Abstract
Protection of telomeres (POT1) binds chromosome ends, recognizing single-strand telomeric DNA via two oligonucleotide/oligosaccharide binding folds (OB-folds). The Arabidopsis thaliana POT1a and POT1b paralogs are atypical: they do not exhibit telomeric DNA binding, and they have opposing roles in regulating telomerase activity. AtPOT1a stimulates repeat addition processivity of the canonical telomerase enzyme, while AtPOT1b interacts with a regulatory lncRNA that represses telomerase activity. Here, we show that OB1 of POT1a, but not POT1b, has an intrinsic affinity for telomeric DNA. DNA binding was dependent upon a highly conserved Phe residue (F65) that in human POT1 directly contacts telomeric DNA. F65A mutation of POT1aOB1 abolished DNA binding and diminished telomerase repeat addition processivity. Conversely, AtPOT1b and other POT1b homologs from Brassicaceae and its sister family, Cleomaceae, naturally bear a non-aromatic amino acid at this position. By swapping Val (V63) with Phe, AtPOT1bOB1 gained the capacity to bind telomeric DNA and to stimulate telomerase repeat addition processivity. We conclude that, in the context of DNA binding, variation at a single amino acid position promotes divergence of the AtPOT1b paralog from the ancestral POT1 protein.
INTRODUCTION
POT1 (Protection of Telomeres) is one of the most highly conserved constituents of the telomere complex, and is widely dispersed across eukarya. POT1 promotes chromosome end-protection, telomere length regulation and is increasingly implicated in stem cell disease and cancer (1,2). The critical role of POT1 is mediated in large part through its interaction with telomeric DNA. Specifically, POT1 contacts the 3′ G-rich single-strand (ss) protrusion on the chromosome end via two N-terminal oligosaccharide/oligonucleotide-binding folds (OB-folds) (3,4). OB-fold domains consist of a five-stranded closed β barrel with connecting loops of varying length, sequence and conformation and are capable of recognizing DNA, RNA and protein interaction partners (5,6). The OB-folds of mammalian POT1, for example, bind telomeric DNA with high affinity (Kd ≈ 10 nM) and base specificity (4). A highly conserved aromatic residue within the first OB-fold (OB1), F62, is crucial for telomeric DNA recognition (4,7,8). This residue also discriminates DNA from RNA through interaction with a single deoxythymidine (dT) within the GGTTAGGGTTAG telomeric repeat. Remarkably, substitution of Phe with Tyr at position 62 converts hPOT1 into an RNA binding protein (9).
Humans harbor a single POT1 ortholog, but multiple POT1 paralogs with distinct functions have been reported in some organisms, including mouse (7,10), worms (11), ciliates (12) and Arabidopsis thaliana (13,14). Two independent POT1 duplications are known in land plants, one in the grasses (Poaceae) and the other in mustards (Brassicaceae), the family that includes A. thaliana (15). Functional diversification may have occurred relatively rapidly in rodents since the two POT1 copies in mouse likely duplicated ∼12 million years ago (Mya) (16) and display 75% similarity (10). In contrast, the AtPOT1a and AtPOT1b duplication is more ancient and the two paralogs display only 50% similarity overall (13,15). AtPOT1a and AtPOT1b fail to bind telomeric DNA in vitro (17). This finding was unexpected as the single copy POT1 gene from the early diverging land plant Physcomitrella patens binds telomeric DNA and is essential for chromosome end protection like its counterparts in vertebrates and fission yeast (18).
AtPOT1a and AtPOT1b can also be distinguished from other POT1 orthologs because they assemble into ribonucleoprotein (RNP) complexes with different telomerase subunits. AtPOT1a interacts with the canonical telomerase RNA (TER1) (19) and positively regulates telomerase activity by stimulating repeat addition processivity (20). Conversely, AtPOT1b associates with a regulatory lncRNA termed TER2, which represses telomerase activity in response to DNA damage (21). Over-expression of a dominant negative allele of AtPOT1b leads to massive telomere erosion and profound genome instability (13), suggesting this protein may play a role in chromosome end protection. AtPOT1b cannot complement a pot1a null mutation in vivo (15), highlighting the distinct contributions of the two paralogs.
The molecular basis for the functional divergence of AtPOT1a and AtPOT1b is unknown. However, recent molecular evolutionary studies reveal that the Brassicaceae POT1a lineage, but not POT1b, has been subjected to positive selection. The three residues with the strongest signatures of selection (E35, S212, E293) enhance the interaction of AtPOT1a with CTC1, a core component of the telomere replication complex, CST and are required for AtPOT1a function in vivo (15).
Here, we examine the nucleic acid binding properties of OB1 from AtPOT1a and AtPOT1b to gain insight into their unique functions. We report that AtPOT1aOB1, as well as single-copy POT1 genes throughout the plant kingdom, bears an evolutionarily conserved Phe that in hPOT1 is required for recognition of ss-telomeric DNA. Interestingly, this residue was not identified as a target of selection in molecular evolutionary analyses of the Brassicaceae POT1b lineage, despite the fact that AtPOT1bOB1 lacks Phe at the homologous position. We demonstrate that AtPOT1aOB1 is sufficient to bind telomeric DNA, and furthermore that DNA binding is likely to be ancestral for plant POT1. The conserved Phe not only dictates telomeric DNA binding, but also plays a critical role in stimulating telomerase repeat addition processivity. AtPOT1bOB1, on the other hand, does not bind telomeric DNA and cannot stimulate telomerase repeat addition processivity. However, when the conserved Phe is restored, AtPOT1bOB1 acquires both properties. We conclude that a single amino acid substitution in the nucleic acid binding pocket plays a critical role in defining the divergent functions of AtPOT1a and AtPOT1b. This finding, in concert with previous positive selection analyses, brings sharper focus to the events that resulted in the evolution of two telomerase RNPs in Brassicaceae.
MATERIALS AND METHODS
In vitro transcription
All the RNAs used in the study were transcribed in vitro from a T7 promoter on either a polymerase chain reaction (PCR) or linearized plasmid template. One or two extra Gs were added at the 5′ end to enhance transcription efficiency (22). Transcription was performed using an Ampliscribe T7-FLASH transcription kit (Epicentre) for at least 2 h at 37°C followed by DNase digestion for 30 min at 37°C. RNAs were then separated by gel electrophoresis (6% polyacrylamide, 7M urea), detected by UV shadowing and eluted in RNA elution buffer (300 mM sodium acetate, pH 5.2, 1 mM EDTA and 0.25% SDS) for at least 2 h at 37°C or overnight at 4°C. The supernatant was filtered using a 0.2 μm filter (VWR International) and ethanol precipitated. RNAs were resuspended in DNase/RNase free ultrapure distilled H2O (Invitrogen), aliquoted and stored at −80°C.
