Abstract
The K15 capsule determinant of uropathogenic Escherichia coli strain 536 (O6:K15:H31) is part of a novel 79.6-kb pathogenicity island (PAI) designated PAI V536 that is absent from the genome of nonpathogenic E. coli K-12 strain MG1655. PAI V536 shows typical characteristics of a composite PAI that is associated with the pheV tRNA gene and contains the pix fimbriae determinant as well as genes coding for a putative phosphoglycerate transport system, an autotransporter protein, and hypothetical open reading frames. A gene cluster coding for a putative general secretion pathway system, together with a kpsK15 determinant, is localized downstream of a truncated pheV gene (′pheV) also present in this chromosomal region. The distribution of genes present on PAI V536 was studied by PCR in different pathogenic and nonpathogenic E. coli isolates of various sources. Analysis of the 20-kb kps locus revealed a so far unknown genetic organization. Generally, the kpsK15 gene cluster resembles that of group 2 and 3 capsules, where two conserved regions (regions 1 and 3) are located up- or downstream of a highly variable serotype-specific region (region 2). Interestingly, recombination of a group 2 and 3 determinant may have been involved in the evolution of the K15 capsule-encoding gene cluster. Expression of the K15 capsule is important for virulence in a murine model of ascending urinary tract infection but not for serum resistance of E. coli strain 536.
The production of extracellular polysaccharides is a common feature of many bacteria (58). Polysaccharides are also components of the bacterial capsule, which often constitutes the outermost layer of the cell and mediates the interaction between the bacterium and its immediate environment. Additionally, capsules play a crucial role for the survival of bacteria in hostile environments. More than 80 chemically and serologically distinct capsules have been described for Escherichia coli (25). According to their genetic organization, regulation of expression, and biosynthetic mechanism, four capsule groups (groups 1 to 4) have been distinguished (57).
Group 1 and 4 capsules resemble those expressed by Klebsiella and Erwinia spp., and their determinants are located within a highly polymorphic region at 45 min of the E. coli K-12 chromosome, which contains genes required for biosynthesis of a number of cell surface-associated polysaccharides (6).
The organization of gene clusters coding for group 2 and 3 capsules resembles those of Neisseria meningitidis and Haemophilus influenzae, respectively. They exhibit a conserved modular genetic organization consisting of three regions (Fig. 1). A highly variable antigen-specific region (region 2) that encodes the enzymes required for polysaccharide biosynthesis is flanked by two conserved regions (region 1 and 3) coding for proteins involved in the secretion of capsule components. Translocation across the cytoplasmic membrane is mediated by the KpsC, -M, -S, and -T proteins, whereas translocation across the periplasm and the outer membrane involves the KpsD and -E proteins (6, 8, 45, 57). In the case of group 2 capsules, two additional genes, kpsF and kpsU, belong to the gene cluster. Whereas the role of KpsF is unknown, the KpsU protein is a CMP-3-keto-3-deoxy-mannooctulosonate synthetase, which is a ligase necessary for successful translocation of sugars through the inner membrane (45). The majority of the known group 2 and 3 capsule-encoding gene clusters are located within the genome at a position corresponding to 64 min of the E. coli K-12 chromosome and are associated with the tRNA genes serA and pheV, respectively (41, 42, 55). tRNA loci frequently serve as chromosomal insertion sites of pathogenicity islands (PAIs). This type of genetic element has been recently reviewed (20, 21). For uropathogenic E. coli (UPEC) strain 536 (O6:K15:H31), the genetic organization and gene content of four PAIs have been characterized in detail. Important virulence factors of UPEC are encoded on these PAIs, e.g., alpha-hemolysin, P- and S-fimbrial adhesins, and the siderophore system yersiniabactin (16). In addition to these PAI-borne virulence-associated genes, smaller chromosomal DNA regions within the genome of strain 536 have been characterized that code for virulence-associated factors that are also important for pathogenesis, e.g., the hemin receptor (ChuA) (38), the enterobactin siderophore system, and a lipopolysaccharide (LPS) of serogroup O6. The K15 capsule locus of strain 536, however, has not been characterized so far.
FIG. 1.
Structural similarities between the K15 capsule determinant and group 2 and 3 capsule gene clusters. (A) Genetic organization of group 2 and 3 capsule gene clusters. Two conserved regions (regions 1 and 3) encode enzymes required for capsule transport through the inner membrane and are localized up- and downstream of a highly variable serotype-specific region (region 2). (B) In UPEC strain 536, the genes kpsFEDUC′K5 of region 1 are highly homologous to region 1 of group 2 capsules, whereas the kpsS and kpsCK10 genes exhibit a higher homology to the corresponding genes of group 3 capsules. The nonfunctional ORF65 that exhibits a high homology to transposase-encoding ORFs and interrupts region 1 is indicated by a hatched arrow.
To identify so far unknown genomic regions of UPEC strain 536 which are absent in nonpathogenic E. coli strain MG1655, the suppression subtractive hybridization (SSH) approach has been used before (24). One of the SSH fragments obtained showed homology to the capsule export protein-encoding gene kpsC of K10 capsules. Southern blot hybridization of genomic fragments of UPEC strain 536 separated by pulsed-field gel electrophoresis with a kpsCK10-specific probe indicated that this SSH fragment was associated with the pheV tRNA-encoding gene. The fact that PAIs carrying capsule gene clusters have also been mapped near the tRNA genes serA and pheV in other E. coli strains (6, 11, 52) suggested that the K15 capsule determinant of strain 536 was localized on a PAI, designated PAI V536, as well. One aim of this study was the characterization and sequence determination of this PAI. Beside the structural analysis, functional studies have also been performed to elucidate the role of PAI V536 and the K15 capsule in uropathogenesis.
In addition, the prevalence of PAI V536-specific sequences was investigated with different pathogenic and nonpathogenic E. coli strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A collection of 136 E. coli strains was used in this study and includes the so-called IMIB (Institut für Molekulare Infektionsbiologie) collection which has already been used for the investigation of the distribution of PAI I536 to IV536 of UPEC strain 536 (15, 16). Bacterial mutants, plasmids, and cosmids used or constructed in this study are listed in Table 1. The strains were grown in Luria-Bertani (LB) broth (49). When necessary, the antibiotics ampicillin (100 μg/ml), tetracycline (5 μg/ml), kanamycin (40 μg/ml), chloramphenicol (20 μg/ml), and streptomycin (100 μg/ml) or sucrose (7% wt/vol) were added to the medium at the concentrations indicated in parentheses.
TABLE 1.
