Abstract
Little is known about the defensive mechanisms induced in epithelial cells by pathogenic versus probiotic bacteria. The aim of our study was to compare probiotic bacterial strains such as Escherichia coli Nissle 1917 with nonprobiotic, pathogenic and nonpathogenic bacteria with respect to innate defense mechanisms in the intestinal mucosal cell. Here we report that E. coli strain Nissle 1917 and a variety of other probiotic bacteria, including lactobacilli—in contrast to more than 40 different E. coli strains tested—strongly induce the expression of the antimicrobial peptide human beta-defensin-2 (hBD-2) in Caco-2 intestinal epithelial cells in a time- and dose-dependent manner. Induction of hBD-2 through E. coli Nissle 1917 was further confirmed by activation of the hBD-2 promoter and detection of the hBD-2 peptide in the culture supernatants of E. coli Nissle 1917-treated Caco-2 cells. Luciferase gene reporter analyses and site-directed mutagenesis experiments demonstrated that functional binding sites for NF-κB and AP-1 in the hBD-2 promoter are required for induction of hBD-2 through E. coli Nissle 1917. Treatment with the NF-κB inhibitor Helenalin, as well as with SP600125, a selective inhibitor of c-Jun N-terminal kinase, blocked hBD-2 induction by E. coli Nissle 1917 in Caco-2 cells. SB 202190, a specific p38 mitogen-activated protein kinase inhibitor, and PD 98059, a selective inhibitor of extracellular signal-regulated kinase 1/2, were ineffective. This report demonstrates that probiotic bacteria may stimulate the intestinal innate defense through the upregulation of inducible antimicrobial peptides such as hBD-2. The induction of hBD-2 may contribute to an enhanced mucosal barrier to the luminal bacteria.
Probiotics are live microbes which have been shown to have beneficial effects on human health (18). Conditions susceptible to treatment with probiotics include traveler’s, antibiotic-induced, and childhood diarrhea. Recently, several controlled clinical studies have also proven a role for probiotic therapy in different states of inflammatory bowel diseases (24, 33). A particularly interesting strain is E. coli Nissle 1917, which was equivalent to standard mesalamine in maintaining remission of ulcerative colitis (24, 33; W. Kruis, P. Fric, and M. Stolte, Abstr. 102nd Annu. Meet. Am. Gastroenterol. Assoc., abstr. 127, 2001). The convincing outcome of these clinical trials using probiotic bacteria has encouraged us to further explore the mode of action of these bacteria. Several modes of probiotic action have been considered. Bacterial interference with intestinal pathogens is a well-established mode of action (2, 32) and may be mediated by bacteriocins (1, 21, 22), specific antimicrobial substances that antagonize intestinal pathogens. However, probiotics also appear to directly affect mucosal immune function through modulation of immunoglobulin A (IgA) synthesis, mucus formation, or alterations of the pro- versus anti-inflammatory balance of local cytokines (17). Recent findings raise the possibility that microbe-host cell signaling might be a mode of action by which probiotic bacteria could stabilize intestinal microecology and effectively prevent colonization by enteric pathogens (31). Prompted by studies on defensins in colonic mucosa (7, 8) we hypothesized that probiotics may act through an induction of these endogenous antibiotics.
In addition to acting as a physical barrier, the intestinal epithelium contributes to host defense by producing antimicrobial peptides in order to limit access of enteric bacteria and other microorganisms. One important class of human antimicrobial peptides is the family of defensins. These small (3- to 5-kDa) cationic peptides are distinguished as α- and β-defensins based on the positions of their three intramolecular disulfide bridges (10). The known human α-defensins include the human neutrophil peptides 1 to 4 as well as the epithelial human defensin-5 (HD-5) and HD-6. Human β-defensin-1 (hBD-1), -2, -3, and -4 are expressed in various epithelial cells. Defensins have a broad spectrum of antimicrobial activity against bacteria, fungi, and some enveloped viruses (9). The mechanism is not fully understood. Human neutrophil defensin 1 to 3 perforation of the cell wall through formation of multimeric pores has been described (20, 25). Interestingly, some beta defensins, including hBD-2, may also act as chemokines (46). HD-5 and HD-6 are expressed primarily in Paneth cells of the small intestine (19) but are also expressed by metaplastic Paneth cells in the colon during inflammatory bowel diseases (5, 43). Interestingly ileal involvement during Crohn's disease is associated with low HD-5 and HD-6 expression, especially in the case of the NOD2 mutation (41). The normal colonic mucosa expresses hBD-1 (6, 39, 40), whereas hBD-2 and hBD-3 are expressed only in case of inflammation, especially in ulcerative colitis and (at a lower level or not at all) in Crohn's disease (6, 39, 40). The functional importance of mucosal defensins in vivo has been demonstrated by the resistance of HD-5 transgenic mice to salmonella infection on expression of the human alpha defensin HD-5 (35) and the increased susceptibility to infection of mice with a gene knockout of the protease matrilysin, leading to a block in the processing of prodefensins to defensins (45).
To test our hypothesis that probiotics may stimulate the colonic epithelial chemical defense system, we decided to study the effect of E. coli Nissle 1917 and other probiotic strains on defensin induction in human colonic epithelial cell lines in vitro and to compare this strain with known pathogenic strains of E. coli as well as with apathogenic K-12 and with fecal isolates. The probiotic bacterium E. coli Nissle 1917 was selected because, in its long history, it has been shown to be apathogenic, to effectively colonize the gut (26), to exhibit a semirough phenotype with a well-characterized lipopolysaccharide (LPS) (11), and to be immunomodulatory (14, 15), as well as immunogenic (4).
MATERIALS AND METHODS
Culture and stimulation of epithelial cells.
Caco-2 cells were obtained from the American Type Culture Collection and used from passage 15 to 25. Cell monolayers were cultured in 75-cm2 flasks (BD Biosciences, Heidelberg, Germany) in a humidified atmosphere with 5% CO2. Standard media for feeding cells consisted of Dulbecco's modified essential medium, 10% fetal calf serum (FCS), and 3% nonessential amino acids supplemented with 2 mM glutamine. For stimulation experiments cells were seeded in 12-well tissue culture plates (4 cm2/well; BD Biosciences) and used at 70 to 80% confluence. Cells were stimulated in FCS-free medium with heat-killed bacteria for 3 to 48 h. To test if other cell lines show similar effects, we tested defensin induction in T84 as well as HT29 cells.
The mitogen-activated protein (MAP) kinase inhibitors PD 98059, SB 202190, and SP600125 as well as the NF-κB inhibitor helenalin were purchased from Calbiochem (Darmstadt, Germany) and resuspended in dimethyl sulfoxide. The inhibitors were added directly to the culture medium at a final concentration of 20 μM 1 h prior to stimulation.
Preparation of bacteria.
