Abstract
cAMP response element binding protein (CREB) is a key regulator of glucose metabolism and synaptic plasticity that is canonically regulated through recruitment of transcriptional coactivators. Here we show that phosphorylation of CREB on a conserved cluster of Ser residues (the ATM/CK cluster) by the DNA damage-activated protein kinase ataxia-telangiectasia-mutated (ATM) and casein kinase1 (CK1) and casein kinase2 (CK2) positively and negatively regulates CREB-mediated transcription in a signal dependent manner. In response to genotoxic stress, phosphorylation of the ATM/CK cluster inhibited CREB-mediated gene expression, DNA binding activity and chromatin occupancy proportional to the number of modified Ser residues. Paradoxically, substoichiometric, ATM-independent, phosphorylation of the ATM/CK cluster potentiated bursts in CREB-mediated transcription by promoting recruitment of the CREB coactivator, cAMP-regulated transcriptional coactivators (CRTC2). Livers from mice expressing a non-phosphorylatable CREB allele failed to attenuate gluconeogenic genes in response to DNA damage or fully activate the same genes in response to glucagon. We propose that phosphorylation-dependent regulation of DNA binding activity evolved as a tunable mechanism to control CREB transcriptional output and promote metabolic homeostasis in response to rapidly changing environmental conditions.
INTRODUCTION
cAMP response element binding protein (CREB) is a ubiquitously expressed bZIP family transcription factor (TF) that has served as a paradigm for stimulus-induced transcription for more than 25 years (1). CREB and the closely related proteins Activating Transcription Factor 1 (ATF1) and cAMP response element modulator (CREM), comprise a subfamily of bZIP TFs that share high homology in their intrinsically disordered KID and bZIP DNA-binding domains. CREB binds to palindromic (TGACGTCA) or half-site (TGAC/G) cyclic AMP response elements (CREs) that typically occur within several kb of a transcription start site. Chromatin immunoprecipitation (ChIP) and in silico-based strategies suggest that CREB occupies several thousand genes; though only a fraction of these genes are responsive to CREB-activating stimuli (2,3). Physiologic roles of CREB are best understood in the contexts of metabolic regulation—where CREB promotes hepatic gluconeogenesis (1,4)—and memory formation, where CREB contributes to long-term potentiation in organisms as diverse as Aplysia and mice (2,3,5).
The canonical CREB activation pathway involves its phosphorylation on S133 by protein kinase A (PKA) (6–9), which causes the unstructured KID to fold into orthogonal helices that associate with the KID-interacting (KIX) domain of CREB-binding protein (CBP) (10,11). Recruitment of CBP to CREB-loaded promoters triggers local histone acetylation and transcriptional activation. A second class of non-HAT CREB coactivators, the CRTCs activate CREB independent of S133 phosphorylation (12–15). CRTCs are retained in the cytoplasm as a consequence of phosphorylation by AMPK family kinases, including salt-inducible kinases (SIKs) 1 and 2, and binding to 14-3-3 proteins (1). Cytoplasmic sequestration of CRTC2 is relieved by cAMP, which triggers PKA-dependent phosphorylation and inactivation of SIKs, and the actions of the Ca2+-dependent phosphatase calcineurin, which dephosphorylates the inhibitor S171 residue on CRTC2 (15). Upon entering the nucleus, CRTCs bind the CREB bZIP domain and enhance both its DNA-binding activity and interaction with CBP (16–19). Although two-hit CREB activation by CBP and CRTC coactivators imparts signal specificity and robustness to CREB-mediated gene expression, it lacks inherent tunability. In addition, it remains unclear whether CREB DNA binding activity is autonomously regulated independent of CRTC tethering.
Work from our laboratory has focused on a cluster of conserved phosphorylation sites within the N-terminal portion of the CREB KID (S108, S111, S114, S117 and S121) that is coordinately phosphorylated by the apical DNA damage signaling kinase, ataxia-telangiectasia-mutated (ATM) and constitutively active CK1 and CK2 (20). ATM is a large molecular mass protein kinase best known for its critical participation in cell cycle checkpoint regulation and the signaling and repair of DNA double-strand breaks (DSBs) (21). Mutations in ATM cause ataxia-telangiectasia (A-T), a syndrome of cancer susceptibility and neurodegeneration in which sensitivity to ionizing radiation (IR) is a defining cellular characteristic. Genetic instability and cancer susceptibility in A-T are thought to arise from defects in cell cycle checkpoint regulation and DNA repair. However, other aspects of the A-T phenotype, including cerebellar neurodegeneration, metabolic syndrome and mitochondrial defects are more difficult to rationalize with known roles of ATM in DNA damage signaling (21–23).
ATM phosphorylates CREB in response to genotoxic stimuli through a processive phosphorylation mechanism (24). ATM-dependent phosphorylation of S111 primes the phosphorylation of S108 by CK2 and the phosphorylation of S114 and S117 by CK1. The phosphorylation of the upstream ATM and CK1/2 sites is required for phosphorylation of S121, which is restricted to conditions of moderate to severe DNA damage. Available evidence suggests that the ATM/CK cluster antagonizes CREB activity, potentially through destabilization of the CREB–CBP interaction (20); however, the physiologic impacts of ATM-mediated CREB phosphorylation remain to be determined and other biochemical roles for these modifications are possible. Finally, upstream CK1 and CK2 sites (S108, S111, S114 and S117) but not S121 are phosphorylated independent of ATM during cell growth (25); however, neither the upstream molecular signals nor functional consequences of this phosphorylation are understood.
Here we provide evidence that ATM/CK cluster phosphorylation influences CREB DNA binding activity and CRTC2 loading in a signal and phosphorylation stoichiometry-dependent manner. Under conditions of DNA damage, stoichiometric phosphorylation of CREB by ATM and CK1/2 led to its chromatin eviction and transcriptional attenuation. Paradoxically, substoichiometric phosphorylation of the ATM/CK cluster in response to cAMP contributed to maximal CREB activation and CRTC2 loading. Combined with the fact that mice expressing a non-phosphorylatable CREB allele exhibited both hypermorphic and hypomorphic CREB activity, these findings suggest a mechanism whereby graded phosphorylation of CREB tunes DNA binding and transcriptional output in response to changing environmental conditions.
MATERIALS AND METHODS
Mouse strains and glucagon injection
C57BL/6J-CREB+/+ and C57BL/6J-CREBS111A mice (homozygous for a Ser-111 to Ala mutation) were housed in colony cages with 12 h light/dark cycle in a temperature-controlled environment. About 8–12 week-old mice were injected with glucagon (200 μg/kg, i.p.; Sigma) or saline (control group). Livers were collected 1 h after injection and gene expression levels were determined. CREB+/+ and CREBS111A mice were placed on a high fat diet (HFD; Research Diets, Inc, D12492), which derived 60% of the caloric content from lard, for 17 weeks at weaning. Blood glucose was measured using a One Touch Ultra glucometer at week 16 of the HFD treatment, following a 16-h fast with cage change. Body weight and epididymal fat pad mass were measured in 20 week-old male mice, at the time of euthanasia.
Cell culture and reagents
Mouse embryonic fibroblasts (MEFs) were isolated from 14.5-day CREB+/+ and CREBS111A embryos. Where indicated, primary MEFs were immortalized by transfection with a plasmid encoding with SV40 large T antigen (p129; a gift from Dr Janet Mertz). Primary hepatocytes were isolated from CREB+/+ and CREBS111A 12-weeks female mice using the collagenase perfusion method. Primary MEFs, immortalized MEFs, primary hepatocytes, HeLa and HEK 293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin-streptomycin. Forskolin (10 μM, Sigma) and IBMX (100 μM, Sigma) were treated for 90 min after serum starvation for overnight. PKA inhibitor H89 (10 μM, Sigma) and ATM inhibitor KU-55933 (10 μM, Sigma) were maintained in dimethyl sulfide (DMSO) and added to cells 30 min prior to stimulation. Calicheamicin A (CLM) was a kind gift of the Pfizer Compound Transfer Program and was maintained 4 μM stock solution in DMSO.
