Abstract
Congenital diseases account for a large portion of pediatric illness. Prenatal screening and diagnosis permit early detection of many genetic diseases. Fetal therapeutic strategies to manage disease processes in utero represent a powerful new approach for clinical care. A safe and effective fetal pharmacotherapy designed to modulate gene expression ideally would avoid direct mechanical engagement of the fetus and present an external reservoir of drug. The amniotic cavity surrounding the fetus could serve as an ideal drug reservoir. Antisense oligonucleotides (ASOs) are an established tool for the therapeutic modulation of gene expression. We hypothesize that ASOs administered to the amniotic cavity will gain entry to the fetus and modulate gene expression. Here, we show that an ASO targeting MALAT1 RNA, delivered by transuterine microinjection into the mouse amniotic cavity at embryonic day 13-13.5, reduces target RNA expression for up to 4 weeks after birth. A similarly delivered ASO targeting a causal splice site mutation for Usher syndrome corrects gene expression in the inner ear, a therapeutically relevant target tissue. We conclude that intra-amniotic delivery of ASOs is well tolerated and produces a sustained effect on postnatal gene expression. Transuterine delivery of ASOs is an innovative platform for developing fetal therapeutics to efficaciously treat congenital disease.
INTRODUCTION
Congenital disease and anomalies are estimated to cause 276,000 deaths worldwide within the first month of life (http://www.who.int/mediacentre/factsheets/fs370/en/). In 2015 alone, 5-20% of the 2.6 million global stillbirths can be attributed to genetic disease (1). Prenatal diagnosis early in gestation is becoming more feasible with the advent of increasingly non-invasive methods for obtaining fetal DNA samples (2). Given this ability to detect disease early in development, there is a growing interest in developing therapeutic strategies that can be applied in utero. Although there are some fetal interventions available for congenital structural defects using surgical interventions (3), there are few options for genetic disorders, which may have pathology beginning in utero. One limitation in the progress of in utero therapies is the challenge of managing risk to the fetus and pregnant female. Thus, model systems that can define safe and effective therapeutic strategies are needed. In utero stem-cell or gene therapies are currently being explored for the treatment of a limited number of diseases (4–6). However, testing of pharmacologic agents in utero has been largely unexplored.
Antisense oligonucleotides (ASOs) are emerging as promising therapeutic molecules that meet many of the requirements of an ideal drug candidate including high specificity and stability, relatively low toxicity and ease of delivery to many tissues (7,8). ASOs are short, modified nucleic acids that are designed to target and bind to a specific RNA sequence via complementary base-pairing. Base-pairing to a target RNA can have a number of consequences depending on the binding location and the backbone chemistry of the ASO. For example, ASOs with DNA nucleotides can target an mRNA for degradation by RNAse H, while ASOs with modified RNA bases can affect RNA splicing or translation by creating steric blocks to RNA binding proteins (9).
ASO technology has been used in mammalian cells to alter gene expression for therapeutic benefit (10). Some antisense drugs are in clinical use and many more are in development at various stages of clinical trials (8,11–14). Major advances in ASO chemistry have improved specificity, potency, stability, delivery and biodistribution, and toxic effects have been minimized (14,15). The favorable pharmacologic, pharmacokinetic and toxicological properties of ASOs make them a promising drug platform for disease therapeutics though their safety and efficacy in utero is not known.
ASOs are a particularly attractive drug option for diseases with known genetic components. The ASO mechanism of action is based on specific base-pairing to a gene sequence, and thus can be engineered to target and improve defective gene expression caused by mutations (7,10,11). In many cases, genetic diseases affect developmental processes or are associated with prenatal organ damage, as exemplified by many pediatric neurodegenerative and neurosensory disorders (16–20). Thus, there is a need for administration of therapeutics early in human development at either the embryonic or fetal stages. Such delivery modalities would ideally not engage the embryo or fetus directly and thus carry low risk of adverse events.
The exact mechanism by which ASOs enter cells is not entirely clear, thus it is difficult to predict the transuterine microinjection delivery modality that would be most effective. Previous studies have reported successful injections of ASO directly into mouse embryos (21), and ASOs are known to have widespread distribution in the body via this delivery method. Ideally, however, direct injection into an embryo or fetus would be avoided to minimize any potential risk of adverse outcomes. Studies have also suggested that ASO delivery to a pregnant mouse can result in very low but detectable amount of ASO in fetal liver only (22) and can affect gene expression in the developing embryo (23). However, the effect on postnatal expression was not evaluated. One drawback to prenatal maternal drug administration is that the exposure could lead to drug toxicity. One alternative approach for fetal drug administration that overcomes some of the disadvantages of other approaches is the delivery of ASO into the amniotic fluid contained within the amniotic cavity, which immediately surrounds the embryo. The biodistribution and bioactivity of ASOs delivered to the amniotic fluid has not been evaluated. This work seeks to understand the feasibility of using ASOs to treat pathological conditions early in development in a manner that avoids potential complications associated with intra-embryonic injection.
In this study, we use a well-characterized ASO that targets a non-coding RNA, metastasis associated lung adenocarcinoma transcript 1 (MALAT1), to determine whether deliver of ASOs to the amniotic cavity can efficiently target gene expression. ASO-MALAT1 was selected because MALAT1 is highly expressed in many tissues but is dispensable in mice and the ASO has been shown to effectively lower MALAT1 RNA levels when delivered systemically (24,25). These features make ASO-MALAT1 an ideal tool to test the efficacy of ASO delivery to the amniotic cavity.
The MALAT1-targeted ASO (ASO-MALAT1) is a ‘gapmer’ oligonucleotide, comprised of a central block of 2′-deoxynucleotides that are flanked by 2′-O-methoxyethyl (2′-MOE) modified nucleotides. Upon base-pairing, the 2′-deoxy bases of the ASO render the target mRNA susceptible to cleavage by the ubiquitous cellular enzyme, RNAse H, thereby reducing mRNA abundance. Intraperitoneal injection of ASO-MALAT1 in mice and monkeys causes a reduction in target RNA levels in most tissues and cell types examined (25–27). In order to determine whether this highly active ASO can also downregulate MALAT1 when delivered in utero, we injected the ASO into the amniotic cavity at embryonic day 13-13.5 (E13-13.5) and assessed ASO distribution and MALAT1 RNA expression after birth. Transuterine microinjection of ASOs resulted in a significant reduction in MALAT1 RNA in neonatal mouse kidney, liver, and inner ear, and the effect persisted in the liver for at least 30 days after birth.
