Abstract
The Clustered Regularly Interspaced Short Palindromic Repeats system allows a single guide RNA (sgRNA) to direct a protein with combined helicase and nuclease activity to the DNA. Streptococcus pyogenes Cas9 (SpCas9), a CRISPR-associated protein, has revolutionized our ability to probe and edit the human genome in vitro and in vivo. Arguably, the true modularity of the Cas9 platform is conferred through the ease of sgRNA programmability as well as the degree of modifications the sgRNA can tolerate without compromising its association with SpCas9 and function. In this review, we focus on the properties and recent engineering advances of the sgRNA component in Cas9-mediated genome targeting.
INTRODUCTION
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) loci are present in prokaryotes, including both bacteria and archaea (1,2), and are primarily characterized by direct repeat sequences interspaced by similarly sized variable sequences (3–6). Early investigations into the nature of the repeat and variable sequences revealed that CRISPR and the CRISPR-associated proteins (Cas) work in tandem to recognize and cleave invading foreign DNA (1–8). The characterization of CRISPR-Cas as a type of prokaryotic immune system laid the groundwork for what has now become a powerful tool for various applications well outside the original biological context (9–16).
The CRISPR-Cas systems are diverse in prokaryotes, and as such, are divided into two classes based on whether the effector complex is multimeric (Class 1) or monomeric (Class 2). The two classes are in turn subdivided into six types with types I, III and IV belonging to Class 1, and types II, V and VI belonging to Class 2 (17,18). While the primary function of conferring acquired immunity to invading foreign nucleic acids is conserved between types, the individual components required to carry out this function vary. For the purposes of this review we largely focus on the RNA components from the CRISPR type II-A SpCas9 system.
In prokaryotes, the CRISPR-Cas system functions as a microbial analog to the acquired (adaptive) immune system present in higher organisms (5,19,20). The variable sequences of the CRISPR array, known as spacers, are relics of previous infectious events whereby fragments of invading DNA, or protospacers, have been captured and integrated into the host genome at the CRISPR locus to serve as an immunological memory (8). Once a new protospacer has been integrated into the CRISPR array, the entire array can be transcribed into pre-crRNA and processed into mature crRNA.
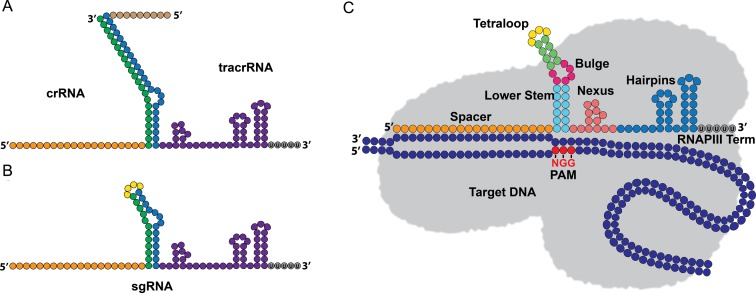
The processing of pre-crRNA into mature crRNA is distinct in type II CRISPR systems in that it relies on the presence of trans-activating crRNAs (tracrRNAs) that hybridize with the pre-crRNA through complementary base pairing to the repeat regions (21–23). RNase III, a dimeric endoribonuclease that cleaves double-stranded RNA, then recognizes the pre-crRNA:tracrRNA hybrid and cleaves individual crRNA:tracrRNA hybrids from the primary CRISPR array transcript (21). Ultimately the crRNA-tracrRNA hybrid spacer sequence (Figure 1A) is trimmed down to 20 nucleotides (21) before tightly associating with the SpCas9 nuclease and forming the catalytically active ribonucleoprotein (RNP) complex used for targeted DNA cleavage (19,20).
Figure 1.
Streptococcus pyogenes CRISPR-SpCas9 guide RNA anatomy. (A) Endogenous CRISPR RNA (crRNA) and transacting crRNA (tracrRNA). The spacer sequence (orange) is 20 nucleotides in length and the repeat sequence (green) is 22 nucleotides that basepairs with tracrRNA complementary region (blue). The 3′ handle region (purple) has functional significance for structure-dependent recognition by SpCas9. (B) The synthetic sgRNA retains dual-tracrRNA:crRNA secondary structure via a fusion of the 3′ end of the crRNA to the 5′ end of the tracrRNA with an engineered tetraloop. (C) Individual functional modules of the sgRNA (sgRNA structure adopted from Briner et al., 2014). The 5′ spacer sequence dictates SpCas9 localization within the genome. The lower stem is formed by the duplex between the CRISPR repeat sequence from the crRNA and the region of complementarity in the tracrRNA. SpCas9 interacts with the upper and lower stems in a sequence-independent manner, whereas the bulge interactions with SpCas9 appear to be sequence-dependent. The nexus contains both sequence and structural features necessary for DNA cleavage and lies at the center of the sgRNA:SpCas9 interactions. The nexus also forms a junction between the sgRNA and both SpCas9 and the target DNA. The terminal hairpins assist in stabilizing the sgRNA and supports stable complex formation with SpCas9. The hairpins can also tolerate large deletion mutations and still exhibit cleavage activity.