Protein expression and purification
Constructs used for Escherichia coli expression of POT1aOB1 (1-158 aa) and POT1bOB1 (1-159 aa) were cloned in a pET28a vector (Novagen). Four amino acids (SISS) and a 6x His tag were added to the C-terminus of POT1aOB1 and POT1bOB1, respectively, to increase the protein solubility as predicted using www.biotech.ou.edu (23). Both constructs also contained a T7 tag at the N-terminus. Site-directed mutagenesis of wild-type POT1aOB1 and POT1bOB1 constructs was performed using Pfu turbo polymerase (Stratagene) following the manufacturer guidelines to generate POT1aOB1-F65A and POT1bOB1-V63F constructs. Constructs were transformed into E. coli BL21 (DE3). Cells were grown at 30°C until the O.D600 nm reached 0.5–0.6. Cells were incubated at 4°C for 30 min followed by induction with 0.5 mM IPTG at 16°C for 18–20 h. Induced cells were centrifuged at 4°C for 20 min at 4,000 g followed by resuspension of cell pellet in lysis buffer containing 50 mM Tris-Cl, pH 7.5, 300 mM NaCl and 1X protease inhibitor cocktail (Sigma). After sonication, the lysate was centrifuged for 45 min at 16,000 rpm at 4°C to remove cell debris. Lysate was loaded on Ni-NTA agarose resin (Qiagen) for affinity purification at room temperature with a flow rate of 6 ml/min. The protein bound Ni-NTA column was washed with 5- to 10-fold excess washing buffer (20 mM Tris-Cl, pH 7.5, 500 mM NaCl and 25–50 mM imidazole) at a flow rate of 6 ml/min. Protein was eluted in 20 mM Tris-Cl, pH 7.5 containing 150 mM NaCl and 250 mM imidazole. DTT (5 mM) was added to the eluted fractions prior to concentration and further purification using a Sephadex G-75 (GE Healthcare) size exclusion column. SDS-PAGE gel analysis was used to monitor protein purity and homogeneity using coomassie staining. LC-MS/MS was used to verify protein identity. A total of 5 mM DTT was added to eluted fractions prior to concentration. The concentrated protein was aliquoted, flash frozen in N2 (l) and stored at −80°C.
Binding assays
Oligonucleotides were acquired from Invitrogen. Double filter binding assays were employed using E. coli expressed POT1aOB1 (0–1 μM) and POT1bOB1 (0–2.5 μM) with either 5′-end labeled or body labeled TER1 and TER2 RNA, respectively. Radioactive labeling of RNA was performed as described (24) and labeled RNAs were purified, eluted and stored as described above. Approximately 10,000 cpm of labeled RNA was used in each reaction. Prior to the binding reaction, each RNA sample was heated at 95°C for 2 min, cooled on ice for 1 min followed by incubation in 1X binding buffer (20 mM Tris-Cl pH 7.5, 200 mM potassium glutamate and 10 mM MgCl2) containing 50 μg/ml BSA, 25 μg/ml yeast transfer RNA, 1 mM DTT and 10% glycerol for 15 min at 37°C. Varying amounts of protein were added and reactions were incubated at 37°C for 30 min. Prior to loading samples, the protein membrane (GE, Amersham Hybond P) was soaked in 100% methanol followed by a quick wash in water. Both the protein and nucleic acid membranes (GE, Amersham Hybond N+) were equilibrated in washing buffer (25 mM Tris-Cl pH 7.5, 200 mM potassium glutamate, 10 mM magnesium acetate, 0.5 mM EDTA, 0.1% NP-40 and 1 mM DTT) until the binding reaction was completed. Samples were filtered under vacuum through Hybond P and N+ membranes using a dot-blot apparatus (Bio-Rad). Membranes were washed with 100 μl washing buffer at least twice prior to drying and exposure to a phosphorimager screen (Bio-Rad). Protein concentration was calculated using a Bradford assay and confirmed by nanodrop (Thermo Scientific).
For electrophoretic mobility shift assays (EMSA), oligonucleotides were 5′ end labeled with (γ-32P) ATP (Perkin Elmer) in 20 μl reactions using T4 Polynucleotide kinase (NEB) for 1 h at 37°C. Labeled oligonucleotides were purified using 16% denaturing PAGE containing 7M urea. Oligonucleotides were eluted in 1X TE buffer pH 8.0 for overnight at 4°C followed by ethanol precipitation. The pellet was washed in 70% ethanol, dried and resuspended in 50–100 μl DNAse/RNAse free Ultrapure distilled water (Invitrogen). Reactions containing ∼10,000 cpm of 5′ labeled DNA oligonucleotide were heated at 95°C for 2 min followed by cooling on ice for 1 min. Binding assays were conducted with oligonucleotides incubated with 1X DNA binding buffer (25 mM Tris-Cl pH 7.5, 50 mM NaCl, 40 mM KCl, 7% glycerol, 1 mM EDTA and 1 mM DTT) containing 10 μg/ml BSA and 50 μg/ml Herring sperm DNA (Invitrogen) for 15 min at 37°C. EMSA with RNA oligonucleotides was performed in 1X RNA binding buffer (20 mM Tris-Cl pH7.5, 50 mM NaCl, 10 mM MgCl2, 1mM DTT and 5% glycerol) containing 10 μg/ml BSA and 50 μg/ml yeast transfer RNA. Varying amounts of protein were added and binding reactions were incubated at 37°C for 30 min. EMSA products were separated on 6–20% gradient polyacrylamide gels (29:1) for 1–2 h at 150 V in 1X TB buffer (pH 7.5) at 4°C. Gels were dried under vacuum at 80°C for 30–60 min prior to exposure to a phosphorimager screen. Data were quantified using Quantity-one software (Bio-Rad).
The bound fraction of RNA was calculated using the (bound signal/ (bound+ unbound)) signal and was plotted against protein concentration. Data were fit to in Origin 6.0 software using the Hill equation, F = Pn/(Pn + Kdn), where n is the “Hill coefficient”, F is the fraction of total bound RNA with protein, P is the total concentration of protein added to the reaction and Kd is an apparent dissociation constant for the binding representing the concentration of protein at which 50% of the RNA was in the bound state. All experiments were performed at least in duplicate, and the calculated Kd (app) values representing the average of the two independent experiments replicates with standard deviation from the mean.
Phylogenetic analysis and structure homology modeling
Nucleotide sequences, including the sequence from Tarenaya hassleriana, were aligned with existing alignments for eudicot POT1 (15). The alignment was translated to produce an amino acid alignment prior to phylogenetic analysis. The most likely tree was inferred using the PROTGAMMAWAG function in RAxML 7.0.4 (25) support for nodes in the tree was assessed by analysis of 100 bootstrap replicate data sets. Structure prediction for AtPOT1aOB1 and AtPOT1bOB1 was performed using Phyre2 (26) based on homology modeling. Superposition of the predicted structures on structure of hPOT1-DNA complex (PDB ID: 1XJV) was carried out using Chimera (27).
