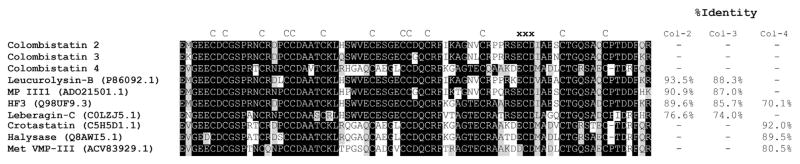Abstract
Crotalid venoms are rich sources of components that affect the hemostatic system. Snake venom metalloproteinases are zinc-dependent enzymes responsible for hemorrhage that also interfere with hemostasis. The disintegrin domain is a part of snake venom metalloproteinases, which involves the binding of integrin receptors. Integrins play an essential role in cancer survival and invasion, and they have been major targets for drug development and design. Both native and recombinant disintegrins have been widely investigated for their anti-cancer activities in biological systems as well as in vitro and in vivo systems. Here, three new cDNAs encoding ECD disintegrin-like domains of metalloproteinase precursor sequences obtained from a Venezuelan mapanare (Bothrops colombiensis) venom gland cDNA library have been cloned. Three different N- and C-terminal truncated ECD disintegrin-like domains of metalloproteinases named colombistatins 2, 3, and 4 were amplified by PCR, cloned into a pGEX-4T-1 vector, expressed in Escherichia coli BL21, and tested for inhibition of platelet aggregation and inhibition of adhesion of human skin melanoma (SK-Mel-28) cancer cell lines on collagen I. Purified recombinant colombistatins 2, 3, and 4 were able to inhibit ristocetin- and collagen-induced platelet aggregation. r-Colombistatins 2 showed the most potent inhibiting SK-Mel-28 cancer cells adhesion to collagen. These results suggest that colombistatins may have utility in the development of therapeutic tools in the treatment of melanoma cancers and also thrombotic diseases.
Keywords: Bothrops colombiensis, Colombistatins, ECD disintegrin-like domain, SK-Mel-28, Tumor
1. Introduction
In the occurrence of the Viperidae snakes, local and systemic hemorrhages are one of the most prevalent and devastating consequences of envenomation (Bjarnason and Fox, 1994). Many of the hemorrhagic toxins in the venom of the mapanare (Bothrops colombiensis), which cause these effects have been isolated and differentiated as zinc metalloproteinases (Girón et al., 2008). In general, metalloproteinases in the mammals play a role in cell growth and proliferation, preservation of the extracellular membrane, synaptogenesis, and angiogenesis. Mutant forms of these metalloproteinases can conduct over activation of cell growth and it has been suspected that they are implicated in tumor growth and metastasis (Ramos and Selistre-De-Araujo, 2006).
cDNA cloning and sequencing of the hemorrhagic toxins of B. colombiensis aside from other Viperidae snakes have pointed out that the metalloproteinases from snake venoms are synthesized as multi-domain precursors (Hite et al., 1992). Snake venom metal-loproteinases (SVMPs) are classified into three major classes (P-I to P-III) based on their structural domains (Fox and Serrano, 2009). The P-I class metalloproteinases, having molecular weights of 20–30 kDa, have pre-, pro-, and metalloproteinase/spacer domains while the bigger forms, P-II (30–60 kDa) and P-III (60–100 kDa), preserve one to three additional carboxy domains (Fox and Bjarnason, 1995). The P-III class is the most hemorrhagic SVMP and has been shown to inhibit collagen-induced platelet aggregation (Moura da Silva et al., 2001; Zigrino et al., 2002).
The P-III class comprises a signal sequence, a pro-domain, a metalloproteinase domain, a spacer peptide, a disintegrin-like domain (non-RGD disintegrin domain), and a cysteine-rich domain (Bjarnason and Fox, 1994). It has been described that class P-III of SVMPs undertakes proteolytic actions to liberate the carboxy disintegrin-like/cysteine rich (DC) domains from the main metalloproteinase using post-translational processing (Shimokawa et al., 1996; Fox and Serrano, 2005, 2009). Several studies reported that both native and recombinant proteins of disintegrin-like domain itself or DC domains of P-III SVMPs show a potent anti-platelet aggregation activity induced by collagen (Higuchi et al., 2011; Tanjoni et al., 2010; Selistre de Araujo et al., 2005; Moura-da-Silva et al., 2001; Zigrino et al., 2002; Shimokawa et al., 1997).
Recently, we have created a Bothrops colombiensis snake venom gland cDNA library (Suntravat et al., 2016). The significant clones were sequenced and functionally explored using cross-species genomic estimation to assess the capacity and operative properties of the constructed library for disintegrins research. The most abundant toxin transcripts were metalloproteinases (35%). The class P-III SVMPs group was the most complex showing five different subclasses with a great diversity of disintegrin-like (D/ SECD motif) and cysteine-rich domains. In this study, three different 5′ and 3′ truncated ECD disintegrin-like domains of the 5′ truncated P-III metalloproteinases from B. colombiensis were cloned and expressed and determined their biological activities. The recombinants proteins, named r-colombistatins 2, 3, and 4, were identified and tested for their abilities to inhibit ristocetin-, ADP-, collagen-induced platelet aggregation in whole human blood and skin melanoma (SK-Mel-28) cell adhesion. These new venom molecules have been documented, and could be utilized as a base for venomic knowledge, evolutionary research and further disintegrin functional and structural studies.
