SUMMARY
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is an aggressive and largely incurable hematologic malignancy originating from plasmacytoid dendritic cells (pDCs). Using RNA interference screening, we identified the E-box transcription factor TCF4 as a master regulator of the BPDCN oncogenic program. TCF4 served as a faithful diagnostic marker of BPDCN, and its downregulation caused the loss of the BPDCN-specific gene expression program and apoptosis. High-throughput drug screening revealed that bromodomain and extra-terminal domain inhibitors (BETi’s) induced BPDCN apoptosis, which was attributable to disruption of a BPDCN-specific transcriptional network controlled by TCF4-dependent super-enhancers. BETi’s retarded the growth of BPDCN xenografts, supporting their clinical evaluation in this recalcitrant malignancy.
INTRODUCTION
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare and aggressive hematologic malignancy that has a characteristic skin tropism but can disseminate widely (Facchetti et al., 2016). Although an initial response to chemotherapy is common, BPDCN prognosis is extremely poor and most patients relapse into a drug-resistant disease with a median overall survival of ~1 year after diagnosis (Garnache-Ottou et al., 2007; Julia et al., 2013; Pagano et al., 2013). Allogenic stem cell transplantation is a viable therapeutic option for BPDCN, but treatment results in only ~40% survival after 3 years (Roos-Weil et al., 2013). Hence, an understanding of the molecular dependencies of BPDCN and the identification of targeted strategies for therapeutic intervention are highly needed. Histologically, BPDCN was first defined as a lineage marker-negative “plasmacytoid T cell lymphoma”, and was later classified as "blastic NK-cell lymphoma" and/or "CD4+CD56+ hematodermic neoplasm" based on the expression of the NK marker CD56. Subsequent studies based on the expression of surface markers (BDCA-2/CD303, IL-3Ra/CD123), signaling molecules (BLNK, CD2AP, TCL1) and transcription factors (BCL11A, SPIB), clearly identified plasmacytoid dendritic cells (pDCs) as the “cell of origin” of BPDCN (Chaperot et al., 2001; Garnache-Ottou et al., 2009; Herling et al., 2003; Jaye et al., 2006; Marafioti et al., 2008; Montes-Moreno et al., 2013; Petrella et al., 2002). Since 2008, this notion has been incorporated into the WHO guidelines for the classification of tumors of hematopoietic and lymphoid tissues, and the BPDCN acronym was established to replace the previous classifiers (S. Swerdlow, 2008).
Recent genomic studies have addressed the molecular basis for BPDCN (Alayed et al., 2013; Dijkman et al., 2007; Jardin et al., 2009; Jardin et al., 2011; Lucioni et al., 2011; Menezes et al., 2014; Sapienza et al., 2014; Stenzinger et al., 2014). Collectively, these studies identified frequent chromosomal losses (5q, 12p13, 13q21, 6q23-ter, 9), inactivation of tumor suppressors (RB1, TP53 and CDKN2A), activation of oncogenes (NRAS, KRAS) and mutations in epigenetic regulators (TET2, TET1, DNMT3A, IDH1, IHD2) that are also frequently mutated in acute myeloid leukemia (Abdel-Wahab and Levine, 2013). However, these studies did not functionally define the molecular pathways that sustain BPDCN viability and did not address how the origin of this malignancy from normal pDCs influences its unique clinical behavior.
pDCs are innate immune cells that counteract viral infections, using Toll-like receptors to sense viral nucleic acids and stimulate the production of anti-viral cytokines, including type I (α, β) interferon. The unique functions of pDCs are fully conserved between humans and mice (Colonna et al., 2004; Liu, 2005; Reizis et al., 2011). pDCs are related to classical (conventional) dendritic cells (cDCs) and develop from common dendritic cell progenitors in the bone marrow (Reizis, 2010). The commitment to the pDC lineage is controlled by the E-box transcription factor TCF4 (E2-2), which is preferentially expressed in both murine and human pDCs (Cisse et al., 2008; Nagasawa et al., 2008). TCF4 also maintains pDC lineage identity and its deletion from pDCs induces trans-differentiation into cDC-like cells (Ghosh et al., 2010). Conversely, the E-box transcription factor inhibitor Id2 is highly expressed in cDCs (Spits et al., 2000), and its loss favors pDC over cDC development (Ghosh et al., 2014). Other transcription factors are required for pDC development (IRF8, BCL11A) or differentiation (SPIB, RUNX2) (Ippolito et al., 2014; Sasaki et al., 2012; Sawai et al., 2013; Schiavoni et al., 2002; Schotte et al., 2004; Tsujimura et al., 2003). All are direct TCF4 targets (Cisse et al., 2008; Ghosh et al., 2010; Sawai et al., 2013) and may in turn regulate TCF4 expression (Ippolito et al., 2014; Nagasawa et al., 2008), thereby constituting a TCF4-orchestrated regulatory network that maintains pDC lineage identity.
Cancer cells often inherit transcription factor programs from their normal cellular counterparts. Such transcription factors are known as "lineage-survival oncogenes" (Garraway and Sellers, 2006), and their inactivation can dissolve the gene expression network of cancer cells, making them an intriguing and relatively unexplored class of therapeutic targets (Rui et al., 2011). The traditional notion that transcription factors cannot be targeted in a clinical setting has recently been challenged by the development of inhibitors of the bromodomain and extraterminal domain (BET) proteins (Filippakopoulos et al., 2010), chromatin readers that bind to acetylated histones and promote transcriptional elongation (Jang et al., 2005). Selective inhibition of key oncogenes has been observed following BET inhibition in multiple tumor types. The ability of BET inhibitors to silence the expression of these oncogenes has been ascribed to their transcriptional control by “super-enhancers” (SEs), defined as large genomic clusters of BET-dependent regulatory regions (Chapuy et al., 2013; Loven et al., 2013).
In an effort to uncover therapeutic options for BPDCN, we have successfully combined RNA interference screening with high-throughput drug screening to identify pathways essential for the proliferation and survival of this cancer.
RESULTS
Loss-of-function RNA interference screening in BPDCN
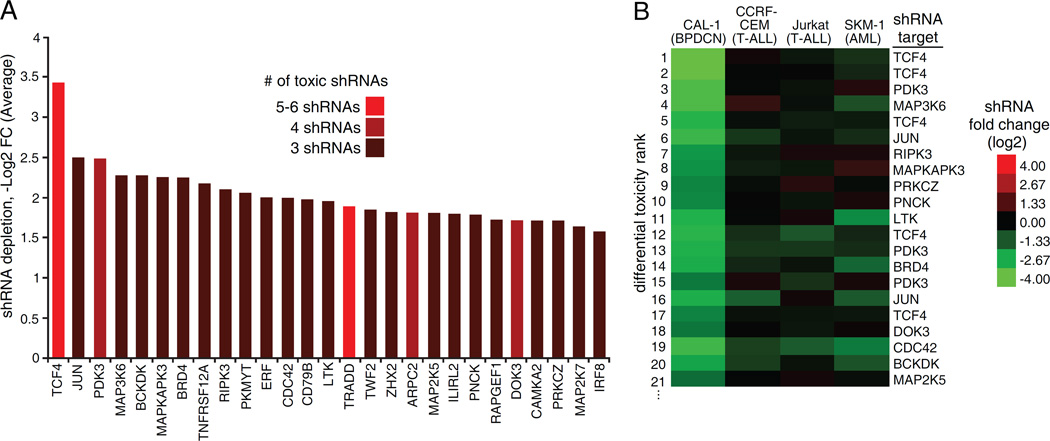
To identify BPDCN molecular dependencies, we performed a loss-of-function RNA interference screen in the BPDCN cell line Cal-1(Maeda et al., 2005) using a 12,515 small hairpin RNA (shRNA) library targeting 1,051 genes encoding kinases, signaling proteins and transcription factors relevant to hematopoiesis. Following induction of shRNA expression for 3 weeks, the depletion of specific shRNAs was monitored by next generation sequencing (Table S1) (Ngo et al., 2006). We identified 27 genes that had at least 3 independent shRNAs depleted by at least 2.5-fold across 4 replicates (p<0.001). The top hit, both in terms of number of toxic shRNAs (6) and average depletion (10.8-fold) was TCF4, an E-box transcription factor known to be an essential regulator of normal pDC development (Figure 1A) (Cisse et al., 2008). Other hits included the transcription factor JUN (3 shRNAs, 5.7-fold depletion), the metabolic enzyme PDK3 (4 shRNAs, 5.6-fold depletion) and the chromatin reader BRD4 (3 shRNAs, 4.8-fold depletion) (Figure 1A). Many shRNAs toxic to Cal-1 cells had no toxicity in other leukemia lines we screened as specificity controls (CCRF-CEM, T-cell acute lymphoblastic leukemia (T-ALL); Jurkat, T-ALL; SKM-1, acute myeloid leukemia (AML)) (Figure 1B, Figure S1A, B, C and D). These included the TCF4 shRNAs, suggesting that TCF4 controls an essential regulatory network specific for BPDCN.
Figure 1.
Genetic dependencies of BPDCN identified by shRNA screen. A) shRNA screen hits: for each gene, the average -Log2 FC depletion of toxic shRNAs is shown. B) The shRNAs toxic to Cal-1 cells were ranked based on differential depletion with respect to the average of 3 control cell lines: CCRF-CEM, Jurkat and SKM-1. The top 20 shRNAs specifically toxic to BPDCN Cal-1 are shown. See also Figure S1 and Table S1.