An indispensable aspect of any immune system is the ability to distinguish self from non-self; in other words, the components of the immune system must be able to recognize molecules that do not originate from the host. The SpCas9 CRISPR system achieves this distinction through the recognition of a protospacer adjacent motif (PAM), which is a short G-rich oligonucleotide sequence downstream of the target DNA (8,24). This feature is crucial for targeted DNA cleavage, as the corresponding spacer in the CRISPR array is identical to the target DNA, and would otherwise be cleaved. It is not until after SpCas9 scans invading foreign DNA for the PAM sequence 5′NGG that complementary base pairing between the target DNA and crRNA can occur and trigger targeted DNA cleavage (25,26).
Cas9, the protein
The high-resolution crystal structure of SpCas9 in complex with a single guide RNA (sgRNA), and its cognate target DNA obtained by Nishimasu et al. identified key functional interactions that govern the molecular mechanism of SpCas9-mediated DNA cleavage. The crystal structure revealed that SpCas9 has a bilobed architecture composed of a Recognition lobe (REC) and a Nuclease lobe (NUC), and the site of heteroduplex formation between the sgRNA and its cognate target DNA is a positively charged cleft at the interface between the two lobes (27,28). The REC lobe is comprised of an α-helical region termed the bridge helix domain that recognizes the ‘seed’ region (the 10–12 PAM-proximal nucleotides of the guide region) of the sgRNA through salt bridges with sgRNA backbone, a REC1 domain that recognizes repeat:anti-repeat duplex of the sgRNA and the REC2 domain that does not interact with the guide:target heteroduplex. The NUC lobe is comprised of an RuvC catalytic nuclease domain that cleaves the non-complementary strand of the target DNA, an HNH catalytic nuclease domain that cleaves the complementary strand of the target DNA, and a PAM-interacting (PI) domain that recognizes the 5′NGG PAM on the non-complementary strand (22,23,27,28).
SpCas9 interacts with the sgRNA in both sequence-dependent and independent manners—the guide region is recognized in a sequence-independent mechanism, whereas SpCas9 recognition of the sgRNA repeat:anti-repeat duplex involves sequence-dependent interactions (27). Additional information on the base pair interactions and details regarding conformational changes due to sgRNA and target DNA/PAM recognition can be found in recent literature (27–30).
CRISPR Class 2, a brief overview
Among Class 2 Type II proteins there is a large number of different Cas9 orthologs (31). Here, we include a brief overview of some of the various Cas9 orthologs used in mammalian genome editing.
The Type II-A Cas9 ortholog Staphylococcus aureus (SaCas9) is a notable alternative to SpCas9. SaCas9 has structural similarity to SpCas9, notwithstanding its 17% sequence identity (32), and was shown to edit genomes with efficiency similar to SpCas9 (33). Less popular are the co-expressed StCas9s from Streptococcus thermophilus located on CRISPR loci 1 or 3. These orthologs are only slightly smaller than SpCas9 and have reduced cleavage activity in human cells (10,34). Type II-C Cas9s have shown poor dsDNA cleavage efficiency, a propensity to cleave off-target, and are not commonly used for genome editing (35). However, NmCas9 from Neisseria meningitides, has successfully been used for genome engineering in human pluripotent stem cells (36–38).
Other types within the Class 2 CRISPR-Cas repertoire include the Type V system Cpf1. Cpf1 targets a 5′ T-rich PAM upstream of the target DNA, it generates a five nucleotide 5′ overhang, and does not require a tracrRNA (39). AsCpf1 from Acidaminococcus sp. recognizes a 5′ TTTN PAM and has shown robust activity in human cells (39). The Class 2 Type VI CRISPR system consists of only one member, C2c2 from Leptotrichia shahii, and cleaves RNA rather than DNA, using a one part guide (similar to Cpf1) (18,40).