2. Materials and methods
2.1. PCR amplification and cDNA cloning of partial ECD disintegrin-like domains
The cloning of the partial ECD disintegrin-like domains was done according to the method of Suntravat et al., 2013. The partial cDNAs encoding the Bothrops colombiensis venom class P-III metalloproteinases identified from the cDNA library (GenBank accession no. JZ880093, JZ880098, and JZ880094) (Suntravat et al., 2016) were used as a template for PCR to subclone their N- and C-terminal truncated ECD disintegrin-like domains, which were designated as colombistatins 2, 3, and 4, respectively. PCRs were used to generate double stranded cDNA, with the following disintegrin-specific primers (colombistatin 2, a forward primer 5′-CGCGAATTCGA-GATGGGAGAAGAATGTGAC-3′ and a reverse primer 5′-GACTCGAGTCACCTTTGGAAGTCATCTGTGG-3′; colombistatin 3, 5′-CGCGAATTCGAGAAGGGAGAAGAATGTGACTG-3′ and a reverse primer 5′-GACTCGAGTCACCTTTTGAAGTCATCTGTGGGA-3′; colombistatin 4, a forward primer 5′-CGCGAATTCGAGGTGGGAGAAGAATGTGAC-3′ and a reverse primer 5′-GACTCGAGTCACCTTTGGAAGCGATCTG-TAC-3′, two restriction enzyme sites (underlined): EcoRI in forward primer and XhoI in reverse primer) as previously described (Suntravat et al., 2015, 2013). PCR amplification consisted of a cycle of 94 °C (3 min), 30 cycles of 94 °C (30 s), 58 °C (30 s), and 72 °C (1 min). A final extension step was performed for 10 min, at 72 °C. The PCR product was digested with EcoRI and XhoI and gel purified. The PCR product was ligated into EcoRI and XhoI sites of pGEX-4T-1 expression vector (GE Healthcare Lifesciences, Uppsala, Sweden). The ligated plasmid was transformed into E. coli Top10 competent cells (Invitrogen, CA, USA). Plasmid was extracted using the Gen-Elute plasmid miniprep kit (Sigma-Aldrich, MO, USA). Plasmids containing inserts of the predicted size for recombinant colombistatins 2, 3, and 4 (r-colombistatins 2, r-colombistatins 3, and r-colombistatins 4) were performed by PCR and further confirmed by sequencing for construction of in-frame.
2.2. Sequence analysis
The cDNA sequences and predicted amino acid sequences were compared to the sequences in the GenBank database using Gen-Bank BLASTN and BLASTX programs (Altschul et al., 1997). Multiple alignments of the amino acid sequences were performed with Clustal W program (Thompson et al., 1994). A phylogenetic tree was generated from a multiple alignment of disintegrin-like domains within MegAlign program using the neighbor-joining method, Lasergene 12 software (DNASTAR, Inc. Madison, WI). The bootstrap test was done using 1000 replications. The sequence of mojastin was used as an outgroup.
2.3. Expression and purification of recombinant colombistatins 2, 3, and 4
Once the sequence was obtained, in-frame r-colombistatins 2-, r-colombistatins 3-, and r-colombistatins 4-pGEX-4T-1 plasmids containing an extra five amino acids from this cloning vector was transformed into E. coli BL21 (DE3) star cells (Invitrogen). BL21 cells harboring recombinant plasmid DNA was first cultured in shaking flasks containing Luria-Bertani (LB) medium overnight. After inoculation of the overnight culture into fresh LB medium, the culture cells were grown at 37 °C with shaking at 225 rpm until the absorbance at 600 nm (OD600) reached 0.6. The culture was induced with a final concentration of 0.1 mM isopropyl β-d-thiogalactoside (IPTG) for 5 h to induce expression of recombinant proteins. Bacterial cells were collected by centrifugation at 10,000 × g for 10 min and resuspended in 1x BugBuster Protein Extraction reagent (Novagen CA, USA) by gentle vortexing, using 5 mL reagent per gram of wet cell paste. Cells were resuspended and incubated on a shaking platform for 30 min at room temperature. The lysate was centrifuged at 16,000 × g for 30 min at 4 °C. The soluble supernatant was purified using a glutathione S-transferase (GST)-binding resin (GE Healthcare, CA, USA) in Econo-Column chromatography column (BIO-RAD, CA, USA), which was previously equilibrated with 1X GST Bind/Wash Buffer (Novagen). Recombinant colombistatins 2, 3 and 4 were cleaved and eluted from GST bound to GST-binding resin by thrombin cleavage. Thrombin was removed from colombistatins using a 5 mL HiTrap™ Benzamidine FF (high sub) column (Amersham Biosciences, NJ, USA) according to the manufacturer’s instruction. The column was equilibrated with 5 column volumes of binding buffer (20 mM sodium phosphate, 0.15 M NaCl, pH 7.5). One milliliter of the sample was loaded into the column and colombistatins were obtained by washing the column with a high salt buffer (20 mM sodium phosphate, 1 M NaCl, pH 7.5). The column was finally washed with 10 column volumes of elution buffer (10 mM HCl, 0.5 M NaCl, pH 2.0) to remove the thrombin bound to the column. Each colombistatin was dialyzed in 1X phosphate buffer saline (PBS), pH 7.4 and concentrated using a 3 kDa Amicon Ultra-15 centrifugal filter (Millipore, Carrigtwohill, Ireland), electrophoresed on SDS-PAGE under non-reducing condition. Protein concentrations were estimated from the absorbance at 280 nm.