TCF4 is required for BPDCN viability
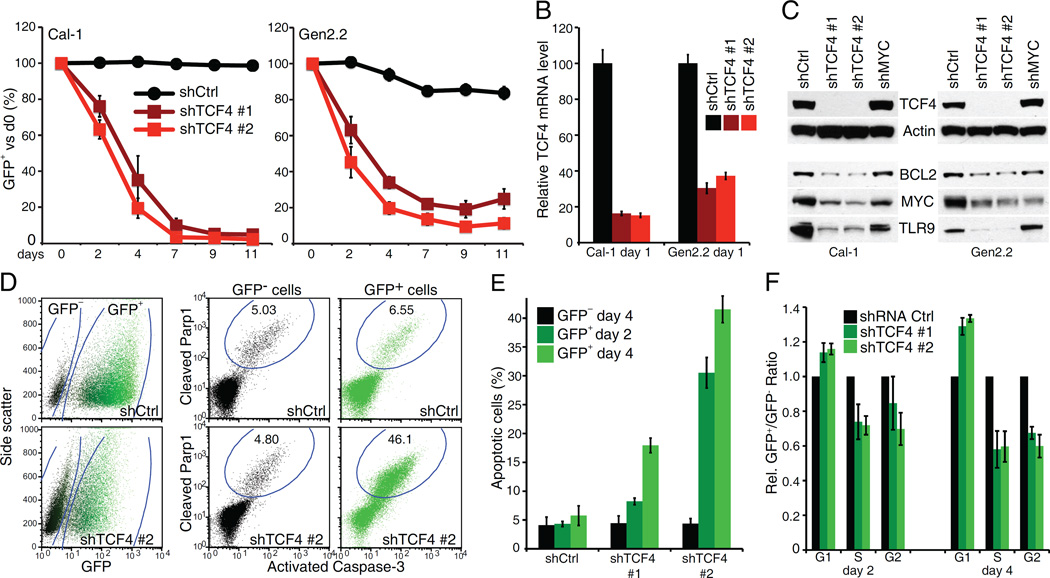
To explore the biological consequences of TCF4 knockdown in BPDCN, we transduced the Cal-1 and Gen2.2 BPDCN lines (Chaperot et al., 2006) with doxycycline-inducible shRNA expression vectors that co-express green fluorescent protein (GFP). Expression of shRNAs targeting the TCF4 3’ UTR resulted in rapid depletion of the shRNA+/GFP+ cell population (Figure 2A, B, C). This toxicity could be rescued by ectopic expression of the TCF4 coding region, supporting on-target effects (Figure S2A). Moreover, the sameTCF4 shRNAs were not toxic in control AML and T-ALL lines, whereas a MYC shRNA was similarly toxic in all the tested lines (Figure S2B). TCF4 knockdown produced a robust, time-dependent and cell-intrinsic increase in apoptosis, as assessed by a dual staining for active Caspase-3 and cleaved Parp1 (Figure 2D, E). TCF4 knockdown also resulted in a minor proliferation arrest, with an increase of G1 cells (~20%) and a decrease of cells in S and G2 phases (Figure 2F, S2C). These findings establish TCF4 as a master regulator of BPDCN viability.
Figure 2.
The E-box transcription factor TCF4 is required for BPDCN viability. A) BPDCN cells were infected with either control or TCF4 shRNAs. Shown is the fraction of live, shRNA expressing (GFP+) cells over time after shRNA induction, compared to the un-induced day 0. B) RT Q-PCR was used to measure the level of TCF4 mRNA in BPDCN cells, after induction of the indicated shRNAs for 1 day. The B2M gene was used as housekeeping control. C) Western-blot analysis of Tcf4 expression 24 hr after the induction of the indicated shRNAs. Actin was used as loading control. The TCF4 targets BCL2, MYC and TLR9 are also shown. D) A representative flow cytometry stain for active Caspase-3 and cleaved Parp1 is shown for Cal-1 cells at day4 post shRNA induction. E) Cal-1 cells were infected with either Ctrl or TCF4 shRNAs. Shown is the fraction of apoptotic cells, for both GFP− and GFP+ (shRNA expressing) populations. Time points refer to days post shRNA induction. F) Cal-1 cells were infected with either Ctrl or TCF4 shRNAs. For each cell cycle phase, the normalized ratio between GFP+ (shRNA expressing) and GFP- cells is shown. Time points refer to days post shRNA induction. Error bars represent SEM of triplicates. See also Figure S2.
The TCF4-dependent transcriptional network in BPDCN
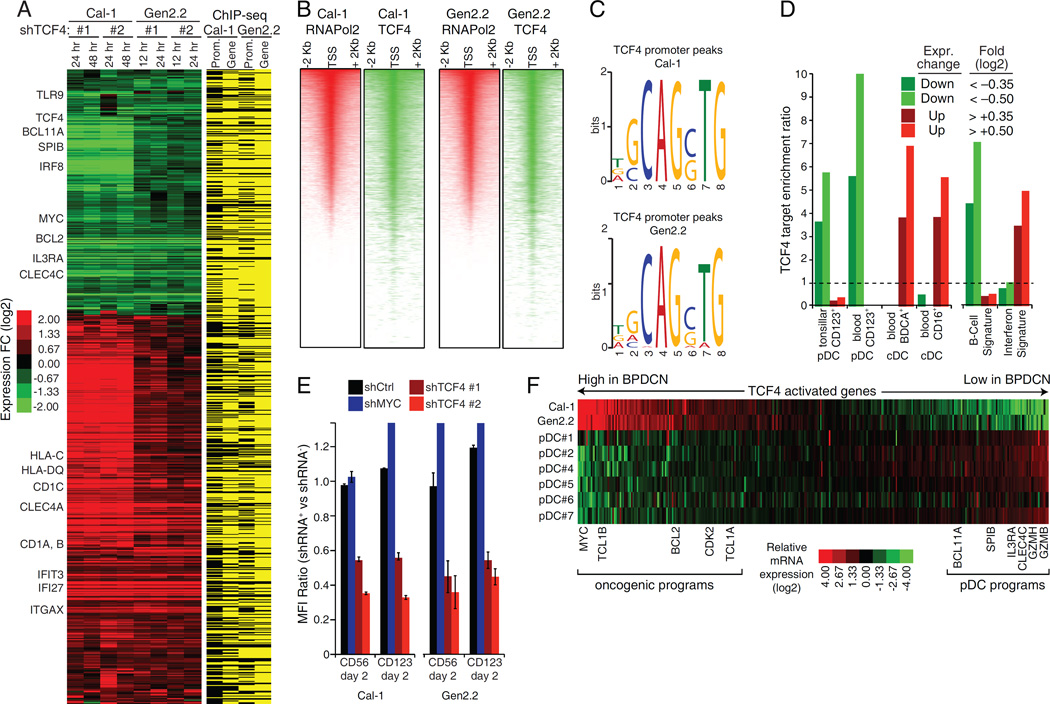
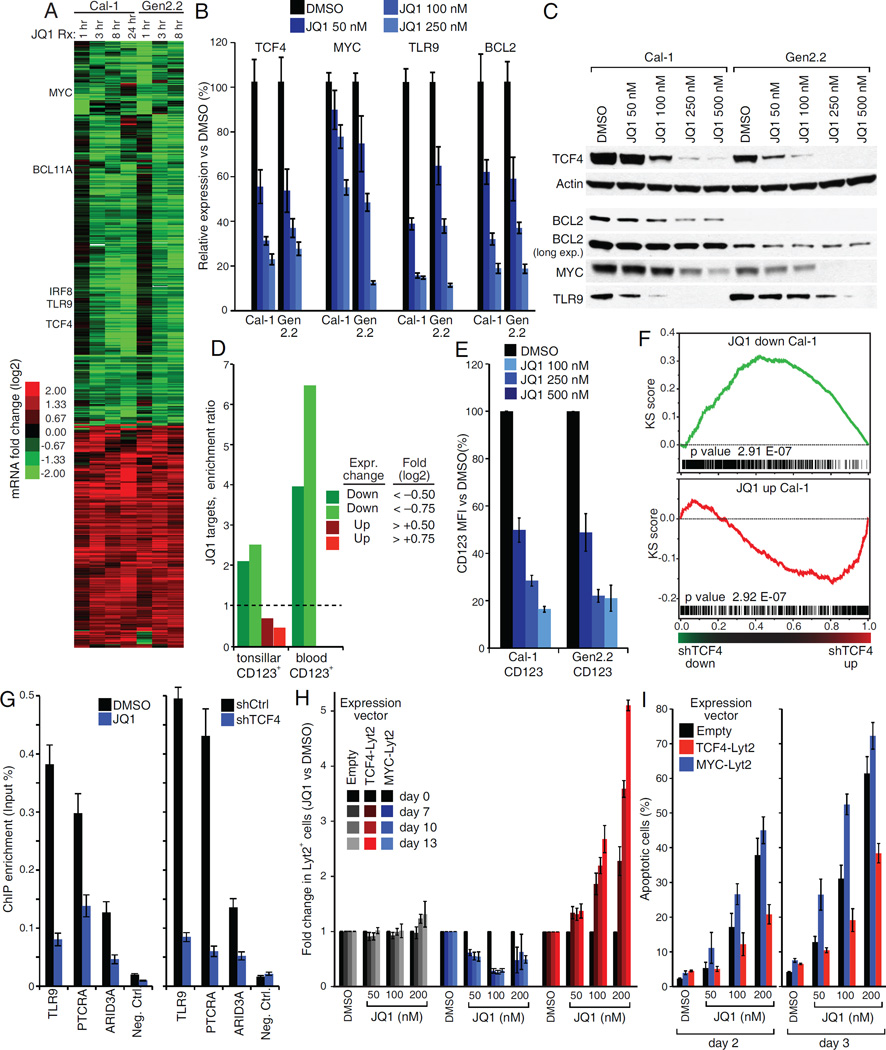
To map the TCF4-dependent regulatory network in BPDCN, we profiled gene expression changes induced by TCF4 shRNAs in Cal-1 and Gen2.2 cells. In parallel, we performed chromatin immunoprecipitation sequencing (ChIP-Seq) to identify TCF4 genomic binding sites, using an anti-TCF4 rabbit monoclonal antibody that we developed for this purpose (Figure S3A, S3B). We identified a total of 399 and 630 genes that were down- or up-regulated following TCF4 knockdown, respectively (Figure 3A, Table S2). Most of them had a TCF4 ChIP-Seq peak in the promoter or gene body. The degree of TCF4 binding at promoters strongly correlated with RNA polymerase-2 (pol2) density, confirming the master regulatory role of TCF4 in BPDCN (Figure 3B). Motif enrichment analysis identified the E-box 5'-CAGCTG-3' as the most enriched motif in TCF4 bound regions in both cell lines, supporting the specificity of our ChIP-Seq assay (Figure 3C, S3C).
