found to promote dimerization and subsequently DNA-binding (34), OmpR-P presumably binds as a dimer and this binding is likely to be essential for control. Unphosphorylated OmpR may bind as well and promote OmpR-P mediated transcription regulation, for instance by activating OmpR-P binding. It is not clear in what form unphosphorylated OmpR would bind, perhaps as a monomer or in complex with another protein, and what promoter elements would allow this binding. An alternative possibility is that OmpR and OmpR-P could form heterodimers able to control gene expression specifically at the Omr promoters.
It is also possible that the observed regulation of Omr synthesis by unphosphorylated OmpR is indirect. Instead, OmpR could control the level of another regulator of Omr transcription; both the nature of this regulator and how its levels would be controlled by OmpR remains to be identified to support this hypothesis. Additional studies are required to discriminate between these possibilities, and identify the promoter elements that make Omr sensitive to both unphosphorylated OmpR and OmpR-P.
Regulating regulators: a recurrent theme in sRNA biology
Synthesis of base-pairing sRNAs is often controlled by transcriptional regulators and, in turn, base-pairing sRNAs commonly target genes encoding transcriptional regulators (60,61 for reviews).
In some cases, sRNAs control expression of their own regulator, thereby creating a feedback circuit, such as the one described here. This has been shown for instance for MicF that negatively controls lrp translation in a double negative feedback circuit (62,63). Another striking example is that of the Vibrio Qrr sRNAs involved in quorum-sensing and that lie at the core of multiple negative feedback circuits (64), which participate in modulating the cell density at which the switch occurs as well as the kinetics of the switch (65–67).
The results presented in this study show that the feedback circuit mediated by OmrA/B on ompR-envZ expression can limit the expression of OmrA/B in steady-state experiments (Figures 5 and 6B). However, this circuit could provide other properties to OmrA/B expression. It could, for instance, restrict Omr induction to a shorter time frame following OmpR activation, allow a surge in Omr levels or provide a faster recovery once the inducing signal(s) for OmpR has disappeared. Another possibility is that this feedback could be involved in ensuring that Omr synthesis occurs only in response to sustained stimulation of OmpR and would filter transient signals, or in limiting cell-to-cell variations among the bacterial population. Interestingly, a recent RNAseq study showed that OmrB, and to a lesser extent OmrA, was strongly induced when Salmonella enters eukaryotic host cells (68). Our findings may thus also be relevant for host-pathogen interactions.
Regulatory network motifs and their properties were first defined by studies of protein transcriptional regulators. However, while many transcriptional regulators are subject to direct autoregulation, examples of protein-based negative feedback circuits reminiscent of the one described in this work are extremely rare in purely transcriptional networks (69). Understanding its properties could thus provide important information regarding not only OmrA/B, but also the role of this specific regulatory circuit in gene expression.
In addition to EnvZ-OmpR TCS, other examples of robustness have been reported such as the phosphorylation of IDH (Isocitrate Dehydrogenase) enzyme by the IDHKP kinase/phosphatase in the glyoxylate bypass pathway (70). Given that prediction of robustness relies on the fact that EnvZ is a bifunctional kinase/phosphatase and that OmpR levels are much greater than those of EnvZ, it is likely that robustness will apply to other TCS as well. In this regard, results obtained with the PhoQ/PhoP TCS suggest that PhoP-P levels might indeed be robust to some changes in total PhoP (71). Our results can thus have implications for these other cases and it is therefore of interest to determine whether these systems also have non-robust targets as well.
Supplementary Material
Acknowledgments
We are grateful to D. Walthers and L. Kenney for providing the OmpR-His overproducing plasmid and the pRLG770 vector, and to L. Morgan for advice regarding the purification procedure. We also thank A. Stock for the anti-OmpR antibody and help with the Phos-Tag technique, T. Silhavy, A. Stock, E. Hobbes and N. Majdalani for providing strains or plasmids, and W. Ross for advice with in vitro transcription. Discussions with T. Silhavy, M. Goulian, L. Kenney, A. Stock, T. Msadek and F. Repoila throughout this work were greatly appreciated. Thanks to H. Kemble and C. Chiaruttini for the ompR transcriptional fusion and the ara::lacIq-KanR allele respectively. We are grateful to M. Springer, J. Jagodnik, C. Chiaruttini and C. Condon for comments on the manuscript.