E. coli K-12 HB101, uropathogenic E. coli (UPEC) W536, and enteropathogenic E. coli (EPEC) were kindly supplied by J. Hacker and G. Blum-Oehler (Institute of Molecular Biology of Infection, Würzburg, Germany). E. coli Nissle 1917 (DSM 6601) and all E. coli strains as well as Lactobacillus fermentum, Lactobacillus gasseri, and three different forms of Lactobacillus acidophilus were provided by Ardeypharm (Herdecke, Germany). Lactobacillus paracasei, Lactobacillus plantarum, Pediococcus pentosaceus, and Leuconostoc mesenteroides, as well as a cocktail containing strains of all four species together, were delivered by Medipharm (Kågeröd, Sweden). An overview of serotypes, origins, and references of selected strains used is given in Tables 1 and 2. All E. coli strains were grown overnight in Trypticase soy broth (TSB) medium at 37°C under permanent shaking. To keep bacteria in a linear growth phase, 200 μl of bacterial suspension was again diluted in 5 ml of fresh TSB medium and bacteria were grown under shaking conditions at 37°C. Lactobacilli were cultured in 10 ml of De Man-Rogosa-Sharpe broth (MRS) under anaerobic conditions at 37°C overnight. As for E. coli strains, 500 μl of the bacterial suspension was then diluted in MRS broth to a final volume of 10 ml and grown again at 37°C under anaerobic conditions. After 5 h lactobacilli and E. coli strains were heat killed in a water bath at 65°C for 60 min and then diluted to the appropriate concentration in FCS-free cell culture medium. For testing of dose dependence, bacteria were used at a concentration between 107 and 109 bacteria per ml. To test whether live E. coli Nissle 1917 (Mutaflor) has the same effect on Caco-2 cells, we used the commercially available E. coli Nissle 1917 suspension and the same suspension without bacteria as controls (provided by Ardeypharm). Mutaflor contains in the commercially available suspension preparation 108 cells of living E. coli Nissle 1917 bacteria/ml. We diluted this suspension 1 to 3 with FCS-free medium for the stimulation experiments. To maintain stable conditions for all other experiments, we used heat-inactivated bacteria at a concentration of 3 × 108 cells/ml.
TABLE 1.
Origins, serotypes, and hemolysis characteristics of some E. coli strains used
| E. coli strain | Type of isolate | Serotypec | Hemolysis | Source or reference |
|---|---|---|---|---|
| Strains known to be nonpathogenic | ||||
| Nissle 1917 (DSM 6601) | Pharmaceutical | O6:K-5:H1 | No | 2a |
| DSM 498 | Reference strain | K-12 | No | DSMZa |
| DSM 1607 (HB101) | Reference strain | K-12 | No | 3a |
| Strains with known pathogenicity | ||||
| W536 (UPEC) | Urinary | O6:K-15:H31 | Yes | 1a |
| E2348/69 (EPEC) | Fecal | O127:H6 | Yes | 5a |
| Strains with unknown pathogenicity | ||||
| PZ 865 | Fecal | O1:H- | Yes | ACS*b |
| PZ 860 | Fecal | O7:H- | No | ACS* |
| PZ 862 | Fecal | O8:H- | No | ACS* |
| PZ 866 | Fecal | O17:H18 | No | ACS* |
| PZ 915 | Fecal | O19:H- | No | ACS |
| PZ 861 | Fecal | O-NT:H- | No | ACS* |
| PZ 864 | Fecal | O-NT:H- | No | ACS* |
| PZ 868 | Fecal | O-NT:H- | No | ACS* |
| PZ 870 | Fecal | O-NT:H- | No | ACS* |
| PZ 863 | Fecal | r:H16 | Yes | ACS* |
| PZ 867 | Fecal | r:H- | No | ACS* |
| PZ 830 | Fecal | O4:H- | No | ACS* |
| PZ 835 | Fecal | O2:H4- | No | ACS* |
DSMZ, Deutsche Sammlung von Mikroorgansimen and Zellkulturen, Baunschweig, Germany.
ACS, Ardeypharm Collection of Strains, fecal isolates from healthy persons, except strains PZ 830 and PZ 835 (fecal isolates from patients with inflammatory bowel disease. *, strain obtained from A. M. Snelling and P. Hawkey, Department of Microbiology, University of Leeds, and The Leeds General Infirmary, Leeds, United Kingdom.
Serotyping of strains was performed by S. Aleksic, Institute of Hygiene, Hamburg, Germany. O-NT, O antigen not typeable; H-, no H antigen (cells nonmotile).
TABLE 2.
Tested lactobacilli and other tested probiotic bacteria
| Species or subspecies (isolate) | Strain designation | Type of isolate | Source and/or referencea |
|---|---|---|---|
| L. gasseri | PZ 1160 | Intestinal isolate | ACS |
| L. acidophilus | PZ 1030 (DSM 20079) | Reference strain | ACS (DSMZ) |
| L. acidophilus | PZ 1129 | Industrial | ACS |
| L. acidophilus | PZ_1130 (JCM 1132) | Reference strain | ACS (GR) |
| L. fermentum | PZ1162 | Intestinal isolate | ACS |
| Pediococcus pentosaceus (16:1) | LMG P-20608 | Intestinal isolate | 24a, BCCM |
| L. plantarum (2362) | LMG P-20606 | Growing rye | 24a, BCCM |
| Leuconostoc mesenteroides (77:1) | LMG P-20607 | Growing rye | 24a, BCCM |
| L. paracasei subsp. paracasei (F19) | LMG P-17806 | Growing rye | 24b, BCCM |
ACS and DSMZ are as defined for Table 1. GR, strain collection of G. Reuter (strain deposited by Mitsuoka at the Japanese Collection of Microorganisms); BCCM, Belgian Coordinated Collection of Microorganisms.
Testing of E. coli LPS.
To look for strain-specific mechanisms of hBD-2 induction, we tested highly purified LPS generated from E. coli Nissle 1917, which was kindly provided by U. Zähringer (Research Center Borstel, Borstel, Germany). As a control we used purified LPS from E. coli serotype O111:B4 (Sigma-Aldrich). The LPS from the control strain and E. coli Nissle 1917 LPS were diluted in FCS-free medium in different concentrations (0.1, 0.5, 1, and 5 μg/ml), and the stimulation was performed for 6 h in Caco-2 cells.
RNA isolation and cDNA synthesis.
After stimulation cells were washed twice with phosphate-buffered saline (PBS) and harvested with TRIzol reagent (Invitrogen, San Diego, Calif.) according to the supplier's protocol. RNA quality and quantity were determined by gel electrophoresis and photometry. Subsequently 1 μg of total RNA was reverse transcribed in cDNA with oligo(dT) primers and 200 U of Superscript II (Invitrogen) according to routine procedure.
Real-time RT-PCR.
Real-time reverse transcription-PCR (RT-PCR) analyses were performed in a fluorescence temperature cycler (LightCycler; Roche Diagnostics GMBH, Mannheim, Germany) as described previously (12, 40). This technique continuously monitors the cycle-by-cycle accumulation of fluorescently labeled PCR product. Briefly, cDNA corresponding to 20 ng of RNA served as a template in a 20-μl reaction mixture containing 4 mM MgCl2, 0.5 μM (each) primer, and 1× LightCycler-FastStart DNA Master SYBR Green I mix (Roche Diagnostics GMBH). Samples were loaded into capillary tubes and incubated in the fluorescence thermocycler (LightCycler). Initial denaturation at 95°C for 10 min was followed by 45 cycles, each cycle consisting of 95°C for 15 s, touchdown of −1°C/cycle from the primer-specific starting to ending annealing temperatures for 5 s, and 72°C for 10 s. For hBD-1 (sense, 5′ ATACTTCAAAAGCAATTTTCCTTTAT 3; antisense, 5′ TTGTCTGAGATGGCCTCAGGTGGTAAC 3′) and hBD-2 (sense, 5′ CCCTTTCTGAATCCGC 3′; antisense, 5′ GAGGGTCTTGTATCTCCT 3′) a touchdown protocol with a primary temperature of 62°C and a target temperature of 58°C was used. For HD-5 (sense, 5′ GCCATCCTTGCTGCCATTC 3′; antisense, 5′ AGATTTCACACACCCCGGAGA 3′), HD-6 (sense, 5′ CCTCACCATCCTCACTGCTGTTC 3′; antisense, 5′ CCATGACAGTGCAGGTCCCATA 3′), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (sense, 5′ CCAGCCGAGCCACATCGCTC 3′; antisense, 5′ ATGAGCCCCAGCCTTCTCCAT 3′) we used a protocol with a primary temperature of 66°C and a target temperature of 60°C. (At the end of each run melting curve profiles were produced by cooling the sample to 65°C for 15 s and then heating slowly at 0.20°C/s to 95°C with continuous measurement of fluorescence to confirm amplification of specific transcripts.) Cycle-to-cycle fluorescence emission readings were monitored and analyzed with LightCycler software (Roche Diagnostics GMBH). The specificity of the amplification products was further verified by subjecting the amplification products to electrophoresis on a 2% agarose gel. The fragments were visualized by ethidium bromide staining, and the specificity of PCR products was verified by sequencing representative samples. Standard curves were obtained for each primer set with serial dilutions of cDNA.