Figure 3.
The TCF4-dependent transcriptional network in BPDCN. A) Left: heat-map of genes down- or up-regulated in both Cal-1 and Gen2.2 cells after TCF4 knockdown. Right: TCF4 regulated genes containing a TCF4 ChIP-Seq peak within the promoter (− +2000bp from the TSS) or the gene body are indicated in yellow. See Table S2. B) Heatmaps of promoter RNA Pol2 (red) and TCF4 (green) density in the Cal-1 and Gen2.2 cells. For each line, representative RefSeq accessions were ranked by RNA Pol2 density and the TCF4 heat-map was displayed accordingly. C) The DNA logos of the top TCF4 binding motif predicted by MEME are shown (Bailey et al., 2009). Please see Figure S3C for the results of the motif prediction tool Clover Figure(Frith et al., 2004). D) The indicated TCF4 gene sets were analyzed by signature enrichment analysis. Enrichment ratios versus the indicated signature are plotted. See Table S3.E) BPDCN cell lines were infected with the indicated shRNAs and the expression of surface CD56 and CD123 was measured by flow cytometry. Shown are mean florescence intensity (MFI) ratios of shRNA+ vs. shRNA− cells. See also Figure S3E. F) Heat-map comparing the RNA-Seq expression of TCF4 activated genes in BPDCN cell lines with that of normal pDCs isolated from 6 healthy donors. Error bars represent SEM of triplicates. See also Figure S3 and Table S2, S3.
Genes downregulated following TCF4 depletion included: 1) regulators of pDC development (BCL11A, IRF8, SPIB); 2) pDC-specific surface receptors (IL3RA, CLEC4C, PTPRS) (Bunin et al., 2015); 3) proteins essential for pDC function (TLR9); 4) oncogenes such as MYC and BCL2 (Figure 2C, S3D). Genes upregulated included: 1) interferon responsive genes, (IFI27, IFIT3); 2) genes encoding adhesion molecules highly expressed in cDCs (ITGAX, CLEC4A); 3) genes encoding proteins involved in antigen presentation by cDCs (CD1) (Figure S3D).
To explore these findings systematically, we performed signature enrichment analysis using a database of gene expression signatures that reflect signaling and regulatory processes in normal and malignant hematopoiesis (Shaffer et al., 2006) (Table S3). Notably, TCF4-activated genes were highly enriched for pDC-specific genes (Figure 3D, CD123+) (Lindstedt et al., 2005). Conversely, TCF4-repressed genes were strongly enriched for cDC genes (Figure 3D, CD16+, BDCA+). Signature enrichment analysis also confirmed upregulation of interferon responsive genes after TCF4 silencing. Notably, knockdown of TCF4, but not MYC, decreased surface expression of CD123 and CD56, which are diagnostic hallmarks of BPDCN (Figure 3E, S3E).
While these findings demonstrate that TCF4 imparts pDC identity on BPDCN cells, we additionally sought genes that were regulated by TCF4 in a BPDCN-specific fashion. To do so, we used global mRNA sequencing (RNA-Seq) to compare the BPDCN lines with normal pDCs isolated from 6 healthy donors, specifically focusing on the TCF4 target genes that we identified in BPDCN (Figure 3F, S3F). Notably, a sizable set of TCF4-activated genes was expressed at higher levels in BPDCN, including several proto-oncogenes (BCL2, MYC, TCL1A and TCL1B). Conversely, TCF4-activated genes that encode important regulators of normal pDC function (BCL11A, SPIB, IL3RA, and CLEC4C) were expressed at lower levels in BPDCN than in pDCs. Thus, the TCF4 regulatory networks in BPDCN cells and normal pDCs are distinct, with TCF4 upregulating genes that may contribute to the BPDCN malignant phenotype.
Diagnostic relevance of TCF4 expression in primary BPDCN cases
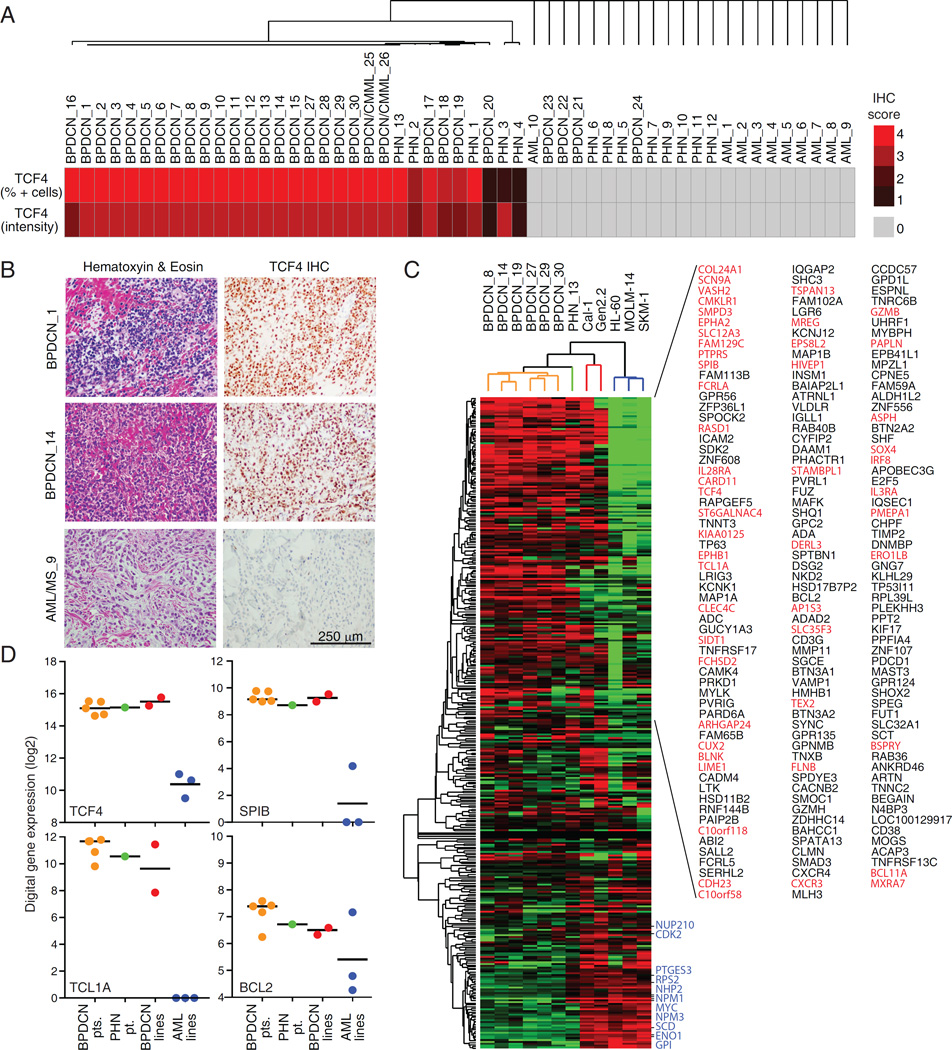
Because BPDCN diagnosis is challenging, we tested whether TCF4 expression could be used to identify BPDCN cases. We performed TCF4 immunohistochemistry (IHC) on a panel of 53 tumors, including BPDCN (n=28), BPDCN mimics such as AML/myeloid sarcoma (n=10) and unclassified primitive hematopoietic neoplasms (PHN; n=13) (Table S4). Consistent with its specificity (Figure S3A), our anti-TCF4 antibody readily identified normal pDCs in human tonsil sections (Figure S4A). We scored each primary tumor based on the distribution and intensity of its IHC signal and perform hierarchical clustering of these data, thereby dividing the primary cases into two major clusters reflecting their TCF4 scores (Figure 4A, B). Cluster #1 was characterized by homogeneous and strong TCF4 signals and included the majority of BPDCN cases (24/28), 2 cases consistent with either BPDCN or chronic myelomonocytic leukemia (CMML) with pDC proliferation (Facchetti et al., 2016), 3 out of 13 unclassified PHN cases, but no AML/Myeloid sarcomas. Cluster #2 included all of the AML/Myeloid sarcoma cases (10/10), the 10 remaining PHN cases and 4 cases initially diagnosed as BPDCN.
Figure 4.
TCF4 expression facilitates BPDCN diagnosis. A) The results of the unsupervised hierarchical clustering performed based on the TCF4 IHC scoring are shown. See Table S4. B) Representative TCF4 and hematoxylin and eosin (H&E) stains are shown at 40X magnification for 2 BPDCN cases and 1 AML case. C) Heat-map view of RNA-Seq expression levels of TCF4-activated genes in the indicated samples, grouped by unsupervised hierarchical clustering. Red indicates genes characteristically expressed in pDCs (DC-1 or DC-4 signatures) whereas blue indicates genes associated with cell proliferation (Prolif-5 signature) (Shaffer et al., 2006). D) The FFPE RNA-Seq digital gene expression values are shown for TCF4 and the indicated TCF4 targets. The black line indicates the mean. See also Figure S4 and Table S4.