Our understanding of other Class 2 CRISPR-Cas systems is growing rapidly, and new systems are being discovered and adapted for genome editing. The sequences of the sgRNAs as well as the PAMs from the Class 2 CRIPSR-Cas proteins mentioned above are listed in Table 1. While this review will focus on guide RNA engineering advances, there are a number of developments in the protein engineering field of Cas9 that cannot be recapitulated by sgRNA engineering alone and warrant mention. For example, human codon optimization has been performed on SpCas9 (9), SaCas9 (41), NmCas9 (42), St1Cas9 (42), St3Cas9 (43) and Cpf1 (39). Likewise, these have been made into catalytically inactive and nicking versions (23,32,42,44–46). Further, mutagenesis of PAM-interacting regions of SpCas9 and SaCas9 has resulted in altered PAM mutants (41,47), and mutational neutralization of key charged residues increased target specificity and decreased off-target effects by SpCas9 (48,49).
Table 1. CRISPR-associated programmable nucleases and cognate sgRNA.
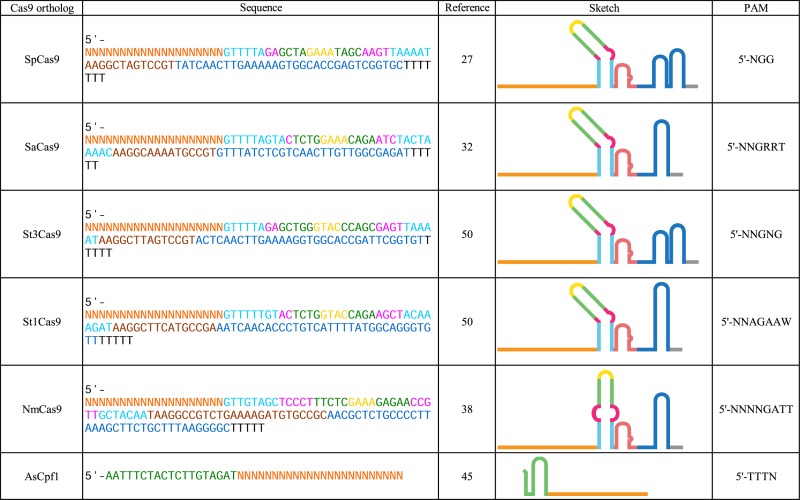
|
Guide RNA production and multiplexing
Endogenous Type II CRISPR RNA components require extensive processing before becoming functional (22,23). The first effort to recapitulate the bacterial CRISPR system in mammalian cells involved the delivery of SpCas9, SpRNase III, the tracrRNA and the pre-crRNA array, which contained the spacer sequence flanked by direct repeats (10). Interestingly, the inclusion of SpRNase III was found to be unnecessary for cleavage of the target DNA sequence in mammalian cells (10).
A key advance in CRISPR programmability came with the engineering of the chimeric sgRNA (23). The chimeric sgRNA (Figure 1B) is a single transcript that retains the dual-tracrRNA:crRNA secondary structure via a fusion of the 3′ end of the crRNA to the 5′ end of the tracrRNA with an engineered tetraloop (23). Results from systematic mutational analysis of 77 sgRNA variants revealed that the sgRNA is composed of six structural modules (Figure 1C): the spacer, the lower stem, the bulge, the upper stem, the nexus and the hairpins (50).
The most common approach to produce sgRNAs in human cells is using the human U6 RNA polymerase III (RNAP III) promoter (9,10,16). This constitutive RNAP III promoter allows the sgRNA transcript to escape post-transcriptional modifications that are coupled to RNAP II transcription (such as 5′ methyl capping and polyadenylation), which would otherwise result in its export out of the nucleus (51,52). Alternatively, the type III RNAP III promoter H1, which requires a purine nucleotide transcriptional start site, can be used (53). Notably, both the U6 and HI promoters can be combined with transcriptional response elements to facilitate inducible sgRNA production (54,55).
RNAP II mediated sgRNA expression has been utilized by placing the sgRNA downstream of a minimal Cytomegalovirus promoter (mCMV) followed by a minimal polyadenylation sequence, with the entire sequence under the inducible control of the tetracycline response element (56). Additionally, RNAP II mediated sgRNA production was also demonstrated by generating an artificial intron in a fluorescent reporter gene by flanking the sgRNA with the appropriate splice sites (56). However, considering that intronic RNAs typically have short half-life (57,58), the stability of the sgRNA product has to be further examined.