Telomerase activity assays
Protein extracts from 5–6 days-old wild type or pot1a null seedlings were prepared (28) for conventional Telomere Repeat Amplification Protocol (TRAP) (28) and telomerase processivity TRAP (TP-TRAP) (29). The protein extract concentration was determined by Bradford assay and confirmed by nanodrop (Thermo Scientific). To assess the effect of recombinant POT1 proteins on telomerase activity, ∼1 μg of the extract was incubated with ∼4–5 μg of E. coli expressed POT1aOB1, POT1bOB1 or their mutant derivatives for 30 min at 4°C followed by extension reaction in 1X primer extension buffer (50 mM Tris-Cl pH 8.0, 50 mM KCl, 2 mM DTT, 3 mM MgCl2 and 1 mM spermidine) at 37°C for 45 min. For immunoprecipitation-TRAP, Dynabeads Protein-G (Invitrogen) were coupled with T7 tag antibody (Novagen) in 1X PBS for 10–20 min at RT. Antibody-coupled beads were incubated with seedling extracts containing POT1 protein for 2 h at 4°C followed by 5X washes with buffer W300 and 2X washes with buffer TMG (30). After the telomerase extension, a non-specific DNA template was added as a TRAP-PCR control, and the reactions were subjected to phenol/chloroform extraction followed by ethanol precipitation and resuspended in DNase/RNase free distilled H2O (Invitrogen) for TRAP and TP-TRAP as described previously (28,29).
RESULTS
A. thaliana POT1aOB1 and POT1bOB1 bind RNA non-specifically
Previous in vitro studies conducted with full-length AtPOT1a expressed in rabbit reticulocyte lysate (RRL) showed that it binds TER1 with higher affinity than TER2 (19). AtPOT1b has the opposite binding specificity, preferring TER2 over TER1 (21). However, binding studies with RRL expressed proteins were not quantitative. To further explore the nucleic acid binding properties of AtPOT1a and AtPOT1b, we attempted to express the full-length A. thaliana proteins in E. coli, yeast and insect cells. None of these expression systems yielded enough soluble protein for analysis. We also tested expression of a construct containing both putative OB folds (OB1 + OB2) in AtPOT1a (13), but again the protein was highly insoluble. Because the N-terminal OB-fold (OB1) of fission yeast and human POT1 is sufficient for recognition of ss-telomeric DNA (31–34), we generated the corresponding constructs for AtPOT1a (1 to 158 aa) and AtPOT1b (1 to 159 aa) encompassing their putative OB1 domain (13). Both protein expression constructs were fused at their N-terminus with 6x His and T7 tag. To increase the protein solubility, four additional amino acids were incorporated on the C-terminus of POT1aOB1 and a 6x His tag at the C-terminus of POT1bOB1. The proteins were expressed in E. coli and purified by affinity chromatography (Supplementary Figure S1A). LC-MS/MS analysis confirmed the identity of the proteins (Supplementary Figure S1B).
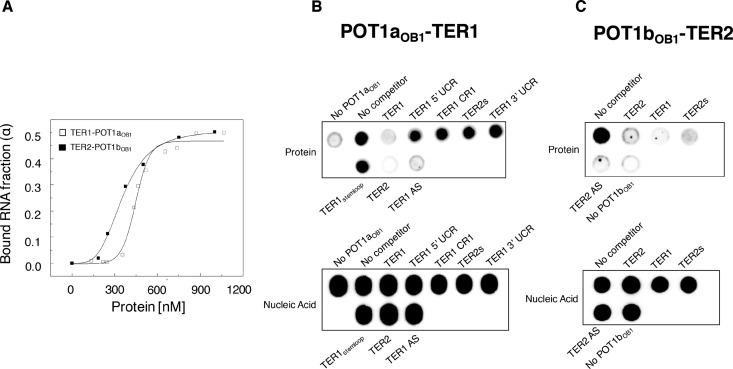
Shown in Supplementary Figure S2A is a schematic representation of three different A. thaliana TER isoforms termed TER1 (748 nt), TER2 (784 nt) and TER2s (219 nt) (19,21). The RNAs share ∼90% sequence identity in Conserved Region 1 and 2 (CR1 and CR2). The template domain is embedded in CR1. In TER2, CR1 and CR2 are interrupted by a 529-nt transposon (19,21,35), which is removed along with the 3′ unique extension to generate TER2s. To assess RNA binding by AtPOT1aOB1, filter binding assays were performed with 5′ radiolabeled TER1 expressed from a T7 promoter. RNA–protein complexes were observed with an apparent binding affinity (Kd) of 450 (±50) nM (Figure 1A). Unexpectedly, competition experiments with TER1, TER2, RNA sequences corresponding to truncated regions of TER1, antisense TER1 and direct binding experiments with U6 snRNA revealed no particular specificity for AtPOT1aOB1 binding to TER1 (Figure 1B; Supplementary Figure S2B). A similar result was obtained in experiments with AtPOT1bOB1 and TER2 (Figure 1C). A Kd of 400 (±50) nM was obtained for POT1b-TER2 complex, but again RNA binding was non-specific (Figure 1A and C). Even at the highest POT1aOB1 and POT1bOB1 concentrations, only 50% of TER1 and TER2 RNA remained bound after filtration (Figure 1A), consistent with formation of weak and/or non-specific RNA–protein complexes. These findings argue that regions outside OB1 promote TER-specific binding by AtPOT1a and AtPOT1b.
Figure 1.
POT1aOB1 and POT1bOB1 interact non-specifically with RNA. (A) Results of filter binding assays using POT1aOB1-TER1 and POT1bOB1-TER2. Radioactive labeled TER1 and TER2 RNA were titrated with increasing concentrations (0–1 μM) of POT1aOB1 and POT1bOB1, respectively. The protein bound fraction of RNA was assessed by a double filter binding assay and results were plotted versus protein concentration. Data were fit using the Hill equation in Origin 6.0 to determine Kd(app) of POT1aOB1 and POT1bOB1 for RNA. Filter binding based competition assay results are shown for (B) POT1aOB1-TER1 and (C) POT1bOB1-TER2 with a fixed protein concentration (∼500 nM) and 100-fold excess of cold RNAs as specified.
