Abstract
Escherichia coli K1 survival in the blood is a critical step for the onset of meningitis in neonates. Therefore, the circulating bacteria are impelled to avoid host defense mechanisms by finding a niche to survive and multiply. Our recent studies have shown that E. coli K1 enters and survives in both monocytes and macrophages in the newborn rat model of meningitis as well as in macrophage cell lines. Here we demonstrate that E. coli K1 not only extends the survival of human and murine infected macrophage cell lines but also renders them resistant to apoptosis induced by staurosporine. Macrophages infected with wild-type E. coli expressing outer membrane protein A (OmpA), but not with OmpA− E. coli, are resistant to DNA fragmentation and phosphatidylserine exposure induced by staurosporine. Infection with OmpA+ E. coli induces the expression of BclXL, an antiapoptotic protein, both at the mRNA level as assessed by gene array analysis and at the protein level as evaluated by immunoblotting. OmpA− E. coli infection of macrophages induced the release of cytochrome c from mitochondria into the cytosol and the activation of caspases 3, 6, and 9, events that were significantly blocked in OmpA+ E. coli-infected macrophages. In addition, OmpA+ E. coli-infected cells were resistant to a decrease in the transmembrane potential of mitochondria induced by staurosporine as measured by the MitoCapture fluorescence technique. Complementation of OmpA− E. coli with a plasmid containing the ompA gene restored the ability of OmpA− E. coli to inhibit the apoptosis of infected macrophages, further demonstrating that E. coli OmpA expression is critical for inducing macrophage survival and thereby finding a safe haven for its growth.
Apoptosis plays a critical role not only during development and homeostasis but also in the regulation of host response during infection with bacteria, viruses, and parasites (26, 39). It has been demonstrated that the apoptosis of infected cells can limit the spread of intracellular microorganisms by provoking inflammatory responses as a complementary mechanism for the removal of these cells through the recruitment of phagocytes (39). Furthermore, induction of target cell apoptosis constitutes an important part of the nonspecific immune responses, and an imbalance in apoptotic responses contributes to several pathological conditions including cancers (23). Therefore, it is essential for intracellular pathogens to develop strategies to inhibit host cell apoptosis.
Escherichia coli K1 is a leading causative agent of neonatal meningitis, and the morbidity and mortality rates due to this pathogen have remained unchanged for the last decade (19, 22). The most important aspect in the pathogenesis of E. coli meningitis is the development of a threshold level of bacteremia, suggesting that the bacteria must survive and propagate in the blood. Our studies have shown that E. coli K1 avoids bactericidal activity in serum by binding via OmpA to C4b-binding protein, a classical complement pathway fluid phase regulator (29). Nevertheless, the circulating bacteria can still be attacked by phagocytes, especially neutrophils, which have the capacity to cleave OmpA by using elastase (4). Besides the role of OmpA in survival in serum, it also plays a role in E. coli entry and survival of the bacteria in human (THP-1) and murine (RAW 264.7) macrophage-like cell lines (33), suggesting that E. coli K1 finds a niche in macrophages to avoid both killing and neutrophil attack in serum. In agreement with the in vitro data, it has also been reported that E. coli enters and survives in both monocytes and macrophages in the newborn rat model of hematogenous meningitis (33). Interestingly, the entry of E. coli into these cells does not require opsonization, suggesting that OmpA of E. coli can directly interact with monocytes and macrophages for entry. In contrast, OmpA− E. coli taken up by the macrophages by an OmpA-independent mechanism, although in small numbers, is killed within an hour. Survival and multiplication of OmpA+ E. coli result in the demise of the macrophages that burst open, releasing the intracellular bacteria by 8 h postinfection. Therefore, it is essential to maintain the integrity of the host cells during E. coli intracellular growth, not only for obtaining nutrients from macrophages for multiplication but also for shielding the intracellular bacteria from host phagocytosis. We therefore speculated that intracellular E. coli might be able to actively inhibit infected host cells from undergoing apoptosis.