2.4. N-terminal sequencing
A 4 μg of each r-colombistatin was transferred from an SDS-PAGE onto a PVDF membrane (Milipore Corporation, MA, USA) using a Semi-Dry Transblot Cell (BIO-RAD) at 125 mA for 1 h. The membrane was stained with Coomassie blue R-250 stain for 5 min and distained with 50% methanol for 5 min. The sample membrane was sent out for N-terminal amino acid sequencing at the Protein Facility, Office of Biotechnology, Iowa State University, Iowa.
2.5. Inhibition of platelet aggregation
Human blood was drawn from healthy donors who had not taken any anticoagulant prescription within two weeks prior to blood donation (Sánchez et al., 2010). Four hundred and fifty microliters of 10% citrated whole human blood was incubated at 37 °C at least 5 min prior to use with equal amounts of 0.15 M sodium chloride. r-Colombistatins at various concentrations (10 μL) were incubated with blood samples in a Chronolog Whole Blood Aggregometer (Chronolog, PA, USA) at 37 °C for 2 min. Platelet aggregation was initiated by adding 8 μL of ristocetin (1 mg/mL), 2 μL of collagen (5 μg/mL), or 10 μL of ADP (10 μM). Percent inhibition of platelet aggregation was calculated using the following equation: [(C-E/C)] ×100, where C is the units of platelet aggregation (ohms) for the control, and E is the unit of platelet aggregation (ohms) for the experimental fraction. The extent of the inhibition of platelet aggregation was assessed by comparison with the maximal aggregation induced by the control dose of agonists (ristocetin, collagen, and ADP). The median inhibitory concentration (IC50) values were determined from dose-response curves generated from various concentrations colombistatins using Microsoft Excel 2011.
2.6. Cell line and culture conditions
The human skin melanoma (SK-Mel-28) cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA). The SK-Mel-28 cell line was sustained with Eagle’s minimum essential medium, supplemented with 10% fetal calf serum and antibiotics (50 units/mL penicillin and 50 μg/mL streptomycin). The cells were cultured in a humidified 5% (v/v) CO2 air incubator at 37°C.
2.7. Cellular adhesion inhibition assay
The Wierzbicka-Patynowski et al. (1999) method was used to measure the inhibition of SK-Mel-28 cell binding to collagen I induced by venom components in triplicate wells of a 96-well plate (Falcon®Tissue Culture Plate), which were coated with 100 μL of collagen I at 10 μg/mL, in 0.01 M phosphate buffer saline (PBS), pH 7.4, and incubated overnight at 4 °C. The plate was blocked by addition of 0.2 mL of PBS in 5% bovine serum albumin (BSA) and incubated at 37 °C for 1 h. Cells were collected with 0.25% Trypsin-EDTA, counted, and resuspended in a medium containing 1% BSA at 5 × 105 cells/mL. Each r-colombistatin at various concentrations (50 μL) was added to the cell suspension (450 μL) and incubated at 37 °C for 1 h. The blocking solution was aspirated, and the cell/ venom fraction suspensions (200 μL) were added to the wells coated with collagen I and incubated at 37 °C for 1 h. The SK-Mel-28 cells incubated with PBS were used as negative control. In these negative control wells, the cells bound to collagen. The wells were washed three times with PBS-5% (w/v) BSA by filling and aspirating. A total of 200 μL of medium in 1% BSA containing 3-[4,5-dimethylthiazol-2-yl] 2,5-diphenltetrazolium bromide (MTT) (5:1, v/v) was added to the wells containing cells and incubated at 37 °C, for 2 h. The MTT was aspirated and 100 μL of dimethyl sulfoxide (DMSO) was dispensed to the wells to lyse the cells. The plate was lightly shaken, and the absorbance read at 570 nm using a Beckman Coulter model AD 340 reader. The percent of inhibition was measured by the next formula: [(absorbance of negative control-absorbance of cell/r-colombistatins)/absorbance of negative control x 100.
3. Results
3.1. cDNA cloning of partial ECD disintegrin-like domains
The cDNA sequences coding for partial metalloproteinases containing ECD disintegrin-like and cysteine rich domains (Gen-Bank accession no. JZ880093, JZ880098, and JZ880094) were used as template to clone and express the 5′ and 3′ truncated ECD disintegrin-like domains, which were designated as colombistatins 2, 3, and 4, respectively. The deduced amino acid sequences of the partial ECD disintegrin-like cDNA colombistatins 2, 3, and 4 presented 231, 231, and 228 bp long, correspondingly, with the deduced sequence containing 77, 77, and 76 amino acids, respectively (Fig. 1). The putative primary structure includes 14 cysteine residues, in that order and the ECD motifs, the molecular masses were estimated as 8.5, 8.5 and 8.2 kDa. Analysis of the predicted amino acid sequence revealed that colombistatins are P-III disintegrin-like proteins with an ECD motif. Fig. 1 shows predicted amino acid sequences and the percent identity in comparison with the partial ECD disintegrin-like domains of colombistatins and other closely related P-III disintegrin-like domains. NCBI protein BLAST analysis showed that the deduced amino acid sequence of the partial ECD disintegrin-like domains of the cloned colombistatins 2 and 3 was homologous to the disintegrin-like domain of leucorolysin-B (P86092.1) from Bothrops leucurus with 93.5% and 88.3% identity, respectively. Colombistatin 4 had a 92% sequence identity with crotastatin (C5H5D1.1) from Crotalus durissus cascavella.