Intrinsic telomeric DNA binding activity within AtPOT1aOB1
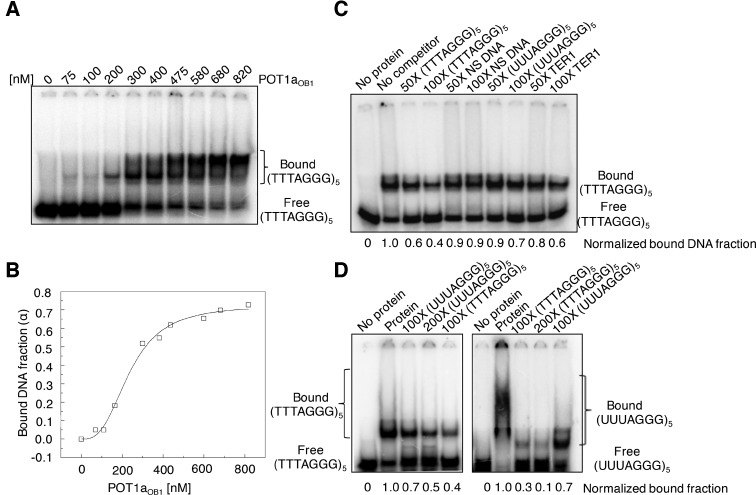
EMSA performed with E. coli expressed POT1aOB1 unexpectedly revealed specificity for ss-telomeric DNA (Figure 2A). AtPOT1aOB1 bound the plant telomere repeat sequence (TTTAGGG)5 with a Kd of 230 (±30) nM (Figure 2B). To assess the binding specificity of POT1aOB1, competition and displacement-based EMSA experiments with non-telomeric oligonucleotides and with telomeric RNA (UUUAGGG)5 were conducted (Figure 2C and D). As shown in Figure 2C, a 50-fold excess of cold telomeric DNA reduced protein binding by 40%, while a 50-fold excess of cold non-telomeric DNA or telomeric RNA reduced protein binding by only 10%. The displacement assay shown in Figure 2D yielded similar results. A 100-fold excess of telomeric DNA disrupted the preformed AtPOT1aOB1-DNA complex by 60%, while telomeric RNA decreased it by only 30%. In contrast, AtPOT1aOB1-RNA complexes were depleted by 70% in the presence of 100X cold telomeric DNA, compared to a decrease of only 30% with cold telomeric RNA. No binding was observed for AtPOT1aOB1 with C-rich ss telomeric DNA or double-stranded telomeric DNA (Supplementary Figure S3A). Taken together, these data argue that POT1aOB1 binds single strand telomeric DNA specifically.
Figure 2.
AtPOT1aOB1 specifically binds telomeric DNA. (A) EMSA assay results for POT1aOB1-(TTTAGGG)5 binding. 5′ labeled (TTTAGGG)5 was titrated with increasing POT1aOB1 (0-820 nM). (B) The Kd (app) of POT1aOB1 for (TTTAGGG)5 was determined as described in Materials and Methods using data from Figure 2A. (C) EMSA based competition assay results for POT1aOB1-(TTTAGGG)5 using a fixed POT1aOB1 concentration (∼500 nM) and 50 to 100-fold excess of the nucleic acids specified. NS DNA stands for non-specific DNA (5′-TTAATTAACCCGGGGATCCGGCTTGATCAACGAATGATCC-3′ (ref. 32)). (D) Results of EMSA based displacement assay using ∼500 nM POT1aOB1 in the absence and presence of 100- to 200-fold excess of different oligonucleotides to assess the specificity of POT1aOB1 binding.
AtPOT1aOB1 interaction with telomeric DNA was length-dependent. The affinity for two telomere repeats (TTTAGGG)2 was decreased relative to (TTTAGGG)5 to an apparent Kd of ∼1 μM (Compare Supplementary Figure S3B with Figure 2A) and no detectable binding was observed for one repeat TTTAGGG (Supplementary Figure S4A; lane 9). EMSA showed that AtPOT1aOB1 has a minimal DNA binding site of 5′-TAGGGTTTAGG-3′ (Supplementary Figure S4A). Because incorporation of an extra 5′ T increased AtPOT1aOB1 binding (Supplementary Figure S4A, lanes 11 and 12), the 12-mer telomere sequence 5′-TTAGGGTTTAGG-3′ was used as a base for nucleotide substitution to determine which nucleotides were critical for interaction. The results indicate that G5, G6, T8, T9, G11 and G12 (5′-TTAGGGTTTAGG-3′) are important for AtPOT1aOB1 binding (Supplementary Figure S4B). We conclude that AtPOT1aOB1 specifically binds ss-telomeric DNA.
A highly conserved phenylalanine residue present in AtPOT1a, but not AtPOT1b, is critical for telomeric DNA binding
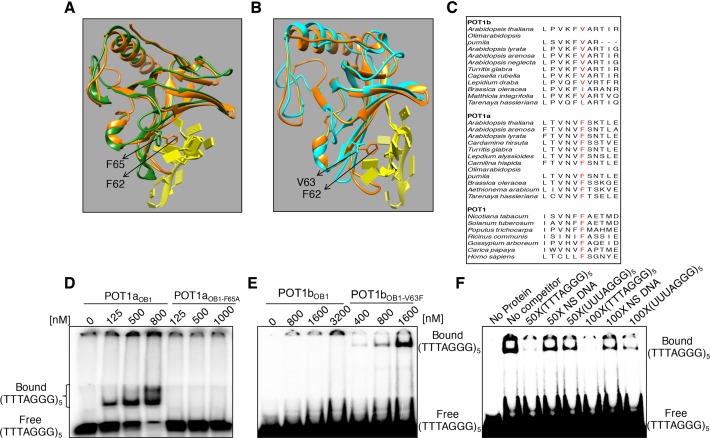
Structure homology analysis and amino acid sequence analysis using Phyre2 (26) and ClustalW2 (36) were performed to compare the predicted nucleic acid binding pocket of OB1 from A. thaliana POT1a and POT1b to the crystal structure of DNA-bound hPOT1 (PDB ID: 1XJV) (Figure 3A and B; Supplementary Figure S5). Human POT1 exhibits nucleotide stacking with several aromatic amino acids in OB1 (F31, F62 and Y89) and OB2 (Y161, Y223, H245 and H266) (4). Strikingly, only two of these amino acids are conserved in AtPOT1a, and none in AtPOT1b (Supplementary Figure S5). Within AtPOT1aOB1 there is a Phe (F65) that directly corresponds to F62 in the OB1 domain of hPOT1 (Figure 3A and C), the residue critical for telomeric DNA binding (4). This residue is retained in all Brassicaceae POT1a proteins as well as all of the single copy POT1 proteins sampled within the plant kingdom (Figure 3C). Notably, F65 was not uncovered as a site under positive selection during our previous analysis of AtPOT1a (15).
Figure 3.