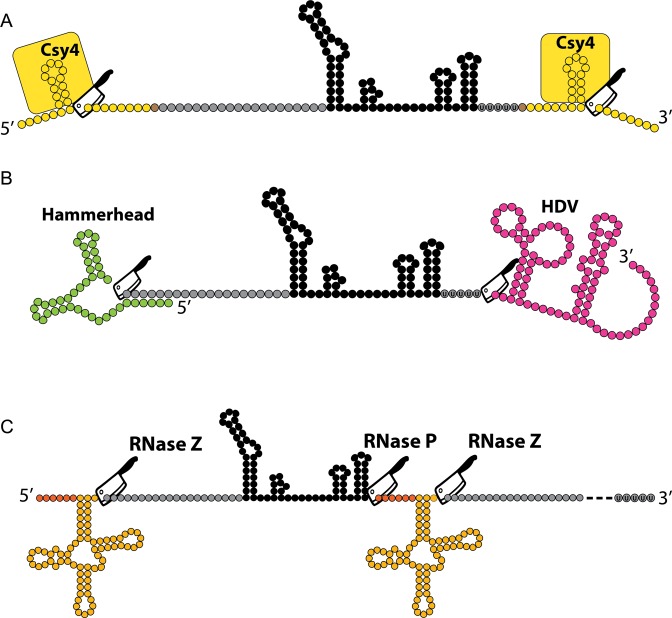
RNAP II based sgRNA production can be combined with strategies that exploit RNA binding proteins and utilize RNA secondary structures for improved efficiency and multiplexed sgRNA production (59). One approach is based on flanking the sgRNA with a 28 nucleotide hairpin that is recognized by the endoribonuclease Cys4. Cys4 cleaves immediately downstream of the hairpin and remains bound to the upstream secondary structure (59–62) (Figure 2A). Critically, it was shown that Csy4 binding can assist with stabilizing intronic sgRNAs (59). The hairpin can also be embedded in the 3′ untranslated region (3′UTR) of a protein coding transcript with an additional RNA module that forms a 3′ triple helical structure (triplex) that stabilizes RNAs lacking poly(A) tails (59,63).
Figure 2.
sgRNA multiplexing strategies. (A) RNA endonuclease Csy4 recognizes a 28 nucleotide sequence flanking the sgRNA sequence and cleaves after the 20th nucleotide while remaining bound to the upstream region. This production strategy allows for RNAP II mediated transcription via a CMV promoter and polyadenylation signal. (B) The cis-acting ribozymes hammerhead ribozyme and HDV ribozyme flanking the 5′ and 3′ of the sgRNA, respectively, allow for self-cleaving production of sgRNAs and are not dependent on the presence of an exogenous protein. This production strategy also allows for RNAP II mediated transcription via a CMV promoter and polyadenylation signal. (C) Polycistronic tRNA–gRNA architecture allows the production of multiple sgRNAs from a single synthetic gene. Endogenous RNases RNaseP and RNase Z cleave the 5′ leader and 3′ trailer sequences at specific sites, respectively. This production strategy relies on the presence of an RNAP III promoter and terminator sequence, but achieves multiple sgRNA production via internal RNAP III promoter elements intrinsic to tRNA genes.
RNAP II sgRNA production can also be carried out with ribozyme flanked sgRNA cassettes, in which the 5′ target sequence is fused to the self-cleaving Hammerhead (HH) ribozyme and 3′end scaffold sequence is fused to the Hepatitis delta virus (HDV) ribozyme (59,64) (Figure 2B). The triplex structure can also be used in stabilizing transcripts originating upstream from the ribozyme-produced sgRNA constructs (59).
Despite these attempts to produce sgRNA from RNAP II promoters, RNAP III remains the predominantly used promoter in recent papers. RNAP III transcribes tRNAs that must undergo processing in the nucleoplasm. By constructing a polycistronic tRNA–gRNA architecture, enzymes used in the endogenous eukaryotic tRNA processing machinery (i.e. RNase P and Z) can cleave the tRNA fragments out of the transcript producing functional and multiplexed sgRNAs (65) (Figure 2C). In Table 2 we include the DNA sequences for the modified sgRNA constructs presented in the Figures 2–4.
Table 2. sgRNA modification sequences.

|
Figure 4.