Fig. 1. Multiple alignment of the partially deduced amino acid sequences of the ECD disintegrin-like domain of metalloproteinases predicted from partially sequenced clones from B. colombiensis with other homologous venom proteins.
Colombistatin 2 (Col-2), colombistatin 3 (Col-3), and colombistatin 4 (Col-4) are aligned with closely related protein in the database, and the % identities are shown in the figure. The alignment was generated with the ClustalW multiple sequence alignment program with manual adjustment and displayed with box shaded. The numbers in parenthesis are the NCBI accession numbers. The tripeptide (ECD) binding motif is marked by the letter “X” above the sequences. All cysteine residues (letter “C” above the sequences) are conserved.
Colombistatin 2 had 90.9% sequence identity with colombistatin 3 and was 64.5% identical to colombistatin 4. Colombistatins 3 and 4 were matched with 61.8% identity (Fig. 1). Colombistatins 2 and 3 were grouped together with other disintegrin domains of P-III class form the genus Bothrops such as leucurolysin-B (P86092.1) from Bothrops leucurus, MP III1 (ADO21501.1) from Bothrops neuwiedi, and HF3 (Q98UF9.3) from Bothrops jararaca. Colombistatin 4 was classified with crotastatin (C5H5D1.1) from Crotalus durissus cascavella, halysase (Q8AWI5.1) from Gloydius halys, Met VMP-III from ACV83929.1) from Agkistrodon contortrix laticinctus.
3.2. Expression and purification of r-colombistatins
Escherichia coli BL21 (DE3) Star cells effectively expressed r-colombistatins. The soluble supernatant was purified using a GST affinity resins in Econo-Column chromatography column. Highly purified colombistatins were eluted with a high salt buffer (20 mM sodium phosphate, 1 M NaCl, pH 7.5). Purified r-colombistatins were identified by N-terminal sequence analysis. The N-terminal end contained an extra five amino acids derived from the vector (GSPEF), which added an additional molecular weight of 0.54 kDa. The total calculated molecular weight of r-colombistatins 2, 3, and 4 were about 9 kDa with the pI values 4.40, 4.76, and 4.73, respectively, by Protein Identification and Analysis Tools on the Expasy Server (Fig. 3).
Fig. 3. SDS-PAGE of r-colombistatins 2, 3, and 4.
Samples (3 μg) were run on 4–12% (w/v) Bis-Tris Gel using an Xcell SureLock Mini-Cell at 200 V for 30 min. The gel was stained with RapidStain. Lane 1: SeeBlue Plus2 Markers; lane 2: purified r-colombistatin 2; lane 3: purified r-colombistatin 3; lane 4: purified r-colombistatin 4. An asterisk (*) represents the N-terminal amino acid sequences of purified r-colombistatins containing the five amino acids from the vector (italicized) before the disintegrin-like sequences, which are shown in bold letters.
3.3. Inhibition of platelet aggregation by r-colombistatins 2, 3 and 4
Table 1 shows the results of inhibition of platelet aggregation of the three recombinant partial ECD disintegrin-like domains from B. colombiensis with three different agonists comprising of ristocetin, collagen, and ADP. Recombinant colombistatins inhibited ristocetin- and collagen-induced platelet aggregation in a dose-dependent manner. No statistical difference in the IC50s for inhibiting ristocetin- and collagen-induced platelet aggregation between r-colombistatins 2 and 3 was observed (p > 0.05). Neither recombinant colombistatins (in this study) inhibited ADP-induced aggregation, even nor at high doses (Table 1).
Table 1.
Inhibition of platelet aggregation by r-colombistatins 2, 3, and 4 from B. colombiensis.
| Agonist | r-Colombistatin-2 | r-Colombistatin-3 | r-Colombistatin-4 |
|---|---|---|---|
| Ristocetin | 0.9 ± 0.1 μM | 1.3 ± 0.2 μM | 13.7 ± 0.4 μM |
| Collagen | 4.2 ± 0.01 μM | 4.1 ± 0.4 μM | 45.9 ± 14.2 μM |
| ADP | NA | NA | NA |
The results are expressed in IC50.
NA: No activity.
3.4. Inhibition of cell adhesion to collagen I
r-Colombistatin 2 inhibited SK-Mel-28 adhesion to collagen I, in a concentration-dependent manner with IC50 value of 22.2 μM (Fig. 4). r-Colombistatin 3 (22 μM) and r-colombistatin 4 (32 μM) inhibited SK-Mel-28 cell adhesion to collagen I by 10.8 ± 6.4% and 6.2 ± 4.3%, respectively.
Fig. 4. Effects of r-colombistatins on adhesion of SK-Mel-28 cancer cell lines on collagen I.
SK-Mel-28 cells were seeded in 96-well plates, which was pre-coated with collagen I in the absence (PBS added), or presence of various concentrations of colombistatins 2, 3 and 4. Cell adhesion was measured by MTT technique and the results were expresses as percent of inhibition. The error bars represent the standard deviation from two independent experiments with n = 3.
4. Discussion
Disintegrins are proteins with low molecular mass ranging from 49 to 84 amino acids in length, which are implicated in cell adhesion ligand recognition, binding expressly to integrin receptors located in the cell surface and in some cases displaying anti-platelet aggregation.