We further demonstrate the general ability of ASOs to target gene expression following injection into the amniotic cavity using another well-characterized ASO, ASO-Ush (previously ASO-29), which targets pre-mRNA generated from a mutated form of the Ush1c gene, Ush1c c.216G>A (28). This mutation creates a de novo splice site that is used preferentially over the authentic splice site, which results in aberrant splicing and Usher syndrome in humans and mice. All of the ASO-Ush bases are 2′-MOE modified, and thus do not induce RNAse H activity upon binding to target RNA, but rather create a steric block to splicing at the targeted position. ASO-Ush has been shown to block recognition of the de novo, aberrant splice site and thereby induce correct splicing of Ush1c216A pre-mRNA in the inner ear and rescue hearing and balance abnormalities in mice following an intraperitoneal injection shortly after birth (28). Here, we show that injection of ASO-Ush to the amniotic cavity corrects Ush1c216A pre-mRNA splicing in the inner ear of postnatal mice. Taken together, the ASO-MALAT1 and ASO-Ush data demonstrate the general ability of ASOs to access embryonic tissue and modulate gene expression after birth. Our results suggest that ASOs may have potential as fetal pharmacotherapeutics to effectively modulate postnatal RNA metabolism.
MATERIALS AND METHODS
Oligonucleotides
A chimeric (gapmer) oligonucleotide (5′-CGGTGCAAGGCTTAGGAATT-3′) targeted to murine MALAT1 RNA (ASO-MALAT1) with a fully-modified phosphorothioate backbone with 2′-MOE substituted nucleotides at positions 1–5 and 16–20, and 2′-deoxynucleotides at positions 6–15, were synthesized as previously described (25). ASO-C, (5′-TTAGTTTAATCACGCTCG-3′) and ASO-Ush (ASO-29) (5′- AGCTGATCATATTCTACC-3′) have a fully-modified phosphorothioate backbone with 2′-MOE modifications at all positions as previously described (28,29).
Animals
All procedures on mice were conducted to minimize pain and discomfort by the use of anesthesia and prophylactic analgesia in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal care protocols governing this study were approved by the Oregon Health & Science University and Rosalind Franklin University Institutional Animal Care and Use Committees.
For experiments with MALAT1-ASO, CD-1 (ICR) female mice were crossed with male C57BL/6J mice and for ASO-Ush experiments, heterozygous Ush1c216AG females were crossed with homozygous Ush1c216AA males (28).
Intra-amniotic cavity injections
ASO-MALAT1, ASO-C or ASO-Ush were microinjected into the amniotic cavity of E13-13.5 conceptuses generated by time-dependent crosses (E0.5 was considered noontime on the day a vaginal plug was detected).
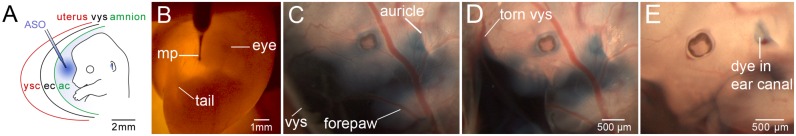
Dams were subjected to transuterine microninjection in the context of mouse survival surgery as described previously (30,31). Aqueous ASOs were mixed with crystalline fast green tracer dye that dissolved fully in solution without appreciably diluting ASO concentration. The ASO-fast green solution was microinjected with a Picospritzer III (source gas nitrogen) using a custom fabricated glass capillary micropipette having an 18–24 μm outer diameter and 20° bevel (32–34). Each pipette was independently calibrated before use so that a known volume and thus a known dose of ASO was delivered per amniotic cavity (34,35). Supplementary Movie S1 and Figure 1 demonstrate the injection paradigm used to deliver ASOs to the amniotic cavity and the embryo line art therein is after Kaufman (36) (Supplementary Movie S1, Figure 1A).
Figure 1.
Transuterine microinjection of antisense oligonucleotide (ASO) into the embryonic day 13 (E13) mouse amniotic cavity. (A) A schematic representation of transuterine microinjection into the amniotic cavity surrounding the E13 embryo. The microinjection pipette (blue) traverses the uterus (red), the yolk sac cavity (ysc), the visceral yolk sac (vys, black), the exocoelomic cavity (ec) and finally the amnion (green) en route to the amniotic cavity (ac). (B) Transillumination of an E13 embryo in utero with light from a fiber optic cable. The left eye and tail are visible. The microinjection pipette (mp) is loaded with ASO tinted with fast green tracer dye and placed in contact with the uterus at the injection site. (C) The E13 embryo injected in (B) after removal of the uterus to enable unobstructed imaging. The blue dye partially obscures the face and digits of the left forepaw. Note the dye accumulation near the nascent pinna or auricle of the external ear. The dye also appears to extend to the boundary of the visceral yolk sac (vys). (D) The vys was torn yet the tracer dye is retained proximal to the embryo in the presumptive amniotic cavity. (E) The amnion was removed from the head and left forepaw allowing most of the dye to diffuse away. Residual dye is retained in the auditory or ear canal. The supplementary movie demonstrates this injection paradigm and the dissection to validate amniotic cavity localization of ASO. The scale bar in (D) applies to (C).
Intraperitoneal injections
Mice were genotyped using ear punch tissue and polymerase chain reaction (PCR) as described previously (28). Mouse pups were injected with 300 mg kg–1 of ASO-Ush at postnatal day 5 (P5) by intraperitoneal injection as previously described (28).
Tissue harvest and handling
Tissue for ASO quantification and RNA isolation was rapidly dissected following euthanasia, distributed to 2.0 ml cryogenic vials, and snap frozen on liquid nitrogen. Tissue was stored at −80°C until shipment on dry ice from Oregon to collaborators in either Illinois or California. Tissue for immunofluorescence was rinsed in phosphate buffered saline (PBS) (pH 7.2-7.4, calcium and magnesium free), fixed overnight in 4% paraformaldehyde in PBS with gentle rotation and rinsed three times in PBS. Tissue was stored at 4°C until shipment in PBS at 4°C to Louisiana.
Quantification of ASO tissue concentration
ASO concentration was determined by HPLC with mass spectrometry detection as previously described (25).
RNA analysis
Frozen tissues were homogenized in TRIZOL solution (ThermoFisher Scientific) using a PowerGen 1000 Homogenizer (ThermoFisher Scientific). Total RNA was purified from TRIZOL reagent following manufacturer's recommendations. RNA was quantitated and purity assessed (A260/280) using a Biophotometer (Eppendorf). The RNA (1 μg) was reverse transcribed using oligo-dT primer and GoScript reverse transcriptase (Promega) following manufacturer's recommendations.
Real time PCR
RNA was quantitated and analyzed by real-time quantitative PCR using TaqMan Gene Expression Master Mix and Primer/Probe set for mouse MALAT1 (Mn01227912_s1) or β-actin (ACTB, Applied Biosystem). Amplification was performed in 96-well plates using the standard condition (50°C for 2 min, and then two steps over 40 cycles at 95°C for 15 s and 60°C for 1 min) for the VIIA7 AB Thermocycler (Applied Biosystem). Each sample was analyzed in duplicate or triplicate. Comparative Ct analysis (ΔΔCt) using ACTB as a normalizer transcript was applied to calculate the relative amount (RQ) of MALAT1 mRNA (37,38) in samples from mice treated with ASO-MALAT1 compared to mice treated with ASO-C. Values that were more than two standard deviations from the mean were not considered, which applied to one sample from the ear at 80 μg and one sample from the spinal cord at 80 μg.