To determine if the TCF4 regulatory network was maintained in primary BPDCN, we performed RNA-Seq on 6 FFPE BPDCN biopsies from which the RNA was not excessively degraded. Because only one PHN and no AML cases passed quality control for RNA-Seq library generation, we used RNA-Seq data from AML lines (HL-60, MOLM-14 and SKM-1) and BPDCN lines that had been formalin-fixed and paraffin-embedded as controls. Unsupervised hierarchical clustering based on the expression of TCF4 activated genes was sufficient to group all the primary BPDCN and the BPDCN lines in a separate cluster from the AML lines (Figure 4C). 40% of the TCF4 targets were consistently expressed at higher levels in primary BPDCN samples than in the AML lines (p<0.05; fold > 1.5; Figure 4C, D), including many that are characteristically expressed in normal blood pDCs (Figure 4C, red). Interestingly, the PHN #13 sample clustered together with primary BPDCNs, consistent with its strong TCF4 IHC scoring, suggesting that this case may be a BPDCN that could not be classified as such using current pathological methods. Together, our findings demonstrate that the expression of TCF4 and its regulatory network are maintained in primary BPDCN tumors and suggest that TCF4 IHC could aid in the differential diagnosis of BPDCN.
Chemical and genetic inhibition of BRD4 reduces BPDCN viability
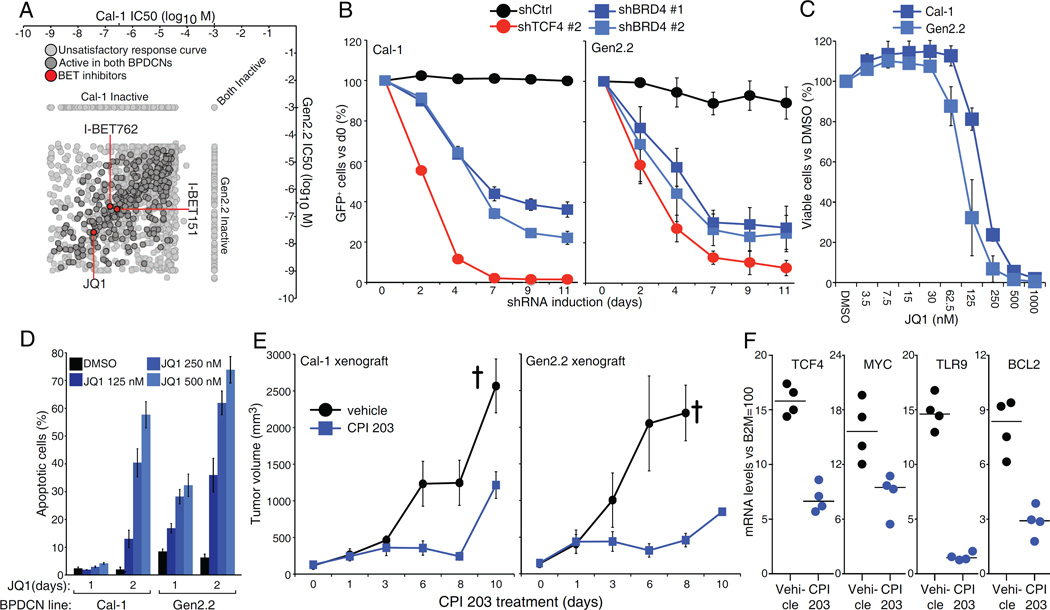
To explore therapeutic strategies in BPDCN, we performed a high-throughput drug screen using a library of 1,910 small molecules that are either approved or in early stages of development for cancer therapy (Ceribelli et al., 2014; Mathews Griner et al., 2014). A plot of the concentrations needed for 50% loss of cell viability (IC50) revealed that all 3 BET inhibitors (BETi's) present in the library (JQ1, I-BET151 and I-BET762) were highly toxic for both BPDCN lines (Figure 5A, Table S5). This was notable since the BET family member BRD4 was among the hits from our shRNA screen (Figure 1A). Inducible expression of BRD4 shRNAs in BPDCN lines resulted in the depletion of shRNA+/GFP+ cells (Figure 5B, S5A). Treatment with the BETi JQ1 decreased the viability of both BPDCN lines in a dose-dependent manner (Figure 5C)) and produced a time- and dose-dependent increase in apoptosis (Figure 5D, S5B).
Figure 5.
The bromodomain and extra-terminal domain BRD4 is required for BPDCN survival. A). Log10 molar IC50 plots comparing Cal-1 and Gen2.2 cells. Of the 1910 compounds screened, 314 showed activity in both BPDCN lines, after excluding inactive and poorly fitting compounds. BETi’s are highlighted in red. See Table S5. B) BPDCN cells were infected with Ctrl, TCF4 or BRD4 shRNAs. Shown is the fraction of live, shRNA expressing (GFP+) cells over time after shRNA induction, compared to the day 0 un-induced value. C) BPDCN cells were treated with either DMSO or the indicated amount of BETi JQ1. Cell viability was assessed by MTS assay at day 3 post-treatment. D) BPDCN cells were treated with either DMSO or the indicated amount of JQ1. The percentage of apoptotic cells (active Caspase-3+, cleaved Parp1+) is shown at day 1 and day 2 post-treatment. E) Human BPDCN xenograft models were established by subcutaneous injection of Cal-1 and Gen2.2 cells in NOD/SCID mice and treated with either vehicle or the BET Inhibitor CPI 203 (5 mg/kg) for the indicated time points. Tumor growth was measured as a function of tumor volume. The cross indicate the mice euthanized because of excessive tumor growth. F) Relative mRNA levels of TCF4 and 3 of its targets in Cal-1 xenografts from mice treated for 5 days with CPI 203 or vehicle control. Error bars represent SEM of triplicates. In E), Error bars represent SEM, n=4 mice for the vehicle group, n=6 mice for the CPI 203 group. See also Figure S5 and Table S5.
To investigate the effect of BETi's in vivo, we established BPDCN xenografts by subcutaneous injection of Cal-1 and Gen2.2 cells into NOD/SCID mice. Treatment with the BETi CPI 203 was effective as a single agent in reducing tumor growth in both xenograft models (Figure 5E). The CPI 203 regimen was well tolerated in vivo, causing only modest weight loss compared to vehicle-treated controls (Figure S5C). Quantitative RT-PCR performed on tumors harvested after 5 days of CPI 203 treatment revealed decreased mRNA expression of TCF4 and three of its targets (MYC, TLR9, BCL2), suggesting that the BETi blocked TCF4 function in vivo (Figure 5F, see below). Interestingly, the anti-tumor activity of CPI 203 correlated with the extent of TCF4 inhibition observed over time and was reduced at the endpoint of our xenograft experiment (Figure S5D).
A TCF4 and BRD4 dependent transcriptional network in BPDCN
To define the molecular mechanism of BETi toxicity in BPDCN, we analyzed gene expression changes after JQ1 treatment in the 2 BPDCN lines. We identified a total of 437 and 251 genes down- or up-regulated, respectively (Figure 6A, Table S6). Interestingly, TCF4 itself was among the genes consistently downregulated by JQ1 in both cell lines. The same was true for multiple TCF4 targets such as TLR9, BCL11A, MYC, IRF8 and BCL2 (Figure 6A, B, C). As we had observed after TCF4 knockdown, pDC-specific signatures were enriched among JQ1 downregulated genes (Figure 6D). JQ1 treatment reduced surface expression of the pDC-specific marker CD123 (Figure 6E, S6A). Gene set enrichment analysis (GSEA) confirmed that JQ1 treatment systematically recapitulates TCF4 silencing in BPDCN, as genes downregulated by JQ1 treatment in were strongly enriched for TCF4-activated genes (Figure 6F, S6B). These findings support the view that the TCF4 program is a central component of the BET-dependent phenotype in BPDCN. Indeed, JQ1 treatment strongly reduced TCF4 binding to its target genes, to an extent similar to that observed after knockdown of TCF4 itself (Figure 6G).
Figure 6.
A TCF4 and BRD4 dependent transcriptional network in BPDCN. A) Heat-map of genes down- or up-regulated following JQ1 treatment in both BPDCN cell lines. B) BPDCN cells were treated with either DMSO or the indicated JQ1 amount for 24 hr. The mRNA levels of TCF4 and its targets BCL2, MYC and TLR9 were evaluated by RT QPCR. The B2M gene was used as housekeeping control. C) Western-blot analysis of TCF4 expression in BPDCN cell lines 24 hr after treatment with the indicated JQ1 amounts. Actin was used as loading control. The TCF4 targets BCL2, MYC and TLR9 are also shown. D) The indicated JQ1 gene sets were analyzed by signature enrichment analysis. Enrichment ratios versus the indicated signature are plotted. E) BPDCN cell lines were treated with either DMSO or the indicated JQ1 amounts and expression of CD123 was measured by flow cytometry 24 hr post treatment. Shown are MFI values, normalized to the DMSO control. A representative stain is shown in Figure S6A. F) Gene Set Enrichment Analysis (GSEA) comparing JQ1 treatment and TCF4 shRNA gene expression datasets in the Cal-1 BPDCN cell line. Genes were first ranked based on the gene expression changes induced by TCF4 shRNAs and the distribution of JQ1 dependent genes was then analyzed. See Figure S5B for Gen2.2 cells. G) Bona fide TCF4 targets were analyzed by single locus ChIP Q-PCR after either inducible expression of TCF4 shRNA #1 (right panel, 24 hr induction) or JQ1 treatment (left panel, 16 hr, 100 nM). H) Cal-1 cells were infected with the indicated rescue vectors and the fraction of Lyt2+ cell was monitored over 2 weeks of treatment with either DMSO or the indicated JQ1 amounts. For each rescue construct, shown is the fraction of Lyt2+ cells normalized to the corresponding DMSO control. See Figure S6C for a representative stain. I) Cal-1 cells were infected with the indicated rescue vectors. Apoptosis induction after JQ1 treatment was monitored by flow cytometry for active Caspase-3 and cleaved Parp1. Error bars represent SEM of triplicates. For G), error bars represent SD of technical triplicates. One of two representative experiments is shown. See also Figure S6 and Table S6.