The first described colombistatin isolated from the venom of B. colombistatin was RGD-disintegrin containing 72 amino acids with a mass of 7.778 kDa as determined by mass spectrometry. This native RGD disintegrin inhibited ADP-induced platelet aggregation (IC50 of 210 nM), human urinary (T24) and skin melanoma (SK-Mel-28) cancer cell adhesion to fibronectin, and cell migration (Sánchez et al., 2009). Recently, our group (Suntravat et al., 2016) provided transcriptomic profiles of SVMPs of B. colombiensis showing a great diversity of P-III SVMPs containing ECD-DC domains. In addition to the transcriptome studies, proteomic techniques (Calvete et al., 2009) also demonstrated that both P-III SVMPs and a few DC domains of P-III SVMPs were isolated from the venom.
In the current study, we cloned and expressed the N- and C-terminal truncated ECD disintegrin-like domains named r-colombistatins 2, 3, and 4 from the B. colombiensis cDNA library to evaluate their biological activities. The deduced amino acid sequence comparison of the colombistatins with other closely related P-III disintegrin-like domains revealed that 14 conserved cysteine residues were observed among disintegrin-like domains (Fig. 1). They share high sequence identity (70%–93.5%) with the previous reported ECD disintegrin-like domain from P-III class SVMPs.
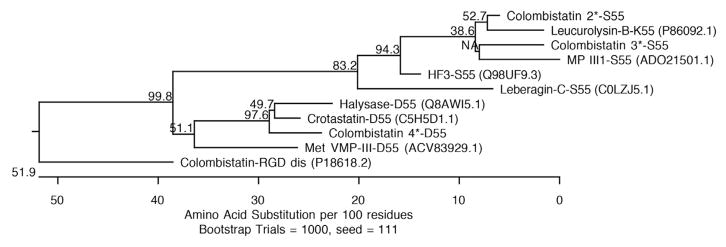
A number of different amino acids in disintegrin-like coding regions were observed among the predicted amino acid sequences of colombistatins 2, 3, and 4, illustrating the diversity of disintegrin-like domains of metalloproteinases in this snake. The phylogenetic relationships of the 5′ and 3′ truncated disintegrin-like domains within P-III class metalloproteinase precursors are based on their predicted amino acid sequences (Fig. 2). Colombistatins 2 and 3 were grouped together with other disintegrin domains of P-III class from the genus Bothrops such as leucurolysin-B (P86092.1) from Bothrops leucurus, MP III1 (ADO21501.1) from Bothrops neuwiedi, and HF3 (Q98UF9.3) from Bothrops jararaca. Colombistatin 4 was clustered in a clade, which was constituted by the P-III disintegrin-like domains from the several genera including the South American rattlesnake (genus Crotalus durissus), the Asian ground pit vipers (genus Gloydius), and the Copperhead (genus Agkistrodon). Interestingly, colombistatins 2 and 3 were clustered in a clade, which was constituted by the P-III disintegrin-like domains with S/K55ECD58, while colombistatin 4 was classified in the lineage of the disintegrin-like domains with an Asp (D) substitution at position 55 (Fig. 2). Colombistatin, an RGD disintegrin isolated from B. colombiensis venom (Sánchez et al., 2009) was defined as an out-group.
Fig. 2. Phylogenetic tree analysis of snake P-III disintegrin-like domains based on their predicted amino acid sequences.
The colombistatins identified in the cDNA library of the Venezuelan mapanare (colombistatins 2, 3, and 4) are marked with an asterisk (*). The tree was constructed using the neighbor-joining methods with a bootstrap for 1000 replications. The number at the branches represents the bootstrap probability. Colombistatin, a RGD disintegrin reported by Sánchez et al. (2009) was used as outgroup.
Edman degradation is even now the typical and most widely used method for sequencing unidentified proteins and has the benefit of providing a simple interpretation of long amino acid sequences. The N-terminal amino acid sequences of 9-kDa proteins were r-colombistatins 2, 3, and 4 containing an additional five amino acids from the vector at the N-terminus end. The total molecular mass and hypothetical pI values of r-colombistatins were shown in Fig. 3. They contain some differences in the amino acid sequence of the ECD-flanking region, RSECDIAE sequence in colombistatins 2 and 3, but KDECDMADL sequence in colombistatin 4 (Fig. 1).
Since disintegrin-like proteins have a particular inhibitory activity on platelet aggregation induced by some agonists, such as collagen, in our study, we examined the inhibitory activity of platelet aggregation by ADP (integrin αIIbβ3 agonist), ristocetin (von Willebrand factor, VWF agonist), and collagen (integrin α2β1 and platelet glycoprotein (GP) VI agonist). Recombinant colombistatins 2, 3, and 4 were not able to inhibit ADP-induced platelet aggregation but showed inhibition when induced by ristocetin and collagen (Table 1). The inhibition of ristocetin- and collagen-induced platelet aggregation of r-colombistatins 2 and 3 was about 13 and 11 times, respectively, more efficient than that of r-colombistatin 4, suggesting that flanking amino acid residues might significantly influence the ECD loop structure. In disintegrins including ECD disintegrins, the loop is important for ligand binding to the integrins (Muniz et al., 2008; McLane et al., 1998). The structural difference may have a role for the biological activity of the ECD motif.
In this study, we showed that recombinant colombistatins had the most potent inhibition effect on platelet aggregation induced by ristocetin, which was about 3–4 times more efficient than that induced by collagen. In addition, several studies reported that ECD distegrins inhibit collagen-induced platelet aggregation and cell adhesion by binding to integrin α2β1 (Kamiguti et al., 1997; Zigrino et al., 2002; Souza et al., 2000). It has been described that ECD disintegrin-like domain of jararhagin, a P-III class SVMP can bind to collagen and appears to be sufficient to inhibit collagen-induced platelet aggregation (Tanjoni et al., 2010). Since collagen is a main initiator for platelet adhesion and aggregation mediated by the interaction to the platelet membrane glycoprotein VI (GPVI) and integrin α2β1, while ristocetin induces the binding of VWF to platelet glycoprotein Ibα (GPIbα) (Scott et al., 1991), it is possible that recombinant colombistatins may possibly act on GPIb either alone or in complex with VWF or on collagen receptors, integrin α2β1, and/or GPVI. However, other integrin specificity for recombinant colombistatins should be further investigated.