Semiquantitative PCR
One microliter of cDNA was used in PCR reactions with GoTaq Green (Promega) supplemented with primers and α–32P–dCTP. Primers specific for human Ush1c exon 3 (5′-GAATATGATCAGCTGACC-3′) and mouse exon 5 (5′-TCTCACTTTGATGGACACGGTCTTC-3′) were used to specifically amplify only mRNA generated from the knocked-in allele of the human Ush1c.216A gene, which is only present in correctly spliced mRNA (28). Mouse Gapdh primers (5′-GTGAGGCCGGTGCTGAGTATG-3′) and (5′-GCCAAAGTTGTCATGGATGAC-3′) were used to detect and measure endogenous murine Gapdh mRNA. Products were separated on a 6% non-denaturing polyacrylamide gel and quantitated using a Typhoon 9400 phosphorimager (GE Healthcare). Ush1c values were divided by Gapdh values and represented graphically following normalization to the Ush1c / Gapdh ratio of one intraperitoneal sample.
Immunohistochemistry
Tissues were harvested from P7 pups and fixed in 4% paraformaldehyde in PBS, cryoprotected in an ascending sucrose gradient (10, 20, 30%), frozen in optimal cutting temperature media and sectioned at 30–60 μm thick frozen sections. Tissue sections were washed in 0.1% Triton X-100 in PBS, blocked in 4% normal goat serum (NGS) in PBS and incubated overnight at 4°C with a polyclonal rabbit antibody to 2′-MOE ASOs (1:1000, Ionis Pharmaceuticals) (25). For brain, a polyclonal chicken anti-glial fibrillary acidic protein primary antibody (Millipore AB5541; 1:250, EMD Millipore) was also used to label astrocytes. Tissues were subsequently washed (0.1% Triton X-100 in PBS) and incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 488, 1:400, A11008; donkey anti-rabbit Alexa Fluor 555, 1:400, A31572; or goat anti-chicken Alexa Fluor 488, 1:400, A11039, ThermoFisher) in 4% NGS in PBS for 2–3 h at room temperature. For liver and kidney, 1% bovine serum albumin was added with rhodamine-phalloidin (1:40, Molecular Probes R415; ThermoFisher) to stain F-actin. Nuclei were counterstained with DAPI (1 μg/ml, D9542, Sigma). Tissues were mounted and stored at room temperature in Prolong Gold (P36934, ThermoFisher) or DAKO (S3023, DAKO). Sections were imaged using a Zeiss LSM710 confocal laser scanning microscope. Files were imported into ImageJ and/or Adobe Photoshop for processing and analysis.
Statistical analyses
Statistical analyses were performed as described in the text and figure legends using GraphPad Prism 6 v6.0h. One sample or two sample t-tests were used to determine significant differences between the test and control data sets, as indicated in figure legends. One-way analysis of variance (parametric) or Kruskal–Wallis tests (non-parametric) were used to compare three or more groups after data sets were checked for normality with the Kolmogorov–Smirnov test to determine whether parametric tests or non-parametric tests should be used. Correction for multiple comparisons was used (Bonferroni's for parametric; Dunn's for non-parametric) as indicated in the text. Significance is reported with an α-level of 0.05.
RESULTS
Transuterine microinjection of ASO-MALAT1 into the amniotic cavity of mouse embryos
We previously showed effective gene transfer to the developing mouse inner ear by electroporation- or virus-mediated approaches (30,32). We adapted our gene transfer approach to test a fetal pharmacotherapy paradigm in which an ASO targeting MALAT1 (ASO-MALAT1), or a control ASO (ASO-C) lacking cellular targets, was delivered by transuterine microinjection into the amniotic cavity surrounding the E13-13.5 mouse embryo. We hypothesized that the amniotic cavity would serve as a biological reservoir for sustained ASO delivery to the developing fetus from the organogenesis stage until birth. ASO-MALAT1 was selected as a test ASO because it has been well-characterized and is very potent in vivo following systemic delivery to adult mice (25).
A micropipette containing aqueous ASO tinted with fast green tracking dye was advanced through the uterus, visceral yolk sac and amnion where it drapes over the forebrain and left forelimb (Figure 1A, B and Supplementary Movie S1). In a representative embryo dissected free of the uterus immediately after microinjection of ASO into the amniotic cavity, tracking dye obscured facial features and collected around the pinna or auricle of the external ear (Figure 1C). Dissection of the visceral yolk sac did not alter dye localization (Figure 1D), indicating that the exocoelomic cavity between the amnion and visceral yolk sac did not retain appreciable dye (Figure 1A). Dissection of the amnion, however, allowed dye to rapidly diffuse away from the embryo permitting a clear view of the face and left forelimb digits (Figure 1E). The presence of residual dye in the lumen of the ear canal validated that the ASO was successfully targeted to the amniotic cavity (Figure 1E). The ASO injection paradigm was well tolerated, with 94% (89/95) of the ASO-MALAT1 and ASO-C injected embryos surviving, none of which had any obvious phenotypic differences compared to uninjected controls.
ASO-MALAT1 is detected in postnatal tissue following ASO microinjection into the amniotic cavity
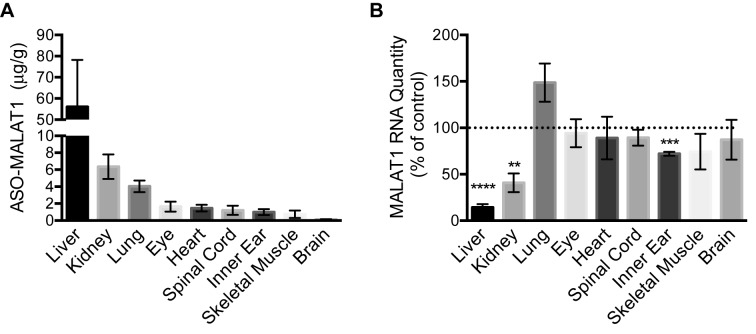
We next assessed whether ASOs that are injected into the amniotic cavity at E13-13.5 gain access to the embryo and are subsequently detectable in neonatal tissue after birth. We measured the ASO concentration in nine different tissues from P7 mice following exposure to 80 μg of ASO via amniotic cavity microinjection at E13-13.5. ASO-MALAT1 was most abundant in the liver tissue (56 μg/g) (Figure 2A and Table 1). All other tissues measured had detectable amounts of ASO including kidney, lung, eye, heart, spinal cord, inner ear, skeletal muscle and brain. These data indicate that ASO is present in diverse P7 tissues after E13-13.5 amniotic cavity loading, and that there are broad differences in ASO abundance among tissues.