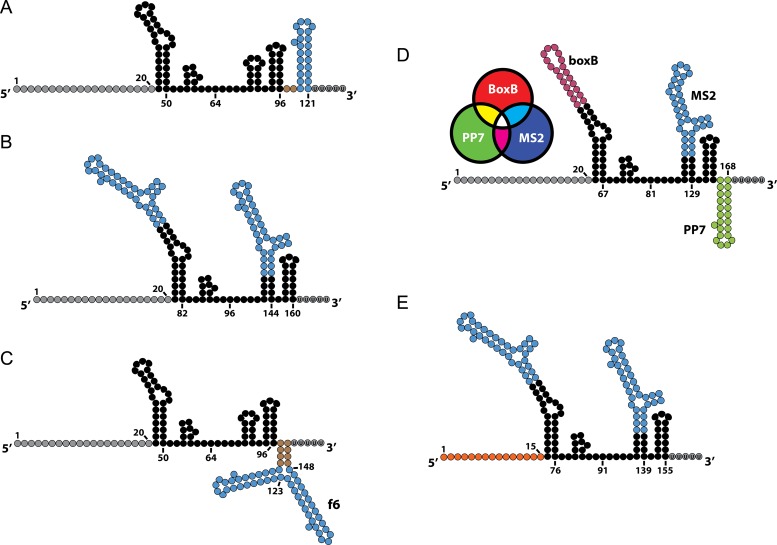
Incorporating RNA aptamer sequences into sgRNA. (A) MS2 loops that selectively bind MCP incorporated into the sgRNA 3′ end. (B) MS2 loops that selectively bind MCP incorporated into both the sgRNA tetraloop and the first hairpin. (C) MS2 loop incorporated in the 3′end in conjunction with a second aptamer hairpin f6 that has been selected to bind MCP. (D) CRISPRainbow sgRNA that utilizes three RNA binding protein-aptamer systems. The N22 peptide is fused to red fluorescent protein that binds the BoxB aptamer, the MCP peptide is fused to blue fluorescent protein and binds the MS2 aptamer and the PCP peptide fused to green fluorescent peptide binds the PP7 aptamer. Using different aptamers to bind red, green and blue fluorescent proteins the CRISPRainbow system creates seven different scaffolds that can be imaged as individual combinations of the primary colors. (E) sgRNA variant similar to the structure shown in Figure 4B, but also utilizes a truncated spacer sgRNA to achieve sgRNA multiplexing schemes that allow for both gene knockout and activation with catalytically active SpCas9.
sgRNA production can also be achieved in vitro by appending a T7 RNA polymerase promoter to the 5′ end of the spacer region, and similar to transcription by the human RNAP III U6 promoter, transcription by T7 RNAP requires 1–2 guanine residues directly upstream of the spacer sequence (66). In vitro-transcribed (IVT) sgRNA can be microinjected into embryos along with mRNA encoding SpCas9 ORF (67), or IVT sgRNAs can be transfected with purified SpCas9 protein (68). Additionally, chemical synthesis of sgRNAs and chemically modified sgRNA nucleotides transfected into human primary cells with either SpCas9 mRNA or purified protein has shown to enhance target specificity (69).
Guide RNA sequence modification and function
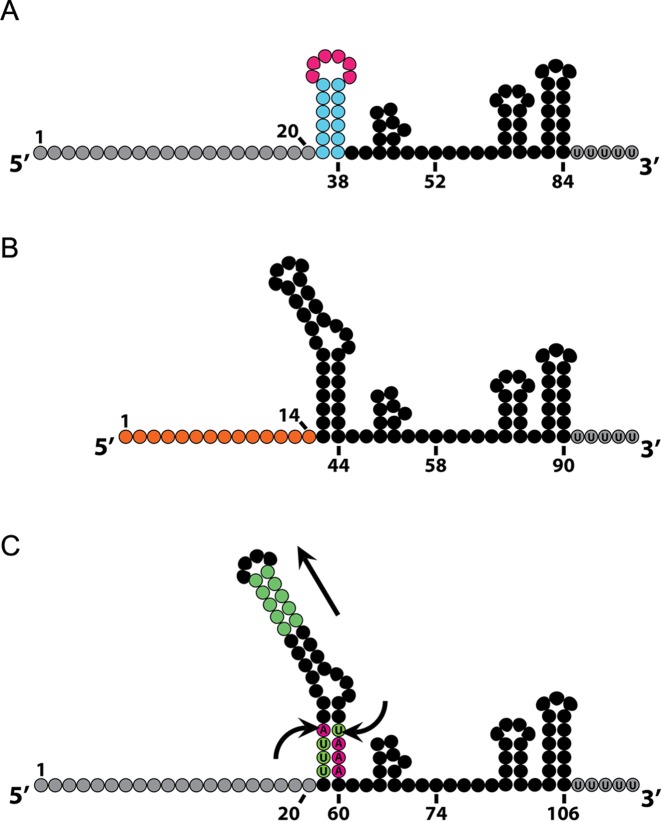
A systematic mutational analysis of the sgRNA (50) revealed that the bulge and nexus are the most sensitive to disruption and are necessary for DNA cleavage. Replacement of the bulge with perfectly complementary base pairing abrogates DNA cleavage. Also, substituting a pair of guanine nucleotides that form the base of the nexus structure with two cytosine nucleotides completely abolished cleavage activity. Interestingly, the upper stem can withstand large deletion mutations and still exhibit DNA cleavage activity (23,27,50) (Figure 3A). Alternatively, extensions to the stemloop were found to increase sgRNA stability and enhance its assembly with catalytically inactive SpCas9 (dSpCas9) (70–72).