Integrin-mediated cellular adhesion functions have been estimated to have an important participation in various biological activities, including the malignant growth of tumors (Humphries et al., 1988; Plantefaber and Hynes, 1989; Boukerche et al., 1989). Previous studies have demonstrated a likely role of integrins in tumor metastasis (Humphries et al., 1986; Saiki et al., 1988; Suntravat al., 2015; Lucena et al., 2011, 2015). In the current paper, we have considered the capability of recombinant colombistatins in inhibiting SK-MEL-28 melanoma cells adhesion to collagen I. In human patients, the primary melanoma propagates either horizontally or vertically in the skin. The vertical growth phase characteristically directs to widespread metastatic disease (Felding-Habermann et al., 1992). Some authors (Albelda et al., 1990) studying normal skin melanocytes and nevi, and comparing with the expression of a number of integrins on primary and metastatic melanoma tissues, established that the vitronectin receptor, integrin αvβ3, was likely detected on the vertical propagation phase primary melanoma in addition to metastatic lesions, and was not noticed on nevi, or horizontal primary melanoma. These findings mean that the expression of this integrin shows a relationship with the metastatic form of human melanoma. Principally, tumor melanoma starts in the skin as proliferative and invasive expansion of the primary site. This process finally guides to expand metastasis dissemination, which is distinguished by a varied development that necessitate not only cell proliferation but also demands the tumor cells capacity of proteases secretion, cell migration, angiogenesis activation and resistance against host immune activities (Poste et al., 1981: Nicolson, 1984). Authors (Chan et al., 1991) many years ago had established that expression of α2β1, a collagen/laminin receptor on rhabdomyosarcoma, amplified the metastatic ability of those cells without affecting the growth rate of the primary tumor.
Sánchez et al. (2009) found that colombistatin, RGD-disintegrin had an extremely potent inhibitory effect on the adhesion of SK-Mel-28 cells to fibronectin with an IC50 of 33 nM. As it is known, different disintegrins have the ability to interact with many integrins, resulting in the inhibition of cell membrane attachment. It is plausible that r-colombistatins 2, 3 and 4 recognize different integrin or multiple integrins, other than the existing on fibronectin; thus, explaining the good reactivity with the SK-Mel-28 cell line.
5. Conclusion
This study reports on the cloning, expression, and the biologic activities of the first three different recombinant ECD disintegrin colombistatins 2, 3, and 4 cloned from B. colombiensis. They can inhibit collagen- and ristocetin-induced platelet aggregation and also inhibits an adhesion effect of SK-Mel-28 cells to collagen I.
Acknowledgments
This work was supported by funds from the NIH/Biological Materials Resource Grant, Viper Resource Grant #s 5P40OD010960-12 (NNTRC, Texas A & M University-Kingsville), International Centre for Genetic Engineering and Biotechnology (Universidad Central de Venezuela, Grant # CRP/VEN13-03), FONACIT (Venezuela) Grant: N2014000490, and the Texas A&M University-Kingsville (TAMUK) Presidential Undergraduate Research Program (PURP Scholars are funded by the President’s Circle and Provost). We want to thank Nora Diaz DeLeon, Mark Hockmuller, Juan Salinas and our NNTRC personnel for their assistance. We also thank the TAMUK Office of Student Access for their support.
Footnotes
Ethical statement
The investigation complied with the bioethical standards taken from “Principles of Laboratory Animal Care” (Anonymous, 1985). All animal work was approved by the Texas A&M University-Kingsville Institutional Animal Care and Use Committee (IACUC Approval # 2015-12-09-A3).
Competing interests
The authors declare that they have no competing interests.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.toxicon.2016.09.007.