Mutants, plasmids, and cosmids used in this study
| Bacterial strain, cosmid, or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Bacterial strains | ||
| 536 | UPEC isolate (O6:K15:H31), Smr | 7 |
| 536ΔkpsSC′K5 | 536, partial deletion of capsule region 1, Smr, Kmr | This study |
| 536ΔR2 | 536, deletion of capsule region 2 mutant, Smr, Cmr | This study |
| 536ΔkpsMT | 536, deletion of capsule region 3 mutant, Smr, Cmr | This study |
| 536ΔkpsC′K5 | 536, deletion of kpsC′K5, Smr, Cmr | This study |
| 536ΔkpsCK10 | 536, deletion of kpsCK10, Smr, Cmr | This study |
| 536ΔkpsK15 | 536, deletion of the complete K15 kps locus, Smr, Cmr | This study |
| 536Δgsp | 536, deletion of gspLKJIHGFED genes, Smr, Cmr | This study |
| IMI402 | Enterotoxigenic E. coli isolate 34344f (O6:K15:H−) | IMIB strain collection |
| IMI403 | Enterotoxigenic E. coli isolate C9221a (O6:K15:H16) | 15 |
| IMI415 | E. coli isolate E642 (O6:K15) | IMIB strain collection |
| SM10λpir | thi-1 thr-1 leuB6 supE44 tonA21 lacY1 recA::RP4-2-Tc::Mu Kmr | 35 |
| SY327 | F−araD Δ(lac pro) argE (Am) recA56 RifrnalA λpir | 35 |
| Plasmids and cosmids | ||
| pKD46 | Arabinose-inducible λ Red recombinase expression plasmid (oriR101 repA101ts ParaB-gam-bet-exo Apr) | 14 |
| pKD3 | Template plasmid for amplification of a cat cassette flanked by FLP recognition sites (FRT) (oriRγ, Apr Cmr) | 14 |
| pKD4 | Template plasmid for amplification of a kan cassette flanked by FLP recognition sites (FRT) (oriRγ Apr Kanr) | 14 |
| pCP20 | Plasmid with temperature-sensitive replication for temporal production of the FLP recombinase (FLP+λ c1857− λ pR Repts Cmr Apr) | 10 |
| pBeloBAC11 | Bacterial artificial chromosome cloning vector | 29 |
| pCVD442 | Suicide vector for gene inactivation (sacB ori ColE1 Apr) | 18 |
| pCos6b38 | Cosmid clone harboring partial sequences of PAI V536, Apr | 24 |
| pCos25/52 | Cosmid clone harboring partial sequences of PAI V536, Apr | This study |
| pCos24/23 | Cosmid clone harboring partial sequences of PAI V536, Apr | This study |
| pCos2/13 | Cosmid clone harboring downstream sequences of PAI V536, Apr | This study |
DNA techniques.
Qiagen products (Hilden, Germany) were used to isolate plasmid and cosmid DNA and to purify DNA fragments or PCR products. For cloning experiments, the Expand Long template PCR system (Roche Diagnostics, Mannheim, Germany) and the Dap Goldstar DNA polymerase (Eurogentec, Seraing, Belgium) were used. For the screening of bacterial strains or the genomic cosmid library of strain 536 and for the verification of mutants, Taq DNA polymerase (Qiagen) was used. Primers were obtained from Sigma-Genosys (Taufkirchen, Germany) while restriction enzymes were purchased from New England Biolabs (Frankfurt am Main, Germany) or Promega (Mannheim, Germany). For Southern blot and colony blot hybridization, DNA or bacterial colonies were transferred to Nytran Supercharge nylon membranes (Schleicher & Schuell BioSciences, Dassel, Germany). Hybridization with and detection of horseradish peroxidase-labeled probes was performed with the ECL labeling and signal detection system (Amersham Biosciences, Freiburg, Germany). Inactivation of PAI V536-localized genes was carried out by allelic exchange by using the suicide vector pCVD442 as described before (35, 36) or by using linear DNA fragments as previously described (14). The complete list of primers used for gene inactivation is available as supplemental material at the homepage of the Enterobacteria research group of the Institute for Molecular Biology of Infectious Diseases (http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm).
RNA techniques.
A small quantity of total RNA was isolated from E. coli by using the RNeasy purification kit (Qiagen). Large quantities of total RNA were extracted from 10 ml of bacterial cultures which were immediately mixed with an equal volume of crushed ice and harvested by centrifugation at 4°C. The cells were resuspended in 0.6 ml of an ice-cold buffer containing 10 mM KCl, 5 mM MgCl2, and 10 mM Tris (pH 7.4) and then immediately added to 0.6 ml of hot lysis buffer (0.4 M NaCl, 40 ml EDTA, 1% sodium dodecyl sulfate) containing 1% β-mercaptoethanol and 100 μl of water-saturated phenol. This mixture was incubated at 100°C for 40 s. Cell debris was removed by centrifugation for 10 min at room temperature, and the supernatant was then extracted five times with phenol-chloroform, ethanol precipitated, washed, and dried. The dried RNA pellet was dissolved in sterile diethyl pyrocarbonate-treated water.
For reverse transcription-PCR (RT-PCR), 15 μg of RNA was digested with 0.5 U of RNase-free DNase I (Roche Diagnostics)/μl for 1 h at 37°C. After purification with RNeasy mini columns (QIAGEN), 2 to 5 μg of RNA was mixed with 3 μg of random hexamer primers (Amersham Biosciences) and heated for 10 min at 70°C. After cooling down on ice for 5 min, cDNA synthesis was performed with the Super Script II RNase H reverse transcriptase system (Gibco BRL, Eggenstein, Germany) at 46°C. After 1 h, the reaction was stopped by alkaline hydrolysis of RNA with 0.25 M NaOH (15 min, 65°C). The neutral pH of the sample was restored by the addition of 50 mM Tris-HCl (pH 7.5) before the cDNA was used as a template for PCRs.
DNA sequencing and sequence analysis.
Overlapping clones of a genomic cosmid library were screened by PCR and colony blot hybridization. Cosmid clones (pCos6b38, pCos24/23, pCos25/52, and pCos2/13) covering the entire PAI V536 and its flanking regions of E. coli strain 536 were sequenced as follows. Small insert libraries (2 to 2.5 kb) were generated by mechanical shearing of cosmid DNA (40). After end repair with T4 polymerase, the fragments were ligated into the prepared pTZ19R vector. Isolated plasmids were sequenced from both ends by using dye-terminator chemistry and analyzed on ABI-377 automated DNA sequencers (Applied Biosystems, Weiterstadt, Germany). After assembly, the remaining gaps were closed by primer walking on the plasmid clones. The Phrap software implemented in the STADEN software package was used for assembling and editing the sequence data (51). The nucleotide sequence of the complete PAI V536 was submitted to the EMBL database. Homology searches were performed with the BLASTN, BLASTX, and PSI- and PHI-BLAST programs of the National Center for Biotechnology Information (3). The percent G+C plot of the kpsK15 determinant was calculated with a window size of 150 bp with VectorNTI (InforMax Inc., Frederick, Md.).