Microarray gene expression data analysis
Microarrays were carried out using Agilent SurePrint G3 Mouse Gene Expression 8 × 60K Microarray 8-plex slides (n = 6 embryos per genotype/per condition). RNA samples were labeled using the Agilent Low Input Quick Amp Labeling Kit, One-Color. Hybridizations were carried out using an Agilent Technologies DNA microarray model #G2565 CA. The pixel intensity data acquired from the microarrays were quantile normalized and log base 2 transformed. Since the arrays were processed at two different times (A and B), the data were corrected for this batch effect using ComBat and an empirical Bayes (26), non-parametric model of the data was performed with genotype and treatment as covariates. The data was then filtered by removing probes if 40 of the 48 arrays had values less than the fifth percentile and missing values subsequently imputed by flooring them at the fifth percentile. Differentially expressed genes were detected by analyzing the pre-processed data with a two-way MicroArray ANalysis Of Variance (MAANOVA) model (27,28) whereby genotype, treatment and the interaction of the two were terms in the model. Contrasts (t-tests) between treated samples and their respective mock (untreated) controls were performed and p-values estimated from 1000 permutations of the samples being compared. Multiple comparisons correction was employed using the Benjamini and Hochberg procedure (29) and statistically significant differentially expressed genes selected based on an absolute fold change cut off of >1.5 and false discovery rate ≤ 0.05. Microarray data have been deposited in the NCBI Gene Expression Omnibus (GSE83891).
Quantitative RT-PCR
RNA analysis was carried out as described (30). The gene-specific primers for qPCR were used: mCREM (5′-TGATTCGCATAAACGTAGAGAAATTC-3′; 5′-CCATGGTAGCAATGTTAGGTGG-3′), mAREG (5′- CACCATAAGCGAAATGCCTTC-3′; 5′-TCTTGGGCTTAATCACCTGTTC-3′), mNR4A1 (5′-GCCTAGCACTGCCAACTTG-3′; 5′-TCTGCCCACTTTCGGATAAC-3′), mNR4A2 (5′- GCGGAGACTTTAGGTGCATGT-3′; 5′- TTGTTTATGTGGCTTGCGC-3′), mHAS2 (5′-CAGACAGGCGGAGGACGA-3′; 5′-AGAAACCTCTCACAATGCATCTTG-3′), mG6P (5′-TCACTTCTACTCTTGCTATCTTTCG-3′; 5′-CCCAGAATCCCAACCACAAG-3′), mPEPCK1 (5′-CCATCCCAACTCGAGATTCTG-3′; 5′-CTGAGGGCTTCATAGACAAGG-3′), mGAPDH (5′-GCCTTCCGTGTTCCTACC-3′; 5′-CCTCAGTGTAGCCCAAGATG-3′) and mPGK1 (5′-ACAGAGGATAAAGTCAGCCATG-3′; 5′-TTGACTTAGCACAGGAAC-3′).
Chromatin immunoprecipitation (ChIP)
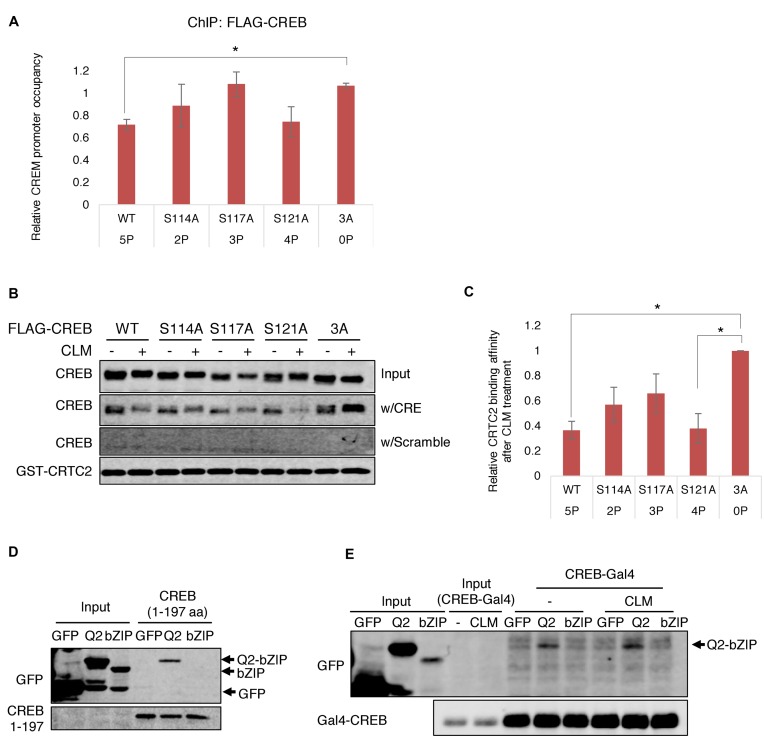
Primary MEFs were cross-linked using 3% formaldehyde (Sigma) for 15 min, lysed and sonicated, and chromatin was immunoprecipitated with α-CREB (Millipore NL-904), α-CBP (Santa Cruz, SC-369) and α-CTRC2 (Calbiochem, ST1099) or α-rabbit IgG. After washing, the immunoprecipitated chromatin was reverse cross-linked and the liberated DNA fragments were purified using Qiagen spin columns. ChIP samples were measured by quantitative real-time polymerase chain reaction (PCR) using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) and SYBR Green. Primer sequences for the promoters: ChIP-mCREM (5′-TGCTAGTTCTTTCCTCCTGCC-3′; 5′-CTCGGAGCTGACGTCAATGT-3′), ChIP-mG6P (5′-GAGGCCTCCCAACATTCAT-3′; 5′-CTCAGAGCGTCTCGCCGGA-3′), ChIP-mPepck1 (5′- AGCCTTTGGTCAACAACTGG-3′; 5′-TGCCTTCGGGGTTAGTTATG-3′). The results were expressed as the relative fold occupancy of the target precipitation as compared to the input. HeLa cells expressed with FLAG-CREBWT, CREBS114A, CREBS117A, CREBS121A or CREB3A were treated with/without 2 nM CLM for 1 h and then crosslinked with 1% formaldehyde for 10 min. After immunoprecipitated with α-FLAG (Sigma, F3165) antibodies, ChIP samples were measured by qPCR using primer sequences for the promoter: ChIP-hCREM (5′-CGCGTGTTATGGATGTTTGTGTGCCT-3′; 5′-GGAACTGACTAAATCACAACATGACTCGGA-3′). The relative FLAG-CREB occupancy on the CREM promoter (Figure 4A) was calculated as a ratio of promoter occupancy in the presence or absence of CLM.
Figure 4.
Phosphorylation of the ATM/CK cluster inhibits DNA binding and CREB–CRTC2–DNA ternary complex formation in graded fashion. (A) ChIP analysis of CREB phosphosite mutants. HeLa cells transfected with FLAG-tagged CREBWT, CREBS114A, CREBS117A, CREBS121A or CREB3A were treated with 2 nM CLM for 1 h and subjected to ChIP-qPCR using α-FLAG antibodies and primers spanning the CREB binding site in the CREM/ICER promoter. *P < 0.05 (Student's t-test). (B) Effects of CREB phosphosite mutations on CREB–CRTC2–DNA ternary complex formation. HEK 293T cells transfected with CREBWT, CREBS114A, CREBS117A, CREBS121A or CREB3A were treated with 2 nM CLM for 1 h and cell lysates incubated with GST-CRTC2CBD and either CRE-containing or scrambled oligonucleotides in the presence of GSH-agarose beads. Immobilized CREB–CRTC2–DNA ternary complexes were washed under indicated buffer conditions analyzed by western blotting with α-CREB and α-GST antibodies. (C) Densitometric analysis of CREB binding to GST–CRTC2CBD–DNA complexes. *P < 0.05 (Student's t-test). (D) Intramolecular interaction between Q2-bZIP and amino-terminal regions of CREB. Recombinant 6×-His-CREB lacking the bZIP domain (CREB 1–197) was incubated with cell lysates from HEK 293T cells transfected with GFP or GFP fused to a minimal CREB bZIP domain (GFP-bZIP) or an extended bZIP domain containing a portion of the adjacent Q2 domain (GFP-Q2-bZIP). The 6×- His tag pull-downs were analyzed by immunoblotting with α-GFP antibody. (E) Interaction between Gal4-CREB and Q2-bZIP is unaffected by DNA damage. GFP, GFP-Q2-bZIP or GFP-bZIP were immunoprecipitated (IP) from transfected HEK 293T cells with α-GFP antibody. The IP samples were incubated with cell lysates from HEK 293T cells separately transfected with Gal4-CREB with or without treatment of 2 nM CLM. Co-immunoprecipitated proteins were analyzed by western blotting with α-GFP or α-CREB antibodies.