Figure 2.
ASO-MALAT1 and RNA content in tissues of P7 mice dosed with ASO-MALAT1 by amniotic cavity injection at E13-13.5. (A) ASO-MALAT1 concentration in P7 mouse tissues measured by HPLC coupled with tandem mass spectrometry detection (mean ± s.e.m., n = 5 except brain and spinal cord, n = 4). (B) The relative quantity (RQ) of MALAT1 RNA normalized to β-actin RNA in the indicated tissues from P7 mice whose amniotic cavities were injected with 80 μg of ASO-MALAT1 (n = 6; except liver and inner ear, n = 5) or ASO-C (n = 3) at E13-13.5. The dotted line represents the normalized control value of 1. Error bars represent s.e.m, **P < 0.01, ***P < 0.001, ****P < 0.0001, One-sample t-test.
Table 1. ASO-MALAT1 tissue concentration.
| Organ | Concentration (μg/g; mean ± s.e.m.) | Ratio to liver | Tissue total (μg; mean ± s.e.m.) |
|---|---|---|---|
| Liver | 56.1 ± 22.2 | 1 | 6.821 ± 0.5125 |
| Kidney | 6.35 ± 1.44 | 0.136 | 0.1345 ± 0.0074 |
| Lung | 4.04 ± 0.69 | 0.105 | 0.3100 ± 0.0170 |
| Eye | 1.64 ± 0.59 | 0.036 | 0.0009 ± 0.0005 |
| Heart | 1.46 ± 0.39 | 0.031 | 0.0354 ± 0.0019 |
| Spinal cord | 1.21 ± 0.54 | 0.027 | 0.0235 ± 0.0020 |
| Inner ear | 1.01 ± 0.35 | 0.023 | 0.0010 ± 0.0009 |
| Skeletal muscle | 0.74 ± 0.44 | 0.010 | 0.0014 ± 0.0008 |
| Brain | 0.13 ± 0.03 | 0.003 | 0.0332 ± 0.0025 |
n = 5, except ear and spinal cord where n = 4.
ASO-MALAT1 reduces MALAT1 RNA in postnatal liver, kidney and inner ear following ASO injection into the amniotic cavity
To assess ASO-MALAT1 bioactivity, 80 μg of ASO-MALAT1 or ASO-C was microinjected into the E13-13.5 amniotic cavity and the relative MALAT1 RNA abundance was determined by real-time quantitative PCR analysis of total RNA isolated from nine different P7 tissues. Relative MALAT1 RNA abundance was significantly reduced in the liver, kidney and inner ear tissue of mice exposed to ASO-MALAT1 intra-amniotically compared to mice exposed to ASO-C (Figure 2B). MALAT1 RNA was not significantly reduced in the lung, eye, heart, spinal cord, skeletal muscle or brain.
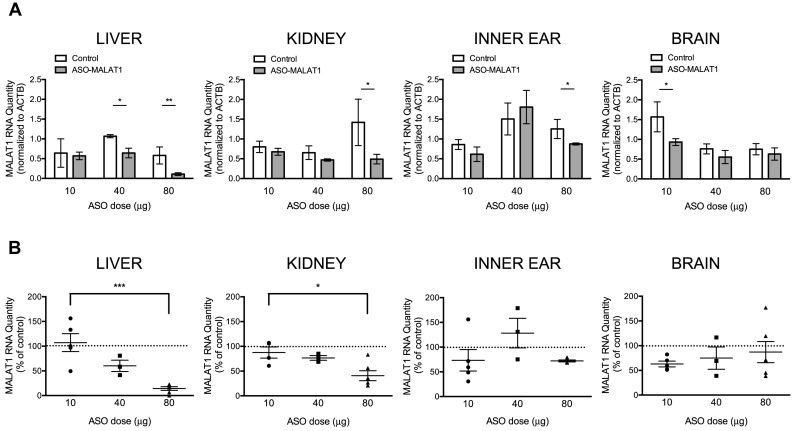
To better understand the functional relationship between amniotic cavity dosing and ASO bioactivity at P7, we quantified MALAT1 RNA abundance following intra-amniotic cavity injection of 10, 40 and 80 μg of ASO-MALAT1. We measured MALAT1 RNA in the liver, kidney, and inner ear, the three tissues found to be most sensitive to ASO-mediated MALAT1 RNA reduction (Figure 3). Brain tissue was also examined as a control in which MALAT1 RNA reduction was not predicted given the lack of an effect of ASO-MALAT1 on MALAT1 RNA following the 80 μg dose (Figure 2A). MALAT1 RNA was reduced significantly in the liver following injection of either 40 or 80 μg of ASO-MALAT1, and in the kidney and inner ear with an 80 μg dose (Figure 3A). The effect of ASO-MALAT1 on MALAT1 RNA was dose-dependent as the reduction in MALAT1 was greater with an 80 μg dose of ASO-MALAT1 than with a 10 μg dose in the liver and kidney (Figure 3B).
Figure 3.
MALAT1 RNA in P7 liver, kidney, inner ear and brain after intra-amniotic cavity dosing with ASO-MALAT1. (A) Quantitation of the average relative quantity (RQ) of MALAT1 RNA normalized to β-actin RNA (ACTB) in the liver, kidney, inner ear and brain from P7 mice subjected to intra-amniotic cavity microinjection of 10, 40 or 80 μg (n = 5, 3, 6, respectively) of ASO-MALAT1 or ASO-C (n = 3). Error bars represent s.e.m., *P < 0.05, **P < 0.01, ****P < 0.0001, one-tail, two-sample t-test (B) The same data as shown in (A) with ASO-MALAT1-treated samples normalized to ASO-C-treated samples which are represented as 100% (dotted line). Error bars represent s.e.m, *P < 0.05, ***P < 0.001 one-way ANOVA with Bonferroni's Multiple Comparison test.