Two major pathways leading to apoptosis have been described. One pathway involves apoptosis mediated by death receptors, such as CD95 (Fas) and tumor necrosis factor alpha (TNF-α) receptors (2, 42). The binding of Fas ligand to Fas receptor activates caspase 8, which processes effector caspases (caspases 3, 6, and 7), thereby inducing apoptosis. In the other pathway, various proapoptotic signals converge at the mitochondrial level, provoking the translocation of cytochrome c from the mitochondria to the cytosol (15). The released cytochrome c binds to Apaf-1 and activates caspase 9, which in turn activates caspase 3. Activation of the caspase cascade family leads to the cleavage of a variety of target proteins with structural or regulatory function, including poly(ADP-ribose) polymerase (PARP), nuclear lamins, protein kinase C, and others leading to destruction of the cell. Furthermore, many proteins of the Bcl-2 family with either antiapoptotic (e.g., Bcl-2, BclXL, or Mac1) or proapoptotic (Bax, Bak, or Bik) functions reside in the outer membrane of the mitochondria (1, 16). The antiapoptotic molecules Bcl-2 and BclXL prevent the translocation of cytochrome c from the mitochondria, while the induced expression or enforced dimerization of Bax results in dysfunction, leading to cytochrome c release.
Although phagocytes upon ingestion kill most bacteria, infection with some bacteria that are capable of surviving inside phagocytes eventually leads to the death of the host cell (8, 10, 11, 14, 18, 23, 25, 32). However, a variety of bacteria and viruses that survive inside host cells have developed strategies to interfere with the host cell apoptotic process (6, 28, 31, 41). Here we show that E. coli K1 interferes with cytochrome c release from the mitochondria and subsequent down-regulation of caspase activity by inducing the expression of BclXL as an antiapoptotic mechanism in macrophages. Interestingly, expression of OmpA in E. coli is important to induce the antiapoptotic mechanism, which was further confirmed by complementation of OmpA− E. coli with a plasmid expressing ompA. This is the first report to show the induction of BclXL in host cells by a bacterial pathogen.
MATERIALS AND METHODS
Reagents and cells.
Staurosporine (ST) and phorbol-12-myristate-13-acetate were obtained from Sigma Chemical Co. (St. Louis, Mo.). A MitoCapture apoptosis detection kit was obtained from BioVision Labs (Mountain View, Calif.). Antibodies to caspases 3, 6, and 9 and to BclXL, Bax, and β-actin were purchased from Cell Signaling Inc. (Beverly, Mass.). Anti-cytochrome c antibody was obtained from eBiosciences (San Diego, Calif.). All these antibodies cross-react with both human and mouse proteins except caspase 9, in which case individual antibodies were used for immunoblotting. The following cell lines used in this study were obtained from American Type Tissue Culture: THP-1 cells were differentiated with and phorbol-12-myristate-13-acetate (300 ng/ml) for 48 h prior to infection and cultured in RPMI medium with 10% fetal calf serum (Omega Scientific Inc.). These cells were designated as THP-M cells. RAW 264.7 cells were cultured in Dulbecco's modified Eagle medium with 10% fetal calf serum. All studies described were carried out by using both these cell lines.
E. coli strains.
E. coli E44 is a rifampin-resistant mutant of E. coli K1 strain RS 218 (serotype O18:K1:H7), which was isolated from the cerebrospinal fluid of a neonate with meningitis, and invades human brain microvascular endothelial cells in vitro (30). A mutant lacking the entire ompA gene (E91) was generated from strain E44 in two steps. First, a tetracycline resistance marker was mobilized from DME 558, which contains the marker neo ompA, by P1 transduction to E. coli K-12 strain BRE51 in which the ompA gene had been deleted (6). Then, a P1 lysate was used to transduce E. coli K1 strain E44. Tetracycline-resistant colonies were screened for lack of OmpA expression by Western blot analysis to identify mutant E91. Bacteria were grown in brain heart infusion broth (Difco Laboratories) with appropriate antibiotics.
E. coli infection.
THP-M or RAW 264.7 cells were grown in 24-well culture dishes until they reached confluence and then were infected with E. coli strains separately at a multiplicity of infection (MOI) of 10 for 1 h. For longer periods of incubation, the cells were washed to remove unbound bacteria and further incubated with medium containing gentamicin (20 μg/ml). The minimum concentration of gentamicin to inhibit the growth of E. coli is 8 μg/ml. Several investigators have used gentamicin in these cells with no significant effect on the intracellular bacteria for more than 48 h (37). In some experiments, host cells were infected with E. coli for various periods of time, washed, and then treated with 1 μM ST for 3 h in medium containing gentamicin. Control cells were treated with dimethyl sulfoxide alone.