Chromatin fractionation
Chromatin fractionation of cells was modified from our previous publication (31). MEFs, HeLa or HEK 293T cell pellets derived from ∼5 × 106 cells were separated to nuclear/cytoplasmic fractions using cytoplasm extraction buffer (10 mM HEPES, 60 mM KCl, 1 mM ethylenediaminetetraacetic acid, 0.1% NP40, 1 mM DTT and 1 mM PMSF). The nuclear fractions were extracted with CSK buffer (10 mM PIPES, 100 mM NaCl, 300 mM sucrose, 1 mM MgCl2, 1 mM DTT, 1 mM EGTA, 0.1% Triton X-100) with protease and phosphatase inhibitors, and centrifuged at 3000 × g for 10 min. The supernatant was collected as the nuclear soluble protein fraction. The remaining cell pellets (chromatin) were washed once with CSK buffer and then boiled in sample loading buffer prior to analysis by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoretic mobility shift assay (EMSA)
The double-stranded oligonucleotides CRE-Forward: 5′-GATCTGCGTCAGAGT-3′ and CRE-Reverse: 5′-ACTCTGACGCAGATC-3′ were labeled with [α-32P] dATP and polynucleotide kinase. Cell lysates or purified CREB proteins were incubated with the probe in a buffer containing 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 7.5% glycerol and 75 ng/μl poly(dI-dC) for 20 min at room temperature. The reaction mixture was separated with 6% native polyacrylamide gel. The [32P]-labeled protein–DNA complexes were visualized by autoradiography.
Recombinant DNA and protein analysis
Gal4-CREB (pFA2-CREB; Agilent) encodes the 147 amino acids Gal4 DNA-binding domain fused to CREB amino acids 1–280. KCREB was a gift from Dr Richard Goodman. GFP-CREB (249-341) and GFP-CREB (282–341) were subcloned into pQCXIN for GFP-Q2-bZIP and GFP-bZIP, respectively. 6×-His-CREB 1–197 was generated by digesting full-length CREB:pET-28a with KpnI followed by Klenow end polishing and religation. The GST-CRTC2CBD plasmid was a kind gift of Dr Marc Montiminy and has been described (12). FLAG-CREB expression plasmids in pFLAG-CMV and pcDNA3.1-based vectors have been described (20,24,25). Purified CREB protein was a kind gift from Dr Jennifer Nyborg (Colorado State University). Protein was extracted from cultured cells in high salt lysis buffer (25 mM HEPES (pH 7.4), 300 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 0.1% Triton X-100) supplemented with protease inhibitors and phosphatase inhibitors for 10 min on ice and proteins were separated on a SDS-PAGE. Antibodies used in this study include: α-pCREB-Ser-108/Ser-111/Ser-114 (24), α-pCREB-Ser-121 (20), α-pCREB-Ser-133 (Millipore 06–519 and Cell Signaling 87G3), α-FLAG (Sigma F7425), α-FUS (Santa Cruz 4H11), α-MCM3 (Bethyl laboratory A300-192A), α-Gal4 (Santa Cruz RK5C1), α-GFP (Santa Cruz B-2), α-β-tubulin (Millipore AA2) and α-HSP90 (Cell Signaling C45G5).
RESULTS
CREB is phosphorylated on CK1/2 sites in response to cAMP
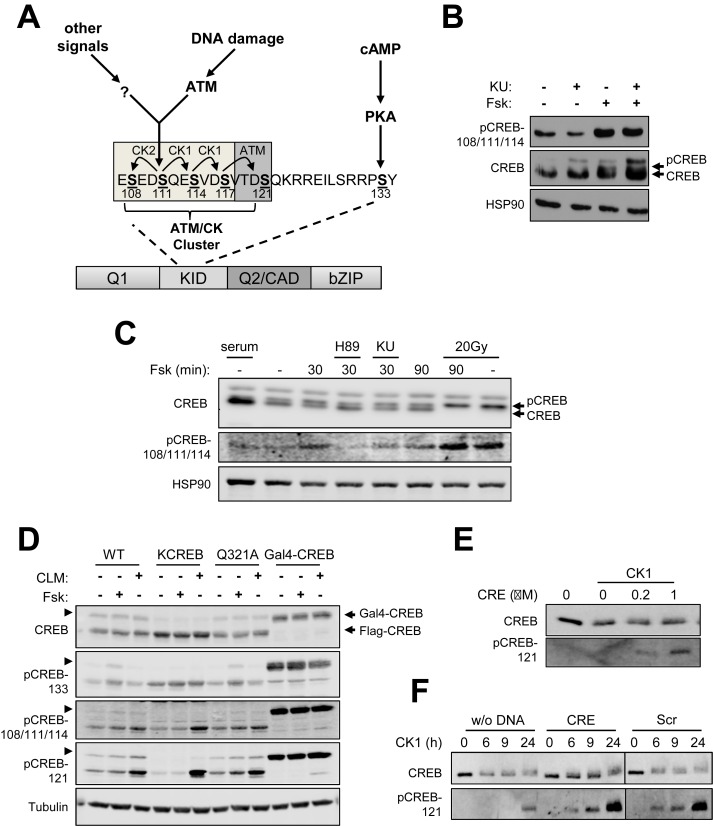
Previous work from our laboratory established that the ATM/CK cluster is processively phosphorylated by ATM and CK1/2 in response to DNA damage and phosphorylated independent of DNA damage and ATM during cell growth (20,24,25) (Figure 1A); however, the upstream signals controlling DNA damage-independent CREB phosphorylation are not known. During the course of experiments using the adenylate cyclase agonist forskolin (Fsk) we noticed that CREB isolated from MEFs treated with Fsk exhibited an electrophoretic mobility shift highly characteristic of ATM/CK cluster phosphorylation (Figure 1B). Immunoblotting with an antibody that detects CREB triply phosphorylated on S108, S111 and S114 (α-CREB-pS108/111/114) confirmed these residues were phosphorylated upon Fsk exposure. Although ATM phosphorylates CREB on S111 in response to IR (20), the ATM inhibitor KU-55933 had no impact on S111 phosphorylation by Fsk (Figure 1B). On the other hand, Fsk did not induce immunoreactivity with an antibody that detects CREB phosphorylated on S121, which is consistent with our previous findings that phosphorylation of this site is DNA damage dependent (data not shown). Fsk-induced CREB S111 phosphorylation was observed within 30 min of treatment and was blocked by the PKA inhibitor, H-89 (Figure 1C, top and middle panels). Thus, even though S111 is not a consensus PKA site and is unlikely to be directly phosphorylated by PKA, PKA activity is upstream of S111 phosphorylation in response to Fsk. One possibility that could account for these findings is that phosphorylation of CREB on S133 renders it more susceptible to phosphorylation by CK1/2, which are abundant and constitutively active kinases. Arguing against this, however, Fsk-induced phosphorylation of the ATM/CK cluster comparably between CREB+/+ MEFs and CREBS133A MEFs generated by Naqvi et al. (32) (Supplementary Figure S1). We conclude that cAMP-induced phosphorylation of the ATM/CK cluster occurs downstream of PKA but independent of S133 phosphorylation.
Figure 1.
cAMP-induced phosphorylation of the CREB ATM/CK cluster. (A) Schematic depiction of the CREB ATM/CK phosphorylation cluster. (B) CREB+/+ MEFs were pretreated with KU-55933 (KU) for 30 min and then the cells were treated with Fsk for 90 min. Cell extracts were immunoblotted with α-pCREB-108/111/114, α-CREB or α-HSP90 antibodies. (C) CREB+/+ MEFs were pretreated with either H-89 (PKA inhibitor), KU-55933 (ATM inhibitor) or 20 Gy IR followed by Fsk for 30 or 90 min. Cell extracts were analyzed using α-CREB, α-pCREB-108/111/114 or α-HSP90 antibodies. (D) Phosphorylation of CREB was analyzed by western blotting using cell lysates prepared from HEK 293T cells transfected with plasmids encoding CREBWT, KCREB, CREBQ321A (CRTC2 binding mutant) or Gal4-CREB. The transfected cells were treated with Fsk or 2 nM CLM for 60 min. CREB expression and phosphorylation were determined by immunoblotting with indicated antibodies. Arrowheads (lane 1–9) indicate presumed Sumo-CREB detected by CREB antibodies. (E) Recombinant CREB was incubated with CK1 and ATP with the indicated amount of CRE oligonucleotide. CREB phosphorylation was analyzed by western blotting using α-CREB or α-pCREB-121 antibodies. (F) Recombinant CREB was phosphorylated with CK1 in the presence or absence of CRE or scrambled DNA (Scr) for the indicated times. CREB phosphorylation was analyzed by western blotting using α-CREB or α-pCREB-121 antibodies.
cAMP-induced CREB phosphorylation requires a functional DNA-binding domain
We used transiently expressed CREB proteins to explore determinants of CREB phosphorylation by cAMP. Interestingly, a R301L mutation (KCREB) that abolishes CREB DNA binding diminished constitutive and Fsk-induced phosphorylation of S111 in HEK 293T cells (Figure 1D), whereas DNA damage-induced phosphorylation of KCREB was largely intact. A CREBQ321A mutant that retains partial DNA binding activity but is defective for CRTC2 binding (16) exhibited an intermediate S111 phosphorylation phenotype (Figure 1D). On the other hand, a C-terminal fusion of CREB to the Gal4 DNA binding domain (Gal4-CREB) showed high levels of both S111 and S121 phosphorylation when expressed in HEK 293T cells even though it lacks a bZIP domain. Thus, DNA binding activity—but not CRE-specific DNA binding—is required for maximal CREB phosphorylation on CK1/2 sites in cells.