To link TCF4 loss to the toxicity of BETi's in BPDCN, we tested whether ectopic expression of TCF4 could rescue BPDCN cells from JQ1-induced toxicity. BPDCN cells were infected with either an empty vector control or a TCF4 expression vector the co-expresses mouse CD8a (Lyt2), allowing transduced cells to be tracked by cell sorting. A mixed population of uninfected (Lyt2−) and infected (Lyt2+) cells was exposed to increasing JQ1 amounts over the course of two weeks. Notably, ectopic TCF4 expression caused a dose-dependent increase in the Lyt2+ fraction over time, indicating rescue of cells from JQ1 toxicity (Figure 6H, S6C). No rescue was observed with an empty vector or a MYC expression vector. Consistently, JQ1-induced apoptosis was reduced in cells ectopically expressing TCF4, but not MYC (Fig 6I, S6D, E). Thus, a significant and specific component of BETi toxicity in BDPCN is due to TCF4 inhibition.
BRD4-dependent super-enhancers in BPDCN
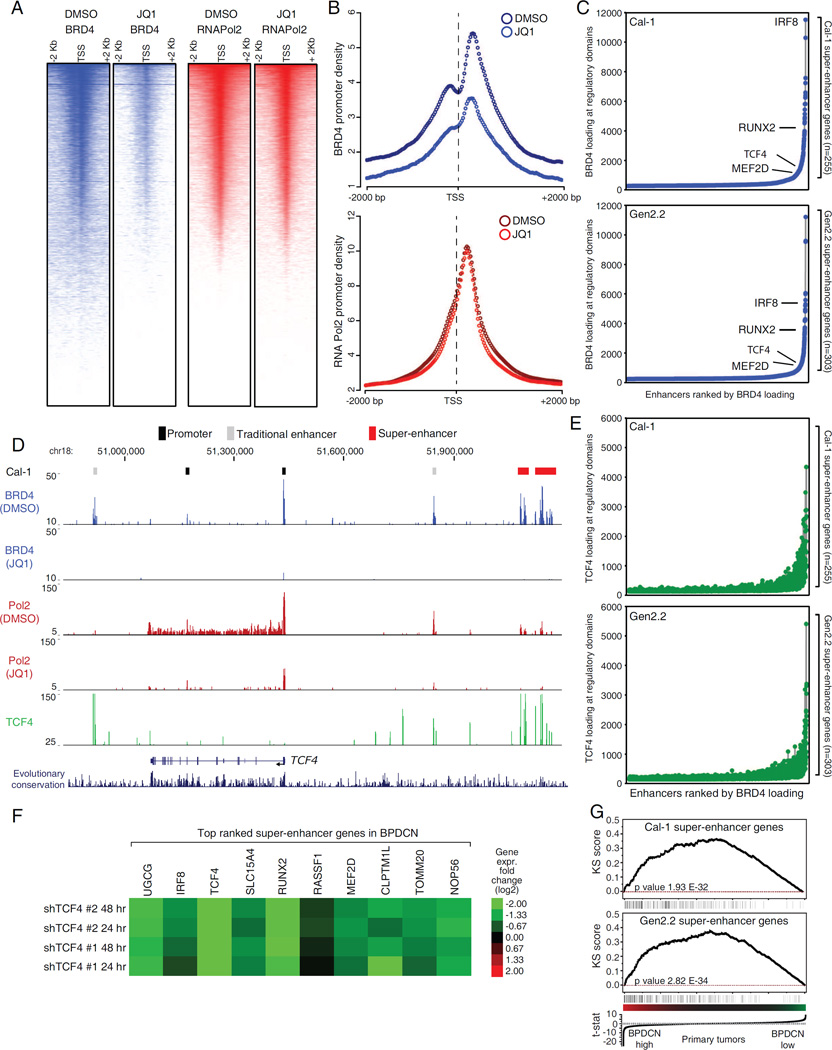
The toxicity of BET inhibitors in multiple cancer types has been linked to a dominant, context-dependent effect on the expression of tumor-specific oncogenes that are regulated by super-enhancers (SEs) (Loven et al., 2013). Because TCF4 behaves as a BPDCN “lineage-survival oncogene” (Garraway and Sellers, 2006) we sought to identify the BRD4-dependent enhancers that sustain TCF4 expression in BPDCNs. To do so, we performed BRD4 and RNA Pol2 ChIP-Seq before and after JQ1 treatment in the 2 BPDCN lines. BRD4 binding at promoters correlated with RNA Pol2 density, as expected (Figure 7A, S7A). JQ1 treatment strongly reduced BRD4 promoter binding but had little effect on promoter-bound RNA Pol2 (Figure 7B, S7B). Conversely, BET inhibition induced pronounced depletion of elongating RNA Pol2 on the majority of expressed genes (Figure S7C). These results are in line with the well-established role of BET proteins in promoting transcriptional elongation.
Figure 7.
Mapping the BRD4 dependent super-enhancers in BPDCN. A) Density heat-maps of promoter BRD4 (blue) and RNA Pol2 (red) in Cal-1 cells, after 12 hr treatment with either DMSO or 250 nM JQ1. Representative RefSeq accessions were ranked based on decreasing RNA Pol2 density in the DMSO treated cells and all the heat-maps were displayed accordingly. See Figure S7A for Gen2.2 cells. B) Meta-promoter profiles of BRD4 (blue) and RNA Pol2 (red) ChIP-Seq data in Cal-1 cells. See Figure S7B for Gen2.2 cells. C) Enhancers were ranked based on increasing BRD4 loading. Relevant SE containing genes are highlighted in the plot. D) The TCF4 locus ChIP-Seq tracks for BRD4 (blue), RNA Pol2 (red) and TCF4 (green) are shown for Cal-1 cells. See Fig S7E for Gen2.2 cells. E) Enhancers were ranked based on increasing BRD4 loading and the corresponding signal from TCF4 ChIP-Seq was then displayed. F) Heat-map of gene expression changes (Log2 FC) observed after TCF4 knockdown in the BPDCN Cal-1 line. G) Gene Set Enrichment Analysis (GSEA) showing the enrichment of SE genes among genes highly expressed in primary BPDCN samples. See also Figure S7 and Table S7.
To identify BPDCN SEs, we ranked BRD4-bound regulatory regions by increasing BRD4 ChIP-Seq occupancy. These plots revealed an obvious inflection point, enabling us to define SEs in both BPDCN lines (Figure 7C). RNA Pol2 loading correlated with BRD4 binding at SEs, supporting their active state (Figure S7D). Altogether, we identified 255 and 303 SE genes in Cal-1 and Gen2.2 cells, respectively (Table S7). Of these, 75 were shared. To identify functionally relevant SEs, we developed a non-parametric ranking based on both the depletion of SE-bound BRD4 and the reduction of elongating RNA Pol2 after JQ1 treatment. Notably, TCF4 itself was among the genes containing a SE in both BPDCN lines and ranked third in our combined SE scoring (Figure 7D, Figure S7E and Table S7). Other top-ranking SE genes included the pDC regulators IRF8 and RUNX2, and SLC15A4, a gene required to sense TLR ligands (Blasius et al., 2010) (Figure S7F, Table S7). These observations support the view that SE scoring identifies genes that are central to BPDCN biology.
Consistent with its master regulator function, TCF4 was detected at the majority of BPDCN SEs, and TCF4 binding SEs positively correlated with both BRD4 and RNA Pol2 loading (Figure 7E, 7D). Interestingly, the TCF4 SE itself was bound by TCF4, identifying a positive auto-regulatory loop that defines BPDCN identity (Figure 7D, S7E). In line with these findings, top ranking SE genes were strongly down-regulated following TCF4 knockdown suggesting that TCF4 is directly responsible for their expression (Figure 7F). Finally, GSEA showed that SE genes were significantly enriched among genes highly expressed in primary BPDCN cases, indicating that the TCF4-dependent regulatory architecture (regulome) sustains the gene expression identity of primary BPDCN tumors (Figure 7G).
The TCF4-dependent regulome in normal pDCs and primary BPDCN
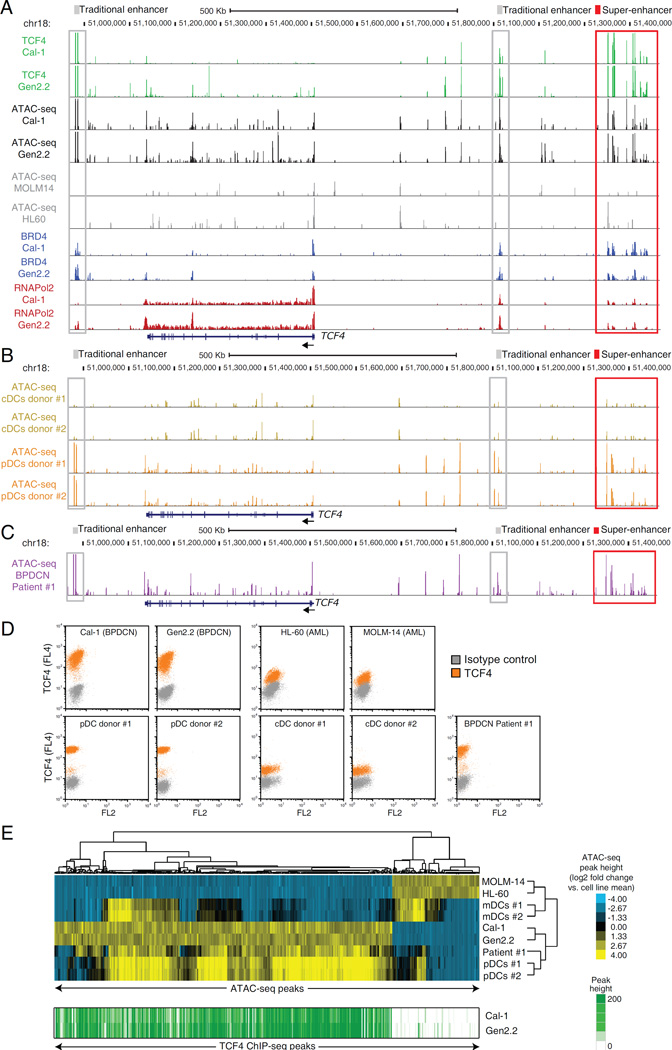
To expand the characterization of the TCF4-dependent regulome, we performed ATAC-Seq (Buenrostro et al., 2013) to map chromatin accessibility in BPDCN lines (Cal-1, Gen2.2) and BPDCN cells purified from the blood a patient with leukemic disease. As controls, we profiled chromatin accessibility in normal blood pDCs and cDCs purified from healthy donors, along with AML lines. As shown in Figure 8A, the TCF4 locus SE was demarcated by highly accessible chromatin in both BPDCN lines (Figure 8A, black). Notably, the ATAC-Seq signal mirrored TCF4 binding at the same locations (green). This was true for the TCF4 SE itself (Figure 8A, red box) as well as for other traditional enhancers (TE) present within the locus (Figure 8A, gray boxes). Remarkably, little or no ATAC-Seq signal was present in the 2 AML lines at these chromosomal regions (Figure 8A, gray). These findings demonstrate that ATAC-Seq can identify BPDCN-specific regulatory elements and suggest that TCF4 binding at these regions is a likely determinant of their open chromatin state.