Persistent reduction in MALAT1 RNA after ASO delivery to the amniotic cavity
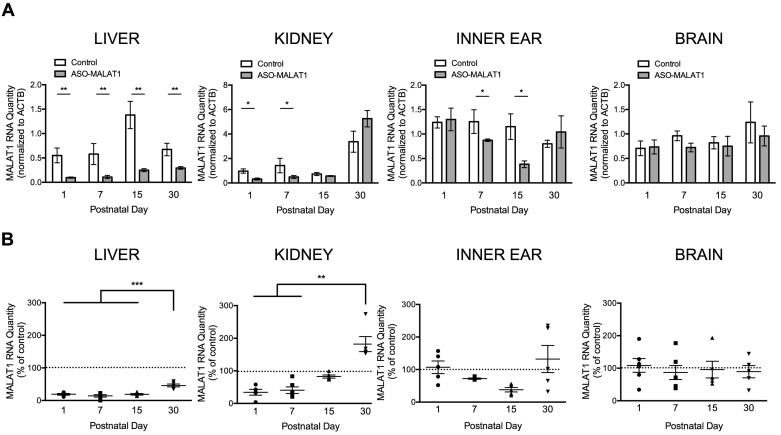
ASOs have a long residence time in a number of tissues (39). In order to determine the duration of the effect of ASOs on MALAT1 RNA abundance following delivery to the amniotic cavity, 80 μg of ASO-MALAT1 or ASO-C was injected into the amniotic cavity at E13-13.5 and liver, kidney, inner ear and brain were collected at P1, P7, P15 and P30 for MALAT1 RNA quantitation. MALAT1 RNA was significantly reduced at all four time points in the liver; at P1 and P7 in kidney tissue; and at P7 and P15 in the inner ear (Figure 4A). There was not a significant difference among the P1, P7 and P15 time points in liver, but there was a significant difference in MALAT1 RNA abundance in P1, P7 and P15 compared to P30 tissue, indicating that the reduction in MALAT1 RNA in the liver diminished over time (Figure 4B).
Figure 4.
MALAT1 RNA in liver, kidney, inner ear and brain at P1, P7, P15 and P30 after intra-amniotic cavity injection of ASO-MALAT1. (A) The average quantity of MALAT1 RNA normalized to β-actin RNA (ACTB) in indicated tissues from mice treated with 80 μg ASO-MALAT1 or ASO-C. Error bars represent s.e.m., *P < 0.05, **P < 0.01, one-tailed, two sample t-test. (B) Graphs depict the average relative quantity (RQ) of MALAT1 RNA normalized to ACTB shown relative to control samples in the liver, kidney, inner ear and brain from P1, P7, P15 and P30 mice subjected to intra-amniotic cavity injection of 80 μg of ASO-MALAT1 (n = 5, 6, 5, 5, respectively) or ASO-C (n = 3,3,4,4, respectively). The dotted lines represent the normalized control value of 1. Error bars represent s.e.m. **P < 0.01, ***P < 0.001 Kruskal–Wallis test with Dunn's Multiple Comparison test (kidney) or one-way ANOVA with Bonferroni's Multiple Comparison test (liver, inner ear, brain).
The effect of ASO-MALAT1 on MALAT1 RNA abundance in the kidney was less stable, with MALAT1 RNA nearing control levels by P15 (Figure 4A). A comparison between the time-points in kidney tissue revealed a significant decrease in MALAT1 RNA abundance in P1 and P7 tissue compared to P30 tissue (Figure 4B). In the inner ear, there was a trend towards an increase in MALAT1 RNA between P15 and P30, suggesting a decline in ASO targeting during this timeframe (Figure 4B). MALAT1 RNA was not significantly reduced in the brain at any of the time-points evaluated (Figure 4A, B).
ASO-MALAT1 distribution in liver, kidney and brain after delivery to the amniotic cavity at E13-13.5
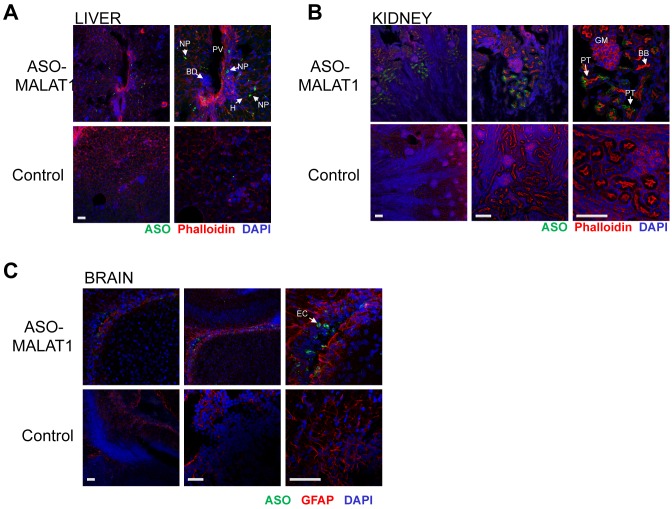
To assess the presence and distribution of ASO in the tissues of postnatal mice that received ASO in the amniotic cavity, immunofluorescence labeling of ASO-MALAT1 was performed using an antibody that specifically recognizes the ASO phosphorothioate backbone (25). We assessed ASO localization in the tissues of P7 mice following an E13-13.5 intra-amniotic cavity injection of 80 μg of ASO (Figure 5 and Table 2). In the liver, ASO was broadly distributed throughout the tissue in both non-parenchymal cells and some hepatocytes. In the kidney, ASO was detected in proximal epithelial cells in some tubules. In the brain, ASO was localized to ventricular ependymal cells. These results show that ASO delivered to the amniotic cavity localizes to the P7 liver, kidney and the brain ventricle.
Figure 5.
Distribution of ASO in P7 liver, kidney and brain after microinjection of ASO-MALAT1 into the E13-13.5 amniotic cavity. Immunofluorescence detection of ASO-MALAT1 in (A) liver (B) kidney and (C) brain. Control panels represent tissues from age-matched uninjected mice. F-actin (red, phalloidin) was labeled in the kidney and liver and glial cells in the brain were immunolabeled with glial-fibrillary acidic protein (GFAP). Nuclei are counterstained with DAPI (blue). BB: brush border; BD: biliary duct; EC: ependymal cells; GM: glomerulus; H: hepatocyte; NP: non-parenchymal cell; PT: proximal tubule cells; PV: portal vein. Scale bars = 100 μm and apply to panels in each respective column.
Table 2. Qualitative summary of immunofluorescence staining of ASO-MALAT1 in P7 mouse tissues.
| Tissue/cell type | IF |
|---|---|
| Liver | |
| Non-parenchymal cells | ++ |
| Hepatocyte | + |
| Kidney | |
| Proximal epithelial cell | ++ |
| Brain | |
| Ventricular ependymal cell | +/++ |
IF, immunofluorescence.
+, weakly positive.
++, moderately positive.
+++, strongly positive.
Correction of Ush1c.216A mRNA splicing following injection of ASO-Ush into the amniotic cavity
We have shown previously that treatment of neonatal mice by intraperitoneal injection of a single dose of an ASO targeting an aberrant splice site created by the mutation Ush1c c.216G>A redirects splicing to the authentic splice site and partially corrects Ush1c.216A splicing in a therapeutically relevant target tissue, the inner ear (28). Because delivery of MALAT1-ASO into the amniotic cavity resulted in a significant downregulation of MALAT1 RNA in the inner ear, we tested whether the ASO targeting Ush1c c.216A (ASO-Ush) could correct splicing of the RNA transcript in the inner ear when delivered by transuterine microinjection into the amniotic cavity.