A highly conserved Phe in the nucleic acid binding pocket of AtPOT1aOB1 is critical for telomeric DNA binding. (A and B) Structure homology modeling. Overlay of AtPOT1aOB1 and AtPOT1bOB1 on the crystal structure of hPOT1-telomeric DNA (PDB ID: 1XJV) using Chimera software (27) is shown. Modeling was performed with AtPOT1aOB1 and AtPOT1bOB1 using Phyre2 (26). AtPOT1aOB1, AtPOT1bOB1 and hPOT1 are highlighted in forest green, cyan and orange, respectively, while the DNA chain is highlighted in yellow. (C) Amino acid sequence alignment. Multiple amino acid sequence alignment of POT1 proteins from different plant species and humans are shown. The highly conserved Phe in OB1 from hPOT1, single copy plant POT1 proteins and Brassicaceae POT1a is indicated in red. AtPOT1b and other POT1b orthologs from the Brassicaceae harbor non-aromatic amino acid such as Val except B. oleracea, which contains Ile at the same position. POT1b from T. hassleriana, a member of Cleomaceae (sister to Brassicaceae) also contains a non-aromatic amino acid, Leu at the corresponding position. (D–F) EMSA results for AtPOT1aOB1 and AtPOT1bOB1 with telomeric DNA. 5′ labeled (TTTAGGG)5 was titrated with increasing concentrations of (D) POT1aOB1 (0–800 nM) and POT1aOB1-F65A (0–1.0 μM) or (E) POT1bOB1 (0–3.2 μM) and POT1bOB1-V63F (0–1.6 μM). (F) Results of EMSA based competition assays for POT1bOB1-V63F -(TTTAGGG)5 using POT1bOB1-V63F (∼900 nM) and a 50 to 100-fold excess of different nucleic acids as specified. NS DNA stands for non-specific DNA (5′-TTAATTAACCCGGGGATCCGGCTTGATCAACGAATGATCC-3′ (ref. 32)).
Since F62 discriminates telomeric DNA from RNA for human POT1 (9), we asked if the corresponding residue in AtPOT1aOB1 (F65) is required for telomeric DNA recognition by substituting Phe with Ala to create AtPOT1aOB1-F65A (Supplementary Figure S1A; lane 2). Telomeric DNA binding was abolished in POT1aOB1-F65A (Figure 3D). Several lines of evidence argue that the loss of DNA binding is not due to gross protein mis-folding. The mutant protein accumulates to the same extent as wild-type AtPOT1aOB1 in E. coli and is recovered at the same level following two rounds of column chromatography. In addition, like AtPOT1aOB1, AtPOT1aOB1-F65A retains non-specific TER1 binding (Supplementary Figure S6). Finally, we previously showed by Western blot analysis that full-length AtPOT1a bearing this same F65A mutation accumulates to the same level as wild-type AtPOT1a in transgenic A. thaliana (15).
Strikingly, AtPOT1b lacks the critical Phe and instead harbors Val (V63) at the analogous position (Figure 3B and C). All of the Brassicaceae POT1b orthologs contain non-aromatic amino acids Val or Ile, instead of Phe at this site (Figure 3C). We asked if AtPOT1bOB1 interacts with telomeric DNA using EMSA. No telomeric DNA binding was detected (Figure 3E). Nevertheless, by substituting Val with Phe at position 63 (POT1bOB1-V63F) (Supplementary Figure S1A, lane 5), specific telomeric DNA binding was acquired (Figure 3E and F). The complex formed by POT1bOB1-V63F bound to telomeric DNA migrates at a position that is markedly different than the POT1aOB1-DNA complex. It is unlikely that the POT1bOB1-V63F complex reflect non-specific aggregation of the mutant protein, because the competition experiments in Figure 3F show specificity and robust telomeric DNA binding. One possibility is that POT1bOB1-V63F forms an oligomeric complex in the presence of telomeric DNA. Taken together, our data indicate that the difference in telomeric DNA recognition by AtPOT1a and AtPOT1b is mediated by a single amino acid substitution.
To determine the timing of the acquisition of alternative functions for POT1a and POT1b, we included sequence data from the Tarenaya hassleriana genome (37), a member of the Cleomaceae and sister to Brassicaceae, permitting a re-examination of the timing of the POT1 duplication (15). We recovered two copies of POT1 from T. hassleriana, suggesting the duplication that produced POT1a and POT1b predates the divergence of Cleomaceae and Brassicaceae. Cleomaceae is characterized by a whole genome duplication (WGD) that is independent of the Brassicaceae WGD (37), raising the possibility that the T. hassleriana paralogs duplicated after the split of the two families. To distinguish between these alternatives, we added POT1 data from T. hassleriana to our existing alignment of eudicot POT1 sequences (15) and inferred phylogeny. Our phylogenetic results indicate that POT1a and POT1b duplicated prior to the divergence of Cleomaceae and Brassicaceae (Supplementary Figure S7), ∼65 Mya (38). Consistent with this finding, ThPOT1aOB1 carries the conserved Phe, but ThPOT1bOB1 contains a hydrophobic residue (Leu) at the corresponding position (Figure 3C). This pattern of duplication along with our functional data support the conclusion that the ancestral function of telomeric DNA binding is conserved in AtPOT1aOB1, but not in AtPOT1bOB1.
AtPOT1aOB1, but not AtPOT1bOB1, stimulates telomerase repeat addition processivity in vitro
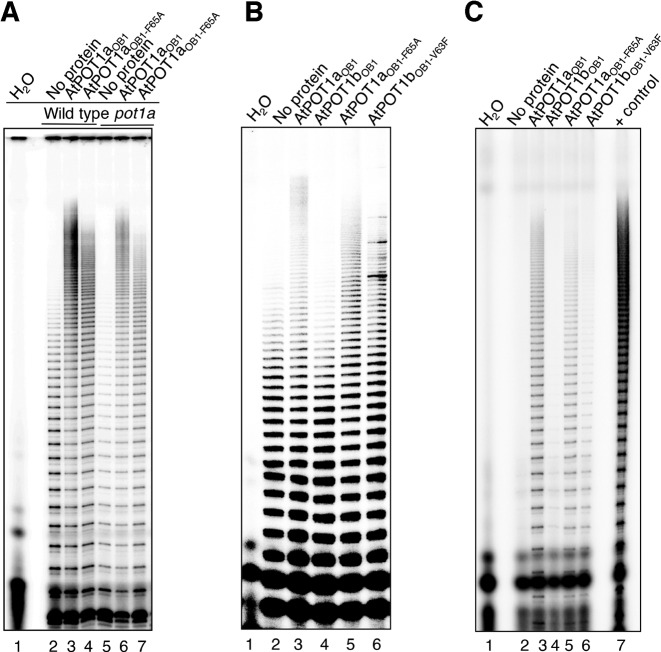
AtPOT1a enhances the repeat addition processivity of telomerase. Therefore, we asked if AtPOT1aOB1 is sufficient to stimulate telomerase in vitro. Extracts prepared from wild type or null for POT1a (pot1a-/-) A. thaliana seedlings were supplemented with recombinant AtPOT1aOB1 and the conventional TRAP was performed. Markedly longer products were obtained in reactions with AtPOT1aOB1 compared to the control (Figure 4A, lanes 2 and 3). The effect was specific: bovine serum albumin did not stimulate telomerase (Supplementary Figure S8A, lane 3) and neither did AtPOT1bOB1 (see below). To test whether the change in telomerase extension products reflects increased repeat addition processivity, we performed TP-TRAP reactions with POT1aOB1. Unlike conventional TRAP, TP-TRAP employs two reverse primers, one of which incorporates a tag on telomerase extension products, allowing their length to be measured accurately (29). The results of TP-TRAP showed that addition of AtPOT1aOB1 caused longer telomerase extension products, consistent with elevated repeat addition processivity (Figure 4B, lanes 2 and 3). Notably, recombinant AtPOT1aOB1 was able to largely rescue the telomerase activity defect of pot1a null extracts (Figure 4A, lanes 5 and 6), indicating that the OB1 domain of POT1a is sufficient to stimulate telomerase repeat addition processivity.