Figure 8.
ATAC-Seq mapping of the regulatory landscape of BPDCN. A) ATAC-Seq was used to map the open chromatin landscape of the TCF4 locus in BPDCN cell lines (black) and in control AML cell lines (gray). TCF4, BRD4 and RNA Pol2 ChIP-Seq in the BPDCN cell lines are also shown in green, blue and red, respectively. B) Normal pDCs and CD1c+ cDCs were isolated from 2 healthy donors .The regulatory landscape of the TCF4 locus is shown in orange and yellow for pDCs and cDCs, respectively. C) ATAC-Seq was used to map the open chromatin regulatory landscape of the TCF4 locus in PBMCs isolated from a patient with leukemic BPDCN (purple). D) TCF4 intracellular flow was performed for the indicated cell lines and primary samples. E) Heat-map results of the unsupervised hierarchical clustering based on the ATAC-Seq predictor regions. Cell lines and primary samples are indicated on the right dendrogram. The corresponding TCF4 ChIP-Seq density is shown in green for the two BPDCN cell lines. See also Figure S8 and Table S8.
The regulatory landscape of the TCF4 locus in normal pDCs was very similar to that observed in BPDCN cell lines, with open chromatin observed at all of the TCF4-bound enhancers (Figure 8B, orange tracks, Figure S8A). Conversely, no ATAC-Seq signal was detected at these same regions in cDCs (Figure 8B, yellow). A similar picture emerged for other TCF4-dependent regulatory elements, such as the RUNX2 SE (Figure S8B). These finding confirmed the pDC-derived nature of BPDCN, and suggested that the TCF4 regulatory network of pDCs is largely maintained in BPDCN lines. Finally, we studied primary BPDCN cells purified from a patient with leukemic disease (Figure S8C). The regulatory architecture of the TCF4 locus in these primary BPDCN cells mirrored that of normal pDCs and BPDCN lines (Figure 8C, compare with Figure 8A, B), consistent with the fact that these cells highly expressed TCF4 by intracellular flow cytometry (Figure 8D, Figure S8D).
We next analyzed the ATAC-Seq data globally to assess similarities and differences among the samples. First, we identified 82,873 ATAC-Seq regions of open chromatin ("peaks") using data from the BPDCN and AML cell lines. We then selected peaks that best distinguished BPDCN from AML, resulting in 497 BPDCN-specific and 129 AML-specific peaks. We used these peaks to perform unsupervised hierarchical clustering of all the ATAC-Seq samples (Figure 8E). Primary cells isolated from the BPDCN patient and BPDCN cell lines clustered together with normal blood pDCs, whereas primary cDCs clustered with the AML cell lines. These findings demonstrated that primary cells and BPDCN lines retain major regulatory features of their cell-oforigin, the pDC.
Notably, 89% of the BPDCN-specific ATAC-Seq peaks overlapped with a TCF4 ChIP-Seq peak identified in both Gen2.2 and Cal-1 cells, whereas none of the AML-specific ATAC-Seq regions satisfied the same criteria (p value =1.34E-90; Figure 8E, green). This striking overlap suggests that TCF4 plays a dominant role in shaping the regulatory and phenotypic differences between BPDCN and AML.
DISCUSSION
BPDCN is an aggressive malignancy with no curative therapeutic options outside of allogeneic bone marrow transplantation (Riaz et al., 2014). Using functional genomics coupled with chemical screening we have uncovered essential survival pathways in BPDCN that are amenable to therapeutic attack. RNAi screening identified TCF4 as a master transcriptional regulator that sustains BPDCN viability. The RNAi screen also uncovered the essential role of the BET protein BRD4 in BPDCN cells, which was consistent with the sensitivity of BPDCN cells to BETi's in the small molecule screen. We were able to link these two observations mechanistically by showing that BRD4 is required for TCF4 expression and that ectopic TCF4 expression blunts the toxic effect of BETi's in BPDCN cells. BET inhibition also attenuated tumor growth in two BPDCN xenograft models, suggesting a targeted option for the therapy of this lethal cancer.
The E-box transcription factor TCF4 is an established determinant of normal pDC development (Cisse et al., 2008; Ghosh et al., 2010). We identified TCF4 targets in BPDCN by combined analysis of TCF4 ChIP-Seq data and gene expression changes following TCF4 knockdown. These analyses confirmed the similarity between pDCs, BPDCN lines and primary BPDCN, and readily distinguished BPDCN from cDCs and AML lines. In parallel, we defined regions of open chromatin by ATAC-Seq and showed that pDCs resembled both BPDCN lines and a primary BPDCN patient sample. Importantly, the vast majority of these BPDCN-specific regions of open chromatin overlapped with peaks of TCF4 binding, raising the possibility that TCF4 acts as a pioneer factor to establish key aspects of the regulomes of BPDCN and normal pDCs. Nonetheless, TCF4 function appears to be somewhat different in BPDCN and pDCs. A subset of TCF4 target genes was more highly expressed in BPDCN than in pDCs, including proto-oncogenes such as BCL2, TCL1A/B and MYC. Conversely, the TCF4 target genes that were more highly expressed in pDCs than in BPDCN included several regulators of pDC function such as BCL11A, SPIB, IL3RA and CLEC4C. Thus, the pDC-specifying function of TCF4 is attenuated in BPDCN in favor of oncogenic gene expression programs. This fits the lineage survival oncogene paradigm in which transcriptional programs inherited from the cell-of-origin are rewired to execute the malignant programs of cancer cells (Garraway and Sellers, 2006).
TCF4 addiction in BPDCN has important clinical implications, both for diagnosis and for therapy. A key to the deployment of targeted agents for cancer treatment is an accurate molecular diagnosis, and this remains a challenge in BPDCN and in the myeloid cancers that can resemble BPDCN histologically (Facchetti et al., 2016). (Assaf et al., 2007; Munoz et al., 2001). In this study, we developed a TCF4 antibody that effectively detected TCF4 protein expression in FFPE biopsy samples. This IHC assay was positive in 85% of cases with a documented diagnosis of BPDCN and conversely, TCF4 was not detected in myeloid BPDCN-mimics, such as cutaneous AML.
Unexpectedly, 4 BPDCN cases scored negative in the TCF4 IHC. Two of these were also weak or negative for bona-fide BPDCN markers such as CD4 and CD123, suggesting that these cases may have been misdiagnosed using histopathological current methods. Finally, 3 of 13 (23%) unclassified primitive hematopoietic malignancies were strongly positive in the TCF4 IHC assay, suggesting that they may be BPDCN cases that could not be diagnosed using current methods. Indeed, one of these cases clustered with primary BPDCN samples based on RNA-Seq expression of TCF4 targets. Altogether, we suggest that TCF4 IHC could improve the routine histopathological diagnosis of BPDCN.
Although initial chemotherapy can produce complete remissions in BPDCN, the cancer typically recurs quickly and the majority of patients succumb, highlighting the need for new treatment options (Pagano et al., 2013; Riaz et al., 2014). Our work addresses this challenge by identifying BETi's as a drug class with therapeutic potential in BPDCN. Three structurally distinct BETi's were toxic for BPDCN cells in our small molecule screen, which fits well with the genetic dependency of these cells on BRD4. Several strands of evidence point to TCF4 as the major downstream target of BETi's BPDCN cells. First, BETi's downregulated several pDC-related genes that are direct targets of TCF4. Second, TCF4 itself was strongly downregulated by BETi's, likely due to the fact that that the TCF4 locus contains a BPDCN-specific SE that is bound by BRD4. Third, TCF4 localized to the majority of SEs in BPDCN, and TCF4 knockdown reduced the expression of these SE-regulated genes. Finally, and crucially, ectopic TCF4 expression was sufficient to rescue BPDCN cells from the toxicity of BETi's, underscoring the functionally dominant, master regulatory role of TCF4 in BPDCN.
Like most cancers, BPDCN tumors acquire a variety of genetic alterations (Alayed et al., 2013; Dijkman et al., 2007; Jardin et al., 2009; Jardin et al., 2011; Lucioni et al., 2011; Menezes et al., 2014; Sapienza et al., 2014; Stenzinger et al., 2014), which could confound agents targeting any one dysregulated pathway. The attractive feature of lineage survival oncogenes such as TCF4 is that therapies targeting this class of factors are likely to cross boundaries of genetic subtypes. Moreover, therapies blocking oncogenic signaling pathways frequently select for the outgrowth of rare subpopulations with compensatory mechanisms that circumvent the targeted agent. By contrast, we show that TCF4 controls a pleiotropic network of target genes in BPDCN, making it less likely that a genetic alteration targeting a single TCF4 target could overcome BETi toxicity. For example, MYC is a direct TCF4 target in BPDCN, but deliberate overexpression of MYC failed to rescue BPDCN cells from BETi-induced death. In all, our genomic and chemical investigations of BPDCN provide a strong mechanistic rationale for the clinical evaluation of BETi's in this lethal cancer that is inadequately treated by conventional chemotherapy and lacks alternative targeted therapies.