Western blot analysis.
Following infection with E. coli strains as described above, cells were washed with ice-cold phosphate-buffered saline (PBS) and placed on ice. The cells in each 60-mm dish were harvested by scraping into 2-ml ice-cold cell homogenization buffer consisting of 20 mM Tris (pH 7.5), 0.25 M sucrose, 10 mM EGTA, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, leupeptin (10 μg/ml), and 2 mM dithiothreitol. Cells were subjected to mild sonication, and a portion of the resulting lysate was centrifuged at low speed (5,000 × g) to remove debris. The protein concentrations were determined by using a Bio-Rad protein assay. Total protein (25 μg) was subjected to polyacrylamide gel electrophoresis on either 10 or 12% gels by using a Bio-Rad Mini PROTEAN II apparatus. The gels were transferred to nitrocellulose by using a Bio-Rad semidry blotting apparatus as recommended by the manufacturer. The blot was incubated with 5% skim milk in PBS containing 0.2% Tween at room temperature for 1 h. The blots were then incubated with primary antibodies (dilution, 1:1,000) in PBS-0.2% Tween at room temperature for 2 h, followed by washing three times with PBS. The blots were further incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (dilution, 1: 5,000). Following three washes with PBS, the immunoreactive bands were visualized by using enhanced chemiluminescence reagent (Pierce) and were exposed to X-ray film.
Annexin V labeling.
THP-M or RAW 264.7 cells were seeded in 8-well chamber slides and cultured to confluence. Appropriate wells were either left uninfected or infected with E. coli strains at an MOI of 10 for 1 h at 37°C. Apoptosis was induced by treating the cells with 1 μM ST for 30 min, followed by washing with tissue culture medium. The cells were then rinsed three times with 1× binding buffer and further incubated with binding buffer containing 5 μl of annexin V for 15 min at room temperature in the dark. The cells were washed with PBS and then fixed in 2% paraformaldehyde in PBS for 15 min at room temperature in the dark. After cells were washed with PBS again, the chambers were removed, and the slides were mounted in Vectashield (Vector Laboratories) antifade solution containing 4′, 6-diamidino-2-phenylindole (DAPI). Cells were viewed with a Leica (Wetzlar, Germany) DMRA microscope with Plan-apochromat 40× (numerical aperture of 1.25) and 63× (numerical aperture of 1.40) oil immersion objective lenses. For quantitative analysis, the number of fluorescent cells per field was counted and expressed as the percent positive of total cells. Images were acquired with a SkyVision-2/VDS digital charge-coupled device camera (12-bit; 1,280 by 1,024 pixels) in unbinned or 2-by-2 binned models into EasyFISH software, saved as 16-bit monochrome images, and merged as 24-bit RGB TIFF images (Applied Spectral Imaging Inc., Carlsbad, Calif.). The images were processed by using Adobe Photoshop 6.0.
DNA fragmentation assay.
The DNA fragmentation assay was performed as described earlier (9). Briefly, macrophage monolayers in 100-mm dishes were treated with ST either uninfected or infected with E. coli strains for various periods of time. The cells were lysed in 2 ml of 7 M guanidine hydrochloride. Genomic DNA was isolated by using a Miniprep DNA purification kit according to the manufacturer's protocol (QIAGEN, Inc). After samples were eluted from the miniprep columns, contaminating RNA was digested by incubation with 1 μg of RNase A (Sigma) for 30 min at 37°C. DNA (20 μg) was then electrophoretically separated in a 1% agarose gel, visualized by ethidium bromide staining, and photographed.
Preparation of mitochondrial fractions.
Cell samples either uninfected or infected with E. coli strains were collected by scraping into ice-cold PBS and centrifuged at 600 × g for 10 min at 4°C; pellets were resuspended in buffer A (20 mM HEPES-KOH [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride) containing 250 mM sucrose on ice for 15 min. The cells were homogenized with 15 to 20 strokes by using a Kontes douncer. The homogenates were centrifuged twice at 750 × g for 10 min at 4°C and then at 10,000 × g for 15 min at 4°C, and the resulting pellets containing mitochondria (designated the mitochondrial fractions) were resuspended in buffer A containing 250 mM sucrose. The supernatants were further cleared of mitochondrial contamination by centrifugation at 100,000 × g for 1 h at 4°C, and the resultant supernatants were designated the cytosolic fractions. Proteins (30 μg) from these two fractions were separated on 12% polyacrylamide gel and subjected to Western blotting.