References
- Albelda SM, Mette SA, Elder DE, Stewart R, Damjanovich L, Herlyn M, Buck CA. Integrin distribution in malignant melanoma: association of the j93 subunit with tumor progression. Cancer Res. 1990;50:6757–6764. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous. Principles of Laboratory Animal Care. National Institute of Health; USA: 1985. Pub, 85 No. 23. [Google Scholar]
- Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venom. Pharmacol Ther. 1994;62:325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Boukerche HO, Berthier-Vergnes M, Bailly JF, Dore LK, Leung JL, McGregor A monoclonal antibody (LYP18) directed against the blood platelet glycoprotein Ib/IIIa complex inhibits human melanoma growth in vivo. Blood. 1989;74:909–912. [PubMed] [Google Scholar]
- Calvete JJ, Borges A, Segura A, Flores-Díaz M, Alape-Girón A, Gutiérrez JM, Diez N, De Sousa L, Kiriakos D, Sánchez E, Faks JG, Escolano J, Sanz J. Snake venomics and antivenomics of Bothrops colombiensis, a medically important pitviper of the Bothrops atroxasper complex endemic to Venezuela: contributing to its taxonomy and snakebite management. J Proteomics. 2009;72:227–240. doi: 10.1016/j.jprot.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Chan BM, Matsuura N, Takada Y, Zetter BR, Hemler ME. In vitro and in vivo consequences ofVLA-2 expression on rhabdomyosarcoma cells. Sci (Wash DC) 1991;251:1600–1602. doi: 10.1126/science.2011740. 2022 B. [DOI] [PubMed] [Google Scholar]
- Felding-Habermann B, Mueller BM, Romerdahl CA, Cheresh DA. Involvement of integrin alpha V gene expression in human melanoma tumorigenicity. J Clin Investig. 1992;89:2018–2022. doi: 10.1172/JCI115811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Bjarnason JB. Proteolytic enzymes. In: Barrett AJ, editor. Methods in Enzymology. Vol. 248. Academic Press; San Diego, CA: 1995. pp. 368–387. [Google Scholar]
- Fox JW, Serrano SM. Timeline of key events in snake venom metalloproteinase research. J Proteomics. 2009;72:200–209. doi: 10.1016/j.jprot.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Girón ME, Salazar AM, Aguilar I, Pérez JC, Sánchez EE, Arocha-Piñango CL, Rodríguez-Acosta A, Guerrero B. Hemorrhagic, coagulant and fibrino(geno)lytic activities of crude venom and fractions from mapanare (Bothrops colombiensis) snakes. Comp Biochem Physiol C Toxicol Pharmacol. 2008;147:113–121. doi: 10.1016/j.cbpc.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Higuchi DA, Almeida MC, Barros CC, Sanchez EF, Pesquero PR, Lang EA, Samaan M, Araujo RC, Pesquero JB, Pesquero JL. Leucurogin, a new recombinant disintegrin cloned from Bothrops leucurus (white-tailed-jararaca) with potent activity upon platelet aggregation and tumor growth. Toxicon. 2011;58:123–129. doi: 10.1016/j.toxicon.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Hite LA, Shannon JD, Bjarnason JB, Fox JW. Sequence of a cDNA clone encoding the zinc metalloproteinase hemorrhagic toxine from Crotalus atrox: evidence for signal, zymogen, and disintegrin-like structures. Biochemistry. 1992;31:6203–6211. doi: 10.1021/bi00142a005. [DOI] [PubMed] [Google Scholar]
- Humphries MJ, Olden K, Yamada KM. A synthetic peptide from fibronectin inhibits experimental metastasis of murine melanoma cells. Sci (Wash DC) 1986;233:467–470. doi: 10.1126/science.3726541. [DOI] [PubMed] [Google Scholar]
- Humphries MJ, Yamada KM, Olden K. Investigation of the biological effects of the anti-cell adhesion synthetic peptides that inhibit experimental metastasis of B16-FIO murine melanoma cells. J Clin Investig. 1988;81:782–790. doi: 10.1172/JCI113384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguti AS, Moura-da-Silva AM, Laing GD, Knapp T, Zuzel M, Crampton JM, Theakston RD. Collagen-induced secretion-dependent phase of platelet aggregation is inhibited by the snake venom metalloproteinase jararhagin. Biochim Biophys Acta. 1997;1335:209–217. doi: 10.1016/s0304-4165(96)00140-7. [DOI] [PubMed] [Google Scholar]
- Lucena S, Sanchez EE, Perez JC. Anti-metastatic activity of the recombinant disintegrin, r-mojastin 1, from the Mohave rattlesnake. Toxicon. 2011;57:794–802. doi: 10.1016/j.toxicon.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena S, Castro R, Lundin C, Hofstetter A, Alaniz A, Suntravat M, Sánchez EE. Inhibition of pancreatic tumoral cells by snake venom disintegrins. Toxicon. 2015;93:136–143. doi: 10.1016/j.toxicon.2014.11.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane MA, Marcinkiewicz C, Vijay-Kumar S, Wierzbicka-Patynowski I, Niewiarowski S. Viper venom disintegrins and related molecules. Proc Soc Exp Biol Med. 1998;219:109–119. doi: 10.3181/00379727-219-44322. [DOI] [PubMed] [Google Scholar]
- Moura-da-Silva AM, Marcinkiewicz C, Marcinkiewicz M, Niewiarowski S. Selective recognition of alpha2beta1 integrin by jararhagin, a Metalloproteinase/disintegrin from Bothrops jararaca venom. Thromb Res. 2001;102:153–159. doi: 10.1016/s0049-3848(01)00216-x. [DOI] [PubMed] [Google Scholar]
- Muniz JR, Ambrosio AL, Selistre-de-Araujo HS, Cominetti MR, Moura-da-Silva AM, Oliva G, Garratt RC, Souza DH. The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: insights for a new classification of snake venom metalloprotease subgroups. Toxicon. 2008;52:807–816. doi: 10.1016/j.toxicon.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Nicolson GL. Cell surface molecules and tumor metastasis. Regulation of metastatic phenotypic diversity. Exp Cell Res. 1984;150:3–22. doi: 10.1016/0014-4827(84)90696-7. [DOI] [PubMed] [Google Scholar]