All quantifications were normalized to the housekeeping GAPDH gene, which showed a very stable expression in Caco-2 cells. Relative expression is given as a ratio between target gene expression and GAPDH gene expression.
Purification and identification of hBD-2 from Caco-2 culture supernatants.
Supernatants of approximately 2 × 107 Caco-2 colon epithelial cells treated for 24 h with 3 × 108 cells of heat-inactivated E. coli Nissle 1917/ml were applied to a heparin affinity column (1 ml; HiTrap; Pharmacia). Bound material was then eluted with 10 ml of 2 M NaCl at a flow rate of 1 ml/min. Eluted material was diafiltered against 0.1% trifluroacetic acid (TFA), pH 3, and applied to a C2/C18 micro-reversed-phase high-performance liquid chromatography (HPLC) column (μRPC C2/C18, SC 2.1/10, Smart-Micro-HPLC apparatus; Amersham Biosciences, Uppsala, Sweden) that was previously equilibrated with 0.1% (vol/vol) TFA in HPLC grade water. Proteins were eluted with a gradient of increasing concentrations of acetonitrile containing 0.1% (vol/vol) TFA (flow rate: 100 μl/min). Fractions were then analyzed for the presence of hBD-2 by enzyme-linked immunosorbent assay (ELISA) or Western blotting.
For ELISA with HPLC fractions, 96-well immunoplates (MaxiSorp; Nunc, Roskilde, Denmark) were coated at 37°C for 1 h with 10 μl of each HPLC fraction and serial dilutions of natural skin-derived hBD-2 in 100 μl of 0.1 M sodium bicarbonate buffer, pH 10.6. Wells were washed with PBS-0.05% Tween and blocked with 1% bovine serum albumin in PBS for 1 h at room temperature. Plates were washed twice with PBS-0.05% Tween, and wells were incubated with 100 μl of hBD-2 rabbit antiserum (Peptide Institute, Osaka, Japan) diluted 1:40,000 in PBS for 1 h at room temperature. After three washes with PBS-0.05% Tween, 100 μl of a 1:20,000 dilution of goat anti-rabbit IgG horseradish peroxidase (HRP) conjugate (Dianova, Hamburg, Germany) per well was incubated for 1 h at room temperature. Plates were washed again three times with PBS-0.05% Tween and filled with 100 μl of peroxidase substrate (o-phenylenediamine dihydrochloride, 0.01 M in 0.1 M citrate-phosphate buffer, pH 5.0, containing 0.001% [vol/vol] H2O2)/well. The enzymatic reaction was stopped with 2 M sulfuric acid, and absorbance at 492 nm was determined in a multichannel photometer (Sunrise; Tecan, Crailsheim, Germany). The ELISA was sensitive to 0.3 ng of hBD-2/well.
For Western blot analysis C2/C18 fractions eluting like natural hBD-2 at 43 to 47% acetonitrile were pooled and loaded onto a sodium dodecyl sulfate-16.5% Tricine polyacrylamide gel containing 8 M urea (36). Proteins were transferred to a Protran-nitrocellulose membrane (Schleicher & Schuell BioScience, Dassel, Germany), blocked for 1 h in blocking buffer (5% [wt/vol] nonfat powdered milk in PBS-0.05% Tween), and then incubated for 18 h at 4°C in 3% (wt/vol) nonfat powdered milk in PBS-0.05% Tween containing 1:40,000-diluted hBD-2 rabbit antiserum (Peptide Institute). The membrane was washed with PBS-0.05% Tween six times for 5 min each, and then incubated for 1 h in 3% (wt/vol) nonfat powdered milk in PBS-0.05% Tween containing a 1:200,000 dilution of goat anti-rabbit IgG HRP conjugate (Dianova). After six washes as described above, the membrane was incubated for 5 min with chemiluminescent peroxidase substrate (Sigma, Taufkirchen, Germany) and visualized with a Diana III cooled charge-coupled device camera imaging system (Raytest, Straubenhardt, Germany). Densitometric quantifications were done with AIDA evaluation software (Raytest).
hBD-2 reporter plasmid construction.
To analyze hBD-2 promoter activity, 2,338 bp of the human hBD-2 promoter was amplified with the primers 5′ CAG TAC AGC AGC AGT GAT AG 3′ and 5′ GGG GAG GAC ATC AAG CCT T 3′. The amplification product was subcloned into the promoterless pGL3-basic firefly luciferase vector (Promega, Madison, Wis.) to generate reporter plasmid hBD-2-2338-luc.
The functional role of putative binding sites for the transcription factor NF-κB as well as for the transcription factor AP-1 in the hBD-2 promoter region was studied by site-directed mutagenesis. The QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used as indicated by the supplier for the introduction of mutations in the hBD-2-2338-luc plasmid. The two putative NF-κB binding sites at positions −205 to −186 and −596 to −572 were mutated with the mutant NF-κB primers to introduce NF-κB mutations as described by Tsutsumi-Ishii and Nagaoka (37). For the first position (HBD2-NF-κB-mut1, positions −205 to −186) we used the following primers: sense, 5′ CTGGTCCCAAGAGCAGGAGGAAGACGCGTTCTGGTACCTCCTGAGTCCAGATTTGCATAAGATC 3′; antisense, 5′ GATCTTATGCAAATCTGGACTCAGGAGGTACCAGAACGCGTCTTCCTCCTGCTCTTGGGACCAG 3′ (boldface letters indicate mutant nucleotides). For the second position (HBD2-NF-κB-mut2, positions −596 to −572) we used the following primers: sense, 5′ CTCTCTCCCGTGGGGAAGATGCTAGCTTTCAGACGCGTTTTCACATAAATTTCACCAGCTCAG 3′; antisense, 5′ CTGAGCTGGTGAAATTTATGTGAAAACGCGTCTGAAAGCTAGCATCTTCCCCACGGGAGAGAG 3′.
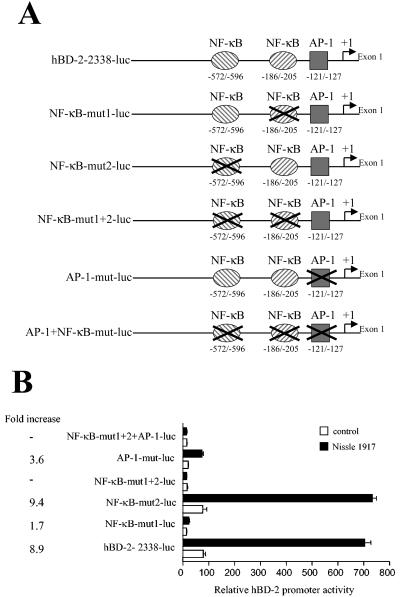
The putative AP-1 binding site at position −127 to −121 was mutated with mutant AP-1 primers (sense, 5′ CATTGTTCTTTGAGGACCATTCTCAGGCTCACCAGGTAAGTGGCTGAAT 3′; antisense, 5′ ATTCAGCCACTTACCTGGTGAGCCTGAGAATGGTCCTCAAAGAACAATG 3′). The resulting hBD-2-pGL3 plasmids containing mutated NF-κB and AP-1 binding sites were termed NF-κB-mut1-luc (containing one mutated NF-κB binding site at positions −205 to −186; see Fig. 6A), NF-κB-mut2-luc (containing one mutated NF-κB binding site at position −596 to −572; see Fig. 6A), NF-κB-mut1+2-luc (containing two mutated NF-κB binding sites; see Fig. 6A), AP1-mut-luc (containing a mutated AP-1 binding site; see Fig. 6A), and AP-1+NF-κB-mut-luc (containing two mutated NF-κB binding sites and a mutated AP-1 binding site; see Fig. 6A).