Gene array analysis.
The GEArray Original Series (hGEA990502N) cDNA expression array contains up to 23 pairs of cDNA fragments of apoptosis-related genes, including caspases and other mediators of apoptosis. β-Actin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) genes are included as controls. Total RNA from control, OmpA+ E. coli-infected, or OmpA− E. coli-infected THP-M cells were prepared by using an RNeasy minikit (QIAGEN). The integrity of the RNA preparation was assessed by agarose gel electrophoresis, followed by ethidium bromide staining. Biotin-dUTP-labeled cDNA probes were prepared according to the manufacturer's protocol. The array membrane was washed with deionized water and incubated with prehybridization buffer containing 100 μg of heat-denatured salmon sperm DNA per ml for 2 h at 68°C with continuous agitation. The prehybridization buffer was replaced with the same buffer containing denatured biotin-labeled probe and incubated overnight at 68°C with continuous agitation. The membrane was then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer containing 1% sodium dodecyl sulfate (SDS), followed by 0.1× SSC containing 0.5% SDS for 20 min each. After the final wash was discarded, the membrane was incubated with GEA blocking solution for 40 min at room temperature, followed by incubation with streptavidin peroxidase (dilution, 1:5,000) in binding buffer for 15 min. The membrane was washed four times with the buffer and exposed to CDP-Star chemiluminescent substrate. The membrane was exposed to X-ray film, and image acquisition was performed by using Adobe Photoshop 6.0. Densitometric analysis was carried out with ImageJ software from the National Institutes of Health (NIH; http://rsb.info.nih.gov/ij).
Detection of changes in mitochondrial transmembrane potential.
RAW 264.7 cells were seeded in 8-well chamber slides and cultured to confluence. The cells were infected with OmpA+ or OmpA− E. coli alone or followed by treatment with ST for 1 h at 37°C. After washing with PBS, cells were incubated in diluted MitoCapture solution (BioVision Research Products) at 37°C in a 5% CO2 incubator for 20 min. The monolayers were washed with the incubation buffer three times and mounted with coverslips with antifade solution. Cells were observed under a fluorescence microscope, and images were captured by using EasyFISH software and merged by Adobe Photoshop 6.0.
RESULTS
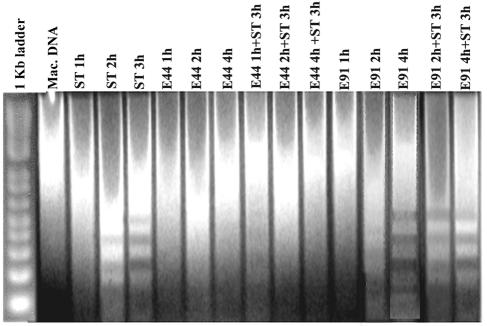
Infection of macrophages with OmpA+E. coli induces antiapoptotic signals in the host cells. Earlier studies showed that OmpA+ E. coli enters macrophages and survives over time inside the cells without inducing apoptosis (33). In contrast, a small number of OmpA− E. coli organisms were also taken up by macrophages, presumably by a different mechanism, and were killed within a short time. Unlike cells invaded by OmpA+ E. coli, those infected with OmpA− E. coli eventually undergo apoptosis as evident by transmission electron microscopy, suggesting that OmpA+ E. coli organisms may be inducing survival strategies in macrophages to enable their own growth. To verify this possibility, both THP-M and RAW 264.7 cells were infected with OmpA+ or OmpA− E. coli for 1, 2, and 4 h at an MOI of 10. Changes in nuclear organization are caused by the activation of specific nucleases and the advent of apoptosis, resulting in DNA fragmentation (42). Therefore, the integrity of infected macrophage cell lines was analyzed by agar gel electrophoresis of total genomic DNA. As shown in Fig. 1, the total DNA from RAW 264.7 cells infected with OmpA+ E. coli showed no signs of DNA laddering even after 4 h, whereas OmpA− E. coli infection induced the degradation of DNA into multimers of 180 to 200 bp by 2 to 4 h postinfection.
FIG. 1.