Production of an antiserum directed against the K15 capsule.
Serum against the K15 capsule was produced according to the method described by Kiesewalter and Seltmann (28). Briefly, bacteria (E. coli strain 536) were incubated overnight in 50% ethanol at 4°C to denature protein antigens. After washing, bacteria were diluted to 2 × 108 CFU/ml in phosphate-buffered saline (PBS). Rabbits were immunized with 0.2, 0.5, 1.0, and 2.0 (twice) ml of bacterial suspensions at 4-day intervals. Four days following the last inoculation, serum samples were taken and tested by tube agglutination. Afterwards, the rabbits were sacrificed. Their sera were collected and diluted 1:10 in sterile saline containing 0.3% phenol as a preservative. The removal of nonspecific antibodies (e.g., against LPS or flagellar sequential epitopes) was achieved by absorption with live and boiled cells of the encapsulated but K15 capsule-negative UPEC strain RZ532 (O6:K+:H31). In brief, a 0.5-g aliquot of wet bacteria was suspended in 1 ml of the diluted serum and the suspension was incubated for 1 h at 37°C and overnight at 4°C. The next day, bacterial cells were harvested, resuspended in saline, and boiled for 1 h. Following centrifugation, the pellet was resuspended in the same serum and suspensions were incubated again for 1 h at 37°C and subsequently overnight at 4°C. After centrifugation, aliquots of serum were stored at −20°C. Due to this procedure, the resulting sera exhibited only minimal immunogenicity to minor protein antigens. The efficient detection of the K15 antigen was proven by enzyme-linked immunosorbent assay (ELISA).
ELISA.
96-well plates (CML-CEB, Nemours, France) were coated overnight with 0.2 ml of bacterial suspension (109 CFU/ml) at 4°C. The following day, plates were washed with PBS plus 0.5% Tween 20 (washing buffer) and then blocked with PBS containing 2% bovine serum albumin (Sigma) for 1 h at 37°C. Anti-K15 serum was diluted in PBS plus 0.5% bovine serum albumin and incubated with antigen-coated plates for 90 min. Serial dilutions were conducted across the plates. After three washes, plates were probed with commercial anti-rabbit immunoglobulin conjugated with horseradish peroxidase (Dako) dissolved in citric acid buffer containing H2O2. The optical density was measured at 490 nm in a conventional ELISA plate reader. Duplicates were used for each strain and dilution, and assays were repeated twice.
Immunofluorescence microscopy.
One milliliter of overnight cultures was washed once with saline and resuspended in water, and 20 μl was dropped on a glass slide. After drying (37°C for 20 min), a 1:4 dilution of the absorbed K15 antiserum was added. After 1 h of incubation at 37°C, the glass slide was carefully washed twice with PBS. Texas Red-labeled anti-rabbit immunoglobulin G (Dianova, Hamburg, Germany) was added, and the slide was incubated for 1 h at 37°C. After incubation, the slides were washed in PBS and analyzed by epifluorescence microscopy.
Serum bactericidal test.
Bacteria grown in LB medium were washed with saline and diluted to 106 cells/ml. One hundred-microliter aliquots were mixed with an equal volume of fresh human serum and incubated at 37°C for 4 h in microtiter plates. Samples were taken at 0-, 0.5-, 1-, 2-, 3-, and 4-h time points. The viable cell count was determined by plating aliquots of the sample onto LB plates and overnight incubation at 37°C. Assays were performed with normal and heat-inactivated serum (56°C for 30 min). Triplicates were used for each strain, and every assay was performed 3 times.
Ascending urinary tract infection (UTI) model.
Intravesical infections of 2- to 3-day-old suckling CFLP mice (Gödöllö, Hungary) were performed as previously described (39). Six to 14 infant mice were injected per strain, and the assays were repeated four times. Bacteria were grown overnight at 37°C in LB medium, harvested by centrifugation, washed once with saline, and normalized to the required inoculum density (107/ml) in PBS. This bacterial suspension (0.025 ml) containing 0.05% Pontamin sky blue dye (Searle Pharmaceuticals, High Wybcombe, United Kingdom) was introduced into the bladder directly through the abdominal wall. The stain, which had no toxic or antibacterial effects, served as an indicator of successful inoculation. Mice that survived infection were sacrificed at 21 days postinfection. The bladder and both kidneys were removed and homogenized in PBS, and aliquots were plated onto agar plates containing a selective antibiotic. Colonies were counted after overnight incubation. Additionally, bacterial counts were determined from the urine and blood. To prove that in this model mortality is due to urosepsis originating from an ascending infection which starts from the bladder and reaches the blood system through the kidneys, 3-day-old mice were infected as described above with wild-type strain 536. At different time points postinfection (1, 2, 4, 6, 8, 10, 12, 18, and 24 h), 2 mice were sacrificed. Following sterilization of the skin with iodine solution, the heart, kidneys, and bladder were removed in this order and the organs were homogenized separately (in 2 ml of 0.9% NaCl) and plated onto LB agar plates containing streptomycin. Viable counts were determined following a 24-h incubation at 37°C. According to this experiment, bacteria were permanently detectable in growing numbers in the bladder. They could only be exceptionally detected in the kidneys earlier than 4 h postinfection. From this time point on, their number increased constantly. In the hearts (blood) of the sacrificed mice, bacteria appeared as late as 12 h postinfection. This is indicative of a primary ascending route of infection with a subsequent entry of bacteria into the bloodstream in this model system.
Nucleotide sequence accession number.
The nucleotide sequence of PAI V536 has been deposited in GenBank under accession number AJ617685.
RESULTS
Genetic features of PAI V536.