FIG. 6.
The relative importance of the NF-κB and AP-1 binding sites in the hBD-2 promoter. (A) The hBD-2 promoter constructs used are diagrammed. Nucleotide positions are marked relative to the hBD-2 transcription start. Two NF-κB sites and one AP-1 site in the hBD-2 promoter (bp −2338 to −1), linked to the luciferase gene, were mutated in different combinations. (B) Caco-2 epithelial cells were transfected with the wild-type (−2338-luc) or mutated hBD-2 promoter luciferase plasmids together with an internal-control Renilla luciferase expression plasmid. After transfection cells were incubated with E. coli Nissle 1917 (3 × 108 cells/ml) for 6 h and hBD-2 promoter activity was determined as a ratio between firefly and Renilla luciferase activities. Data represent means ± SEM of triplicate samples.
Transfection and determination of promoter activity.
For hBD-2 promoter studies Caco-2 cells were seeded in 12-well plates (BD Biosciences) and used for transfection at 60 to 80% confluence. Cells were transfected with 0.5 μg of the indicated hBD-2 reporter plasmids and 0.025 μg of an internal-control Renilla luciferase expression plasmid (phRG-TK; Promega) by using 1 μl of transfection reagent Fugene 6 (Roche Diagnostics GMBH) according to the manufacturer's instructions. Twenty-four hours after transfection cells were stimulated with bacteria for 5 h in FCS-free medium. After stimulation cells were harvested with 500 μl of passive lysis buffer (Promega), and firefly luciferase activity from the hBD-2-pGL3 reporter vector and Renilla luciferase activity were measured by the Dual Luciferase assay system (Promega) on a TD-20/20 luminometer (Turner Design). Promoter activity was reported as the ratio between firefly and Renilla luciferase activities in each sample.
hBD-2 ELISA.
To directly test the presence of the hBD-2 peptide in the supernatants of Caco-2 cells, a sensitive hBD-2 ELISA was established. Ninety-six-well immunoplates (MaxiSorp; Nunc) were coated at 4°C for 20 h with 50 μl of hBD-2 rabbit antiserum (Peptide Institute) diluted 1:5,000 in 0.05 M carbonate buffer, pH 9.6. Subsequently, wells were blocked with 200 μl of 1% bovine serum albumin in PBS for 10 min at room temperature. After three washes with 200 μl of PBS-0.1% Tween 20, 100 μl of cell culture supernatants per well and serial dilutions of natural skin-derived hBD-2 in cell culture medium were incubated for 30 min at room temperature. Plates were washed thrice with PBS-0.1% Tween 20, and wells were incubated for 30 min at room temperature with 50 μl of biotinylated goat anti-hBD-2 antibody (Cell Concepts, Umkirch, Germany) diluted 1:2,500 to 0.2 μg/ml in PBS-0.1% Tween 20. Plates were washed again three times with PBS-0.1% Tween and filled with 50 μl of streptavidin-peroxidase (Roche Diagnostics; 1:10,000 in PBS-0.1% Tween 20)/well. The plates were then incubated for 30 min at room temperature, washed three times as described above, and incubated with 2,2′-azinobis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS; Roche Diagnostics) as the development agent for 15 min at room temperature in the dark. Absorbance was measured at 405 nm with a multichannel photometer (Sunrise; Tecan).
Statistics.
Experiments were performed in triplicate samples. Differences between stimulated and control cells were analyzed by two-tailed, nonpaired t test, with a P value <0.05 indicative of statistical significance.
RESULTS
E. coli Nissle 1917 and other probiotic bacteria are potent activators of hBD-2 gene expression in colonic epithelial cells.
To analyze the influence of various probiotic as well as nonprobiotic E. coli strains on defensin expression in colon epithelial cells, we treated Caco-2 cells with E. coli Nissle 1917, 9 other probiotic bacterial strains including lactobacilli, and over 40 other nonprobiotic heat-inactivated E. coli strains and analyzed α- and β-defensin gene expression by the use of real-time PCR.
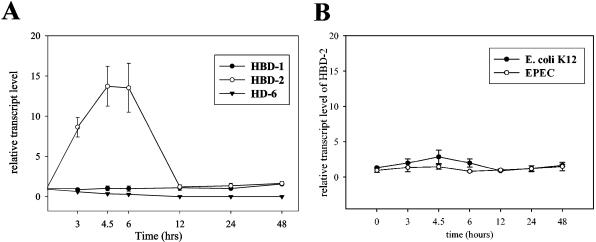
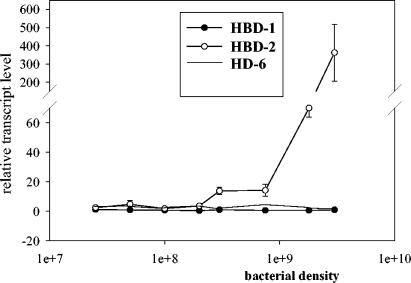
Basal constitutive gene expression of hBD-1 was not affected by E. coli Nissle 1917, either over time (Fig. 1A) or with increasing dose (Fig. 2). Similar results were obtained for the α-defensin HD-6, which was also not influenced by heat-inactivated E. coli Nissle 1917 at any time or any dose (Fig. 1 and 2). HD-5 could be detected only at a very minimal level and inconsistently.
FIG. 1.
E. coli Nissle 1917 but not EPEC or E. coli K-12 induces hBD-2 in Caco-2 cells in a time-dependent manner. Caco-2 cells were stimulated with E. coli Nissle 1917 (A) and EPEC and K-12 (B) with a concentration of 3 × 108 heat-inactivated bacteria per ml. Defensin expression was measured at different time points by real-time PCR. Panel B shows only the level of hBD-2 expression. Data shown represent the means ± SEM of representative results of two to five separate experiments using triplicates.
FIG. 2.
E. coli Nissle 1917 induces hBD-2 in Caco-2 cells in a dose-dependent manner. Caco-2 cells were stimulated for 4.5 h with different concentrations of heat-killed E. coli Nissle 1917. Defensin expression was measured by real-time PCR. Data shown represent the means ± SEM of a representative result of three independent experiments performed in triplicate.