EXPERIMENTAL PROCEDURES
Cell lines
Cal-1, CCRF-CEM, Jurkat, HL-60, MOLM-14 and SKM-1 cells were cultured in RPMI-1640 medium supplemented with penicillin/streptomycin and 10% fetal bovine serum (Tet tested, Atlanta Biologicals). Gen2.2 cells were cultured in RPMI 1640 medium supplemented with non-essential amino acids, sodium pyruvate, penicillin/streptomycin and 10% fetal bovine serum. The mouse MS-5 stromal feeder cell line was used to support growth of Gen2.2 cells.
shRNA library toxicity screening
The shRNA library toxicity screening was performed as previously described (Ngo et al., 2006). Briefly, pools of 1,000 shRNAs were used to retrovirally transduce Cal-1 cells. After puromycin selection, shRNA expression was induced by doxycycline (50 ng/ml). Un-induced cultures were kept in parallel as a control. After 3 weeks, genomic DNA from both un-induced and induced cultures was harvested and depletion of specific shRNAs was quantitatively assessed by next generation sequencing.
Gene expression profiling
For gene expression profiling, total RNA was extracted with TRIzol (Invitrogen) and subsequently cleaned-up with RNeasy mini columns (Qiagen). Gene-expression profiling was performed using two-color human Agilent gene-expression arrays, according to manufacturer’s instructions.
RNA-Seq
For RNA-Seq on the BPDCN cell lines Cal-1 and Gen2.2, total RNA was extracted with the AllPrep DNA/RNA kit (QIAGEN) and libraries were generated with the TruSeq RNA Sample Prep Kit-v2 (Illumina) according to the manufacturer’s instruction. Pair-end sequencing was performed on a HiSeq 2000 sequencer with v3 sequencing reagents (2×101bp reads). For RNA-Seq on the FFPE samples, RNA was extracted with the AllPrep DNA/RNA FFPE Kit (QIAGEN) and libraries were generated with the TruSeq RNA Access kit (Illumina) according to the manufacturer’s instruction. Pair-end sequencing was performed on a NextSeq 500 sequencer with v2 sequencing reagents.
ChIP-Seq
Chromatin immunoprecipitation was performed as described in the supplemental experimental procedures section. ChIP DNA was used to generate ChIP-Seq libraries with the NEXTflex™ Illumina ChIP-Seq Library Prep Kit (Bioo Scientific), according to manufacturer’s instructions. The sequencing of the TCF4 ChIP-Seqs was performed on a GA2x (Illumina) sequencer, with single reads (SR) of 36 bp length. The sequencing of all the other ChIP-Seqs was performed on the High-Output flow-cell of a NextSeq 500 sequencer (Illumina), with single reads (SR) of 76 bp length.
ATAC-Seq
ATAC-Seq was performed essentially as previously described (Buernostro et al.). Briefly, and 50,000 cells were pelleted and were resuspended in room-temperature lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.5% IGEPAL CA-630). Following centrifugation (10’, 500g), isolated nuclei were tagmented in 50 ul transposase reaction mix, for 30 min at 37°C (25 µl 2 × TD buffer, 10 µl transposase (NextEra DNA Library Prep Kit, Illumina) and 15 µl nuclease-free water). Following purification, the tagmented DNA was PCR amplified for 8–10 total cycles. ATAC-Seq libraries were sequenced on the High-Output Next-Seq flow cell (Illumina, 150 cycles, paired end 2 × 75 bp).
Accession numbers
All the genomic datasets have been deposited in GEO with the following accessions: Agilent gene expression: GSE75650. RNA-Seq gene expression: GSE84471.ChIP-Seq: GSE76147. ATAC-Seq: GSE84623.
High-throughput drug screen
The high-throughput drug screen was performed \as previously described (Ceribelli et al., 2014). The screening data have been deposited in PubChem.Cal-1 cells: AID 1224825; Gen2.2 cells: AID 1224824.
Xenograft experiments
All animal experiments were approved by the National Cancer Institute Animal Care and Use Committee (NCI ACUC) and were carried out in accordance with the NCI ACUC guidelines.
Primary samples
This study was approved by the Institutional Review Board (IRB) of the National Cancer Institute, Research Protocol 10-CN-074 C. All the primary samples were archival sample submitted for consultation to the Hematopathology Section of the National Cancer Institute (NCI), National Institutes of Health (NIH). Based on the retrospective nature of the tissue samples, the above mentioned protocol has been approved for a waiver of informed consent by the NCI IRB.
Supplementary Material
SIGNIFICANCE.
We combined loss-of-function RNA interference screening and high-throughput drug toxicity screening to uncover therapeutic options for patients with BPDCN, who have a median overall survival of only ~1 year with current therapy. The E-box transcription factor TCF4 emerged as the master transcriptional regulator of the BPDCN oncogenic program, a finding that can be exploited for the accurate molecular diagnosis of BPDCN. TCF4 functions as a BPDCN lineage-survival oncogene that can be targeted by BETi drugs, supporting their clinical evaluation in this lethal cancer.
Acknowledgments
This research was supported by the Intramural Research Programs of the NIH, National Cancer Institute, Center for Cancer Research and the National Human Genome Research Institute; the Frederick National Laboratory for Cancer Research, National Institutes of Health including contract HHSN261200800001E and grant # U54CA143930; the Division of Preclinical Innovation, National Center for Advancing Translational Sciences; the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research; and by NIH grant AI072571 and the Feinberg Lymphoma Research Pilot award to B.R. We thank Kathleen Meyer for help with the GEO submission.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, M.C., S.P., C.J.T., B.R. and L.M.S.; Investigation, M.C. Z.R.H., P.N. K., K.G., M.O.E., S.P., A.L.S. and X.Z.; Formal Analysis, D.W.H, G.W., W.X. and R.G.; Resources, G.M, S.J.F., M.F., L.C., J.P., E.S.J., C.J.T., B.R. and L.M.S.; Writing, M.C., B.R. and L.M.S.; Visualization: M.C. and L.M.S.; Supervision, C.J.T, B.R. and L.M.S.
See Supplemental Information for all experimental procedures details.
REFERENCES
- Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563–3572. doi: 10.1182/blood-2013-01-451781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alayed K, Patel KP, Konoplev S, Singh RR, Routbort MJ, Reddy N, Pemmaraju N, Zhang L, Shaikh AA, Aladily TN, et al. TET2 mutations, myelodysplastic features, and a distinct immunoprofile characterize blastic plasmacytoid dendritic cell neoplasm in the bone marrow. American journal of hematology. 2013;88:1055–1061. doi: 10.1002/ajh.23567. [DOI] [PubMed] [Google Scholar]
- Assaf C, Gellrich S, Whittaker S, Robson A, Cerroni L, Massone C, Kerl H, Rose C, Chott A, Chimenti S, et al. CD56-positive haematological neoplasms of the skin: a multicentre study of the Cutaneous Lymphoma Project Group of the European Organisation for Research and Treatment of Cancer. J Clin Pathol. 2007;60:981–989. doi: 10.1136/jcp.2006.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–w208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, Li X, Smart NG, Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunin A, Sisirak V, Ghosh HS, Grajkowska LT, Hou ZE, Miron M, Yang C, Ceribelli M, Uetani N, Chaperot L, et al. Protein Tyrosine Phosphatase PTPRS Is an Inhibitory Receptor on Human and Murine Plasmacytoid Dendritic Cells. Immunity. 2015;43:277–288. doi: 10.1016/j.immuni.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, Mathews Griner LA, Guha R, Shinn P, Keller JM, et al. Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci U S A. 2014;111:11365–11370. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaperot L, Bendriss N, Manches O, Gressin R, Maynadie M, Trimoreau F, Orfeuvre H, Corront B, Feuillard J, Sotto JJ, et al. Identification of a leukemic counterpart of the plasmacytoid dendritic cells. Blood. 2001;97:3210–3217. doi: 10.1182/blood.v97.10.3210. [DOI] [PubMed] [Google Scholar]
- Chaperot L, Blum A, Manches O, Lui G, Angel J, Molens JP, Plumas J. Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J Immunol. 2006;176:248–255. doi: 10.4049/jimmunol.176.1.248. [DOI] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- Dijkman R, van Doorn R, Szuhai K, Willemze R, Vermeer MH, Tensen CP. Gene-expression profiling and array-based CGH classify CD4+CD56+ hematodermic neoplasm and cutaneous myelomonocytic leukemia as distinct disease entities. Blood. 2007;109:1720–1727. doi: 10.1182/blood-2006-04-018143. [DOI] [PubMed] [Google Scholar]
- Facchetti F, Cigognetti M, Fisogni S, Rossi G, Lonardi S, Vermi W. Neoplasms derived from plasmacytoid dendritic cells. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2016;29:98–111. doi: 10.1038/modpathol.2015.145. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnache-Ottou F, Feuillard J, Ferrand C, Biichle S, Trimoreau F, Seilles E, Salaun V, Garand R, Lepelley P, Maynadie M, et al. Extended diagnostic criteria for plasmacytoid dendritic cell leukaemia. Br J Haematol. 2009;145:624–636. doi: 10.1111/j.1365-2141.2009.07679.x. [DOI] [PubMed] [Google Scholar]