Footnotes
Present address: Anna Korobeinikova, Institute of Protein Research, Russian Academy of Sciences, Pushchino, Moscow Region 142290, Russia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS, ANR [ANR-2010-JCJC-130801 and ANR-14-CE10-0004-01 to M.G.]; ‘Initiative d'Excellence’ program from the French State [‘DYNAMO’, ANR-11-LABX-0011]; Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding for open access charge: ANR (french national research agency) [ANR-14-CE10-0004-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Goulian M. Two-component signaling circuit structure and properties. Curr. Opin. Microbiol. 2010;13:184–189. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pratt L., Silhavy T. Porin regulon of Escherichia coli. In: Hoch J, Silhavy T, editors. Two-Component Signal Transduction. American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 3.Pratt L.A., Hsing W., Gibson K.E., Silhavy T.J. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 4.Kenney L.J. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 2002;5:135–141. doi: 10.1016/s1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- 5.Goh E.B., Siino D.F., Igo M.M. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J. Bacteriol. 2004;186:4019–4024. doi: 10.1128/JB.186.12.4019-4024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin S., Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogasawara H., Yamada K., Kori A., Yamamoto K., Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K., Nagura R., Tanabe H., Fujita N., Ishihama A., Utsumi R. Negative regulation of the bolA1p of Escherichia coli K-12 by the transcription factor OmpR for osmolarity response genes. FEMS Microbiol. Lett. 2000;186:257–262. doi: 10.1111/j.1574-6968.2000.tb09114.x. [DOI] [PubMed] [Google Scholar]
- 9.Dressaire C., Moreira R.N., Barahona S., Alves de Matos A.P., Arraiano C.M. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio. 2015;6 doi: 10.1128/mBio.02352-14. doi:10.1128/mBio.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn H.J., Cameron A.D., Dorman C.J. Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 2014;10:e1004215. doi: 10.1371/journal.pgen.1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stincone A., Daudi N., Rahman A.S., Antczak P., Henderson I., Cole J., Johnson M.D., Lund P., Falciani F. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res. 2011;39:7512–7528. doi: 10.1093/nar/gkr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takayanagi K., Maeda S., Mizuno T. Expression of micF involved in porin synthesis in Escherichia coli: two distinct cis-acting elements respectively regulate micF expression positively and negatively. FEMS Microbiol. Lett. 1991;67:39–44. doi: 10.1016/0378-1097(91)90440-l. [DOI] [PubMed] [Google Scholar]
- 13.Chen S., Zhang A., Blyn L.B., Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillier M., Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 15.Storz G., Vogel J., Wassarman K.M. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Lay N., Schu D.J., Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel J., Luisi B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmqvist E., Reimegard J., Sterk M., Grantcharova N., Romling U., Wagner E.G. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen M.G., Nielsen J.S., Boysen A., Franch T., Moller-Jensen J., Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- 20.Thomason M.K., Fontaine F., De Lay N., Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Lay N., Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mika F., Busse S., Possling A., Berkholz J., Tschowri N., Sommerfeldt N., Pruteanu M., Hengge R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol. Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno T., Chou M.-Y., Inouye M. A unique mechanism regulating gene expression: Translational inhibition by a complementary RNA transcript (micRNA) Proc. Natl. Acad. Sci. U.S.A. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer V., Papenfort K., Lucchini S., Hinton J.C., Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 25.Guillier M., Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 26.Guillier M., Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beisel C.L., Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagodnik J., Thieffry D., Guillier M. Bacterial small RNAs in mixed regulatory circuits. In: de Bruijn FJ, editor. Stress and environmental regulation of gene expression and adaptation in bacteria. Hoboken: Wiley-Blackwell Publishers; 2016. pp. 371–382. [Google Scholar]
- 29.Beisel C.L., Storz G. The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papenfort K., Espinosa E., Casadesus J., Vogel J. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4772–E4781. doi: 10.1073/pnas.1507825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee J.E., Sheng W., Morgan L.K., Nolet R., Liao X., Kenney L.J. Amino acids important for DNA recognition by the response regulator OmpR. J. Biol. Chem. 2008;283:8664–8677. doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walthers D., Li Y., Liu Y., Anand G., Yan J., Kenney L.J. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross W., Gourse R.L. Analysis of RNA polymerase-promoter complex formation. Methods. 2009;47:13–24. doi: 10.1016/j.ymeth.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbieri C.M., Wu T., Stock A.M. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J. Mol. Biol. 2013;425:1612–1626. doi: 10.1016/j.jmb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J.H. A Short Course in Bacterial Genetics. NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 36.Ross W., Thompson J.F., Newlands J.T., Gourse R.L. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai S.J., Inouye M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 2002;277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 38.Vanderpool C.K., Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 39.Hsing W., Russo F.D., Bernd K.K., Silhavy T.J. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J. Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verhoef C., Lugtenberg B., van Boxtel R., de Graaff P., Verheij H. Genetics and biochemistry of the peptidoglycan-associated proteins b and c of Escherichia coli K12. Mol. Gen. Genet. 1979;169:137–146. doi: 10.1007/BF00271664. [DOI] [PubMed] [Google Scholar]