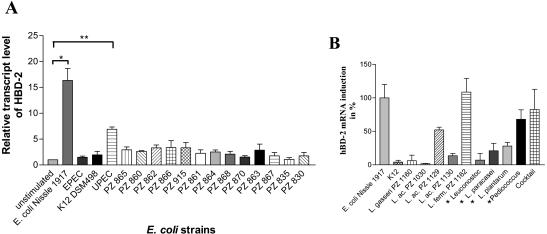
In contrast to that of the hBD-1, HD-5, and HD-6 genes, expression of the hBD-2 gene in Caco-2 cells was clearly induced upon treatment of the cells with E. coli Nissle 1917 and other probiotic bacteria. hBD-2 induction peaked between 3 and 6 h, but expression returned to basal values after 12 h (Fig. 1A). E. coli K-12 and EPEC did not influence any tested defensin expression at any time (Fig. 1B). UPEC showed its maximum induction at the same time as E. coli Nissle 1917 (data not shown). The induction of hBD-2 by E. coli Nissle 1917 was found to be dose dependent, with a maximum of 362-fold at the highest bacterial concentration tested (Fig. 2). From a comparison of all tested E. coli strains, hBD-2 induction by the probiotic E. coli Nissle 1917 (mean, 16-fold, P = 0.0006) and the UPEC W536 (mean, 6-fold, P < 0.0001) was observed exclusively (Fig. 3A). To clarify whether the induction of hBD-2 is a general feature of probiotic bacteria, we tested nine other known probiotic strains. Lactobacillus fermentum and Pediococcus pentosaceus showed an hBD-2 induction which was comparable to that of E. coli Nissle 1917. After stimulation with Lactobacillus acidophilus PZ 1129 and PZ 1130, Lactobacillus paracasei, and Lactobacillus plantarum, the intestinal cells induced a remarkable expression of hBD-2, which was stronger than that after stimulation with E. coli K-12, but lower than that after stimulation with E. coli Nissle 1917 (Fig. 3B). A cocktail containing four different probiotic strains also showed hBD-2 induction without any cumulative effect compared to the different strains added individually (Fig. 3B), and Lactobacillus gasseri, one of three different strains of Lactobacillus acidophilus (PZ 1030), and Leuconostoc showed expression levels comparable to that shown by E. coli K-12 (Fig. 3B). The suspension containing the living form of E. coli Nissle 1917 (as taken by patients) showed a strong induction of hBD-2 after incubation with Caco-2 cells for 4.5 h (68-fold). In contrast the same suspension without bacteria did not induce hBD-2 (data not shown). Since we saw the same effect by living strains, but with the disadvantage of extensive growth while in culture, we decided to use heat-killed bacteria for all other experiments. Heat-inactivated bacteria allowed stable culture conditions and bacterial concentrations. Other E. coli strains, such as the nonpathogenic K-12 reference strains (DSM 498 and HB101) and the EPEC strain, failed to induce hBD-2 (Fig. 3A). In addition, the 13 fecal isolates showed no significant difference in basal expression of hBD-2, with a range of 0.7- to 4-fold induction (Fig. 3A). Further, 30 E. coli strains (not classified, as shown in Table 1) isolated from human feces had a range of 0.7- to 4-fold (mean, 1.5-fold) induction of hBD-2 expression (data not shown). All tested E. coli strains, including the 11 serotyped strains isolated from the feces of healthy volunteers (Table 1), did not influence hBD-1 basal expression in Caco-2 cells (data not shown). Like Caco-2 cells T84 and HT29 cells also showed an induction of hBD-2 after incubation with E. coli Nissle 1917 but not K-12 or EPEC. Compared to those in Caco-2 cells the expression levels were lower, suggesting that Caco-2 cells are a better model to study hBD-2 expression.
FIG. 3.
Stimulation of Caco-2 cells with pathogenic and nonpathogenic E. coli strains, E. coli strains with unknown pathogenicity, and various lactobacilli. Caco-2 cells were stimulated for 4.5 h with a concentration of 3 × 108 cells of heat-inactivated bacteria/ml. Gene expression of hBD-2 was analyzed by real-time PCR. (A) hBD-2 expression after stimulation by different E. coli strains, including probiotic E. coli Nissle 1917. Data represent the means ± SEM normalized to basal expression of controls (set at 1) from one to six separate experiments run in triplicate. (*, P = 0.0006; **, P < 0.0001). (B) hBD-2 gene expression levels (± SEM) after stimulation with various lactobacilli in relation to that for E. coli Nissle 1917, which is set as 100%. *, bacteria which are also part of the cocktail containing a mix of four probiotic bacteria. L. ac., L. acidophilus; L. ferm., L. fermentum.
E. coli Nissle 1917 LPS and control purified LPS did not induce hBD-2.
Highly purified LPS from E. coli Nissle 1917 (0.1 μg, 1- ± 0.1-fold; 0.5 μg, 2.2- ± 0.1-fold; 1 μg, 1- ± 0.1-fold; 5 μg, 1- ± 0.1-fold; values are means ± standard errors of the means [SEM]) and LPS from the control strain (0.5 μg, 1- ± 0.33-fold; 1 μg, 1.4- ± 0.52-fold; 5 μg, 1.6- ± 1.01-fold) did not show any influence on hBD-2 expression at any concentration. These experiments suggest that factors other than LPS are responsible for the hBD-2 induction, and a specific semirough LPS phenotype, which has been described for E. coli Nissle 1917 (11), does not explain the described differences.
Caco-2 cells release hBD-2 upon contact with E. coli Nissle 1917.
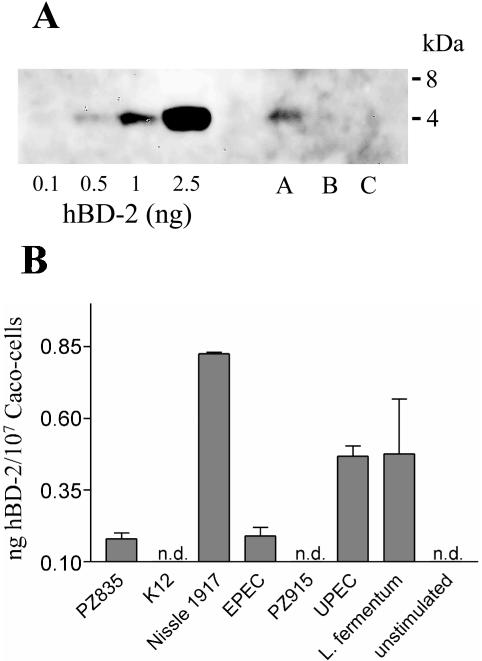
To investigate whether E. coli Nissle 1917 induces the release of the hBD-2 peptide from Caco-2 cells, we analyzed supernatants of untreated as well as E. coli Nissle 1917- or E. coli K-12-treated Caco-2 cells for the presence of the hBD-2 peptide. hBD-2 peptide released in the culture supernatants was bound to a heparin affinity column. Heparin-bound material was further purified by C2/C18 reversed-phase HPLC, and the resulting fractions were analyzed by ELISA and Western blotting for the presence of the hBD-2 peptide. Western blot analysis (Fig. 4A) and ELISA (not shown) detected approximately 0.5 to 1 ng of peptide in the culture supernatants of approximately 107 Caco-2 cells stimulated with E. coli Nissle 1917. In the supernatants of untreated cells and K-12-treated cells no hBD-2 peptide could be detected (Fig. 4A). Furthermore, we used an hBD-2 ELISA to directly analyze supernatants of cultured Caco-2 cells treated with various strains of E. coli as well as L. fermentum. As shown in Fig. 4B, only treatment with E. coli Nissle 1917 and, to a lesser extent, L. fermentum and UPEC led to a high release of hBD-2 in the culture supernatants of Caco-2 cells.
FIG. 4.