- Garnache-Ottou F, Feuillard J, Saas P. Plasmacytoid dendritic cell leukaemia/lymphoma: towards a well defined entity? Br J Haematol. 2007;136:539–548. doi: 10.1111/j.1365-2141.2006.06458.x. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Ghosh HS, Ceribelli M, Matos I, Lazarovici A, Bussemaker HJ, Lasorella A, Hiebert SW, Liu K, Staudt LM, Reizis B. ETO family protein Mtg16 regulates the balance of dendritic cell subsets by repressing Id2. J Exp Med. 2014;211:1623–1635. doi: 10.1084/jem.20132121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herling M, Teitell MA, Shen RR, Medeiros LJ, Jones D. TCL1 expression in plasmacytoid dendritic cells (DC2s) and the related CD4+ CD56+ blastic tumors of skin. Blood. 2003;101:5007–5009. doi: 10.1182/blood-2002-10-3297. [DOI] [PubMed] [Google Scholar]
- Ippolito GC, Dekker JD, Wang YH, Lee BK, Shaffer AL, 3rd, Lin J, Wall JK, Lee BS, Staudt LM, Liu YJ, et al. Dendritic cell fate is determined by BCL11A. Proc Natl Acad Sci U S A. 2014;111:E998–E1006. doi: 10.1073/pnas.1319228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jardin F, Callanan M, Penther D, Ruminy P, Troussard X, Kerckaert JP, Figeac M, Parmentier F, Rainville V, Vaida I, et al. Recurrent genomic aberrations combined with deletions of various tumour suppressor genes may deregulate the G1/S transition in CD4+CD56+ haematodermic neoplasms and contribute to the aggressiveness of the disease. Leukemia. 2009;23:698–707. doi: 10.1038/leu.2008.359. [DOI] [PubMed] [Google Scholar]
- Jardin F, Ruminy P, Parmentier F, Troussard X, Vaida I, Stamatoullas A, Lepretre S, Penther D, Duval AB, Picquenot JM, et al. TET2 and TP53 mutations are frequently observed in blastic plasmacytoid dendritic cell neoplasm. Br J Haematol. 2011;153:413–416. doi: 10.1111/j.1365-2141.2010.08556.x. [DOI] [PubMed] [Google Scholar]
- Jaye DL, Geigerman CM, Herling M, Eastburn K, Waller EK, Jones D. Expression of the plasmacytoid dendritic cell marker BDCA-2 supports a spectrum of maturation among CD4+ CD56+ hematodermic neoplasms. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2006;19:1555–1562. doi: 10.1038/modpathol.3800679. [DOI] [PubMed] [Google Scholar]
- Julia F, Petrella T, Beylot-Barry M, Bagot M, Lipsker D, Machet L, Joly P, Dereure O, Wetterwald M, d'Incan M, et al. Blastic plasmacytoid dendritic cell neoplasm: clinical features in 90 patients. The British journal of dermatology. 2013;169:579–586. doi: 10.1111/bjd.12412. [DOI] [PubMed] [Google Scholar]
- Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005;175:4839–4846. doi: 10.4049/jimmunol.175.8.4839. [DOI] [PubMed] [Google Scholar]
- Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucioni M, Novara F, Fiandrino G, Riboni R, Fanoni D, Arra M, Venegoni L, Nicola M, Dallera E, Arcaini L, et al. Twenty-one cases of blastic plasmacytoid dendritic cell neoplasm: focus on biallelic locus 9p21.3 deletion. Blood. 2011;118:4591–4594. doi: 10.1182/blood-2011-03-337501. [DOI] [PubMed] [Google Scholar]
- Maeda T, Murata K, Fukushima T, Sugahara K, Tsuruda K, Anami M, Onimaru Y, Tsukasaki K, Tomonaga M, Moriuchi R, et al. A novel plasmacytoid dendritic cell line, CAL-1, established from a patient with blastic natural killer cell lymphoma. Int J Hematol. 2005;81:148–154. doi: 10.1532/ijh97.04116. [DOI] [PubMed] [Google Scholar]
- Marafioti T, Paterson JC, Ballabio E, Reichard KK, Tedoldi S, Hollowood K, Dictor M, Hansmann ML, Pileri SA, Dyer MJ, et al. Novel markers of normal and neoplastic human plasmacytoid dendritic cells. Blood. 2008;111:3778–3792. doi: 10.1182/blood-2007-10-117531. [DOI] [PubMed] [Google Scholar]
- Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014;111:2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes J, Acquadro F, Wiseman M, Gomez-Lopez G, Salgado RN, Talavera-Casanas JG, Buno I, Cervera JV, Montes-Moreno S, Hernandez-Rivas JM, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2014;28:823–829. doi: 10.1038/leu.2013.283. [DOI] [PubMed] [Google Scholar]
- Montes-Moreno S, Ramos-Medina R, Martinez-Lopez A, Barrionuevo Cornejo C, Parra Cubillos A, Quintana-Truyenque S, Rodriguez Pinilla SM, Pajares R, Sanchez-Verde L, Martinez-Torrecuadrada J, et al. SPIB, a novel immunohistochemical marker for human blastic plasmacytoid dendritic cell neoplasms: characterization of its expression in major hematolymphoid neoplasms. Blood. 2013;121:643–647. doi: 10.1182/blood-2012-08-447599. [DOI] [PubMed] [Google Scholar]
- Munoz L, Nomdedeu JF, Lopez O, Carnicer MJ, Bellido M, Aventin A, Brunet S, Sierra J. Interleukin-3 receptor alpha chain (CD123) is widely expressed in hematologic malignancies. Haematologica. 2001;86:1261–1269. [PubMed] [Google Scholar]
- Nagasawa M, Schmidlin H, Hazekamp MG, Schotte R, Blom B. Development of human plasmacytoid dendritic cells depends on the combined action of the basic helix-loop-helix factor E2-2 and the Ets factor Spi-B. Eur J Immunol. 2008;38:2389–2400. doi: 10.1002/eji.200838470. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Pagano L, Valentini CG, Pulsoni A, Fisogni S, Carluccio P, Mannelli F, Lunghi M, Pica G, Onida F, Cattaneo C, et al. Blastic plasmacytoid dendritic cell neoplasm with leukemic presentation: an Italian multicenter study. Haematologica. 2013;98:239–246. doi: 10.3324/haematol.2012.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella T, Comeau MR, Maynadie M, Couillault G, De Muret A, Maliszewski CR, Dalac S, Durlach A, Galibert L. 'Agranular CD4+ CD56+ hematodermic neoplasm' (blastic NK-cell lymphoma) originates from a population of CD56+ precursor cells related to plasmacytoid monocytes. The American journal of surgical pathology. 2002;26:852–862. doi: 10.1097/00000478-200207000-00003. [DOI] [PubMed] [Google Scholar]
- Reizis B. Regulation of plasmacytoid dendritic cell development. Curr Opin Immunol. 2010;22:206–211. doi: 10.1016/j.coi.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz W, Zhang L, Horna P, Sokol L. Blastic plasmacytoid dendritic cell neoplasm: update on molecular biology, diagnosis, and therapy. Cancer control : journal of the Moffitt Cancer Center. 2014;21:279–289. doi: 10.1177/107327481402100404. [DOI] [PubMed] [Google Scholar]
- Roos-Weil D, Dietrich S, Boumendil A, Polge E, Bron D, Carreras E, Iriondo Atienza A, Arcese W, Beelen DW, Cornelissen JJ, et al. Stem cell transplantation can provide durable disease control in blastic plasmacytoid dendritic cell neoplasm: a retrospective study from the European Group for Blood and Marrow Transplantation. Blood. 2013;121:440–446. doi: 10.1182/blood-2012-08-448613. [DOI] [PubMed] [Google Scholar]
- Rui L, Schmitz R, Ceribelli M, Staudt LM. Malignant pirates of the immune system. Nat Immunol. 2011;12:933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- Swerdlow S, C E, Lee Harris N, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue. 2008 [Google Scholar]
- Sapienza MR, Fuligni F, Agostinelli C, Tripodo C, Righi S, Laginestra MA, Pileri A, Jr, Mancini M, Rossi M, Ricci F, et al. Molecular profiling of blastic plasmacytoid dendritic cell neoplasm reveals a unique pattern and suggests selective sensitivity to NF-kB pathway inhibition. Leukemia. 2014;28:1606–1616. doi: 10.1038/leu.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I, Hoshino K, Sugiyama T, Yamazaki C, Yano T, Iizuka A, Hemmi H, Tanaka T, Saito M, Sugiyama M, et al. Spi-B is critical for plasmacytoid dendritic cell function and development. Blood. 2012;120:4733–4743. doi: 10.1182/blood-2012-06-436527. [DOI] [PubMed] [Google Scholar]
- Sawai CM, Sisirak V, Ghosh HS, Hou EZ, Ceribelli M, Staudt LM, Reizis B. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J Exp Med. 2013;210:2151–2159. doi: 10.1084/jem.20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC, 3rd, Belardelli F, Gabriele L. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte R, Nagasawa M, Weijer K, Spits H, Blom B. The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J Exp Med. 2004;200:1503–1509. doi: 10.1084/jem.20041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, Lam LT, Davis RE, Staudt LM. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunological reviews. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Spits H, Couwenberg F, Bakker AQ, Weijer K, Uittenbogaart CH. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre- DC)2 but not into pre-DC1. Evidence for a lymphoid origin of pre-DC2. J Exp Med. 2000;192:1775–1784. doi: 10.1084/jem.192.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzinger A, Endris V, Pfarr N, Andrulis M, Johrens K, Klauschen F, Siebolts U, Wolf T, Koch PS, Schulz M, et al. Targeted ultra-deep sequencing reveals recurrent and mutually exclusive mutations of cancer genes in blastic plasmacytoid dendritic cell neoplasm. Oncotarget. 2014;5:6404–6413. doi: 10.18632/oncotarget.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura H, Tamura T, Gongora C, Aliberti J, Reis e Sousa C, Sher A, Ozato K. ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood. 2003;101:961–969. doi: 10.1182/blood-2002-05-1327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.