A 105,801-bp chromosomal region of E. coli strain 536 comprising the complete PAI V536 and its downstream region was subcloned and sequenced. The nucleotide sequence of PAI V536 is available under the GenBank accession number AJ617685. This PAI is associated with the tRNA-encoding gene pheV located at 64 min of the E. coli K-12 strain MG1655 chromosomal map (Fig. 2). A truncated imperfect copy of this tRNA gene (′pheV) which comprises the last 22 bp of this gene's 3′ end and lacks one G at position 71 of pheV is located 48,708 bp downstream of the intact pheV gene. Similarly to PAI III536, the pheV-associated PAI V536 represents a composite PAI, consisting of 58 open reading frames (ORFs) which are flanked by a 22-bp imperfect direct repeat (DR) structure (pheV and its truncated copy) as well as a 30,987-bp DNA region downstream of ′pheV that also contains the determinants required for K15 capsule biosynthesis and for a type II secretion system. The composite island is 79,695 bp in size and exhibits an overall G+C content of 47.7%. Transition into the chromosomal backbone shared with E. coli K-12 strain MG1655 and UPEC O6 strain CFT073 occurs again upstream of gene b2970. In addition, the conserved E. coli chromosomal backbone is interrupted 17,969 bp further downstream between the E. coli K-12 genes b2981 and b2983. The IS5 transposase-encoding gene (b2982) present in the E. coli K-12 strain MG1655 is replaced by an 8,418-bp DNA stretch described for UPEC strain CFT073 (GenBank accession no. AE016766) and Shigella flexneri 2a (GenBank accession no. AE016988 and AE015315) that contains genes required for glycolate utilization as well as several hypothetical ORFs.
FIG. 2.
Genetic structure of PAI V536 and its downstream region. Important PAI regions are indicated as shown at the bottom of the figure. The localization of tRNA-encoding genes is indicated as well as that of the DNA regions included into the PCR screening approach for the detection of PAI V536-specific sequences.
Several fragments of PAI V536 are highly homologous to chromosomal regions of other extraintestinal pathogenic E. coli (ExPEC) and intestinal pathogenic E. coli strains (e.g., strains CFT073, EDL933, and 239 KH 89), S. flexneri 2a (she and SRL PAIs), and Salmonella enterica serovar Typhimurium LT2. Moreover, PAI V536 also contains fragments found on different virulence plasmids of E. coli (pB171 and pSFO15) and Shigella spp. (pWR501 and pWR301) as well as on the Yersinia pestis plasmids pMT1 and pCD1. PAI V536 exhibits typical features of a PAI, e.g., a mosaic-like structure and the presence of multiple functional and fragmented mobile genetic elements. Several virulence-associated factors are encoded on this PAI, including the K15 capsule (ORF60 to 74), the recently described Pix fimbria-encoding gene cluster (ORF6 to 13) which belongs to the P adhesin family (33), and an autotransporter protein (ORF48) with the highest similarity to Sap, a putative autotransporter adhesin encoded on the she PAI of S. flexneri (GenBank accession no. AF326777 and AAL08472) (32). In addition to the corresponding virulence-associated determinants, a gene cluster (ORF16 to 19) is located on PAI V536 that codes for a putative phosphoglycerate transport system (pgt) also present in S. enterica serovar Typhimurium LT2 and other UPEC strains (19, 56). A complete list of the putative ORFs located on PAI V536 is available as supplemental material.
Transcriptional analysis of pgtC, pixB, and pixC expression by RT-PCR demonstrated that pgtC was transcribed under in vitro conditions (37°C, LB batch culture), as was pixB, a hypothetical negative regulator of the pix operon. Due to the expression of pixB, the structural genes of the pix operon were not transcribed, as no pixC transcript was detected by RT-PCR (data not shown).
Mobile genetic elements present on PAI V536.
Mobile genetic elements play an important role in recombination, resulting in genetic rearrangements and deletions. Insertion sequence (IS) elements and prophages are typical components of PAIs, and their high numbers indicate how frequently recombination occurs within PAIs. As a typical feature of PAIs, a P4-like bacteriophage integrase-encoding gene (ORF1) is located downstream of the pheV gene, the chromosomal insertion site of PAI V536. The same type of integrase gene is reported to be present on different PAIs integrated at the same tRNA-encoding gene of different E. coli strains (1, 30, 43, 53, 56). In addition, several putative transposase-encoding and other IS element-associated ORFs of IS2, IS21, IS100, IS110, and IS911, as well as the R6 transposase-encoding gene of PAI IICFT073, are present on PAI V536.
Characterization of the K15 capsule determinant of E. coli strain 536.
The kpsK15 gene cluster required for K15 capsule biosynthesis was localized about 1 kb downstream of ′pheV. The genetic organization of the capsule determinant of UPEC strain 536 combines features of both group 2 and 3 capsules (Fig. 1 and 3A). Region 1 of the kpsK15 gene cluster encodes enzymes required for the transport of capsule components and consists of five ORFs (kpsF, -E, -D, -U, and -C). This region is highly homologous to those of K1 and K5 capsule determinants. However, the kpsC gene (kpsC′K5) is truncated and the kpsS gene is missing relative to its K1 and K5 counterparts. Downstream of kpsC′K5, an inactivated putative ORF (ORF65) is localized that is highly similar to the putative ORF c1248 of UPEC strain CFT073, coding for a putative transposase (NP_753162).
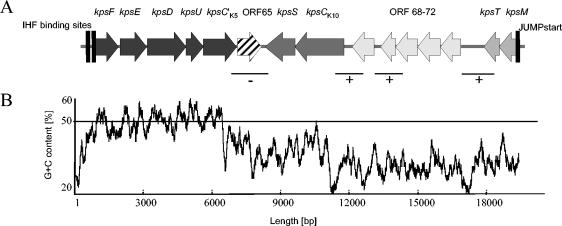
FIG. 3.
Genetic organization of the K15 capsule determinant. (A) Regions 1 to 3 of the kpsK15 gene cluster are indicated by different shades of gray. The nonfunctional ORF65 is indicated by a hatched arrow. The locations of two putative integration host factor (IHF) binding sites within the kpsF promoter region and that of the JUMPstart (just upstream of many polysaccharide-associated gene starts) region located upstream of kpsM are indicated, as are the DNA regions whose transcription has been studied by RT-PCR. Fractions of the kpsK15 gene cluster that could be amplified from the corresponding cDNA are marked with plus signs, and those whose transcripts were not detectable are marked with a minus sign. (B) Percent G+C plot of the kpsK15 gene cluster indicative of the segmental structure of this capsule locus representing DNA regions from different sources. The average percent G+C content of the E. coli chromosomal backbone (50.4%) is indicated by a horizontal line.