To further investigate the relationship between DNA binding and phosphorylation we examined the effect of dsDNA oligonucleotides on CREB phosphorylation by CK1 in vitro. Oligonucleotides containing or lacking an embedded CRE stimulated the CK1-dependent phosphorylation of CREB on the ATM/CK cluster in vitro. The stimulation of CREB phosphorylation by CK1 was DNA concentration dependent and most pronounced for the S121 phosphorylation site (Figure 1E and F). We conclude that targeting to DNA increases the phosphorylation of CREB by CK1/2 in cellulo and that binding to DNA increases the intrinsic phosphorylation potential of recombinant CREB in vitro.
CREB S111 phosphorylation inhibits or enhances CREB target gene expression in a signal dependent manner
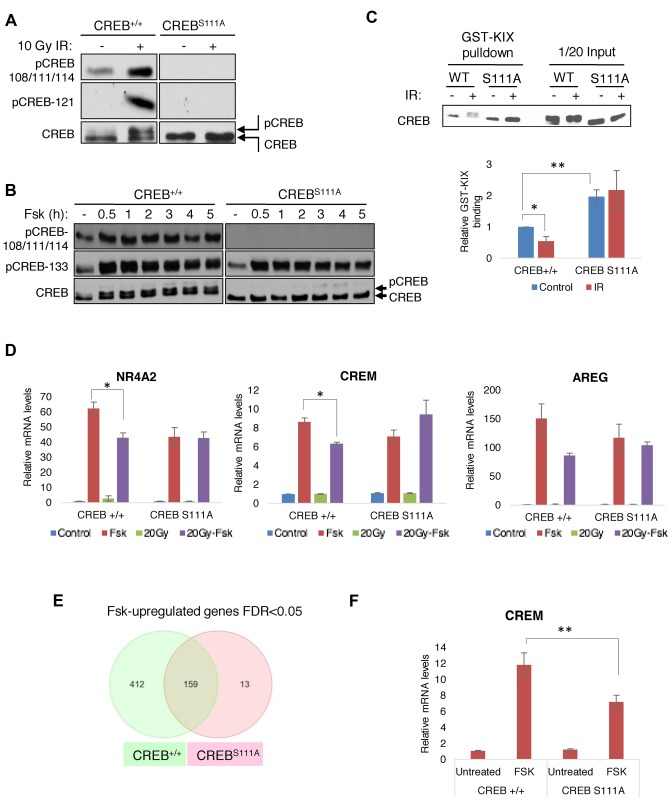
To further explore the physiologic relevance of ATM/CK cluster phosphorylation in response to activating and stress stimuli, we introduced a Ser-111 to Ala knock-in mutation into the mouse germ line that abolishes phosphorylation of the ATM/CK cluster (24) (Supplementary Figure S2). Homozygous CREBS111A/S111A (CREBS111A) knock-in mice were indistinguishable in appearance or activity level from wild-type (WT) littermates. As predicted from previous studies, IR-induced phosphorylation of S121 was undetectable in CREBS111A splenocytes (Figure 2A) and the characteristic IR-induced electrophoretic mobility shift of CREB due to phosphorylation of CK1/2 sites at S108, S111, S114 and S117, was abolished. Fsk-induced phosphorylation of the ATM/CK cluster was also abolished in CREBS111A MEFs (Figure 2B). Finally, given previous work showing that DNA damage-induced phosphorylation of S111 inhibited CREB interaction with the KID-interacting (KIX) domain of CBP (20,24) we tested binding of CREB to a GST-KIX domain fragment in pull-down assays. We found that CREBS111A from spleen extracts bound to the CBP KIX domain more strongly than did CREBWT. IR had a small but reproducible inhibitory effect on the CREBWT/KIX domain interaction, whereas CREBS111A binding to KIX was completely resistant to IR (Figure 2C).
Figure 2.
ATM/CK cluster positively and negatively regulates CREB transcriptional activity. (A and B) A Ser-111 to Ala knock-in mutation abolishes ATM/CK cluster phosphorylation in response to DNA damage (A) and Fsk (B). In panel (A) ex vivo cultured splenocytes were sham treated or exposed to 10 Gy IR and cell extracts were prepared 1 h later for western analysis using the indicated antibodies. In panel (B) CREB+/+ and CREBS111A MEFs were treated with 10 μM Fsk and the cells were collected at the indicated times. Cell extracts were immunoblotted with α-pCREB-108/111/114, α-pCREB-133 or α-CREB antibodies. (C) CREBS111A protein exhibited increased affinity for the CBP KIX domain. GST-KIX pull-downs were performed using spleen extracts from CREB+/+ or CREBS111A mice before or after 10 Gy irradiation. GST-KIX binding activity was measured by densitometric analysis. n = 4. (D) DNA damage-resistant gene expression in CREBS111A MEFs. CREB+/+ and CREBS111A MEFs were treated as in (E) and levels of indicated genes determined by qPCR. Each bar represents averaged results for three biological replicates, assayed three times each. (E) Fsk-inducible genes in CREB+/+ and CREBS111A MEFs determined by microarray analysis. (F) Defective cAMP-induced expression of CREM in CREBS111A MEFs. CREB+/+ and CREBS111A MEFs were treated with Fsk for 90 min and CREM mRNA expression analyzed by qPCR. Each bar represents averaged results, n = 24. Data information: in (C–F), data are presented as mean ± SEM. *P < 0.05, **P < 0.02 (Student's t-test).
Microarray experiments were employed to examine the effects of the S111A mutation on CREB target gene expression in MEFs treated with IR, Fsk or Fsk preceded by IR. The top 50 Fsk-induced genes and their expression under the IR-Fsk condition in CREB+/+ and CREBS111A MEFs are shown in Supplementary Table S1A and B. The majority of Fsk-induced genes in CREB+/+ MEFs showed reduced expression when Fsk was preceded by IR, whereas the same CREB target genes were, for the most part, not attenuated in IR-Fsk-treated CREBS111A MEFs (Table 1). Guided by a publication from Brindle and colleagues (33), we used qPCR to query expression of CBP/p300-dependent and CBP/p300-independent CREB target genes in CREB+/+ and CREBS111A MEFs, reasoning that the former set of genes may be most sensitive to DNA damage and the S111A mutation. The Fsk-induced expression of both CBP/p300-dependent (AREG) and CBP/p300-independent (CREM, NR4A2) was attenuated by IR and, in all instances, attenuation failed to occur in CREBS111A MEFs (Figure 2D). Similar findings were made for additional CBP-dependent and CBP-independent CREB target genes (Supplementary Figure S3). Thus, IR attenuates CREB-mediated gene expression in a Ser-111 phosphorylation-dependent manner; however, because this occurred for both CBP-dependent and CBP-independent genes, diminution of CBP/p300 binding is not the major mechanism through which this occurs.
Table 1. Relative attenuation of Fsk-induced genes by IR in CREB+/+ and CREBS111A MEFs.
| CREB +/+ | CREB S111A | |
|---|---|---|
| Gene symbol | Fold change (log2) Fsk and IR versus Fsk_only and IR only | Fold change (log2) Fsk and IR versus Fsk_only and IR_only |
| Junb | −1.99482 | 1.13155 |
| Areg | −1.80694 | −1.21135 |
| Il6 | −1.80623 | −1.16726 |
| Gem | −1.73148 | −1.02846 |
| Scrt1 | −1.72122 | −1.07231 |
| Nefl | −1.70273 | −1.10401 |
| Crem | −1.68828 | 1.0897 |
| Adrb1 | −1.55724 | −1.06747 |
| Ier3 | −1.54472 | 1.23746 |
The above findings supported an inhibitory role for the ATM/CK cluster in CREB transcriptional activation following IR exposure. Surprisingly, however, a comparison of the MEF microarray data revealed the opposite trend in the context of cAMP signaling, where 571 and 172 genes were significantly upregulated by Fsk in CREB+/+ and CREBS111A MEFs, respectively (Figure 2E). Consistent with a mild activation defect, qPCR experiments revealed blunted CREM induction by Fsk in CREBS111A MEFs, with similar findings made for other CREB target genes (Figure 2F and Supplementary Figure S4A). As a whole, fewer genes were significantly upregulated by Fsk in CREBS111A MEFs versus CREB+/+ MEFs, supporting the notion that Fsk-inducible gene expression was slightly compromised by the S111A mutation (Supplementary Figure S4B). Thus, ATM/CK cluster phosphorylation attenuates target gene expression in the context of DNA damage but paradoxically contributes to maximal gene expression in response to cAMP.