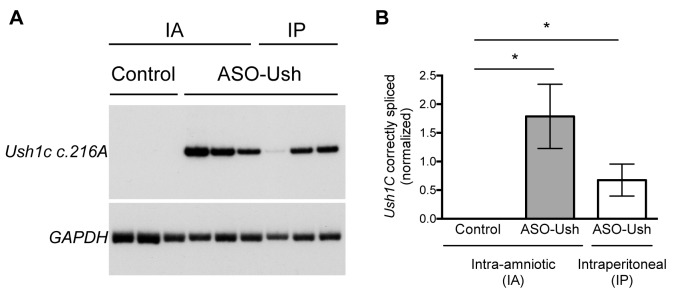
E13-13.5 embryos from a heterozygous Ush1c216GA female crossed with a homozygous Ush1c216AA male were injected with 100 μg of ASO-Ush and inner ear tissue was collected and analyzed at P22. For comparison, neonatal Ush1c216AA mice were treated by intraperitoneal injection of 300 mg/kg (∼600 μg) of ASO-Ush at P5 and tissues were collected at P22. Correct Ush1c splicing was detected in all of the samples from mice injected with ASO-Ush at E13-13.5 indicating successful delivery of the ASO to the inner ear by delivery to the amniotic cavity (Figure 6). The amount of correctly spliced Ush1c216A mRNA that was detected in the inner ear of mice treated by delivery to the amniotic cavity had an average increase in the amount of correctly spliced mRNA detected in the inner ear of Ush1c216AA mice compared to those treated by intraperitoneal injection of ASO-Ush, though the difference between the two delivery methods was not statistically significant (Figure 6).
Figure 6.
Ush1c.216A splicing in the inner ear of P22 mice dosed with ASO-Ush by amniotic cavity injection at E13-13.5. (A) Semi-quantitative radiolabeled RT-PCR analysis with primer sets specific for human USH1C (to specifically amplify only the knocked in human portion of the Ush1c216A allele (28)) and mouse Gapdh was performed on RNA isolated from the inner ear of P22 Ush1c216AA or Ush1c216GA mice treated with ASO-Ush at E13-13.5 by transuterine injection to the amniotic cavity (IA) (n = 3), or neonatal Ush1c216AA mice treated at P5 with ASO-Ush by intraperitoneal (IP) injection (n = 3), or untreated P22 Ush1c216AA (control)(n = 3) mice. (B) Quantification of correctly spliced mRNA from the Ush1c216A allele. Error bars represent s.e.m (n = 3). *P ≤ 0.05, one-tailed, two-sample t-test.
DISCUSSION
Our results demonstrate that ASOs injected into the amniotic cavity at E13-13.5 can alter expression of a targeted RNA transcript in the liver, kidney, and inner ear of postnatal mice. We find that an ASO delivered in this manner to mouse embryos can effectively target RNA for 1 (kidney), 2 to 3 (inner ear) or 4 weeks (liver) after birth. We have demonstrated the efficacy of ASO delivery to the amniotic cavity using an ASO that targets the long non-coding RNA, MALAT1, for RNAse H-mediated degradation and an ASO that binds its target RNA creating a steric block of cryptic splicing of pre-mRNA from the Ush1c.216A gene. We propose that this approach can be applied to the use of ASOs, in general, for modulation of any gene target. Our data suggest that intra-amniotic cavity injection of ASOs represents a relatively non-invasive modality for the treatment of congenital disease or other conditions that adversely affect fetal growth and development.
Fetal approaches for treating genetic disease are desirable for the prevention of antenatal pathology that can cause irreversible damage in utero. In addition, prenatal treatment would be expected to target a rapidly dividing population of stem or progenitor cells that may be more easily accessed by ASOs than post-mitotic differentiated cell types. Injection into the amniotic cavity is likely to have as favorable a benefit/risk balance as amniocentesis, a diagnostic test involving removal of amniotic fluid, which carries a low miscarriage rate of ∼1 in 1000 (0.11%) (40). In utero therapies involving delivery of reagents to the amniotic cavity are not common but the approach is under increasing consideration as new promising therapeutic interventions become available for the treatment of disorders in early development (4,41–43).
In the present study, 94% (89/95) of the ASO-MALAT1 or ASO-C injected embryos survived, all of which were indistinguishable from uninjected controls. These results demonstrate that transuterine microinjection of ASOs into the amniotic cavity is a feasible methodology for fetal drug delivery made possible by the specialized properties of the amnion, a membrane that encases the mammalian embryo. The mouse amnion is a clear, bilayered membrane that develops from the amniochorionic fold during gastrulation (44). The shear resistance and strength of the amnion results from the fusion of a layer of cuboidal ectoderm that faces the surface of the embryo and a layer of squamous mesoderm that faces the exocoelomic cavity (44). This structure is resistant to mechanical perturbation and self-seals after puncture with a 10–40 micron microinjection pipette by immediately clogging with cells present in the amniotic fluid. The amnion further defines a cavity surrounding the embryo that has a comparatively large volume of ∼ 85–95 microliters at E13-13.5 (ref. (45)), allowing sufficient space to load the cavity with a relevant quantity of bioactive ASO for delivery to the embryo. Finally, the diverse solute concentration of the amniotic fluid, consisting of sodium, potassium, chloride, glucose, lactate, bicarbonate and calcium as well as nutrients and factors that promote fetal growth, produces an osmolality of 300–305 mOsm/kg from E13-17 (45) which is similar to the osmolality of mouse blood plasma at 327 mOsm/kg (46), thus ensuring a corollary pharmacokinetic environment for ASO mobilization that is well established from systemic ASO injection studies (47). Our work establishes that the amniotic cavity has the patency, volume and solute concentration to serve as an efficient biological reservoir for ASO delivery to the developing embryo.
The mechanism by which ASOs in the amniotic cavity gain access to the embryo is not clearly understood. ASOs with a phosphorothioate-modified backbone bind to plasma proteins after systemic delivery, which slows their renal clearance and permits broad distribution in tissues, with the highest accumulation seen in the liver and kidney following subcutaneous or intravenous injection (25,47,48). Our study demonstrates that intra-amniotic cavity injection of ASOs results in a similar liver and kidney enrichment. We hypothesize that the ASOs in the amniotic cavity gain entry into the circulatory system of the fetus and subsequently become enriched in hepatic and renal tissues. The ∼25 mOsm/kg differential between the osmolality of the amniotic cavity fluid and mouse blood plasma may establish an osmotic gradient that produces a strong hydrostatic force to efficiently drive water directly into blood vessels of the fetal skin, the umbilical cord and the embryonic component of the placenta (49,50). Remarkably, this so-called intramembranous transport pathway drives 400 ml of water into the sheep fetal circulation per day (30). In the E13-13.5 mouse fetus, the intramembranous transport pathway through the skin is restricted due to keratinization which occurs at approximately E12.5-14 in rodents (51). It is thus likely that the bulk of water transport in the E13-13.5 mouse embryo occurs through the fetal placenta or umbilicus. We predict that the strong hydrodynamic flow of water into the fetal circulation supports ASO translocation from the amniotic cavity to the fetal blood stream. A more complete understanding of fetal ASO transport may enable its modulation to enhance fetal uptake, broaden tissue distribution and heighten therapeutic impact.