Interestingly, a kpsS gene and another complete kpsC gene were identified downstream of the nonfunctional ORF65 on the complementary strand. This kpsC gene (kpsCK10) was homologous to kpsC of the E. coli K10 capsule type, a well-studied representative of group 3 capsules (12). kpsS exhibited significant similarity to the corresponding gene of Aeromonas hydrophila (GenBank accession no. AF375657). Due to the nucleotide sequence homology and gene order of these two genes, this DNA stretch can be considered a fragment of region 3 of a group 3 capsule determinant. The presence of a nonfunctional putative transposase-encoding ORF between kpsC′K5 and kpsCK10 indicates that region 1 of the K15 capsule determinant resulted from recombination between a group 2 and 3 capsule gene cluster involving IS element(s). Upstream of kpsS and kpsCK10, region 2 of the K15 capsule locus was located, consisting of five ORFs (ORF68 to 72), without any homology on the nucleotide level. The deduced amino acid sequences of two genes (ORF71 and 72) were similar to those of a putative glycosyltransferase of Actinobacillus (AAC26630) and a mannosyltransferase B of Aquifex aeolicus strain VF5 (NP_213361), respectively. Region 3 of the K15 capsule determinant was located upstream of region 2. The encoded KpsM and KpsT proteins were shown to be highly homologous (>70% identity) to corresponding proteins of other group 2 capsules.
Analysis of the distribution of PAI V536-specific sequences among various E. coli strains from different sources.
To study the distribution of PAI V536-specific sequences among 137 nonpathogenic and pathogenic E. coli isolates as well as to find out whether an identical repertoire of genetic information can be detected in other E. coli strains, PCRs were designed for the amplification of selected regions of PAI V536. These regions include the pixC, pgtC, and gspD genes as well as parts of regions 1 to 3 of the kpsK15 determinant (Fig. 2). The complete list of PCR screening results is available as supplemental material at the homepage of the Enterobacteria research group of the Institute for Molecular Biology of Infectious Diseases (http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm).
A large group of ExPEC and intestinal pathogenic E. coli strains as well as nonpathogenic, commensal E. coli strains has been screened, including the strains of the IMIB collection. The results obtained (Table 2) demonstrate that, generally, PAI V536-specific sequences were distributed more frequently among ExPEC and, to a lesser extent, in commensal isolates than in diarrheagenic isolates. More than 50% of the ExPEC strains tested carried kpsCK5, indicative of group 2 capsule determinants, whereas kpsCK10, indicative of DNA regions of group 3 capsule-encoding gene clusters, has rarely been detected among the strains included in this study. Within this group of strains, only three strains including UPEC isolate 536 have been shown to carry the K15 capsule determinant. The general secretion pathway determinant, although more frequently distributed among ExPEC strains, was also detectable in at least 50% of the diarrheagenic and commensal isolates. These findings underline the notion that PAI-associated DNA regions are also detectable in commensal isolates, indicating that the commensal flora represents the natural reservoir for ExPEC strains.
TABLE 2.
Detection of PAI V536-specific sequences in various E. coli isolates
| Source or type of E. coli strain (n) | % Presence of PAI V536-specific region (gene):
|
||||||
|---|---|---|---|---|---|---|---|
| 1 (pixC) | 2 (pgtC) | 3 (kpsCK5) | 4 (kpsCK10) | 5 (ORF68 to 69) | 6 (kpsMT) | 7 (gspD) | |
| UTI strains (67) | 8.9 | 47.8a | 53.7 | 17.9 | 3.0 | 4.5 | 74.6 |
| Human and animal MNEC and SEPEC isolatesb (28) | 17.9 | 42.9 | 60.7a | 3.6 | 3.6 | 3.6 | 96.4a |
| Diarrheagenic E. coli pathogens (18) | 0 | 0a | 5.6a | 5.6 | 0 | 5.6 | 50 |
| Nonpathogenic strains (24) | 4.2 | 25.0 | 33.3 | 12.5 | 0 | 0 | 58.3 |
| Total (137) | 8.8 | 36.5 | 45.3 | 12.4 | 2.2 | 3.6 | 73.0 |
Significantly different from the frequency of occurence among nonpathogenic strains (chi-square probe, P < 0.05).
MNEC, meningitis-causing E. coli; SEPEC, sepsis-causing E. coli.
Transcription analysis of the K15 capsule-encoding locus.
To analyze the operon structure of the K15 capsule determinant, RT-PCRs were performed. Both kpsC variants (kpsC′K5 and kpsCK10) were shown to be transcribed (see Fig. 1 in the supplemental material). According to the amplification of cDNAs representing different parts of the K15 capsule determinant, it can be assumed that region 3, region 2, and kpsCK10 and kpsS of region 1 represent a single transcription unit (Fig. 3B). The JUMPstart motif (CAGTGTATTGGTAGCTGTTAAGCCAAGGGCGGTAGCGTA) including the ops element required for efficient transcription of long operons (5) is localized upstream of kpsM. It is already known that region 1 of group 2 capsules is organized as a single transcription unit (57). Consequently, the genes kpsF-kpsC′K5 of the K15 capsule gene cluster may also constitute one transcription unit. Two putative integration host factor binding sites have been detected upstream of kpsF.
Inactivation of the K15 gene cluster and identification of the functional kpsC gene.
To confirm that the detected putative kps determinant represents the functional K15 capsule-encoding gene cluster of strain 536, different mutants have been generated in which one of the regions of the kps determinant has been inactivated. For this purpose, the DNA regions comprising kpsS/kpsCK10, ORF1 to 5, or kpsMT have been deleted as well as the complete kpsK15 determinant of strain 536. Furthermore, to find out which kpsC gene was functional and required for expression of the K15 capsule determinant, either kpsC′K5 or kpsCK10 was inactivated. The mutants generated were tested for K15 capsule production by ELISA (Fig. 4). According to these experiments, partial deletion of one of the regions always resulted in a markedly reduced expression of the K15 capsule. Capsule expression of these mutants was comparable to that of UPEC strain RZ532 (O6:K+:H31), which does not express a K15 capsule, to that of the complete kpsK15 deletion mutant and of the strain 536 rfaH mutant, in which the transcriptional antiterminator required for kps expression has been inactivated (17, 39). Complementation of strain 536 ΔkpsK15 with a bacterial artificial chromosome clone carrying the complete kpsK15 gene cluster restored capsule expression. These results confirm that the investigated gene cluster is the functional capsule determinant of E. coli strain 536. The loss of capsule production upon inactivation of region 2 of the K15 kps gene cluster was also visualized by immunofluorescence microscopy. ELISA with an anti-K15 serum indicated that inactivation of kpsCK10 resulted in reduced K15 capsule production, whereas the kpsC′K5 mutant exhibited the same encapsulated phenotype as wild-type strain 536 (Fig. 4). This demonstrates that kpsCK10 is required for K15 capsule biosynthesis, whereas kpsC′K5 has been inactivated due to truncation.