Figure 4.
AtPOT1aOB1 stimulates telomerase repeat addition processivity. (A) Conventional TRAP results from wild type and pot1a mutant seedling extracts in the presence of POT1aOB1 and POT1aOB1-F65A. (B) TP-TRAP results from wild type seedling extracts in the presence of POT1aOB1, POT1aOB1-F65A, POT1bOB1 or POT1bOB1-V63F. H2O lane is without extract to monitor PCR contamination. (C) IP-TRAP results for wild type extracts supplemented with POT1aOB1, POT1aOB1-F65A, POT1bOB1 or POT1bOB1-V63F.
We next asked if the telomeric DNA binding activity of POT1aOB1 is required for telomerase stimulation. TRAP and TP-TRAP reactions performed using wild type and pot1a extracts supplemented with POT1aOB1-F65A showed some degree of telomerase stimulation, but not as much as with wild type AtPOT1aOB1 (Figure 4A, lanes 4 and 7; Figure 4B, lane 5). On the other hand, wild-type POT1bOB1 did not enhance overall telomerase activity and repeat addition processivity at all (Supplementary Figure S8B, lane 3; Figure 4B, lane 4). POT1bOB1-V63F modestly stimulated repeat addition processivity, albeit not to the same level as wild-type POT1aOB1 (Figure 4B, lanes 3 and 6). We conclude that telomeric DNA binding by OB1 of POT1a and POT1b is necessary, but not sufficient to fully stimulate telomerase repeat addition processivity.
Finally, we tested if POT1aOB1, POT1bOB1 or their mutant variants associate with enzymatically active telomerase RNP. Tagged proteins were pre-incubated with seedling extract then immunoprecipitation (IP) was conducted, followed by coomassie staining to assess protein IP and TRAP to monitor telomerase activity recovered (Supplementary Figure S8C and Figure 4C). Robust enzyme activity was precipitated from the POT1aOB1 reaction (Figure 4C, lane 3). Telomerase activity was also recovered in the POT1aOB1-F65A IP (Figure 4C, lane 5), but at a reduced level compared to wild-type POT1aOB1 (Figure 4C, lane 3). This observation is consistent with diminished telomerase stimulation by POT1aOB1-F65A. Because active telomerase activity is present in the IP with POT1aOB1-F65A, which cannot bind telomeric DNA, the data indicate that either POT1aOB1 interacts with the holoenzyme directly or engages it indirectly through additional telomere proteins.
In marked contrast, telomerase activity was barely detectable in the POT1bOB1 IP (Figure 4C, lane 4). This finding corroborates earlier results showing that endogenous AtPOT1b does not interact with enzymatically active telomerase (21). A higher level of telomerase activity was detected following IP with POT1bOB1-V63F (Figure 4C, lane 6). We postulate that POT1bOB1-V63F weakly interacts with telomerase, and once telomerase begins synthesis, POT1bOB1-V63F binding to the extended telomeric DNA tract can stimulate telomerase repeat addition processivity through transient interactions with the RNP complex. Taken together, our data indicate that the robust stimulation of telomerase by AtPOT1aOB1 requires both telomeric DNA binding and stable association with the telomerase holoenzyme, two functions lacking in POT1bOB1.
DISCUSSION
AtPOT1 proteins challenge the existing paradigm of POT1 as a core component of the shelterin telomere cap. In this study, we reconcile some of the confounding observations pertaining to Arabidopsis POT1 proteins, and identify the molecular basis for the differential regulation of telomerase activity by AtPOT1a and AtPOT1b. There are several key findings from this work. First, we demonstrate that the ancestral function of telomeric DNA binding is retained in AtPOT1aOB1. We suspect that the intrinsic DNA binding capacity of AtPOT1a was masked in the full-length protein, since the two N-terminal OB-folds from fission yeast and human POT1 (hPOT1V2) bind telomeric DNA with substantially higher affinity than the respective full-length proteins (31,32,34). The specificity of AtPOT1aOB1 for telomeric DNA is quite similar to hPOT1. The minimum binding site (MBS) are comparable, 5′-TAGGGTTTAGG-3′ and 5′-(T)TAGGGTTAG-3′ for AtPOT1aOB1 and hPOT1, respectively, (this study; 4 and 33). In addition, mutation of the penultimate G11 nucleotide within the AtPOT1aOB1 MBS dramatically reduced binding, consistent with the importance of the 3′ guanosine residue in the hPOT1-telomeric DNA interaction (4). Most importantly, the critical Phe in hPOT1 OB1 that is required for telomeric DNA binding is not only conserved in AtPOT1a, but is essential for its recognition of telomeric DNA.
Despite these similarities, the affinity of AtPOT1aOB1 for telomeric DNA is substantially weaker than for other POT1 proteins. A Kd of ∼1 μM for AtPOT1aOB1 binding to two TTTAGGG repeats is nearly two orders of magnitude higher than for the S. pombe POT1 N-terminal OB fold (Pot1pN) (Kd= 19 nM) (34). Moreover, no AtPOT1aOB1 binding was detected for one TTTAGGG repeat, while Pot1pN showed a Kd of 83 nM for hexameric GGTTAC (34). Full-length hPOT1 binds TTAGGG with a Kd of ∼450 nM (4), and its affinity for two telomere repeats varies from 9 nM to 70 nM depending on the permutation of the oligonucleotide. One explanation for the decreased telomeric DNA binding affinity of AtPOT1aOB1 is that it adopts a different mode of nucleic acid recognition than the S. pombe or human POT1 proteins. In addition, our results point to unanticipated fluidity between DNA and RNA substrate recognition by the Arabidopsis POT1 proteins. Aside from F62, most of the residues in hPOT1 OB2 that make contact with telomeric DNA (4) are not conserved in AtPOT1a. Moreover, in the case of AtPOT1bOB1, telomeric DNA binding is abolished altogether (see below). Finally, unlike SpPOT1 and hPOT1, full-length AtPOT1a and AtPOT1b specifically bind TER (19,21). Our data indicate that specificity for TER1 or TER2, by AtPOT1 and AtPOT1b, respectively, is conveyed via OB2 and/or the C-terminal domain, since OB1 binds RNA non-specifically. Detailed structural information will be required to investigate precisely how AtPOT1a and AtPOT1b contact nucleic acid binding partners.