S111 dependence of CREB target genes correlates with CRTC2 occupancy
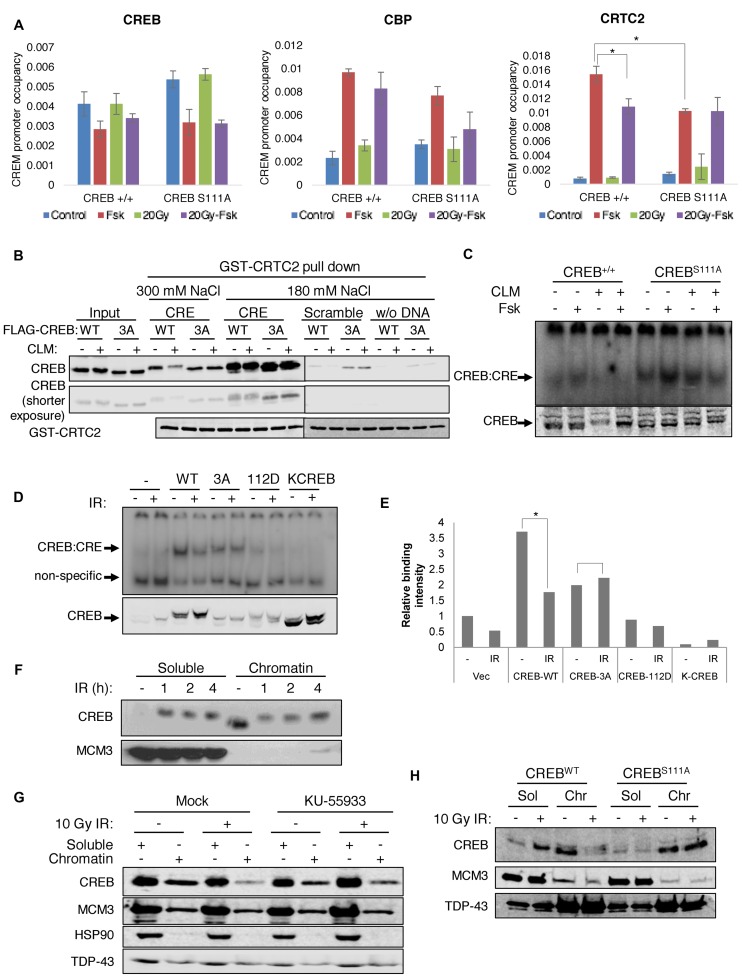
We next sought to determine the underlying mechanisms for S111 phosphorylation-dependent changes in CREB-mediated transcription. ChIP assays were performed to measure the occupancy of CREB and its coactivators, CBP and CRTC2 over CREB target genes in CREB+/+ and CREBS111A MEFs before and after DNA damage. Fsk exposure increased CBP occupancy over AREG and CREM/ICER promoters in CREB+/+ MEFs (Figure 3A and Supplementary Figure S5A); however, CBP recruitment was not significantly inhibited by IR, suggesting that either CBP is not the critical target of DNA damage-dependent regulation or that the CBP ChIP assay lacks sufficient dynamic range to reveal such differences. On the other hand, Fsk-dependent recruitment of CRTC2 to CREM and AREG was reduced in an S111 phosphorylation-dependent manner upon irradiation (Figure 3A and Supplementary Figure S5A). CRTC2 recruitment to CREM was also reduced in immortalized CREB+/+ MEFs, but not CREBS111A MEFs treated with the radiomimetic agent CLM (Supplementary Figure S5B). Reductions in CRTC2 promoter recruitment could be caused by defects in its nuclear translocation; however, CRTC2 nuclear accumulation was comparable between CREB+/+ and CREBS111A MEFs treated with Fsk in the absence or presence of CLM (Supplementary Figure S6A and B). Finally, ChIP experiments also revealed that Fsk-induced CRTC2 recruitment to CREM was significantly lower in CREBS111A MEFs than CREB+/+ MEFs (Figure 3A). The reduced recruitment of CRTC2 plausibly explains the blunted induction of CREB target genes in CREBS111A MEFs.
Figure 3.
Impact of DNA damage and CREB S111 phosphorylation on coactivator recruitment and DNA binding activity. (A) ChIP analysis of CREB, CBP and CRTC2 over the CREM promoter in CREB+/+ and CREBS111A MEFs. Cells were treated with Fsk for 90 min, irradiated with 20 Gy IR for 120 min (20 Gy) or treated with 20 Gy IR 30 min prior to Fsk (20 Gy-Fsk) and processed for ChIP-qPCR using the indicated antibodies. Y-axis represents the CREM promoter occupancy. Each bar represents averaged results, n = 3. Error bars indicate SEM. *P < 0.05 (Student's t-test). (B) DNA damage inhibits CREB–CRTC2–DNA ternary complex formation in an S111 phosphorylation-dependent manner. HEK 293T cells transfected with CREBWT or CREB3A were treated with 2 nM CLM for 1 h and cell lysates incubated with GST-CRTC2CBD and either CRE-containing or scrambled oligonucleotides in the presence of GSH-agarose beads. Immobilized CREB–CRTC2–DNA ternary complexes were washed under indicated buffer conditions analyzed by western blotting with α-CREB and α-GST antibodies. (C) Immortalized CREB+/+ and CREBS111A MEFs were treated with Fsk for 90 min, treated with 2 nM CLM for 120 min (CLM), or treated with CLM for 30 min prior to Fsk (CLM + Fsk). CREB–CRE complexes were analyzed by electrophoretic mobility shift assay (EMSA). (D) DNA-binding activity was analyzed by EMSA using cell lysates prepared from HEK 293T cells transfected with empty vector, or plasmids encoding CREBWT, CREB3A (100A, 111A and 121A), CREBQ112D or DNA-binding defective KCREB. The transfected cells were irradiated with 50 Gy IR for 1 h and CREB expression analyzed by immunoblotting. (E) CRE binding activity was measured by densitometric analysis. *P < 0.05 (Student's t-test). (F) DNA damage caused dissociation of CREB from bulk chromatin. HeLa cells were irradiated with 10 Gy IR, collected at the indicated times, and fractionated into soluble (Sol) or chromatin (Chr) fractions prior to sodium dodecyl sulphate-polyacrylamide gel electrophoresisand immunoblotting with α-CREB or α-MCM3 antibodies. (G) CREB chromatin dissociation is ATM dependent. HeLa cells were pretreated with DMSO or ATM inhibitor (KU-55933) for 30 min, and then irradiated with 10 Gy IR for 1 h. Soluble (Sol) or chromatin (Chr) fractions were analyzed using α-CREB, α-HSP90, α-MCM3 or α-TDP-43 antibodies. (H) The S111A mutation inhibits IR-dependent CREB chromatin eviction. Chromatin fractionation of CREB+/+ and CREBS111A MEFs was carried out 1 h after irradiation of 10 Gy IR.
The CREB bZIP binds to the CREB-binding domain (CBD) of CRTC2 to form a ternary complex with enhanced DNA-binding activity (12,16). To investigate the effects of DNA damage and ATM/CK cluster phosphorylation on CREB–CRTC2–DNA ternary complexes, we incubated cell extracts from control or CLM-treated HEK 293T cells expressing CREBWT or a non-phosphorylatable CREB3A mutant harboring Ala mutations at Thr-100, Ser-111 and Ser-121 with recombinant GST-CBD and dsDNA oligonucleotides with or without an embedded CRE. Consistent with previous findings (16), the interaction of GST-CBD and CREB was enhanced by dsDNA, and CRE-containing DNA fragments stimulated interaction to a greater extent than those lacking a CRE (Figure 3B). Under conditions of isotonic NaCl, CREBWT and CREB3A interacted comparably with GST-CBD/DNA and cellular exposure to CLM had little effect on the interaction. However, ternary complexes containing phosphorylated CREBWT but not CREB3A, were dissociated upon washing with buffer containing 300 mM NaCl (Figure 3B). Thus, DNA damage disrupts CREB–CRTC2–DNA ternary complexes in a phosphorylation-dependent manner.
DNA damage-induced phosphorylation of the ATM/CK cluster inhibits CREB DNA-binding activity
Because CRTC2 binding is stimulated by CREB interaction with DNA, it was possible that DNA damage-dependent reductions in CREB–CRTC2–DNA ternary complexes occurred secondary to reduced association of CREB with DNA. To investigate this we performed electrophoretic mobility shift assays (EMSA) using extracts prepared from CREB+/+ and CREBS111A MEFs. CLM exposure also reduced CREB DNA binding affinity in CREB+/+ MEFs but not CREBS111A MEFs, supporting the notion that ATM-mediated phosphorylation of the ATM/CK cluster reduces CREB DNA-binding affinity (Figure 3C). DSB inducers etoposide and camptothecin also reduced CREB DNA binding affinity in CREB+/+ MEFs but not CREBS111A MEFs (Supplementary Figure S7).