Detection of the hBD-2 peptide in culture supernatants from E. coli Nissle 1917-stimulated Caco-2 cells. (A) Heparin-bound proteins from culture supernatants of approximately 107 Caco-2 colon epithelial cells stimulated for 24 h with 3 × 108 cells of heat-inactivated E. coli/ml were purified by C2/C18 reversed-phase HPLC. Fractions eluting like natural hBD-2 at 43 to 47% acetonitrile were pooled and subjected to Western blot analysis using antibodies directed against hBD-2. The amounts of hBD-2 were calculated by densitometry using the indicated amounts of natural skin-derived hBD-2 as standards. Lane A, E. coli Nissle 1917-treated cells; lane B, E. coli K-12-treated cells; lane C, untreated cells. (B) Caco-2 epithelial cells were stimulated for 6 h with 3 × 108 cells of the indicated heat-inactivated bacterial strains/ml. Detection of hBD-2 released in the culture supernatants was achieved by an hBD-2 ELISA.
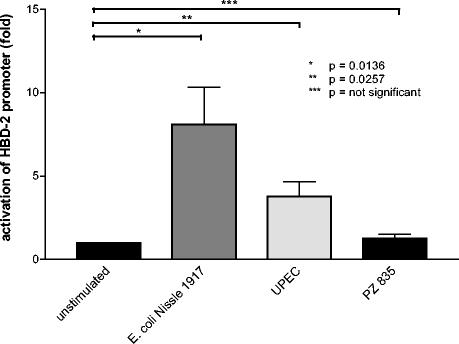
Activation of the hBD-2 promoter in Caco-2 cells by E. coli Nissle 1917.
To test whether hBD-2 gene induction measured by real-time PCR correlates with hBD-2 promoter activation, the hBD-2 promoter was ligated to the firefly luciferase gene and this construct was used to transiently transfect Caco-2 cells. As a result we found that E. coli Nissle 1917 induced the hBD-2 promoter approximately sevenfold compared to promoter levels in nonstimulated Caco-2 cells (Fig. 5). The activation of the hBD-2 promoter by the uropathogenic strain was less pronounced and coincided with the weaker induction of hBD-2 mRNA expression by this UPEC (Fig. 3). Two randomly selected strains (serotypes O4:H- and O2:H4-; Table 1) from patients with inflammatory bowel disease were also tested, but none activated the hBD-2 promoter (Fig. 5; only one representative strain is shown).
FIG. 5.
E. coli Nissle 1917 activates the hBD-2 promoter. Caco-2 cells were transiently transfected with a luciferase gene reporter vector containing the hBD-2 promoter as described in Materials and Methods. Twenty-four hours after transfection cells were stimulated for 4.5 h with pathogenic and nonpathogenic bacteria at a concentration of 3 × 108 heat-inactivated bacteria per ml, and hBD-2 promoter activity was determined as a ratio between firefly and Renilla luciferase activities. Data represent means ± SEM of two to four experiments performed in triplicate. The means were normalized to basal unstimulated luminescence of controls, set at 1.
NF-κB and AP-1 binding sites in the hBD-2 promoter are required for hBD-2 induction by E. coli Nissle 1917.
To elucidate the role of putative binding sites for the transcription factors NF-κB and AP-1 in the hBD-2 promoter region, we generated hBD-2 promoter luciferase expression constructs containing mutations of the two NF-κB sites (positions −205 to −186 and −596 to −572) and the AP-1 site (positions −127 to −121; Fig. 6A). Caco-2 cells transfected with these constructs were treated with E. coli Nissle 1917 and analyzed for hBD-2 promoter activation. Mutation of the two NF-κB sites together with the AP-1 site abolished E. coli Nissle 1917-mediated hBD-2 promoter activation (Fig. 6B). The same result was obtained by mutating only both NF-κB sites (Fig. 6B). Mutation of the proximal NF-κB site (positions −205 to −186) strongly inhibited E. coli Nissle 1917-mediated activation of the hBD-2 promoter (from 8.9-fold to 1.7-fold). In contrast, mutation of the NF-κB site at positions −596 to −572 did not inhibit hBD-2 promoter activation (Fig. 6B). Mutating the AP-1 site reduced E. coli Nissle 1917-mediated hBD-2 promoter activation from 8.9-fold to 3.6-fold (Fig. 6B).
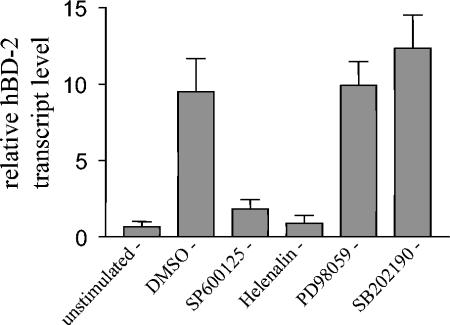
Inhibitors of the JNK and NF-κB pathways block hBD-2 induction by E. coli Nissle 1917.
Since AP-1 activity can be regulated by three MAP kinase cascades (extracellular signal-regulated kinase 1/2 [ERK1/2] pathway, c-Jun N-terminal kinase [JNK] pathway, and p38 pathway) (44), we tested specific inhibitors for all three MAP kinase pathways in relation to E. coli Nissle 1917-mediated hBD-2 gene induction. Treatment with SP600125, a selective inhibitor of JNK, blocked hBD-2 induction by E. coli Nissle 1917 (Fig. 7). SB 202190, a specific p38 MAP kinase inhibitor, and PD 98059, a selective inhibitor of ERK1/2, did not block E. coli Nissle 1917-mediated hBD-2 gene induction (Fig. 7). Furthermore hBD-2 gene induction by E. coli Nissle 1917 was also blocked by treatment of the Caco-2 cells with the NF-κB inhibitor helenalin (Fig. 7). In all experiments no changes of cell morphology upon inhibitor treatment were observed, indicating that the inhibitors exhibited no cytotoxic activity.
FIG. 7.
Inhibition of JNK but not ERK and p38 MAP kinase pathways blocks E. coli Nissle 1917-mediated hBD-2 induction in Caco-2 cells. Caco-2 cells were pretreated with the indicated inhibitors at a final concentration of 20 μM for 1 h and then stimulated with 3 × 108 cells of heat-inactivated E. coli Nissle 1917/ml for 4.5 h. Gene expression of hBD-2 was analyzed by real-time PCR. Data represent means ± SEM of triplicate samples. DMSO, dimethyl sulfoxide.