Inhibition of DNA fragmentation in OmpA+ E. coli-infected RAW 264.7 cells with or without ST treatment. Total cellular DNA was isolated after 3 h from untreated control (Mac. DNA), ST-treated, OmpA+ E. coli-infected, and OmpA− E. coli-infected RAW 264.7 cells and from RAW 264.7 cells treated with ST after OmpA+ and OmpA− E. coli infection. The DNA (20 μg) samples were analyzed by electrophoresis in 1% agarose gel and stained with ethidium bromide. The results of one representative experiment of three performed are shown.
To further examine whether the OmpA+ E. coli-infected macrophages are resistant to external apoptotic stimuli, the infected cells were treated with ST, a kinase inhibitor that induces apoptosis by the mitochondrial pathway (5, 34, 35). RAW 264.7 cells infected with bacteria for various periods of time were washed and incubated with medium containing gentamicin and ST for an additional 3 h. The total DNA from macrophages treated with ST alone showed DNA laddering after 2 h. Interestingly, DNA from macrophages infected with OmpA+ E. coli for 1 h prior to ST treatment showed a slight smearing of DNA, which could be due to the effect of ST on cells not infected by 1 h. But no evidence of chromatin cleavage or degradation was observed in cells infected with OmpA+ E. coli at 2 and 4 h, despite treatment with ST, and infected cells had a DNA pattern similar to that of uninfected cells. In contrast, the RAW 264.7 cells infected with OmpA− E. coli were not protected from apoptosis even when treated with ST. Similar results were also obtained with THP-M cells (data not shown).
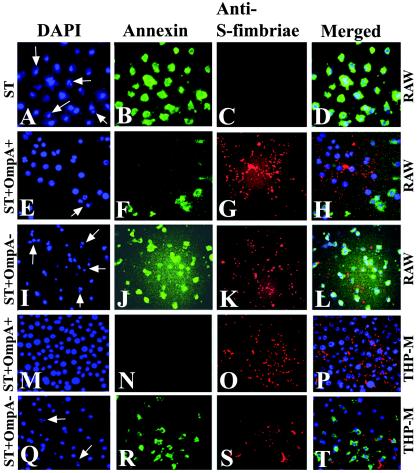
Apoptosis is associated with the loss of plasma membrane asymmetry manifested by the translocation of phosphatidylserine from the inner to the outer leaflet of the plasma membrane (24). This change can be readily detected by the calcium-dependent binding of fluorescently labeled annexin V to the plasma membrane. Since phosphatidylserine exposure is an early event in apoptosis, macrophages were treated with ST for 30 min with or without prior infection with bacteria. As expected, ST treatment of uninfected macrophages showed that almost all cells stained for annexin V binding (Fig. 2B). Nuclear staining of the same cells with DAPI also showed fragmentation of the nucleus in several cells (Fig. 2A). In contrast, significantly fewer annexin V-positive cells were observed with OmpA+ E. coli-infected macrophages treated with ST under similar conditions (95% ± 5% positive cells for both ST and OmpA− E. coli versus 26% ± 7% for OmpA+ E. coli; P < 0.0001 by paired t test) (Fig. 2F). The infected cells showed that a significant number of bacteria associated with the macrophages when cells were stained with anti-S-fimbriae antibody followed by Cy3-conjugated secondary antibody (Fig. 2G). Most of the macrophages infected with OmpA+ E. coli contain at least one bacteria per cell inside, as shown by a differential staining method (33), whereas cells infected with OmpA− E. coli showed fragmented bacteria (data not shown). The cells that showed positive staining for annexin V in the OmpA+ E. coli-infected monolayer were probably not infected, as there were no bacteria observed in the vicinity of these cells (Fig. 2H). On the other hand, 90% of the cells infected with OmpA− E. coli stained positive for annexin V, despite containing several bacteria on the monolayer (Fig. 2J). Moreover, several of these cells showed fragmentation of the nuclei (Fig. 2I), which are similar to the nuclei observed in ST-treated but uninfected cells. Similarly, THP-M cells infected with OmpA+ E. coli also became resistant to ST treatment (Fig. 2M to P); however, OmpA− E. coli-infected cells could not survive (Fig. 2Q to T). Taken together, these data indicate that OmpA+ E. coli infection inhibits the apoptosis of macrophages even after an attempt is made to induce apoptosis by an external stimulus.
FIG. 2.