We also performed EMSA using extracts prepared from HEK 293T cells transiently transfected with plasmids encoding WT and mutant CREB cDNAs. Binding of CREBWT to a double-stranded oligonucleotide probe containing a CRE half-site was reduced following exposure of HEK 293T cells to IR whereas the binding of non-phosphorylatable CREB3A was unaffected (Figure 3D and E). A CREBQ112D mutant that exhibits a reduced threshold for DNA damage-dependent phosphorylation (24) bound weakly to the probe and its binding was further reduced upon DNA damage. A DNA-binding defective CREBR301L ‘KCREB’ mutant (34), failed to bind the CRE probe in these experiments. Finally, we observed a strong decrease in CREB chromatin association following DNA damage (Figure 3F). IR-dependent CREB chromatin dissociation was partially suppressed by the ATM inhibitor KU-55933 and was abolished by the S111A mutation (Figure 3G and H). From the combined EMSA and chromatin fractionation studies we conclude that DNA damage-induced phosphorylation of the ATM/CK cluster diminishes affinity for DNA.
The ATM/CK cluster functions as a DNA binding rheostat
Multisite phosphorylation domains can function as molecular switches (35) to mediate on-off responses to threshold stimuli, or as molecular rheostats to impart graded biochemical behavior proportional to the number of phospho-residues (36). To investigate whether the ATM/CK cluster operates as a switch or rheostat we carried out ChIP experiments using an allelic series of CREB phospho-site mutants, which owing to the processive nature of phosphorylation (24), generate predictable phosphorylation intermediates upon DNA damage. Because initial ChIP experiments in Figure 3A did not reveal different occupancy patterns between endogenous CREBWT and CREBS111A in MEFs, we employed milder formaldehyde crosslinking conditions to reduce handcuffing artifacts that may mask dynamic CREB–DNA interactions. We found that DNA damage reduced CREB occupancy of the CREM promoter, with the magnitude of dissociation roughly correlating with predicted CREB phosphorylation stoichiometry. CREBWT (five phosphorylated residues, or 5P), showed maximal dissociation from the CREM promoter following CLM, whereas CREB3A (no phosphorylated residues or 0P) was completely resistant to CLM (Figure 4A). CREBS114A (2P), CREBS117A (3P) and CREBS121A (4P), showed intermediate levels of dissociation following CLM treatment, with CREBS121A behaving similar to CREBWT. These findings suggest that ATM/CK cluster phosphorylation inhibits CREB DNA binding activity in graded fashion proportional to the number of phosphate groups.
The same allelic series of CREB phosphosite mutants was tested for ternary complex formation in the presence of GST-CBD and dsDNA. As expected, CLM treatment inhibited the interaction between CREBWT and GST-CBD, but had essentially no effect on the interaction between CREB3A and GST-CBD (Figure 4B and C). Binding of GST-CBD to CREBS114A, CREBS117A and CREBS121A, showed intermediate responsiveness to CLM, with the more heavily phosphorylated CREBS121A isoform exhibiting the lowest affinity. Combined with the ChIP findings, these results indicate that the multisite phosphorylation of the ATM/CK cluster imparts graded behavior, as opposed to switch-like properties, to CREB DNA binding and CRTC ternary complex formation.
We next considered how phosphorylation of the distal ATM/CK cluster, which resides in N-terminal portion of the unstructured KID, might modulate CREB DNA binding activity and CREB–CRTC2–DNA ternary complex formation. One possibility was that the hyperphosphorylated ATM/CK cluster competes with the phosphate backbone of DNA for binding to basic residues within the bZIP. To test this we carried out pull-down experiments using a purified, truncated form of His-CREB lacking the bZIP domain (CREB 1–197) and GFP-bZIP-domain fragments (see methods) that had been expressed in HEK 293T cells. As shown in Figure 4D, His-CREB 1–197 interacted with a GFP-tagged CREB fragment containing the bZIP and a portion of the adjacent Q2 domain (Q2-bZIP) but not a minimal GFP-bZIP. Thus, the isolated amino terminal and Q2-bZIP domains interact; however, attempts to phosphorylate CREB 1–197 with CK1/2 in vitro were unsuccessful, possibly because phosphorylation requires a functional DNA binding domain (Figure 1D). To work around this difficulty we employed the Gal4-CREB construct that undergoes robust phosphorylation of the ATM/CK cluster in intact cells (Figure 1D). Gal4-CREB interacted with the GFP-bZIP in co-IP assays, however the interaction was not affected by CLM treatment (Figure 4E). We conclude that CREB is capable of intramolecular association; however, the interaction appears to occur both in the absence and presence of ATM/CK cluster phosphorylation.
CREB S111 phosphorylation contributes to the regulation of gluconeogenic genes
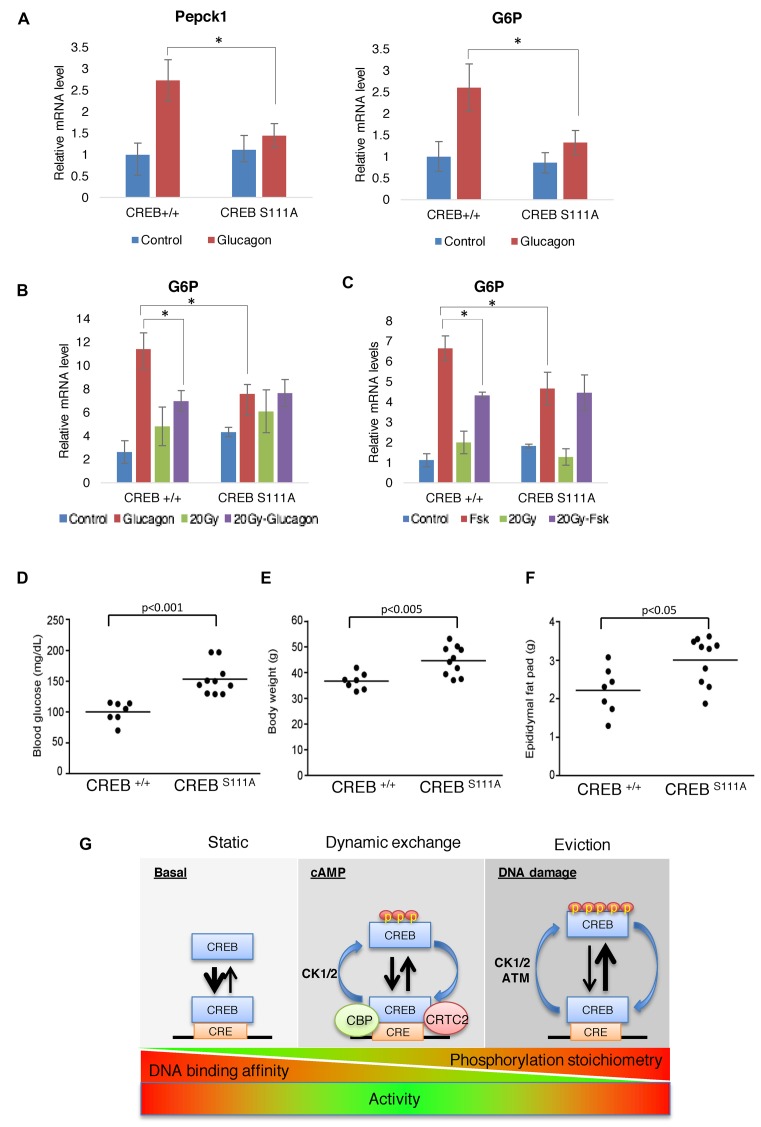
During fasting, CREB works in conjunction with FOXO TFs in the liver to induce gluconeogenic gene expression in response to the peptide hormone glucagon (37). To examine whether ATM/CK cluster phosphorylation modulates CREB-dependent expression of gluconeogenic genes we measured hepatic glucose-6 phosphatase (G6P) and PEPCK expression CREB+/+ and CREBS111A mice following intraperitoneal injection with glucagon. Glucagon-induced expression of hepatic G6P and PEPCK were attenuated in CREBS111A mice versus CREB+/+ mice (Figure 5A), suggesting that S111 phosphorylation is required for maximal induction of gluconeogenic CREB target genes in vivo. To extend these findings we measured G6P expression in CREB+/+ and CREBS111A hepatocytes under conditions of DNA damage and in response glucagon. Preexposure to IR attenuated glucagon-induced expression of G6P in CREB+/+ hepatocytes, but not CREBS111A hepatocytes, with similar findings made for Fsk (Figure 5B and C), supporting an inhibitory role for ATM/CK cluster phosphorylation in the context of these gluconeogenic genes. On the other hand, and consistent with studies using MEFs, Fsk-induced level of G6P was lower in CREBS111A hepatocytes relative to CREB+/+ hepatocytes (Figure 5C). CREBS111A mice also exhibited elevated fasting glucose and increased body mass and adiposity under high fat diet feeding (Figure 5D–F) supporting the notion that phosphorylation of the ATM/CK cluster contributes to glucose metabolism and metabolic homeostasis in vivo.