ASO pharmacokinetics and pharmacodynamics in the pre-natal and neonatal mouse are complex and not fully understood, and thus the amount of ASO in any given tissue or organ and the extent of the anticipated ASO-mediated RNA modulation may not reliably predict phenotypic effects. For example, we have recently shown that only a very low level of ASO targeting was necessary to achieve a complete phenotypic rescue of vestibular dysfunction and partial rescue of hearing in a mouse model of Usher syndrome (28). Considering these previous results, it is particularly encouraging to find that the intra-amniotic cavity delivery of ASO results in a significant effect on the target RNA in the inner ear, which would be the target tissue to treat congenital forms of deafness such as that seen in human Usher syndrome. Indeed, effective ASO delivery via amniotic cavity injection was confirmed using an ASO targeting the aberrant splice site in Ush1c c.216A that is associated with Type 1C Usher syndrome. This ASO, which has been previously shown to correct splicing and rescue vestibular and hearing deficits in the mice when delivered by intraperitoneal injection shortly after birth, caused a greater correction of splicing when delivered by intra-amniotic cavity injection. We are currently refining fetal delivery of ASO-Ush to further enhance targeting of the Ush1c.216A allele and to determine the impact on ameliorating the deleterious sensory phenotypes.
We conclude that ASO delivery to the intra-amniotic cavity modulates neonatal gene expression and may serve as a therapeutic intervention in itself or when paired with a suitable postnatal therapeutic strategy. Further optimization of the method will broaden the potential impact and applicability of this approach. We predict that fetal ASO pharmacotherapy has the potential to safely enable therapeutic strategies for the treatment of fetal and congenital genetic disease.
Supplementary Material
Acknowledgments
We thank Steven Miller for assistance with data analysis and statistics, Viktoriya Georgieva and Anthony Hinrich for experimental assistance and consultation and Monica Oblinger for comments on the manuscript. M.L.H., J.V.B., F.R. and J.J.L. conceived and designed the experiments and analyzed the data. F.F.D., L.W., H.J., F.M.J. and R.F.R. performed the experiments and analyzed the data. F.F.D., M.L.H. and J.V.B. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-DC012596 to M.L.H., R21-DC012916 to J.V.B., R01-DC014160 to J.V.B., P30-DC005983 to J.V.B., 1 U54 GM104940 to J.J.L., P30-GM103340 to J.J.L.]; Foundation Fighting Blindness (to J.J.L., M.L.H.). Funding for open access charge: The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. Frank Rigo is an employee of Ionis Pharmaceuticals. Michelle Hastings receives funding from Ionis Pharmaceuticals.
REFERENCES
- 1.Lawn J.E., Blencowe H., Waiswa P., Amouzou A., Mathers C., Hogan D., Flenady V., Froen J.F., Qureshi Z.U., Calderwood C., et al. Stillbirths: rates, risk factors, and acceleration towards 2030. Lancet. 2016;387:587–603. doi: 10.1016/S0140-6736(15)00837-5. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi D.W. From prenatal genomic diagnosis to fetal personalized medicine: progress and challenges. Nat. Med. 2012;18:1041–1051. doi: 10.1038/nm.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson E.G., Flake A.W. Stem cell and genetic therapies for the fetus. Semin. Pediatr. Surg. 2013;22:56–61. doi: 10.1053/j.sempedsurg.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Ramachandra D.L., Shaw S.S., Shangaris P., Loukogeorgakis S., Guillot P.V., Coppi P.D., David A.L. In utero therapy for congenital disorders using amniotic fluid stem cells. Front. Pharmacol. 2014;5:270–281. doi: 10.3389/fphar.2014.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay T.R., Rahim A.A., Buckley S.M., Ward N.J., Chan J.K., Howe S.J., Waddington S.N. Perinatal gene transfer to the liver. Curr. Pharm. Des. 2011;17:2528–2541. doi: 10.2174/138161211797247541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pergament E. The future of prenatal diagnosis and screening. J. Clin. Med. 2014;3:1291–1301. doi: 10.3390/jcm3041291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigo F., Seth P.P., Bennett C.F. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv. Exp. Med. Biol. 2014;825:303–352. doi: 10.1007/978-1-4939-1221-6_9. [DOI] [PubMed] [Google Scholar]
- 8.Havens M.A., Hastings M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudet A.L., Meng L. Gene-targeting pharmaceuticals for single-gene disorders. Hum. Mol. Genet. 2015;25:R18–R26. doi: 10.1093/hmg/ddv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havens M.A., Duelli D.M., Hastings M.L. Targeting RNA splicing for disease therapy. Wiley Interdiscip. Rev. RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundin K.E., Gissberg O., Smith C.I. Oligonucleotide therapies: the past and the present. Hum. Gene Ther. 2015;26:475–485. doi: 10.1089/hum.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geary R.S., Baker B.F., Crooke S.T. Clinical and preclinical pharmacokinetics and pharmacodynamics of mipomersen (kynamro((R))): a second-generation antisense oligonucleotide inhibitor of apolipoprotein B. Clin. Pharmacokinet. 2015;54:133–146. doi: 10.1007/s40262-014-0224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marafini I., Di Fusco D., Calabrese E., Sedda S., Pallone F., Monteleone G. Antisense approach to inflammatory bowel disease: prospects and challenges. Drugs. 2015;75:723–730. doi: 10.1007/s40265-015-0391-0. [DOI] [PubMed] [Google Scholar]
- 14.McClorey G., Wood M.J. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr. Opin. Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poretti A., Boltshauser E., Huisman T.A. Pre- and postnatal neuroimaging of congenital cerebellar abnormalities. Cerebellum. 2016;15:5–9. doi: 10.1007/s12311-015-0699-z. [DOI] [PubMed] [Google Scholar]
- 17.Utz J.R., Crutcher T., Schneider J., Sorgen P., Whitley C.B. Biomarkers of central nervous system inflammation in infantile and juvenile gangliosidoses. Mol. Genet. Metab. 2015;114:274–280. doi: 10.1016/j.ymgme.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braverman N.E., Raymond G.V., Rizzo W.B., Moser A.B., Wilkinson M.E., Stone E.M., Steinberg S.J., Wangler M.F., Rush E.T., Hacia J.G., et al. Peroxisome biogenesis disorders in the Zellweger spectrum: an overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol. Genet. Metab. 2016;117:313–321. doi: 10.1016/j.ymgme.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poenaru L., Castelnau L., Dumez Y., Thepot F. First-trimester prenatal diagnosis of mucolipidosis II (I-cell disease) by chorionic biopsy. Am. J. Hum. Genet. 1984;36:1379–1385. [PMC free article] [PubMed] [Google Scholar]