DISCUSSION
Our results demonstrate that active as well as heat-inactivated E. coli Nissle 1917 and several other probiotic bacteria, in contrast to most nonprobiotic E. coli strains, potently induce the antimicrobial peptide hBD-2 in intestinal epithelial cells. The induction of hBD-2 by E. coli Nissle 1917 seems to be a characteristic of this and other probiotic strains, because other E. coli strains (with the exception of UPEC), including several fecal isolates (40 different strains total), failed to induce hBD-2 in Caco-2 cells. The fact that other probiotic bacteria, including several lactobacilli, also induce hBD-2 suggests that the induction of antimicrobial peptides such as hBD-2 may be a common feature of probiotics which enables these bacteria to strengthen barrier function. The induction of hBD-2 through UPEC and Nissle 1917 gives rise to the hypothesis that these bacteria contain similar (or the same?) unique molecular patterns that are responsible for hBD-2 induction and that are not present in other E. coli strains that do not induce hBD-2. But, in contrast to Nissle 1917, UPEC expresses various virulence factors, and many of them are encoded by the pathogenicity islands present in all UPEC strains (2a). It is possible that these virulence factors, which are not expressed by E. coli Nissle 1917, are responsible for the inflammation associated with the pathogenic effects of UPEC. In contrast to hBD-2, hBD-1 remained unaffected by the bacterial strains tested. This is consistent with previous reports demonstrating constitutive hBD-1 gene expression in colon epithelial cells that was not upregulated upon treatment of the cells with E. coli or Salmonella strains (30). Since the promoter region of the hBD-2 gene contains binding sites for NF-κB as well as for AP-1 (13), we analyzed the relevance of these binding sites for E. coli Nissle 1917-mediated hBD-2 induction. Indeed, mutation of the two NF-κB sites in the hBD-2 promoter abolished hBD-2 promoter activation upon treatment with E. coli Nissle 1917, indicating that NF-κB plays a critical role in the regulation of the E. coli Nissle 1917-mediated hBD-2 gene induction. Especially the first proximal NF-κB binding site has a pivotal function for E. coli Nissle 1917-mediated hBD-2 induction, because mutation of this binding site nearly completely abolished the induction of the hBD-2 promoter. In contrast, mutation of the second potential NF-κB binding site at positions −596 to −572 revealed no significant effect, suggesting that this NF-κB binding site plays no role in E. coli Nissle 1917-mediated hBD-2 induction. The involvement of NF-κB in the signal transduction pathway leading to hBD-2 gene induction by E. coli Nissle 1917 was further supported by the observation that the specific NF-κB inhibitor helenalin blocked E. coli Nissle 1917-mediated hBD-2 induction. This fits well with several other reports showing activation of the NF-κB pathway in human epithelial cells upon bacterial challenge. For example, upregulation of hBD-2 in colon epithelial cells infected with enteroinvasive Salmonella enterica serovar Dublin or enteroinvasive E. coli is mediated by NF-κB (30). The S. enterica serovar Enteritidis flagellum filament protein induces hBD-2 expression in Caco-2 cells via NF-κB activation (29), and Helicobacter pylori-mediated hBD-2 induction in gastric mucosal cells, which was recently described in vivo (42), also requires activation of NF-κB (38). However, a recent report from Krisanaprakornkit and coworkers demonstrated that hBD-2 induction by Fusobacterium nucleatum in human gingival epithelial cells was not blocked by NF-κB inhibitors. They found that hBD-2 induction by F. nucleatum is regulated via the p38 and JNK kinase pathways and speculated that the activation of p38 and JNK may regulate gene expression of hBD-2 through transcription factor AP-1 (23). As shown here by site-directed mutagenesis, the proximal AP-1 binding site in the hBD-2 promoter is important for induction of hBD-2 by E. coli Nissle 1917. It is known that AP-1 can be activated by different MAP kinase pathways, including the ERK1/2 pathway, the JNK pathway, and the p38 pathway. Our data indicate that activation of AP-1 through E. coli Nissle 1917 may be regulated by the JNK kinase pathway, because blocking this pathway with the specific JNK kinase inhibitor SP600125 inhibits hBD-2 gene induction by E. coli Nissle 1917 in Caco-2 cells. In contrast, two specific inhibitors for the other MAP kinase pathways, the ERK1/2 and p38 pathways, did not inhibit hBD-2 induction by E. coli Nissle 1917. Although we cannot completely exclude the possibility that the inhibitors used block pathways other than those mentioned, the results of the inhibitor experiments are in full concordance with the data obtained by the gene reporter assays, thus indicating that NF-κB and AP-1 are required for hBD-2 induction in Caco-2 epithelial cells through E. coli Nissle 1917. However, one cannot exclude that possibly that another transcription factor(s) might participate in hBD-2 induction by E. coli Nissle 1917 in Caco-2 cells. The response of the Caco-2 cells to E. coli Nissle 1917 requires specific recognition mechanisms, which should be addressed by future investigations.
It is obvious that some pathogenic strains may suppress defensin release, probably as a self-defense (16, 34), whereas other bacteria, probably including the probiotic E. coli Nissle 1917, are protective to the host organism by inducing defensins. The protective effect documented in several controlled trials (24, 33) may be closely linked to this feature, since in the remission state defensin expression is low (6, 39, 40) and induction may prevent a bacterially induced relapse. Further evidence is given by coculture experiments, where invasion of epithelial cells by enteroinvasive pathogens is greatly reduced in the presence of E. coli Nissle 1917 (3). It remains to be determined whether reduced pathogen growth is caused by increased beta-defensin levels induced by E. coli Nissle 1917.
The present findings of an induction of the innate chemical defense system through E. coli Nissle 1917 may explain various experimental and clinical findings, including the initial hypothesis by the pioneer Alfred Nissle postulating that the presence of E. coli Nissle 1917 recovered from the feces of a soldier immune to salmonellosis prevented this disease (28). Others (26) reported that the load of pathogens recovered from stool samples of newborn infants was reduced when the neonates had been colonized with E. coli Nissle 1917. Experiments with BALB/c mice showed that oral administration of E. coli Nissle 1917 prior to infection with Candida albicans minimized the systemic spread of the pathogen (15). The same effect was observed in Listeria monocytogenes infection experiments (15), as well as in gnotobiotic rats when the Nissle 1917 strain was administered orally prior to a challenge with infective doses of Candida albicans (27). These observations in animal models may well be due to induction of antimicrobial peptides, but more in vivo experiments are required to support this hypothesis.
In conclusion, our data provide the first evidence that the probiotic bacterium E. coli Nissle 1917 and other probiotic strains may exert their beneficial effects through stimulation of the synthesis of endogenous epithelial antimicrobial peptides such as hBD-2.
Acknowledgments
We thank J. Quitzau, K. Schultz, and K. Siegel for excellent technical assistance, and we also thank C. Bevins (University of California, Davis, Davis) for help with the manuscript. We thank U. Zaehringer (Research Center Borstel, Borstel, Germany) for providing highly purified E. coli Nissle 1917 LPS.
This work was supported by the Robert Bosch Foundation, Stuttgart; by Ardeypharm, Herdecke, Germany; and by Deutsche Forschungsgemeinschaft (SFB 617).
Editor: A. D. O'Brien
REFERENCES
- 1.Baquero, F., and F. Moreno. 1984. The microcins. FEMS Microbiol. Lett. 23:117-124. [Google Scholar]
- 1a.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibel, D. J. 1982. Bacterial interference, bacteriotherapy and bacterioprophylaxis, p. 1-12. In R. Aly and H. R. Shinefield (ed.), Bacterial interference. CRC Press, Boca Raton, Fla.