Figure 5.
CREB S111 phosphorylation contributes to the bidirectional regulation of gluconeogenic genes by cAMP and IR in vivo. (A) CREB+/+ and CREBS111A mice were injected with either PBS or 200 μg/kg glucagon and expression levels of G6P and PEPCK at 1 h analyzed by qPCR. Each bar represents averaged results for three biological replicates, assayed three times each. Error bars indicate SEM *P < 0.02 (Student's t-test). (B) CREB+/+ and CREBS111A primary hepatocytes were treated with glucagon for 60 min, irradiated with 20 Gy IR for 120 min (20 Gy) or treated with glucagon for 60 min after 20 Gy IR for 30 min (20 Gy-glucagon). *P < 0.05 (Student's t-test). (C) CREB+/+ and CREBS111A primary hepatocytes were treated with Fsk for 90 min, irradiated with 20 Gy IR for 120 min (20 Gy) or treated with Fsk for 90 min after 20 Gy IR for 30 min (20 Gy-Fsk). *P < 0.05 (Student's t-test). (D) CREBS111A mice exhibit hyperglycemia. Fasting blood glucose levels were measured in 19-week-old male mice (n = 7 CREB+/+ and 10 CREBS111A, respectively) under fasted (16 h) conditions. (E and F) CREBS111A mice maintained on a high fat diet exhibit increased body weight (E) and adiposity (F). (G) Working model for phosphoregulation of CREB DNA binding activity in response to cAMP and DNA damage.
DISCUSSION
The current model for CREB-mediated transcription invokes a two-hit mechanism in which the independent recruitment of CBP and CRTC coactivators is required for maximal CREB target gene induction (1). Our studies expand the CREB regulation paradigm to include autonomous, phosphorylation-dependent control of CREB DNA-binding activity. We propose that phospho-dependent regulation enhances the dynamic range of CREB-mediated transcription and speculate that such regulation may engender fine control at the level of individual CREB target genes and binding sites.
Previous experiments in the setting of DNA damage revealed that phosphorylation of the ATM/CK cluster diminished CBP/p300 binding (20), which is compatible with the location of the ATM/CK cluster near the KIX-interacting surface of the CREB KID. Although endogenously expressed CREBS111A exhibited elevated binding to the CBP KIX domain relative to WT CREB (Figure 2C), the finding that both CBP-dependent (AREG) and CBP-independent (CREM) genes underwent S111 phosphorylation-dependent attenuation following IR makes it unlikely that inhibition of CBP binding is the primary mechanism of CREB target gene attenuation. Instead, chromatin fractionation, DNA pull-down, ChIP and EMSA experiments point toward reductions in DNA binding activity and CRTC2 association as the major functional consequence of ATM/CK cluster phosphorylation. Regulation of CREB DNA binding activity through post-translational modification has been implied through a handful of studies (38–41). Most notably, Horiuchi and et al. demonstrated that phosphorylation of positionally conserved CK1/2 sites in Drosophila CREB inhibited its DNA-binding affinity in vitro (40). However, neither the regulatory mechanisms nor functional impacts of these phosphorylation sites were identified.
ATM/CK cluster phosphorylation inhibited CREB DNA binding and CRTC2 association in graded fashion proportionate to the number of phosphorylated residues. Under conditions of high DNA damage and maximal ATM activity, the ATM/CK cluster is stoichiometrically phosphorylated on all five sites and DNA binding at its minimum. Exposure to lower levels of DNA damage or cAMP (discussed below) leads to intermediate phosphorylation stoichiometries and intermediate DNA-binding affinities (Figure 5G). By preventing the phosphorylation of all five sites, the S111A mutation ostensibly abolished DNA damage-dependent attenuation of CREB DNA-binding activity, whereas S114A, S117A and S121A mutations—which have successively smaller impacts on ATM/CK cluster phosphorylation—had increasingly smaller impacts on DNA binding activity (Figure 5A). Although structural information is lacking, attenuation may involve allosteric communication between the intrinsically disordered KID and bZIP domain. Intrinsically disordered domains are increasingly recognized for their ability to form folded structures upon interaction with cognate binding partners, and these disorder to order transitions are often regulated through post-translational modification (42). Precedent for such a mechanism exists for Ets-1, where multisite phosphorylation of a Ser-rich region allosterically modulates the Ets-1 DNA-binding domain (43–45). An N-terminal fragment of CREB containing the ATM/CK cluster interacted with an extended CREB bZIP domain in vitro, though the phospho-dependence of this interaction could not be assessed (Figure 4D and E).
An unexpected finding from this study is that cAMP-induced phosphorylation of the ATM/CK cluster contributes to maximal CREB activation (Figure 1B). cAMP-induced phosphorylation of the ATM/CK cluster required PKA, but not S133 phosphorylation and was coupled to a functional DNA-binding domain (Figure 2B). Although the precise phosphorylation mechanism remains to be determined, it is plausible that DNA binding leads to a change in the ATM/CK cluster that renders it more accessible to CK1/2, and possibly, additional CREB kinases. That DNA binding strongly enhanced CREB phosphorylation lends credence to the idea that the KID and bZIP domains engage in bidirectional communication. How can cAMP-induced phosphorylation of the ATM/CK cluster promote transcriptional activation given its negative impacts on DNA binding? One parsimonious explanation for this paradox is that CREB proteins harboring a substoichiometric phosphorylation profile—and thus an intermediate affinity for DNA—are transcriptionally more potent than CREB proteins that are either unphosphorylated or fully phosphorylated. Whereas full phosphorylation of the ATM/CK cluster leads to overt chromatin eviction, intermediate CREB phosphorylation stoichiometries may reduce sequence non-specific CREB DNA binding affinity, thereby increasing its sliding rate, and/or enable dynamic promoter exchange, which has been implicated in transcriptional activation by other TFs (46–48). Given CRTC2 recruitment defects in CREBS111A cells (Figure 3A), dynamic CREB-DNA interactions may also facilitate CRTC2 loading at CREB target gene promoters. Future studies will address whether the effects of ATM/CK cluster phosphorylation and CRTC2 loading can be genetically separated.
Physiologic importance of cAMP-induced ATM/CK cluster phosphorylation is supported by the blunted induction of gluconeogenic CREB target genes in CREBS111A livers and primary hepatocytes (Figure 5A–C). At the same time, CREBS111A mice exhibited elevated fasting glucose, suggesting that ATM/CK cluster phosphorylation is required for long-term glucose homeostasis. Conditional knockout of CREB in the liver did not lead to overt changes in gluconeogenesis or circulating glucose level (49), suggesting that glucose handling defects in CREBS111A mice occur through a dominant mechanism. Because CREBS111A was expressed in all tissues it is possible that hyperglycemia in CREBS111A mice is due to increased gluconeogenesis in the liver, defective pancreatic insulin secretion, global insulin resistance or a combination of mechanisms.
How CREB phosphorylation contributes to the ATM-dependent DNA damage response is presently unclear. Cell cycle checkpoint activation, proliferation rates, apoptosis and gross DBS repair were essentially normal in CREBS111A MEFs or hematopoietic tissues, suggesting that CREB phosphorylation does not make a major contribution to these established endpoints of ATM signaling. CREBS111A mice also did not exhibit elevated rates of tumorigenesis on the C57BL/6 background. Although we can only speculate, additional collaborating genetic insults may be required to unmask phenotypes associated with defective ATM-CREB signaling.
On the other hand, given that A-T patients and ATM knockout mice develop features of metabolic syndrome (23), it is conceivable CREB phosphorylation selectively plays a role in ATM-dependent metabolic regulation. Future studies using ATM−/− and CREBS111A mice will be needed to measure the activation and functional consequences of ATM-CREB signaling in liver under chronic conditions of HFD and other stress insults. Finally, it is worth considering that a role in transcriptional activation constitutes the primordial function of the ATM/CK cluster and that sequence differences between the ATM/CK clusters of mammalian CREB paralogs contribute to functional uniqueness. ATF1 is heavily phosphorylated on CK1/2 sites and undergoes additional phosphorylation by ATM in response to DNA damage, whereas CREM lacks ATM sites altogether (25). CK1/2 site are also present diverse metazoan CREB proteins from Caenorhabditis elegans to Drosophila whose regulation by DNA damage is uncertain. We speculate that cAMP-induced phosphorylation of CK1/2 sites evolved to enhance dynamic CREB–CRTC2–DNA interactions and transcriptional activation, whereas DNA damage-dependent regulation of this domain arose later during vertebrate evolution, potentially as an additional barrier to neoplastic growth.