- 20.Kral A., Kronenberger W.G., Pisoni D.B., O'Donoghue G.M. Neurocognitive factors in sensory restoration of early deafness: a connectome model. Lancet Neurol. 2016;15:610–621. doi: 10.1016/S1474-4422(16)00034-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soucy N.V., Riley J.P., Templin M.V., Geary R., de Peyster A., Levin A.A. Maternal and fetal distribution of a phosphorothioate oligonucleotide in rats after intravenous infusion. Birth Defects Res. B Dev. Reprod. Toxicol. 2006;77:22–28. doi: 10.1002/bdrb.20067. [DOI] [PubMed] [Google Scholar]
- 23.Driver S.E., Robinson G.S., Flanagan J., Shen W., Smith L.E., Thomas D.W., Roberts P.C. Oligonucleotide-based inhibition of embryonic gene expression. Nat. Biotechnol. 1999;17:1184–1187. doi: 10.1038/70724. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B., Arun G., Mao Y.S., Lazar Z., Hung G., Bhattacharjee G., Xiao X., Booth C.J., Wu J., Zhang C., et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung G., Xiao X., Peralta R., Bhattacharjee G., Murray S., Norris D., Guo S., Monia B.P. Characterization of target mRNA reduction through in situ RNA hybridization in multiple organ systems following systemic antisense treatment in animals. Nucleic Acid Ther. 2013;23:369–378. doi: 10.1089/nat.2013.0443. [DOI] [PubMed] [Google Scholar]
- 26.Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L., et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lentz J.J., Jodelka F.M., Hinrich A.J., McCaffrey K.E., Farris H.E., Spalitta M.J., Bazan N.G., Duelli D.M., Rigo F., Hastings M.L. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 2013;19:345–350. doi: 10.1038/nm.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinrich A.J., Jodelka F.M., Chang J.L., Brutman D., Bruno A.M., Briggs C.A., James B.D., Stutzmann G.E., Bennett D.A., Miller S.A., et al. Therapeutic correction of ApoER2 splicing in Alzheimer's disease mice using antisense oligonucleotides. EMBO Mol. Med. 2016;8:328–345. doi: 10.15252/emmm.201505846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H., Wang L., Beier K.T., Cepko C.L., Fekete D.M., Brigande J.V. Lineage analysis of the late otocyst stage mouse inner ear by transuterine microinjection of a retroviral vector encoding alkaline phosphatase and an oligonucleotide library. PLoS One. 2013;8:e69314. doi: 10.1371/journal.pone.0069314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Jiang H., Brigande J.V. Gene transfer to the developing mouse inner ear by in vivo electroporation. J. Vis. Exp. 2012;64:3653. doi: 10.3791/3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gubbels S.P., Woessner D.W., Mitchell J.C., Ricci A.J., Brigande J.V. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amaral D.G., Price J.L. An air pressure system for the injection of tracer substances into the brain. J. Neurosci. Methods. 1983;9:35–43. doi: 10.1016/0165-0270(83)90107-3. [DOI] [PubMed] [Google Scholar]
- 34.Sakai M., Swartz B.E., Woody C.D. Controlled micro release of pharmacological agents: measurements of volume ejected in vitro through fine tipped glass microelectrodes by pressure. Neuropharmacology. 1979;18:209–213. doi: 10.1016/0028-3908(79)90063-7. [DOI] [PubMed] [Google Scholar]
- 35.Brigande J.V., Seyfried T.N. Glycosphingolipid biosynthesis may not be necessary for vertebrate brain development. Ann. N.Y. Acad. Sci. 1998;845:215–218. doi: 10.1111/j.1749-6632.1998.tb09673.x. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman M. The Atlas of Mouse Development. London: Academic Press Limited; 1992. [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Havens M.A., Reich A.A., Hastings M.L. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet. 2014;10:e1004312. doi: 10.1371/journal.pgen.1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigo F., Chun S.J., Norris D.A., Hung G., Lee S., Matson J., Fey R.A., Gaus H., Hua Y., Grundy J.S., et al. Pharmacology of a central nervous system delivered 2′-o-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J. Pharmacol. Exp. Ther. 2014;350:46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akolekar R., Beta J., Picciarelli G., Ogilvie C., D'Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2015;45:16–26. doi: 10.1002/uog.14636. [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin E.S., Schlosser B.A., Border W.L. Fetal diagnostics and fetal intervention. Clin. Perinatol. 2016;43:23–38. doi: 10.1016/j.clp.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Hermes K., Schneider P., Krieg P., Dang A., Huttner K., Schneider H. Prenatal therapy in developmental disorders: drug targeting via intra-amniotic injection to treat X-linked hypohidrotic ectodermal dysplasia. J. Investig. Dermatol. 2014;134:2985–2987. doi: 10.1038/jid.2014.264. [DOI] [PubMed] [Google Scholar]
- 43.Ross M.G., Brace R.A., National Institute of Child, H. and Development Workshop, P. National Institute of Child Health and Development Conference summary: amniotic fluid biology–basic and clinical aspects. J. Matern. Fetal Med. 2001;10:2–19. doi: 10.1080/714904292. [DOI] [PubMed] [Google Scholar]
- 44.Pereira P.N., Dobreva M.P., Graham L., Huylebroeck D., Lawson K.A., Zwijsen A.N. Amnion formation in the mouse embryo: the single amniochorionic fold model. BMC Dev. Biol. 2011;11:48–60. doi: 10.1186/1471-213X-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheung C.Y., Brace R.A. Amniotic fluid volume and composition in mouse pregnancy. J. Soc. Gynecol. Investig. 2005;12:558–562. doi: 10.1016/j.jsgi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Bekkevold C.M., Robertson K.L., Reinhard M.K., Battles A.H., Rowland N.E. Dehydration parameters and standards for laboratory mice. J. Am. Assoc. Lab. Anim. Sci. 2013;52:233–239. [PMC free article] [PubMed] [Google Scholar]
- 47.Geary R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 48.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Brace R.A. Progress toward understanding the regulation of amniotic fluid volume: water and solute fluxes in and through the fetal membranes. Placenta. 1995;16:1–18. doi: 10.1016/0143-4004(95)90077-2. [DOI] [PubMed] [Google Scholar]
- 50.Brace R.A. Physiology of amniotic fluid volume regulation. Clin. Obstet. Gynecol. 1997;40:280–289. doi: 10.1097/00003081-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 51.Hayward A.F. The permeability of the epithelium of the skin of fetal rats demonstrated with a lanthanum-containing solution. J. Anatomy. 1983;136:379–388. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.