FIG. 4.
Detection of K15 capsule expression of E. coli strain 536 and various derivatives. (A) Detection of K15 capsule by ELISA with a K15 capsule-specific antiserum. The binding of a preabsorbed polyclonal K15 capsule-specific antibody was visualized by using an anti-rabbit antibody conjugated with horseradish peroxidase. The optical density was measured at 490 nm. Values marked with an asterisk are statistically significant (P < 0.001) compared to the values of the wild type as calculated by the Student t test. (B) Detection of K15 capsule expression by immunofluorescence microscopy. The binding of a preabsorbed polyclonal K15 capsule-specific antibody was visualized by a Texas Red-labeled secondary antibody and examined by epifluorescence microscopy. Upper row, phase contrast images; lower row, fluorescence images; left column, UPEC strain 536 (K15, encapsulated); middle column, UPEC strain 536 ΔkpsK15 (nonencapsulated); right column, E. coli K-12 strain MG1655 (negative control).
Role of the K15 capsule and the PAI V536-encoded general secretion pathway for serum resistance and urovirulence.
Serum resistance assays demonstrated that nonencapsulated mutants of strain 536 in which one of the regions of the kpsK15 determinant has been partially deleted were able to grow in 50% human serum as well as the encapsulated wild-type strain. In contrast, the strain 536 rfaH mutant does not express K15 capsule and LPS and was not able to grow in human serum (Fig. 5). This indicates that, in contrast to LPS, the K15 capsule is not required for serum resistance of strain 536. Serum resistance of the PAI V536-encoded general secretion pathway mutant strain 536 Δgsp was not impaired relative to that of the wild type.
FIG. 5.
Influence of the K15 capsule on serum resistance of UPEC strain 536 and various capsule-negative derivatives. Bacteria were incubated in 50% serum obtained from a healthy individual. The graph shows means ± standard errors of the means of values originated from three similar assays. The values obtained for the rfaH-negative mutant are significantly different (P < 0.01) between the time points from 1 to 4 h (Student's t test).
To analyze whether the K15 capsule contributes to virulence of UPEC strain 536, different kpsK15 mutants that have been tested for K15 capsule expression before (Fig. 4) were examined in a murine model of experimental ascending UTI. According to the results of the experiments, the virulence of the different nonencapsulated mutants was markedly reduced relative to that of the encapsulated wild-type strain and the encapsulated kpsC′K5 mutant (Table 3). Consequently, the K15 capsule contributes to the urovirulence of strain 536. Inactivation of the general secretion pathway determinant localized on PAI V536 did not affect capsule expression (see Fig. 2 in the supplemental material) and the virulence of the corresponding mutant (Table 3).
TABLE 3.
Contribution of K15 capsule and PAI V536-encoded general secretion pathway (gsp) to urovirulence of E. coli strain 536
| Strain | Characteristics | No. of infected mice | Mortality rate (%) |
|---|---|---|---|
| 536 wild type | K15 capsule+, GSP+ | 21 | 100 |
| 536 ΔkpsSC | K15 capsule−, GSP+ | 27 | 14.8a |
| 536 ΔR2 | K15 capsule−, GSP+ | 20 | 10.0a |
| 536 ΔkpsMT | K15 capsule−, GSP+ | 24 | 8.3a |
| 536 ΔkpsK15 | K15 capsule−, GSP+ | 21 | 19.0a |
| 536 ΔkpsC′K5 | K15 capsule+, GSP+ | 12 | 91.7 |
| 536 ΔkpsCK10 | K15 capsule−, GSP+ | 13 | 15.4a |
| 536 Δgsp | K15 capsule+, GSP− | 12 | 91.7 |
Significant difference from wild-type strain 536 (chi-square probe, P < 0.001).
DISCUSSION
Four PAIs have so far been described in detail for UPEC strain 536 (O6:K15:H31) (16). In this study, the genetic organization of PAI V536 was characterized and prominent gene clusters of this PAI have been subjected to functional characterization. The composite PAI V536 exhibits typical features of PAIs, such as (i) the association with a tRNA-encoding gene, (ii) the presence of different virulence-associated genes (the pix fimbria determinant [33] and the K15 capsule-encoding gene cluster), and (iii) the presence of DR structures formed by the last 22 bp of the 3′ end of pheV and its truncated copy ′pheV. Similarly, PAI III536 also represents a composite island with flanking DR structures formed by the last 47 bp of the 3′ end of thrW and its truncated copy ′thrW (16). In the case of PAI V536, the G+C content of 47.7% and the fact that this DNA region is absent in nonpathogenic E. coli strain MG1655 underline the fact that it has been acquired by horizontal gene transfer. In addition, several regions of PAI V536 are highly homologous to regions of (formerly) mobile DNA elements, i.e., virulence plasmids or PAIs, bacteriophages, and other accessory DNA elements such as IS elements. In UPEC strain 536, a bacteriophage P4-like integrase gene is localized immediately downstream of the pheV tRNA gene. P4-like integrase genes seem also to be the typical integrase genes present on pheV-associated PAIs of other E. coli strains (1, 30, 43, 52, 53, 56).
Analysis of the gene content of PAI V536 revealed that the putative K15 capsule determinant is located downstream of ′pheV. Several other kps gene clusters have been reported to be localized on pheV-associated PAIs (52, 56) or next to pheV (11), and it is anticipated that group 2 and 3 capsule loci are chromosomally inserted between pheV and yghD (6). Comparison of the genetic structure of the PAI V536-localized kps determinant with that of three other E. coli K15 strains present in the IMIB strain collection (IMI402, IMI403, and IMI415) by overlapping PCRs demonstrated that all K15 isolates tested share a kps locus with an identical genetic organization (data not shown). This underlines the notion that the kps determinant localized on PAI V536 is indeed the functional K15 capsule-encoding gene cluster.