- Plantefaber LC, Hynes RO. Changes in integrin receptors on oncogenically transformed cells. Cell. 1989;56:281–290. doi: 10.1016/0092-8674(89)90902-1. [DOI] [PubMed] [Google Scholar]
- Poste G, Doll J, Fidler IJ. Interaction among clonal subpopulations affect stability of the metastatic phenotype in polyclonal populations of B16 mela-noma cells. Proc Natl Acad Sci U S A. 1981;78:6226–6230. doi: 10.1073/pnas.78.10.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos OHP, Selistre-De-Araujo HS. Snake venom metalloproteases -structure and function of catalytic and disintegrin domains. Comp Biochem Physiol Part C Toxicol Pharmacol. 2006;142:328–346. doi: 10.1016/j.cbpc.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Saiki I, Murata J, Lida J, Sugimura K, Azuma I. The inhibition of murine lung metastasis by synthetic polypeptides [poly RGD] and poly [YIGSR]with a core sequence of cell adhesion molecules. Br J Cancer. 1988;59:194–197. doi: 10.1038/bjc.1989.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez EE, Rodríguez-Acosta A, Palomar R, Lucena SE, Bashir S, Soto JG, Pérez JC. Colombistatin: a disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch Toxicol. 2009;83:271–279. doi: 10.1007/s00204-008-0358-y. [DOI] [PubMed] [Google Scholar]
- Sánchez EE, Lucena SE, Reyes S, Soto JG, Cantu E, Lopez-Johnston JC, Guerrero B, Salazar AM, Rodríguez-Acosta A, Galán JA, Tao WA, Pérez JC. Cloning, expression, and hemostatic activities of a disintegrin, r-mojastin 1, from the Mohave rattlesnake (Crotalus scutulatus scutulatus) Thrombosis Res. 2010;126:e211–e219. doi: 10.1016/j.thromres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JP, Montgomery RR, Retzinger GS. Dimeric ristocetin flocculates proteins, binds to platelets, and mediates von Willebrand factor-dependent agglutination of platelets. J Biol Chem. 1991;266:8149–8155. [PubMed] [Google Scholar]
- Selistre-de-Araujo HS, Cominetti MR, Terruggi CH, Mariano-Oliveira A, De Freitas MS, Crepin M, Figueiredo CC, Morandi V. Alternagin-C, a disintegrin-like protein from the venom of Bothrops alternatus, modulates alpha2beta1 integrin-mediated cell adhesion, migration and proliferation. Braz J Med Biol Res. 2005;38:1505–1511. doi: 10.1590/s0100-879x2005001000007. [DOI] [PubMed] [Google Scholar]
- Shimokawa K, Shannon JD, Jia LG, Fox JW. Sequence and biological activity of catrocollastatin-C: a disintegrin-like/cysteine-rich two-domain protein from Crotalus atrox venom. Arch Biochem Biophys. 1997;343:35–43. doi: 10.1006/abbi.1997.0133. [DOI] [PubMed] [Google Scholar]
- Shimokawa K, Jia LG, Wang XM, Fox JW. Expression, activation, and processing of the recombinant snake venom metalloproteinase, pro-atrolysin E. Arch Biochem Biophys. 1996;335:283–294. doi: 10.1006/abbi.1996.0509. [DOI] [PubMed] [Google Scholar]
- Souza DH, Iemma MR, Ferreira LL, Faria JP, Oliva ML, Zingali RB, Niewiarowski S, Selistre-de-Araujo HS. The disintegrin-like domain of the snake venom metalloprotease alternagin inhibits alpha2beta1 integrin-mediated cell adhesion. Arch Biochem Biophys. 2000;384:341–350. doi: 10.1006/abbi.2000.2120. [DOI] [PubMed] [Google Scholar]
- Suntravat M, Jia Y, Lucena SE, Sánchez EE, Pérez JC. cDNA cloning of a snake venom metalloproteinase from the eastern diamondback rattlesnake (Crotalus adamanteus), and the expression of its disintegrin domain with anti-platelet effects. Toxicon. 2013;64:43–54. doi: 10.1016/j.toxicon.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntravat M, Barret HS, Jurica CA, Lucena SE, Perez JC, Sánchez EE. Recombinant disintegrin (r-Camdis) from Crotalus adamanteus inhibits adhesion of human pancreatic cancer cell lines to laminin-1 and vitronectin. J Venom Res. 2015;6:1–10. [PMC free article] [PubMed] [Google Scholar]
- Suntravat M, Uzcátegui NL, Atphaisit C, Helmke TJ, Lucena SE, Sánchez EE, Rodríguez-Acosta A. Gene expression profiling of the venom gland from the Venezuelan mapanare (Bothrops colombiensis) using expressed sequence tags (ESTs) BMC Mol Biol. 2016;17(1):17–7. doi: 10.1186/s12867-016-0059-7. Erratum BMC Mol Biol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanjoni I, Evangelista K, Della-Casa MS, Butera D, Magalhães GS, Baldo C, Clissa PB, Fernandes I, Eble J, Moura-da-Silva AM. Different regions of the class P-III snake venom metalloproteinase jararhagin are involved in binding to alpha2beta1 integrin and collagen. Toxicon. 2010;55:1093–1099. doi: 10.1016/j.toxicon.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I, Niewiarowski S, Marcinkiewicz C, Calvete JJ, Marcinkiewicz MM, McLane MA. Structural requirements of echistatin for the recognition of alpha(v)beta(3) and alpha(5)beta(1) integrins. J Biol Chem. 1999;274:37809–37814. doi: 10.1074/jbc.274.53.37809. [DOI] [PubMed] [Google Scholar]
- Zigrino P, Kamiguti AS, Eble J, Drescher C, Nischt R, Fox JW, Mauch C. The reprolysin jararhagin, a snake venom metalloproteinase, functions as a fibrillar collagen agonist involved in fibroblast cell adhesion and signaling. J Biol Chem. 2002;277:40528–40535. doi: 10.1074/jbc.M202049200. [DOI] [PubMed] [Google Scholar]