Inhibition of apoptotic signals induced by ST in OmpA+ E. coli-infected macrophages. Confluent macrophages (RAW 264.7 and THP-M) in 8-well chamber slides either uninfected (A to D) or infected with OmpA+ E. coli (E to H and M to P) or OmpA− E. coli (I to L and Q to T) were treated with ST for 30 min. The cells were then fixed with 2% paraformaldehyde for 15 min, followed by staining with annexin V and fluorescein isothiocyanate. The monolayers containing the bacteria were later incubated with anti-S-fimbriae antibody (dilution, 1:1,000) for 1 h at room temperature, followed by incubation with Cy3-conjugated secondary antibody (dilution, 1:5,000). The cells were then washed and mounted with a coverslip with antifade solution containing DAPI. These experiments were carried out two times in triplicate. The cells were viewed by using a microscope equipped with both red and green filters. Images were acquired by EasyFISH software, converted into TIFF files, and merged by using Adobe Photoshop 6.0. Arrows indicate fragmented nuclei.
OmpA+ E. coli infection of macrophages induces the expression of antiapoptotic protein BclXL.
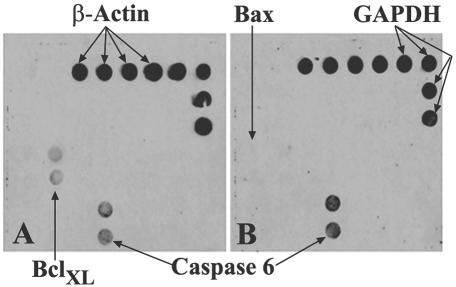
In an attempt to determine the mechanisms by which OmpA+ E. coli exerts its antiapoptotic effects in macrophages, gene array analysis was performed by using RNA isolated from THP-M cells infected with either OmpA+ or OmpA− E. coli. The human gene array membrane obtained from SuperArray Inc. contains 23 genes related to apoptosis and includes the two housekeeping genes β-actin and GAPDH. The films were scanned, and the density of each gene was estimated by normalizing to GAPDH. The RNA from control uninfected cells showed no hybridization with any of the apoptotic genes but hybridized with housekeeping genes under the conditions employed (data not shown). Increased expression of BclXL was observed in THP-M cells infected with OmpA+ E. coli (Fig. 3A). In contrast, macrophages infected with OmpA− E. coli revealed an increase in the expression of the caspase 6 gene (Fig. 3B) and of the Bax genes when the membrane was exposed to the film for longer periods of time (data not shown). The induction of BclXL gene expression was threefold greater than that of the gene expression in macrophages infected with OmpA− E. coli. The level of caspase 6 expression in OmpA+ E. coli-infected cells, though up-regulated, was reduced in signal by more than 50% compared to the levels of caspase 6 expression in THP-M cells infected with OmpA− E. coli. The expression levels of the housekeeping genes were similar in all three macrophages (control and OmpA+ and OmpA− E. coli-infected cells). These results suggest that OmpA+ E. coli induces significantly greater gene expression of an antiapoptotic protein, BclXL, for macrophage survival, which may in turn utilize the macrophage environment for its own propagation.
FIG. 3.
Apoptotic gene array analysis with RNA from THP-M cells infected with E. coli. Gene array membranes containing 23 apoptotic genes including β-actin and GAPDH are screened by using the RNA isolated from THP-M cells infected with either OmpA+ (A) or OmpA− (B) E. coli for 1 h; a nonradioactive detection method was used, and the blots were exposed to X-ray film and scanned. The densities of the spots were calculated by using the NIH software Image J.