A second important observation from our study is that OB1 from AtPOT1a can act in trans to promote telomerase repeat addition processivity, and further this domain is sufficient to rescue the telomerase activity defect in extracts from pot1a null mutants. We found that POT1aOB1 contacts telomeric DNA as well as telomerase to increase repeat addition processivity. Loss of telomeric DNA binding in POT1aOB1-F65A reduced telomerase repeat addition processivity, but did not completely abrogate it. One plausible explanation is that POT1aOB1-F65A retains its interaction with telomerase, a prediction supported by our IP results.
AtPOT1a exhibits striking parallels with the Tetrahymena telomerase processivity factor p82 (Teb1) (39–42). Teb1 harbors three OB-folds that increase telomerase repeat addition processivity in different ways. The two central OB-folds of Teb1 bind telomeric DNA and suppress folding of the nascent telomerase elongation products, while the C-terminal OB-fold promotes telomerase repeat addition processivity through its association with the TASC (p75-p45-p19) sub-complex of telomerase (39,40). Recent studies reveal TASC to be the Tetrahymena CST complex (43,44). We hypothesize that AtPOT1a bridges telomerase and components of CST at telomeres in a similar fashion as Teb1. AtPOT1aOB1 interacts directly with CTC1 and STN1, and further CTC1 and STN1 associate with enzymatically active telomerase (20). Notably, vertebrate POT1 also interacts with CST components, suggesting that CST association may be another ancestral function of POT1.
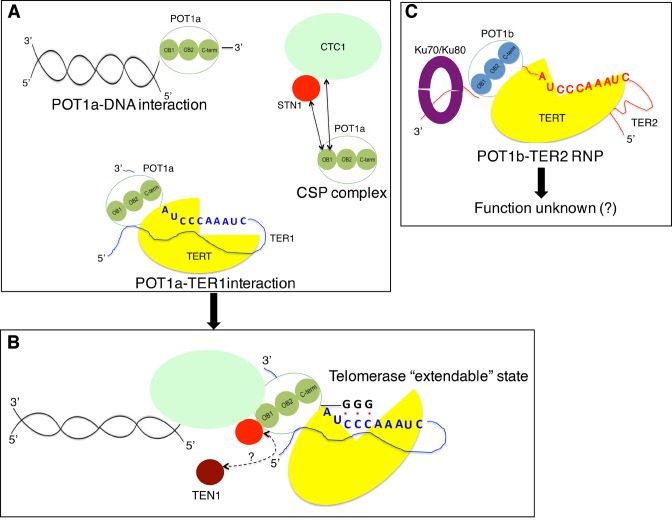
We propose that POT1a is at the nexus of coordinating telomere elongation in A. thaliana via a network of interactions with telomerase through contacts with TER1 (19) and possibly TERT (14), with CTC1 and STN1 (20) and with telomeric DNA (this study) (Figure 5A). AtPOT1a is enriched at telomeres only during S phase (45), where it can stimulate telomerase repeat addition processivity (20) (Figure 5B). Interestingly, the TEN1 subunit of Arabidopsis CST negatively regulates telomerase repeat addition processivity (29) and can displace POT1a from STN1 in vitro (20). Thus, dynamic exchange of POT1a for TEN1 in late S/G2 could play a role in converting telomerase from the extendable to the un-extendable state (20).
Figure 5.
Different modes of interaction for Arabidopsis POT1 proteins. (A) AtPOT1aOB1 has the capacity to bind telomeric DNA (this study) and two components of the CST complex, CTC1 and STN1 (20). The full-length protein can interact specifically with TER1 (19). (B) During S phase, POT1a is proposed to be the bridging factor that engages telomeric DNA, the telomerase RNP as well as CTC1 and STN1 to promote telomere synthesis and specifically to enhance telomerase repeat addition processivity. The interaction of STN1 with POT1aOB1 and TEN1 is mutually exclusive (20). Thus, TEN1 may be dislodged from STN1 to facilitate the telomerase extendable state. (C) AtPOT1b cannot interact with telomeric DNA. It assembles into an alternative RNP complex containing TER2/TER2s, Ku70/80 and perhaps TERT (21). The function of the AtPOT1b RNP complex is unknown.
A final key contribution from this work is elucidation of the molecular basis for the functional divergence between AtPOT1a and AtPOT1b in telomerase regulation. We discovered that substitution of a single highly conserved amino acid in the nucleic acid binding pocket of AtPOT1bOB1 prevents it from binding telomeric DNA and, as a consequence, stimulating telomerase repeat addition processivity. DNA binding can be restored to AtPOT1bOB1 by simply swapping V63 with Phe. Although AtPOT1bOB1-V63F did not elevate telomerase repeat addition processivity to the same extent as wild-type AtPOT1aOB1, this can be explained by the failure of AtPOT1bOB1 to stably associate with the telomerase holoenzyme. In this view, residues in OB1 that lie outside the nucleic acid-binding site would mediate contact with telomerase.
As with most telomere-related components, retention of duplicated POT1 genes is rare. Dosage of individual constituents within multi-subunit complexes, such as those that modulate telomere metabolism, is undoubtedly strictly controlled. As a consequence, evolutionary pressure is exerted to eliminate duplicate copies or diversify them to prevent toxic-genic imbalance. In the case of the Brassicaceae POT1 paralogs, reduced affinity for telomeric DNA and the rise of RNA binding are consistent with evolutionary transition. More importantly, our data argue that telomeric DNA binding was completely ablated in the POT1b lineage, profoundly altering its function. The diversification of POT1b relative to POT1a is not limited to the loss of DNA binding. AtPOT1b assembles into RNP complexes with RNA and protein composition distinct from the AtPOT1a-TER1 RNP (21) (Figure 5C). In addition, AtPOT1a and AtPOT1b display different affinities for CTC1 (15). The altered interaction with CTC1 can be traced to the three residues under positive selection within the Brassicaceae POT1a lineage that specifically enhance POT1a binding to CTC1 relative to POT1b (15). Whether the increased interaction between CTC1 and AtPOT1a stimulates telomere elongation by telomerase is still an open question.
Finally, our analyses support the idea that the functional changes that characterize AtPOT1a and AtPOT1b evolved following the duplication event that produced them. The results indicate that the paralogs were generated as part of the β whole genome duplication event that characterizes Brassicaceae and Cleomaceae genomes, but not Carica papaya (46). Notably, the amino acid residues under positive selection in Brassicaceae POT1a are shared with T. hassleriana POT1a and additionally all POT1a orthologs exhibit the highly conserved Phe within the first OB fold of the protein. Taken together, these data indicate that, post-duplication, POT1a evolved enhanced CTC1 binding via positive selection, reinforcing its role in telomere elongation. Not surprisingly, F65 is under strong purifying selection in AtPOT1a, consistent with its conservation of the ancestral role of telomeric DNA binding. Conversely, the homologous site in AtPOT1b is more variable, indicating that it experienced relaxed selection, consistent with a departure of its role in telomere elongation. This scenario is consistent with escape from adaptive conflict (47), which predicts partitioning of conflicting functions following duplication. A more detailed understanding of how evolutionary pressure has molded AtPOT1a, AtPOT1b and a third, uncharacterized AtPOT1 paralog, POT1c, may yield fascinating new insight into telomere–telomerase engagement.