Supplementary Material
Acknowledgments
The authors wish to thank Qijing Xie for technical contributions and Dr Shigeki Miyamoto (UW-Madison Department of Oncology) and Dr Jerry Yin (UW-Madison Department of Genetics) for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [R01CA180765-01 to R.S.T.]; National Institute for Neurological Disorders and Stroke [1R21NS090313-01A1 to R.S.T.]; National Institute of Diabetes and Digestive and Kidney diseases [R01DK090249 to C.R.J.]; Intramural Research Program of the National Institutes of Health [NIH]; National Institute of Environmental Health Sciences [NIEHS]. Funding for open access charge: National Cancer Institute [R01CA180765-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X., Odom D.T., Koo S.-H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh K.-J., Han H.-S., Kim M.-J., Koo S.-H. Transcriptional regulators of hepatic gluconeogenesis. Arch. Pharm. Res. 2013;36:189–200. doi: 10.1007/s12272-013-0018-5. [DOI] [PubMed] [Google Scholar]
- 5.Kida S., Serita T. Functional roles of CREB as a positive regulator in the formation and enhancement of memory. Brain Res. Bull. 2014;105:17–24. doi: 10.1016/j.brainresbull.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez G.A., Montminy M.R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez G.A., Yamamoto K.K., Fischer W.H., Karr D., Menzel P., Biggs W., Vale W.W., Montminy M.R. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K.K., Gonzalez G.A., Biggs W.H., Montminy M.R. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 9.Mayr B., Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 10.Radhakrishnan I., Pérez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 11.Chrivia J.C., Kwok R.P., Lamb N., Hagiwara M., Montminy M.R., Goodman R.H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 12.Conkright M.D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J.B., Montminy M. TORCs: transducers of regulated CREB activity. Mol. Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Iourgenko V., Zhang W., Mickanin C., Daly I., Jiang C., Hexham J.M., Orth A.P., Miraglia L., Meltzer J., Garza D., et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bittinger M.A., McWhinnie E., Meltzer J., Iourgenko V., Latario B., Liu X., Chen C.H., Song C., Garza D., Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Screaton R.A., Conkright M.D., Katoh Y., Best J.L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J.R., Takemori H., et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Luo Q., Viste K., Urday-Zaa J.C., Senthil Kumar G., Tsai W.-W., Talai A., Mayo K.E., Montminy M., Radhakrishnan I. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20865–20870. doi: 10.1073/pnas.1219028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Weaver I.C.G., Gauthier-Fisher A., Wang H., He L., Yeomans J., Wondisford F., Kaplan D.R., Miller F.D. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein-Taybi syndrome brain. Dev. Cell. 2010;18:114–125. doi: 10.1016/j.devcel.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 18.Ravnskjaer K., Kester H., Liu Y., Zhang X., Lee D., Yates J.R., Montminy M. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W., Kasper L.H., Lerach S., Jeevan T., Brindle P.K. Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. EMBO J. 2007;26:2890–2903. doi: 10.1038/sj.emboj.7601734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Y., Venkataraman S.L., Dodson G.E., Mabb A.M., LeBlanc S., Tibbetts R.S. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5898–5903. doi: 10.1073/pnas.0307718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 22.Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C., Cleveland J.L., Green D.R., Kastan M.B. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider J.G., Finck B.N., Ren J., Standley K.N., Takagi M., Maclean K.H., Bernal-Mizrachi C., Muslin A.J., Kastan M.B., Semenkovich C.F. ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell Metab. 2006;4:377–389. doi: 10.1016/j.cmet.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Shanware N.P., Trinh A.T., Williams L.M., Tibbetts R.S. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J. Biol. Chem. 2007;282:6283–6291. doi: 10.1074/jbc.M610674200. [DOI] [PubMed] [Google Scholar]
- 25.Shanware N.P., Zhan L., Hutchinson J.A., Kim S.H., Williams L.M., Tibbetts R.S. Conserved and distinct modes of CREB/ATF transcription factor regulation by PP2A/B56gamma and genotoxic stress. PLoS One. 2010;5:e12173. doi: 10.1371/journal.pone.0012173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 27.Kerr M.K., Martin M., Churchill G.A. Analysis of variance for gene expression microarray data. J. Comput. Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- 28.Kerr M.K., Churchill G.A. Statistical design and the analysis of gene expression microarray data. Genet. Res. 2001;77:123–128. doi: 10.1017/s0016672301005055. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing on JSTOR. J. R. Stat. Soc. B. 1995;57:300–289. [Google Scholar]
- 30.Kim S.H., Shanware N.P., Bowler M.J., Tibbetts R.S. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y., Dodson G.E., Mukhopadhyay P.S., Shanware N.P., Trinh A.T., Tibbetts R.S. Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies. J. Biol. Chem. 2007;282:9236–9243. doi: 10.1074/jbc.M609256200. [DOI] [PubMed] [Google Scholar]
- 32.Naqvi S., Martin K.J., Arthur J.S.C. CREB phosphorylation at Ser133 regulates transcription via distinct mechanisms downstream of cAMP and MAPK signalling. Biochem. J. 2014;458:469–479. doi: 10.1042/BJ20131115. [DOI] [PubMed] [Google Scholar]
- 33.Kasper L.H., Lerach S., Wang J., Wu S., Jeevan T., Brindle P.K. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walton K.M., Rehfuss R.P., Chrivia J.C., Lochner J.E., Goodman R.H. A dominant repressor of cyclic adenosine 3′, 5′-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol. Endocrinol. 1992;6:647–655. doi: 10.1210/mend.6.4.1350057. [DOI] [PubMed] [Google Scholar]
- 35.Nash P., Tang X., Orlicky S., Chen Q., Gertler F.B., Mendenhall M.D., Sicheri F., Pawson T., Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 36.Lee C.W., Ferreon J.C., Ferreon A.C.M., Arai M., Wright P.E. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh K.-J., Han H.-S., Kim M.-J., Koo S.-H. CREB and FoxO1: two transcription factors for the regulation of hepatic gluconeogenesis. BMB Rep. 2013;46:567–574. doi: 10.5483/BMBRep.2013.46.12.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimes C.A., Jope R.S. CREB DNA binding activity is inhibited by glycogen synthase kinase-3 beta and facilitated by lithium. J. Neurochem. 2001;78:1219–1232. doi: 10.1046/j.1471-4159.2001.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bullock B.P., Habener J.F. Phosphorylation of the cAMP response element binding protein CREB by cAMP-dependent protein kinase A and glycogen synthase kinase-3 alters DNA-binding affinity, conformation, and increases net charge. Biochemistry. 1998;37:3795–3809. doi: 10.1021/bi970982t. [DOI] [PubMed] [Google Scholar]
- 40.Horiuchi J., Jiang W., Zhou H., Wu P., Yin J.C.P. Phosphorylation of conserved casein kinase sites regulates cAMP-response element-binding protein DNA binding in Drosophila. J. Biol. Chem. 2004;279:12117–12125. doi: 10.1074/jbc.M212839200. [DOI] [PubMed] [Google Scholar]
- 41.Masson N., John J., Lee K.A. In vitro phosphorylation studies of a conserved region of the transcription factor ATF1. Nucleic Acids Res. 1993;21:4166–4173. doi: 10.1093/nar/21.18.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desjardins G., Meeker C.A., Bhachech N., Currie S.L., Okon M., Graves B.J., McIntosh L.P. Synergy of aromatic residues and phosphoserines within the intrinsically disordered DNA-binding inhibitory elements of the Ets-1 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2014;111:11019–11024. doi: 10.1073/pnas.1401891111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pufall M.A., Lee G.M., Nelson M.L., Kang H.-S., Velyvis A., Kay L.E., McIntosh L.P., Graves B.J. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 45.Lee G.M., Pufall M.A., Meeker C.A., Kang H.-S., Graves B.J., McIntosh L.P. The affinity of Ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J. Mol. Biol. 2008;382:1014–1030. doi: 10.1016/j.jmb.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNally J.G., Müller W.G., Walker D., Wolford R., Hager G.L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 47.Bosisio D., Marazzi I., Agresti A., Shimizu N., Bianchi M.E., Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 49.Lee D., Le Lay J., Kaestner K.H. The transcription factor CREB has no non-redundant functions in hepatic glucose metabolism in mice. Diabetologia. 2014;57:1242–1248. doi: 10.1007/s00125-014-3203-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.