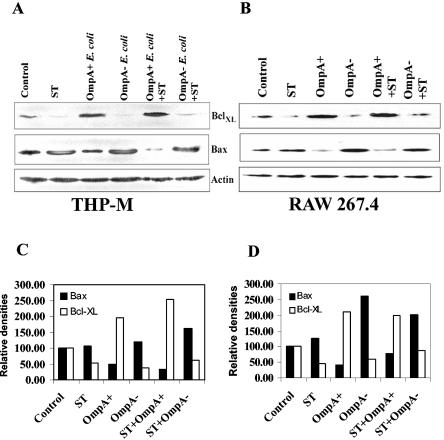
Next we examined the expression of BclXL and Bax at the protein level by Western blot analysis. In agreement with the gene array analysis, the protein expression of BclXL increased significantly in both THP-M and RAW 264.7 cells infected with OmpA+ E. coli (Fig. 4A and B). In contrast, macrophages treated with either ST or infected with OmpA− E. coli showed decreased levels of BclXL compared to levels in control uninfected cells. In addition, the decrease in levels of BclXL usually observed in ST-treated macrophages was inhibited when cells were infected with OmpA+ E. coli, but not with OmpA− E. coli, prior to ST treatment. The expression level of Bax, a proapoptotic protein, increased in ST-treated and OmpA− E. coli-infected cells compared to expression in OmpA+ E. coli-infected cells. In addition, the increase in Bax expression was significantly blocked when cells were infected with OmpA+ E. coli prior to ST treatment. The blot was reprobed with anti-β-actin antibody to determine the equality of protein loading. Densitometric analysis of all bands that were normalized to that of β-actin revealed that BclXL expression was twofold higher in OmpA+ E. coli-infected macrophages compared to levels in control cells (Fig. 4C and D). In contrast, Bax expression was two- to threefold lower than that observed in ST-treated cells. Taken together, these results demonstrate that OmpA+ E. coli induces the expression of BclXL at the protein level in both THP-M and RAW 264.7 cells.
FIG. 4.
Increased expression of BclXL in OmpA+ E. coli-infected macrophage cell lines. Confluent monolayers of either THP-M (A) or RAW 264.7 (B) cells were treated with ST with or without prior infection with either OmpA+ or OmpA− E. coli. In separate experiments the cells were infected with bacteria only. Total cell lysates were prepared, separated by SDS-10% polyacrylamide gel electrophoresis, and subjected to immunoblotting with anti-BclXL, anti-Bax, or anti-β-actin antibodies. Total cell lysates from untreated cells were used as controls. (C and D) The blots were scanned and plotted by using arbitrary units of the densities that are normalized to the densities of the respective β-actin bands. The densities of bands in the control are taken as 100%.
Mitochondrial cytochrome c release is blocked by OmpA+ E. coli infection of macrophages.
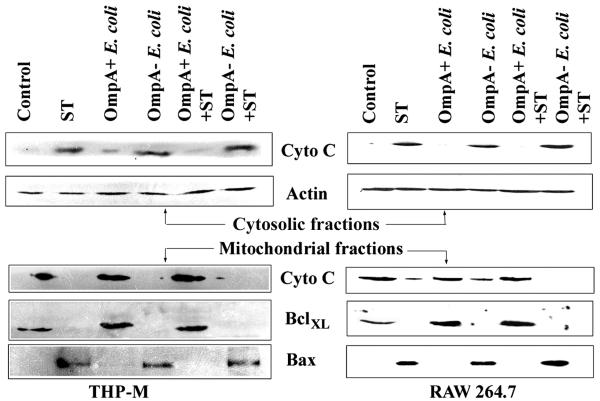
It has been suggested that mitochondria play an important role in cellular apoptosis (15). Alteration of the ratio of BclXL to Bax expressed on the mitochondrial outer membrane induces changes in the membrane potential, which results in the release of cytochrome c into cytosol (7). The released cytochrome c then gives signals for the activation of caspase 9. Since we observed an increase in the expression of BclXL in OmpA+ E. coli-infected macrophages, we tested whether OmpA+ E. coli infection prevents cytochrome c release in these cells treated with ST. Cytosolic and mitochondrial fractions were prepared from both THP-M and RAW 264.7 cells after various treatments, which were then analyzed by immunoblotting. In untreated control cells, cytochrome c was exclusively present in the mitochondrial fraction and was not detectable in the cytosolic fraction (Fig. 5). OmpA+ E. coli-infected macrophage fractions also showed similar results to those of uninfected cells. In contrast, ST treatment induced the release of cytochrome c from mitochondria into the cytosol, as indicated by a significant decrease in expression in the mitochondrial fraction and a simultaneous increase of expression in the cytosolic fraction. However, macrophages infected with OmpA+ E. coli prior to ST treatment significantly blocked the release of cytochrome c from the mitochondria to the cytosol. OmpA− E. coli infection of macrophages could not protect these cells from ST-induced cytochrome c release. The blots containing the cytosolic fractions when reprobed with anti-β-actin antibody showed the presence of equal quantities of protein in all the lanes.