Supplementary Material
Acknowledgments
The authors are grateful to Callie Kobayashi, Anthony Andrade, Chang Shu, Pingwei Lei and Boris Novikov for discussions and technical assistance. The authors also thank Andrew Nelson and members of the Shippen lab for insightful comments on the manuscript.
Author contributions: A.A. designed and performed experiments. M.E.B analyzed the phylogenetic data, and edited the manuscript. D.E.S. gave conceptual advice. A.A and D.E.S wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01 GM065383 to D.E.S.]; NSF [MCB 1540273 to M.A.B.]. Funding for open access charge: NIH [R01 GM065383 to D.E.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Stewart J.A., Chaiken M.F., Wang F., Price C.M. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage S.A. Human telomeres and telomere biology disorders. Prog. Mol. Biol. Transl. Sci. 2014;125:41–66. doi: 10.1016/B978-0-12-397898-1.00002-5. [DOI] [PubMed] [Google Scholar]
- 3.Lei M., Podell E.R., Baumann P., Cech T.R. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 4.Lei M., Podell E.R., Cech T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 5.Theobald D.L., Mitton-Fry R.M., Wuttke D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn R.L., Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L., Multani A.S., He H., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 8.He H., Multani A.S., Cosme-Blanco W., Tahara H., Ma J., Pathak S., Deng Y., Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nandakumar J., Podell E.R., Cech T.R. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hockemeyer D., Daniels J.-P., Takai H., de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 11.Raices M., Verdun R.E., Compton S.A., Haggblom C.I., Griffith J.D., Dillin A., Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 12.Jacob N.K., Lescasse R., Linger B.R., Price C.M. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol. Cell. Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shakirov E.V., Surovtseva Y.V., Osbun N., Shippen D.E. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossignol P., Collier S., Bush M., Shaw P., Doonan J.H. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- 15.Beilstein M.A., Renfrew K.B., Song X., Shakirov E.V., Zanis M.J., Shippen D.E. Evolution of the telomere-associated protein POT1a in Arabidopsis thaliana is characterized by positive selection to reinforce protein-protein interaction. Mol. Biol. Evol. 2015;32:1329–1341. doi: 10.1093/molbev/msv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura Y., Hawkins M.T.R., McDonough M.M., Jacobs L.L., Flynn L.J. Corrected placement of Mus-Rattus fossil calibration forces precision in the molecular tree of rodents. Sci. Rep. 2015;5:14444. doi: 10.1038/srep14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shakirov E.V., McKnight T.D., Shippen D.E. POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 2009;58:1004–1015. doi: 10.1111/j.1365-313X.2009.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shakirov E.V., Perroud P.-F., Nelson A.D., Cannell M.E., Quatrano R.S., Shippen D.E. Protection of Telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell. 2010;22:1838–1848. doi: 10.1105/tpc.110.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cifuentes-Rojas C., Kannan K., Tseng L., Shippen D.E. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Renfrew K.B., Song X., Lee J.R., Arora A., Shippen D.E. POT1a and components of CST engage telomerase and regulate its activity in Arabidopsis. PLoS Genet. 2014;10:e1004738. doi: 10.1371/journal.pgen.1004738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cifuentes-Rojas C., Nelson A.D.L., Boltz K.A., Kannan K., She X., Shippen D.E. An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 2012;26:2512–2523. doi: 10.1101/gad.202960.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz A.A., Tomba E., Lennarson R., Richard R., Bagajewicz M.J., Harrison R.G. Prediction of protein solubility in Escherichia coli using logistic regression. Biotechnol. Bioeng. 2010;105:374–383. doi: 10.1002/bit.22537. [DOI] [PubMed] [Google Scholar]
- 24.Huang C., Yu Y.T. Synthesis and labeling of RNA in vitro. Curr. Protoc. Mol. Biol. 2013 doi: 10.1002/0471142727.mb0415s102. Ch. 4, Unit 4.15, doi:10.1002/0471142727.mb0415s102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 26.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald M.S., McKnight T.D., Shippen D.E. Characterization and developmental patterns of telomerase expression in plants. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leehy K.A., Lee J.R., Song X., Renfrew K.B., Shippen D.E. MERISTEM DISORGANIZATION1 encodes TEN1, an essential telomere protein that modulates telomerase processivity in Arabidopsis. Plant Cell. 2013;25:1343–1354. doi: 10.1105/tpc.112.107425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryan T.M., Goodrich K.J., Cech T.R. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 31.Baumann P., Cech T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 32.Baumann P., Podell E., Cech T.R. Human Pot1 (Protection of Telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loayza D., Parsons H., Donigian J., Hoke K., de Lange T. DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 34.Lei M., Baumann P., Cech T.R. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry (Mosc.) 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 35.Xu H., Nelson A.D.L., Shippen D.E. A transposable element in the non-canonical telomerase RNA of Arabidopsis thaliana modulates telomerase activity in response to DNA damage. PLoS Genet. 2015;11:e1005281. doi: 10.1371/journal.pgen.1005281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Cheng S., van den Bergh E., Zeng P., Zhong X., Xu J., Liu X., Hofberger J., de Bruijn S., Bhide A.S., Kuelahoglu C., et al. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell. 2013;25:2813–2830. doi: 10.1105/tpc.113.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beilstein M.A., Nagalingum N.S., Clements M.D., Manchester S.R., Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min B., Collins K. Multiple mechanisms for elongation processivity within the reconstituted tetrahymena telomerase holoenzyme. J. Biol. Chem. 2010;285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng Z., Min B., Huang J., Hong K., Yang Y., Collins K., Lei M. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20357–20361. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong K., Upton H., Miracco E.J., Jiang J., Zhou Z.H., Feigon J., Collins K. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol. Cell. Biol. 2013;33:3962–3971. doi: 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Upton H.E., Hong K., Collins K. Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol. Cell. Biol. 2014;34:4200–4212. doi: 10.1128/MCB.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan B., Tang T., Upton H., Shuai J., Zhou Y., Li S., Chen Brunzelle J. J.S., Zeng Z., Collins K., et al. The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat. Struct. Mol. Biol. 2015;22:1023–1026. doi: 10.1038/nsmb.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang J., Chan H., Cash D.D., Miracco E.J., Ogorzalek Loo R.R., Upton H.E., Cascio D., O'Brien Johnson R., Collins Loo K. J.A., et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surovtseva Y.V., Shakirov E.V., Vespa L., Osbun N., Song X., Shippen D.E. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., Saw J.H., Senin P., Wang W., Ly B.V., Lewis K.L.T., et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Des Marais D.L., Rausher M.D. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.