Figure 3.
SpCas9 sgRNA mutational variants. (A) sgRNA variant in which the entire upper stem is removed and the bulge is replaced by a tetraloop that retains cleavage activity, suggesting that the upper stem may be dispensable. (B) sgRNA variant in which the spacer sequence is truncated from the canonical 20 nucleotides down to 14–15 nucleotides that allows catalytically active SpCas9 to still bind its target DNA without cleaving the target DNA. (C) sgRNA variant in which a putative RNAP III terminator sequence is removed from the lower stem by an A-U base pair flip and the upper stem is extended that increase sgRNA stability and enhance its assembly with SpCas9.
sgRNA targeting efficacy has shown to vary between guides (67,73–78). There are many factors that can contribute to off-target effects such as chromatin accessibility (78), nucleotide composition of the guide (79,80) and length of the guide (81). Indeed, deletions to the sgRNA have proved to be beneficial, including decreased off-target effects by truncation of the 5′ end of the sgRNA such that they possess 17 or 18 nucleotides of complementarity to the target sequence. Utilization of truncated sgRNAs (tru-sgRNAs) by RNA-guided nucleases (RGNs) such as SpCas9 (tru-RGNs) decreased undesired cleavage at known off-targets sites by several orders of magnitude (81). Truncations of the spacer sequence down to 15 nucleotides abolish SpCas9 cleavage activity, though the enzyme still retains genomic targeting function (81,82) (Figure 3B). Notably, 5′ mismatches and truncations as low as 11 nucleotides have shown not to significantly compromise dSpCas9 binding activity (82).
The truncated sgRNA can be used in conjunction with a catalytically active SpCas9 fused to a transactivation domain such as VPR, for multiplexed sgRNA schemes that require both targeted activation and targeting cleavage with a single SpCas9 enzyme (83). Moreover, truncated sgRNAs were utilized for genomic imaging and found to outperform their longer spacer counterparts when targeting repetitive elements (70,72). Other sgRNA augmentations that increased genomic labeling efficiency include mutations that disrupt the putative Pol-III terminator (four consecutive U's) in the sgRNA stemloop, as well as the previously mentioned stemloop extension (70,72) (Figure 3C). Both modifications helped to improve sgRNA-dSpCas9 assembly and increase the sgRNA stability.
The versatility of sgRNA is exemplified by the ability to incorporate separate RNA secondary structures into the sgRNA scaffold without compromising its association of SpCas9. Indeed, Shechner et al. established a platform dubbed CRISPR-Display that utilizes catalytically inactive SpCas9 for targeted localization of large RNA cargos (84). The CRISPR-Display platform enabled the incorporation of large non-coding RNA (lncRNA) domains such as the repressive Xist A-repeat domains (85), and enhancer-transcribed RNAs (86) up to 4.8 kb (87) into the sgRNA. While the effects of lncRNA-mediated transcriptional repression and activation were modest, the CRISPR-Display platform introduced a novel approach for studying lncRNA function. MS2 bacteriophage coat proteins (MCP) can dimerize and selectively bind a specific RNA hairpin-forming aptamer (88,89). Effector domains such as transcriptional activators, transcriptional repressors and epigenetic demethylators have been fused to the MCP protein. By engineering MS2 loops onto the sgRNA, dCas9 can localize to a specific locus and recruit the corresponding effector fusions (32,42,46,56,82,90–92). MS2 loops have been added to the 3′ end of the sgRNA (Figure 4A), as well as engineered into the tetraloop and 1st hairpin with successful transactivation of target genes (84,90,91) (Figure 4B). Conversely, 5′ additions of single MS2 or PP7 loops were degraded and failed to achieve transactivation (91), but 5′ additions of multiple tandem MS2 and PP7 loops were found to be intact and achieved modest activation (84). For 3′ appended loops, 2–20 bp long linkers have been shown to function, with increasing linker length correlated to decreased stability and recruitment efficiency. Additions of 3X MS2 loops to the 3′ end have been attempted, but increasing the number of MS2 loops corresponded to a decrease in transactivation and stability (91). To address this problem, a double stranded linker was developed to enhance the stability of the scaffold, which resulted in a notable increase in MCP-VP64-mediated transactivation for sgRNAs bearing two MS2 loops (91). The design was further improved by replacing the second MS2 loop for an in vitro selection-derived aptamer that also binds to MCP so as to reduce misfolding between the aptamers (91,93) (Figure 4C). It is worth noting that this construct was developed and tested in yeast, but such transcriptional control should be achievable in mammalian cells.