The K15 capsule locus exhibits a so far unknown genetic organization and combines structural features of group 2 and 3 capsule determinants (Fig. 1 and 3A). Group 2 and 3 capsule gene clusters have a segmental gene organization with two conserved regions (1 and 3) flanking the serotype-specific region 2 (12, 45). The most striking difference between the kpsK15 gene cluster and a typical group 2 capsule locus is the inversion of the functional kpsS and kpsCK10 genes relative to the rest of the genes of region 1. kpsS and kpsCK10, which more closely resemble corresponding genes of group 3 capsule operons, are separated from the truncated kpsC′K5 gene by a nonfunctional ORF with homology to the putative transposase-encoding ORF c1248 of UPEC strain CFT073. The encoded gene product is similar to a transposase of the IS630 family (NP_929067). This indicates that truncation of kpsC′K5 and the presence of an inversed orientation of kpsS and kpsCK10 relative to kpsC′K5 is due to recombination between group 2 and 3 capsule gene clusters and involved IS element(s). The concomitant loss of functional kpsCK5 and kpsSK5 genes has been complemented by the horizontally acquired kpsS and kpsCK10 genes. The KpsS and KpsC proteins are involved in capsule maturation when the synthesized polysaccharide is ligated to phosphatidyl-3-keto-3-deoxy-mannooctulosonate (45). Clarke and coworkers demonstrated that KpsC and KpsS of group 2 and group 3 capsules are homologous and functionally interchangeable (12). The fusion of segments of two unrelated kps determinants is further corroborated by a dramatic decrease of the G+C content immediately downstream of kpsC′K5. The overall G+C content of the ORF65-kpsS-kpsCK10 region (37.8%) is markedly lower than that of the DNA stretch comprising the kpsFEDUC′K5 genes (50.9%). Region 2 exhibits the lowest G+C content (30.9%) within the kpsK15 determinant (Fig. 3B). This is typical for genes encoding enzymes involved in polysaccharide biosynthesis (44). It has been hypothesized that acquisition of different region 2 sequences by homologous recombination between flanking regions 1 and 3 of two group 2 capsule gene clusters contributes to the diversity of group 2 capsules and that the insertion of new capsule genes into an existing group 2 gene cluster may lead to the generation of group 3 capsule determinants (6). This is supported by the genetic structure of the kpsK15 locus indicative of genetic rearrangements and recombination between group 2 and 3 capsule determinants that result in the generation of a new capsule type.
The KpsM and KpsT proteins are similar to ABC (ATP-binding cassette) transporters (8, 22) that are believed to be the cytoplasmic membrane component of the capsule transport system. Further steps of capsule polysaccharide translocation through the outer membrane still remain to be elucidated. It has been speculated that this process may also involve a general mechanism such as the type II general secretion pathway (GSP). Interestingly, a GSP determinant was identified 5 kb downstream of the kpsK15 locus on PAI V536. To find out whether this gene cluster in the vicinity of the kpsK15 determinant codes for proteins involved in the transport of capsule polysaccharides across the outer membrane, the GSP determinant has been inactivated. However, this inactivation had no effect on K15 capsule expression, serum resistance, and virulence of E. coli strain 536 (Table 3 and Fig. 5; see also Fig. 2 in the supplemental material).
It has been amply reported that group 2 capsules are more frequently distributed among uropathogenic isolates than among fecal isolates (26, 27, 50). This led to the speculation that group 2 capsules contribute to urovirulence according to their ability to impede phagocytosis (13, 34, 54). The capsule has also been reported to confer resistance to the bactericidal activity of serum (2, 13, 31, 47). Furthermore, a signature-tagged mutagenesis screen has previously suggested the role of the K2 capsule of UPEC strain CFT073 in UTI (4). However, other findings do not support a role of group 2 capsules as a virulence factor of UPEC and favor a role in systemic infections rather than in an ascending UTI (47, 48). It has been suggested that the capsule protects the organism by masking the cell surface and by serving as a physical barrier to prevent bacterial attachment to phagocytic cells like polymorphonuclear leukocytes and monocytes and, consequently, to protect the cells from internalization (9, 13, 23, 37, 54). To analyze whether the K15 capsule contributes to pathogenesis, the virulence of capsule-deficient mutants of UPEC strain 536 have been compared to that of the corresponding wild-type strain in an experimental ascending UTI model with infant mice. The data obtained demonstrate that the loss of the K15 capsule is associated with a dramatic attenuation. It has been proven that, in the experimental infection model used in this study, mortality is due to urosepsis originating from an ascending UTI which starts from the bladder and reaches the blood system through the kidneys. This indicates that the K15 capsule contributes to the urovirulence of strain 536 (Table 3) and not only to a systemic infection as reported before (46, 48). However, the K15 capsule is not required for serum resistance, as the wild-type strain 536 and the different kpsK15 mutants used in this study multiplied for several hours in human serum (Fig. 5).
In summary, the study of the genetic organization of PAI V536 and, in particular, that of the K15 capsule gene cluster of E. coli strain 536 supports the model that horizontal gene transfer and recombination play an important role for acquisition of additional genetic information and the generation of new variants of already existing determinants. Whereas the function of the K15 capsule has been successfully analyzed, the transport and assembly pathway of the capsule polysaccharides across the outer membrane as well as the functional characterization of other PAI V536-localized genes will have to be further elucidated.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB479 and TPA1) and OTKA grants T037833 and ETT 086/2001. The Göttingen Genomics Laboratory received support from the Forschungsmittel des Landes Niedersachsen. G.S. was supported by a grant from the Bayerische Forschungstiftung.
We thank H. Merkert (Würzburg) for immunofluorescence microscopy and B. Plaschke (Würzburg) for excellent technical assistance.
Editor: J. T. Barbieri
REFERENCES
- 1.Al-Hasani, K., K. Rajakumar, D. Bulach, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Allen, P. M., I. Roberts, G. J. Boulnois, J. R. Saunders, and C. A. Hart. 1987. Contribution of capsular polysaccharide and surface properties to virulence of Escherichia coli K1. Infect. Immun. 55:2662-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 6.Barrett, B., L. Ebah, and I. S. Roberts. 2002. Genomic structure of capsular determinants, p. 137-155. In J. Hacker and L. B. Kaper (ed.), Pathogenicity islands and the evolution of pathogenic microbes, vol. 264. Springer, Berlin, Germany. [PubMed] [Google Scholar]
- 7.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bliss, J. M., C. F. Garon, and R. P. Silver. 1996. Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology 6:445-452. [DOI] [PubMed] [Google Scholar]
- 9.Burns, S. M., and S. I. Hull. 1999. Loss of resistance to ingestion and phagocytic killing by O− and K− mutants of a uropathogenic Escherichia coli O75:K5 strain. Infect. Immun. 67:3757-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 11.Cieslewicz, M., and E. Vimr. 1997. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol. Microbiol. 26:237-249. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, B. R., R. Pearce, and I. S. Roberts. 1999. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol. 181:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross, A. S., K. S. Kim, D. C. Wright, J. C. Sadoff, and P. Gemski. 1986. Role of lipopolysaccharide and capsule in the serum resistance of bacteremic strains of Escherichia coli. J. Infect. Dis. 154:497-503. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Fünfstück, and J. Hacker. 2001. S-Fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt, U., L. Emödy, I. Gentschev, W. Goebel, and J. Hacker. 2002. Efficient expression of the alpha-haemolysin determinant in the uropathogenic Escherichia coli strain 536 requires the leuX-encoded tRNA(5) (Leu). Mol. Genet. Genomics 267:370-379. [DOI] [PubMed] [Google Scholar]
- 18.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldrick, D., G. Q. Yu, S. Q. Jiang, and J. S. Hong. 1988. Nucleotide sequence and transcription start point of the phosphoglycerate transporter gene of Salmonella typhimurium. J. Bacteriol. 170:3421-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker, J., G. Blum-Oehler, I. Mühldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 21.Hacker, J., and J. B. Kaper (ed.). 2002. Pathogenicity islands and the evolution of pathogenic microbes, vol. 264. Springer, Berlin, Germany.