- 41.Hall M.N., Silhavy T.J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J. Mol. Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita-Kikuta E., Aoki Y., Kinoshita E., Koike T. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell Proteomics. 2007;6:356–366. doi: 10.1074/mcp.T600044-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Slauch J.M., Silhavy T.J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 1989;210:281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- 44.Tran V.K., Oropeza R., Kenney L.J. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J. Mol. Biol. 2000;299:1257–1270. doi: 10.1006/jmbi.2000.3809. [DOI] [PubMed] [Google Scholar]
- 45.Aiba H., Nakasai F., Mizushima S., Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J. Biol. Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- 46.Mizuno T., Kato M., Jo Y.L., Mizushima S. Interaction of OmpR, a positive regulator, with the osmoregulated ompC and ompF genes of Escherichia coli. Studies with wild-type and mutant OmpR proteins. J. Biol. Chem. 1988;263:1008–1012. [PubMed] [Google Scholar]
- 47.Hall M.N., Silhavy T.J. Transcriptional regulation of Escherichia coli K-12 major outer membrane protein 1b. J. Bacteriol. 1979;140:342–350. doi: 10.1128/jb.140.2.342-350.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batchelor E., Goulian M. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U.S.A. 2003;100:691–696. doi: 10.1073/pnas.0234782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siryaporn A., Perchuk B.S., Laub M.T., Goulian M. Evolving a robust signal transduction pathway from weak cross-talk. Mol. Syst. Biol. 2010;6:452. doi: 10.1038/msb.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsing W., Silhavy T.J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delihas N., Forst S. MicF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- 52.Rentschler A.E., Lovrich S.D., Fitton R., Enos-Berlage J., Schwan W.R. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology. 2013;159:316–327. doi: 10.1099/mic.0.059386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L.C., Morgan L.K., Godakumbura P., Kenney L.J., Anand G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foo Y.H., Gao Y., Zhang H., Kenney L.J. Cytoplasmic sensing by the inner membrane histidine kinase EnvZ. Progr. Biophys. Mol. Biol. 2015;118:119–129. doi: 10.1016/j.pbiomolbio.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsui P., Huang L., Freundlich M. Integration host factor binds specifically to multiple sites in the ompB promoter of Escherichia coli and inhibits transcription. J. Bacteriol. 1991;173:5800–5807. doi: 10.1128/jb.173.18.5800-5807.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang L., Tsui P., Freundlich M. Positive and negative control of ompB transcription in Escherichia coli by cyclic AMP and the cyclic AMP receptor protein. J. Bacteriol. 1992;174:664–670. doi: 10.1128/jb.174.3.664-670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinar G., Milo R., Martinez M.R., Alon U. Input output robustness in simple bacterial signaling systems. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19931–19935. doi: 10.1073/pnas.0706792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dahl M.K., Msadek T., Kunst F., Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- 59.Latasa C., Garcia B., Echeverz M., Toledo-Arana A., Valle J., Campoy S., Garcia-del Portillo F., Solano C., Lasa I. Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol. 2012;194:3708–3722. doi: 10.1128/JB.00361-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gopel Y., Gorke B. Rewiring two-component signal transduction with small RNAs. Curr. Opin. Microbiol. 2012;15:132–139. doi: 10.1016/j.mib.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Mandin P., Guillier M. Expanding control in bacteria: interplay between small RNAs and transcriptional regulators to control gene expression. Curr. Opin. Microbiol. 2013;16:125–132. doi: 10.1016/j.mib.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 62.Corcoran C.P., Podkaminski D., Papenfort K., Urban J.H., Hinton J.C., Vogel J. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012;84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 63.Holmqvist E., Unoson C., Reimegard J., Wagner E.G. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol. Microbiol. 2012;84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
- 64.Rutherford S.T., van Kessel J.C., Shao Y., Bassler B.L. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svenningsen S.L., Waters C.M., Bassler B.L. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev. 2008;22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tu K.C., Waters C.M., Svenningsen S.L., Bassler B.L. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol. Microbiol. 2008;70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tu K.C., Long T., Svenningsen S.L., Wingreen N.S., Bassler B.L. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell. 2010;37:567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westermann A.J., Forstner K.U., Amman F., Barquist L., Chao Y., Schulte L.N., Muller L., Reinhardt R., Stadler P.F., Vogel J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 69.Alon U. Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Chapman & Hall/CRC; 2006. [Google Scholar]
- 70.LaPorte D.C., Thorsness P.E., Koshland D.E., Jr Compensatory phosphorylation of isocitrate dehydrogenase. A mechanism for adaptation to the intracellular environment. J. Biol. Chem. 1985;260:10563–10568. [PubMed] [Google Scholar]
- 71.Shin D., Groisman E.A. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J. Biol. Chem. 2005;280:4089–4094. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.