Alternatively, Konermann et al. have engineered MS2 loops into the tetraloop and 1st hairpin of the sgRNA separately (sgRNA 1.1 and 1.2, respectively) and within the same construct (sgRNA 2.0), showing substantially increased activity when using both modifications. Another study was able to demonstrate epigenetic editing through utilization of the sgRNA 2.0 construct (90) in which the MCP was fused to the catalytic domain of the Tet1 dioxygenase protein via a flexible linker that enables site directed demethylation of endogenous promoters in mammalian cell cultures (92).
Similar to the MCP-MS2 system are other RNA binding protein-aptamer systems (e.g. PCP-PP7, Com-com and NN2-BoxB) that have been used in sgRNA scaffold design. By utilizing several sgRNAs in a multiplexed targeting scheme, combinatorial sets of transcriptional control sgRNAs at endogenous loci can be used to engineer complex regulatory networks (10,59,94). Moreover, RNA aptamer systems with distinct effector fusions can be appended to the same sgRNA molecule and function in parallel without crosstalk (71,72,91). This was exemplified in the CRISPRainbow system in which multiple fluorescent proteins were fused to distinct RNA aptamer-binding proteins with their respective RNA aptamer sequence incorporated into a combination of either the tetraloop, 1st hairpin and/or appended to the 3′ end resulting in a total of seven different sgRNA species for multiplexed imaging of genomic loci (72) (Figure 4D).
Notably the multiplexing efforts described here have largely been performed with the catalytically inactive dSpCas9. As mentioned previously, the catalytically active SpCas9 has been shown to bind but not cleave its target DNA sequence when guided by truncated sgRNAs (82,83). Therefore, orthogonal control can be conferred through the length and the specific sequence of the sgRNA spacer, as well as the type of RNA aptamer incorporated onto the sgRNA secondary structure. This allows for the concurrent transactivation and site-specific DNA cleavage at desired genomic loci using the catalytically active SpCas9 (82,83) (Figure 4E).
CONCLUSION
The Cas9:sgRNA technology has revolutionized our ability to probe and edit the human genome in vitro and in vivo. The ability to reliably control and modify the human genome is expected to be instrumental toward unraveling disease properties and developing novel therapeutics. Engineering increasingly sophisticated functions in cells will rely on the continued expansion of the CRISPR toolkit and the rational engineering of novel guide RNAs. Novel engineered sgRNA constructs will not only point to a specific genomic address but also define the desired function (82,91). As more diverse CRISPR-associated proteins appear in literature (17,39,40) we expect that current advances and observations on the SpCas9 sgRNAs to readily inform the RNA engineering for Cas9 orthologs and other CRISPR-associated RNPs.
Acknowledgments
This work was funded by NSF CAREER (1351354) and NSF/NIGMS (1361355) grants.
FUNDING
NSF CAREER [1351354]; NSF/NIGMS [1361355]. Funding for open access charge: NSF CAREER [1351354]; NSF/NIGMS [1361355]
Conflict of interest statement. None declared.
REFERENCES
- 1.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 2.Garneau J.E., Dupuis M.-È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 3.Sorek R., Kunin V., Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 4.Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., Dickman M.J., Makarova K.S., Koonin E. V, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 6.Jansen R., van Embden J.D.A., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 7.Lander E.S. The Heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Deveau H., Barrangou R., Garneau J.E., Labonté J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 12.Hai T., Teng F., Guo R., Li W., Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore R., Spinhirne A., Lai M.J., Preisser S., Li Y., Kang T., Bleris L. CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res. 2015;43:1297–1303. doi: 10.1093/nar/gku1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Mendiratta S., Ehrhardt K., Kashyap N., White M.A., Bleris L. Exploiting the CRISPR/Cas9 PAM Constraint for Single-Nucleotide Resolution Interventions. PLoS One. 2016;11:e0144970. doi: 10.1371/journal.pone.0144970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilton I.B., D'Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J.J., Charpentier E., Haft D.H., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolotin A., Quinquis B., Sorokin A., Dusko Ehrlich S. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 20.Makarova K.S., Grishin N. V, Shabalina S.A., Wolf Y.I., Koonin E. V, Fire A., Hannon G., Cogoni C., Macino G., Bernstein E., et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marraffini L.A., Sontheimer E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 26.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders C., Niewoehner O., Duerst A., Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chylinski K., Makarova K.S., Charpentier E., Koonin E. V. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimasu H., Cong L., Yan W.X., Ran F.A., Zetsche B., Li Y., Kurabayashi A., Ishitani R., Zhang F., Nureki O. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath P., Romero D.A., Coute-Monvoisin A.-C., Richards M., Deveau H., Moineau S., Boyaval P., Fremaux C., Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma E., Harrington L.B., O'Connell M.R., Zhou K., Doudna J.A., Briner A.E., Donohoue P.D., Gomaa A.A., Selle K., Slorach E.M., et al. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol. Cell. 2015;60:398–407. doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou Z., Zhang Y., Propson N.E., Howden S.E., Chu L.-F., Sontheimer E.J., Thomson J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Heidrich N., Ampattu B.J., Gunderson C.W., Seifert H.S., Schoen C., Vogel J., Sontheimer E.J. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C.M., Cradick T.J., Bao G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol. Ther. 2016;24:645-654. doi: 10.1038/mt.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V. V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K., Ren C., Liu Z., Zhang T., Zhang T., Li D., Wang L., Yan Q., Guo L., Shen J., et al. Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell. Mol. Life Sci. 2015;72:383–399. doi: 10.1007/s00018-014-1679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M., et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V., et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V. V, Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F., Cong L., Ran F.A., Cox D., Lin S., et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Briner A.E., Donohoue P.D., Gomaa A.A., Selle K., Slorach E.M., Nye C.H., Haurwitz R.E., Beisel C.L., May A.P., Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 51.Hamm J., Mattaj I.W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 52.McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 53.Ranganathan V., Wahlin K., Maruotti J., Zack D.J. Expansion of the CRISPR–Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat. Commun. 2014;5:4516. doi: 10.1038/ncomms5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin X., Yang J., Chen J., Gunasekera A., Fesik S.W., Shen Y. Development of a tightly regulated U6 promoter for shRNA expression. FEBS Lett. 2004;577:376–380. doi: 10.1016/j.febslet.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 55.Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O'Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Kiani S., Beal J., Ebrahimkhani M.R., Huh J., Hall R.N., Xie Z., Li Y., Weiss R. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods. 2014;11:723–1726. doi: 10.1038/nmeth.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Audibert A., Weil D., Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clement J.Q., Qian L., Kaplinsky N., Wilkinson M.F. The stability and fate of a spliced intron from vertebrate cells. RNA. 1999;5:206–220. doi: 10.1017/s1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haurwitz R.E., Sternberg S.H., Doudna J.A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sternberg S.H., Haurwitz R.E., Doudna J.A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.-D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao Y., Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 65.Xie K., Minkenberg B., Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R., Brown A.J., Fisher D.A., Kouranova E., McCoy A., et al. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hashimoto M., Yamashita Y., Takemoto T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 2016;418:1–9. doi: 10.1016/j.ydbio.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 69.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B., et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao S., Zhang W., Hu H., Xue B., Qin J., Sun C., Sun Y., Wei W., Sun Y. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 2016;44:e86. doi: 10.1093/nar/gkw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma H., Tu L.-C., Naseri A., Huisman M., Zhang S., Grunwald D., Pederson T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016;34:528–530. doi: 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koike-Yusa H., Li Y., Tan E.-P., Velasco-Herrera M.D.C., Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2013;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 76.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.-D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dahlman J.E., Abudayyeh O.O., Joung J., Gootenberg J.S., Zhang F., Konermann S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kiani S., Chavez A., Tuttle M., Hall R.N., Chari R., Ter-Ovanesyan D., Qian J., Pruitt B.W., Beal J., Vora S., et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shechner D.M., Hacisuleyman E., Younger S.T., Rinn J.L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minks J., Baldry S. EL, Yang C., Cotton A.M., Brown C.J., Lyon M., Brown C., Ballabio A., Rupert J., Lafreniere R., et al. XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics Chromatin. 2013;6:23. doi: 10.1186/1756-8935-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peabody D.S. The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 90.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zalatan J.G., Lee M.E., Almeida R., Gilbert L.A., Whitehead E.H., La Russa M., Tsai J.C., Weissman J.S., Dueber J.E., Qi L.S., et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu X., Tao Y., Gao X., Zhang L., Li X., Zou W., Ruan K., Wang F., Xu G., Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hirao I., Spingola M., Peabody D., Ellington A.D. The limits of specificity: an experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Divers. 1999;4:75–89. doi: 10.1023/a:1026401917416. [DOI] [PubMed] [Google Scholar]
- 94.Li J.-F., Norville J.E., Aach J., McCormack M., Zhang D., Bush J., Church G.M., Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]