- 2a.Blum, G., R. Marre, and J. Hacker. 1995. Properties of Escherichia coli strains of serotype O6. Infection 23:234-236. [DOI] [PubMed] [Google Scholar]
- 3.Boudeau, J., A. L. Glasser, S. Julien, J. F. Colombel, and A. Darfeuille-Michaud. 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease. Aliment. Pharmacol. Ther. 18:45-56. [DOI] [PubMed] [Google Scholar]
- 3a.Boyer, H. W., and D. Roulland-Dussoix. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 1969 41:459-472. [DOI] [PubMed]
- 4.Cukrowska, B., R. Lodinova-Zadnikova, C. Enders, U. Sonnenborn, J. Schulze, and H. Tlaskalova-Hogenova. 2002. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand. J. Immunol. 55:204-209. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe, R. N., F. R. A. J. Rose, J. Keyte, L. Abberley, W. C. Chan, and Y. R. Mahida. 2001. Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut 48:176-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahlgren, A., S. Hammarstrom, A. Danielsson, and M. L. Hammarstrom. 2003. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin. Exp. Immunol. 131:90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellermann, K., J. Wehkamp, K. R. Herrlinger, and E. F. Stange. 2003. Crohn′s disease: a defensin deficiency syndrome? Eur. J. Gastroenterol. Hepatol. 15:627-634. [DOI] [PubMed] [Google Scholar]
- 8.Fellermann, K., and E. F. Stange. 2001. Defensins—innate immunity at the epithelial frontier. Eur. J. Gastroenterol. Hepatol. 13:771-776. [DOI] [PubMed] [Google Scholar]
- 9.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 10.Ganz, T., and R. I. Lehrer. 1994. Defensins. Curr. Opin. Immunol. 6:584-589. [DOI] [PubMed] [Google Scholar]
- 11.Grozdanov, L., U. Zaehringer, G. Blum-Oehler, L. Brade, A. Henne, Y. A. Knirel, U. Schombel, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, E. T. Rietschel, and U. Dobrindt. 2002. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 184:5912-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 2001. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 13.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schröder. 2000. Mucoid Pseudomonas aeruginosa, TNF-α, and IL-1β, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell. Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 14.Hockertz, S. 1991. Immunomodulating effect of killed, apathogenic Escherichia coli, strain Nissle 1917, on the macrophage system. Arzneim.-Forsch./Drug Res. 41:1108-1112. [PubMed] [Google Scholar]
- 15.Hockertz, S. 1997. Augmentation of host defence against bacterial and fungal infections of mice pretreated with the non-pathogenic Escherichia coli strain Nissle 1917. Arzneim.-Forsch./Drug Res. 47:793-796. [PubMed] [Google Scholar]
- 16.Islam, D., L. Bandholtz, J. Nilsson, H. Wigzell, B. Christensson, B. Agerberth, and G. Gudmundsson. 2001. Downregulation of bactericidal peptides in enteric infections: an immune escape mechanism with bacterial DNA as a potential regulator. Nat. Med. 7:180-185. [DOI] [PubMed] [Google Scholar]
- 17.Isolauri, E. 2001. Probiotics in human disease. Am. J. Clin. Nutr. 73:1142S-1146S. [DOI] [PubMed] [Google Scholar]
- 18.Isolauri, E., P. V. Kirjavainen, and S. Salminen. 2002. Probiotics: a role in the treatment of intestinal infection and inflammation? Gut 50(Suppl. 3):III54-III59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, D. E., and C. L. Bevins. 1992. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 267:23216-23225. [PubMed] [Google Scholar]
- 20.Kagan, B. L., M. E. Selsted, T. Ganz, and R. I. Lehrer. 1990. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl. Acad. Sci. USA 87:210-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaenhammer, T. R. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337-349. [DOI] [PubMed] [Google Scholar]
- 22.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 23.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-κB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 24.Kruis, W., E. Schutz, P. Fric, B. Fixa, G. Judmaier, and M. Stolte. 1997. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 11:853-858. [DOI] [PubMed] [Google Scholar]
- 24a.Kruszewska, K., J. Lan, G. Lorca, et al. 2002. Selection of lactic acid bacteria as probiotic strains by in vitro tests. Microecol. Ther. 29:37-51. [Google Scholar]
- 24b.Lan, J.-G., and N. Yamagisawa. 2002. Isolation, selection and characteristics of Lactobacillus paracasei ssp. paracasei isolate F19. Microb. Ecol. Health Dis. Suppl. 3:4-6.
- 25.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodinova-Zadnikova, R., and U. Sonnenborn. 1997. Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol. Neonate 71:224-232. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz, A., and J. Schulze. 1996. Establishment of E. coli Nissle 1917 and its interaction with Candida albicans in gnotobiotic rats. Microecol. Ther. 24:45-51. [Google Scholar]
- 28.Nissle, A. 1918. Die antagonistische Behandlung chronischer Darmstörungen mit Colibakterien. Medizinische Klinik No. 2:29-33. [Google Scholar]
- 29.Ogushi, K., A. Wada, T. Niidome, N. Mori, K. Oishi, T. Nagatake, A. Takahashi, H. Asakura, S. Makino, H. Hojo, Y. Nakahara, M. Ohsaki, T. Hatakeyama, H. Aoyagi, H. Kurazono, J. Moss, and T. Hirayama. 2001. Salmonella enteritidis FliC (flagella filament protein) induces hu beta-defensin-2 mRNA production by Caco-2 cells. J. Biol. Chem. 276:30521-30526. [DOI] [PubMed] [Google Scholar]
- 30.O′Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 31.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 32.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 33.Rembacken, B. J., A. M. Snelling, P. M. Hawkey, D. M. Chalmers, and A. T. Axon. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635-639. [DOI] [PubMed] [Google Scholar]
- 34.Salzman, N. H., M. M. Chou, H. de Jong, L. Liu, E. M. Porter, and Y. Paterson. 2003. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect. Immun. 71:1109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman, N. H., D. Ghosh, K. M. Huttner, Y. Paterson, and C. L. Bevins. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522-526. [DOI] [PubMed] [Google Scholar]
- 36.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsumi-Ishii, Y., and I. Nagaoka. 2003. Modulation of human beta-defensin-2 transcription in pulmonary epithelial cells by lipopolysaccharide-stimulated mononuclear phagocytes via proinflammatory cytokine production. J. Immunol. 170:4226-4236. [DOI] [PubMed] [Google Scholar]
- 38.Wada, A., K. Ogushi, T. Kimura, H. Hojo, N. Mori, S. Suzuki, A. Kumatori, M. Se, Y. Nakahara, M. Nakamura, J. Moss, and T. Hirayama. 2001. Helicobacter pylori-mediated transcriptional regulation of the human β-defensin 2 gene requires NF-κB. Cell. Microbiol. 3:115-123. [DOI] [PubMed] [Google Scholar]
- 39.Wehkamp, J., K. Fellermann, K. R. Herrlinger, S. Baxmann, K. Schmidt, B. Schwind, M. Duchrow, C. Wohlschlager, A. C. Feller, and E. F. Stange. 2002. Human beta-defensin 2 but not beta-defensin 1 is expressed preferentially in colonic mucosa of inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 14:745-752. [DOI] [PubMed] [Google Scholar]
- 40.Wehkamp, J., J. Harder, M. Weichenthal, O. Mueller, K. R. Herrlinger, K. Fellermann, J. M. Schroeder, and E. F. Stange. 2003. Inducible and constitutive beta-defensins are differentially expressed in Crohn's disease and ulcerative colitis. Inflamm. Bowel Dis. 9:215-223. [DOI] [PubMed] [Google Scholar]
- 41.Wehkamp, J., J. Harder, M. Weichenthal, M. Schwab, E. Schaeffeler, M. Schlee, A. Stallmach, F. Noack, P. Fritz, J. M. Schroder, C. L. Bevins, K. Fellermann, and E. F. Stange. NOD2 (CARD15) mutations in Crohn′s disease are associated with diminished mucosal α-defensin expression. Gut, in press. [DOI] [PMC free article] [PubMed]
- 42.Wehkamp, J., K. Schmidt, K. R. Herrlinger, S. Baxmann, S. Behling, C. Wohlschlager, A. C. Feller, E. F. Stange, and K. Fellermann. 2003. Defensin pattern in chronic gastritis: HBD-2 is differentially expressed with respect to Helicobacter pylori status. J. Clin. Pathol. 56:352-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wehkamp, J., B. Schwind, K. R. Herrlinger, S. Baxmann, K. Schmidt, M. Duchrow, C. Wohlschlager, A. C. Feller, E. F. Stange, and K. Fellermann. 2002. Innate immunity and colonic inflammation: enhanced expression of epithelial alpha-defensins. Dig. Dis. Sci. 47:1349-1355. [DOI] [PubMed] [Google Scholar]
- 44.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589-607. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, C. L., A. J. Ouellette, D. P. Satchell, T. Ayabe, Y. S. Lopez-Boado, J. L. Stratman, S. J. Hultgren, L. M. Matrisian, and W. C. Parks. 1999. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286:113-117. [DOI] [PubMed] [Google Scholar]
- 46.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schröder, J. M. Wang, O. M. Z. Howard, and J. J. Oppenheim. 1999. β-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]