- 22.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism-an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz, M. A. 1982. Phagocytosis of microorganisms. Rev. Infect. Dis. 4:104-123. [DOI] [PubMed] [Google Scholar]
- 24.Janke, B., U. Dobrindt, J. Hacker, and G. Blum-Oehler. 2001. A subtractive hybridisation analysis of genomic differences between the uropathogenic E. coli strain 536 and the E. coli K-12 strain MG1655. FEMS Microbiol. Lett. 199:61-66. [DOI] [PubMed] [Google Scholar]
- 25.Jann, K., and B. Jann. 1992. Capsules of Escherichia coli, expression and biological significance. Can. J. Microbiol. 38:705-710. [DOI] [PubMed] [Google Scholar]
- 26.Kaijser, B. 1973. Immunology of Escherichia coli: K antigen and its relation to urinary-tract infection. J. Infect. Dis. 127:670-677. [DOI] [PubMed] [Google Scholar]
- 27.Kaijser, B., L. A. Hanson, U. Jodal, G. Lidin-Janson, and J. B. Robbins. 1977. Frequency of E. coli K antigens in urinary-tract infections in children. Lancet i:663-666. [DOI] [PubMed] [Google Scholar]
- 28.Kiesewalter, J., and G. Seltmann. 1968. Herstellung der diagnostischen Hyperimmun-Sera, p. 158-160. In H. Rische (ed.), Enterobacteriaceae-Infektionen. VEB Georg Thieme, Leipzig, Germany.
- 29.Kim, U. J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H. L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 30.Lalioui, L., and C. Le Bouguenec. 2001. afa-8 gene cluster is carried by a pathogenicity island inserted into the tRNAPhe of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leying, H., S. Suerbaum, H. P. Kroll, D. Stahl, and W. Opferkuch. 1990. The capsular polysaccharide is a major determinant of serum resistance in K-1-positive blood culture isolates of Escherichia coli. Infect. Immun. 58:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lügering, A., I. Benz, S. Knochenhauer, M. Ruffing, and M. A. Schmidt. 2003. The Pix pilus adhesin of the uropathogenic Escherichia coli strain X2194 (O2: K−: H6) is related to Pap pili but exhibits a truncated regulatory region. Microbiology 149:1387-1397. [DOI] [PubMed] [Google Scholar]
- 34.Medearis, D. N., Jr., B. M. Camitta, and E. C. Heath. 1968. Cell wall composition and virulence in Escherichia coli. J. Exp. Med. 128:399-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 37.Moxon, E. R., and J. S. Kroll. 1990. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 150:65-85. [DOI] [PubMed] [Google Scholar]
- 38.Nagy, G., U. Dobrindt, M. Kupfer, L. Emödy, H. Karch, and J. Hacker. 2001. Expression of hemin receptor molecule ChuA is influenced by RfaH in uropathogenic Escherichia coli strain 536. Infect. Immun. 69:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, G., U. Dobrindt, G. Schneider, A. S. Khan, J. Hacker, and L. Emödy. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oefner, P. J., S. P. Hunicke-Smith, L. Chiang, F. Dietrich, J. Mulligan, and R. W. Davis. 1996. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 24:3879-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ørskov, I., and K. Nyman. 1974. Genetic mapping of the antigenic determinants of two polysaccharide K antigens, K10 and K54, in Escherichia coli. J. Bacteriol. 120:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ørskov, I., V. Sharma, and F. Ørskov. 1976. Genetic mapping of the K1 and K4 antigens (L) of Escherichia coli. Non-allelism of K(L) antigens with K antigens of O8:K27(A), O8:K8(L) and O9:K57(B). Acta Pathol. Microbiol. Scand. B 84:125-131. [PubMed] [Google Scholar]
- 43.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, I. S. 1995. Bacterial polysaccharides in sickness and in health. The 1995 Fleming Lecture. Microbiology 141:2023-2031. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 46.Russo, T., J. J. Brown, S. T. Jodush, and J. R. Johnson. 1996. The O4 specific antigen moiety of lipopolysaccharide but not the K54 group 2 capsule is important for urovirulence of an extraintestinal isolate of Escherichia coli. Infect. Immun. 64:2343-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo, T. A., M. C. Moffitt, C. H. Hammer, and M. M. Frank. 1993. TnphoA-mediated disruption of K54 capsular polysaccharide genes in Escherichia coli confers serum sensitivity. Infect. Immun. 61:3578-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo, T. A., Y. Liang, and A. S. Cross. 1994. The presence of K54 capsular polysaccharide increases the pathogenicity of Escherichia coli in vivo. J. Infect. Dis. 169:112-118. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Sandberg, T., B. Kaijser, G. Lidin-Janson, K. Lincoln, F. Ørskov, I. Ørskov, E. Stokland, and C. Svanborg-Eden. 1988. Virulence of Escherichia coli in relation to host factors in women with symptomatic urinary tract infection. J. Clin. Microbiol. 26:1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 52.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 54.Timmis, K. N., G. J. Boulnois, D. Bitter-Suermann, and F. C. Cabello. 1985. Surface components of Escherichia coli that mediate resistance to the bactericidal activities of serum and phagocytes. Curr. Top. Microbiol. Immunol. 118:197-218. [DOI] [PubMed] [Google Scholar]
- 55.Vimr, E. R. 1991. Map position and genomic organization of the kps cluster for polysialic acid synthesis in Escherichia coli K1. J. Bacteriol. 173:1335-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 58.Whitfield, C., and M. A. Valvano. 1993. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv. Microb. Physiol. 35:135-246. [DOI] [PubMed] [Google Scholar]