FIG. 5.
OmpA+ E. coli-infected macrophages are protected from cytochrome c release into the cytoplasm from mitochondria induced by ST. Both THP-M and RAW 264.7 cells grown in 100-mm dishes were either untreated (control) or treated with ST for 3 h after infection with OmpA+ and OmpA− E. coli for 1 h as described in Materials and Methods. The mitochondrial and cytosolic fractions were prepared, and the proteins (30 μg) were separated by SDS-10% polyacrylamide gel electrophoresis. The cytosolic proteins were subjected to immunoblotting with anti-cytochrome c antibody (dilution, 1:1,000). The same blot was stripped and reprobed with anti-β-actin antibody (dilution, 1:1,000) to determine the equality of loading. The mitochondrial fractions were subjected to immunoblotting with anti-BclXL, anti-Bax, and anti-cytochrome c antibodies.
We next examined the presence of BclXL and Bax proteins in the mitochondrial fractions of the macrophages. As expected, ST-treated cells completely lost BclXL from the mitochondria compared to untreated cells, whereas Bax was present in significant quantities. In contrast, the amount of BclXL in the mitochondrial fraction of OmpA+ E. coli-infected macrophages was 50% greater than the BclXL quantities in untreated cells, whereas Bax was undetectable in this fraction. In addition, the OmpA+ E. coli-infected cells, but not OmpA− E. coli-infected cells, were resistant to ST-induced loss of BclXL from the mitochondria, suggesting that OmpA+ E. coli inhibits cytochrome c release from mitochondria by increasing the expression of BclXL.
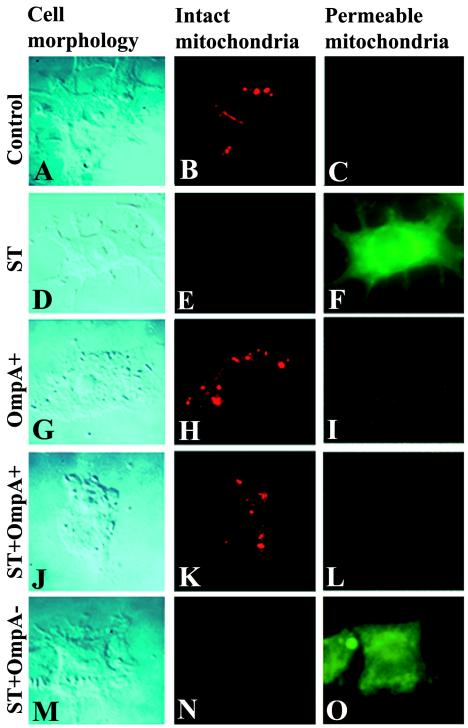
These observations were further confirmed at the single-cell level by immunofluorescence by using a MitoCapture detection kit. Disruption of the mitochondrial transmembrane potential is one of the earliest intracellular events that occur following induction of apoptosis. MitoCapture provides a simple, fluorescence-based method for distinguishing between healthy and apoptotic cells by detecting changes in the mitochondrial transmembrane potential. The kit utilizes a cationic dye that fluoresces differently in healthy cells and in apoptotic cells. In healthy cells MitoCapture aggregates in the mitochondria, giving off a bright red fluorescence. In apoptotic cells, the dye remains in the cytoplasm in its monomeric form, fluorescing green. RAW 264.7 cells were infected with OmpA+ E. coli or OmpA− E. coli and/or treated with ST, followed by staining with the MitoCapture reagent in an 8-well chamber slide, according to the manufacturer's protocols. The control uninfected cells showed no green fluorescence but contained aggregates of red fluorescence in the cell (Fig. 6B and C). Similar staining was also observed when the cells were infected with OmpA+ E. coli (Fig. 6H and I). Both OmpA− E. coli-infected and ST-treated RAW 264.7 cells showed significant green fluorescence in the cytoplasm and no red fluorescence, indicating an increase in mitochondrial permeability (Fig. 6E and F; only one panel of ST-treated cells is presented). Nonetheless, OmpA+ E. coli-infected cells were protected from ST-induced mitochondrial changes (Fig. 6K and L), whereas OmpA− E. coli-infected cells were not (Fig. 6N and O). Infection of THP-M cells with bacteria alone or with ST treatment also revealed similar results (data not shown). These data suggest that OmpA+ E. coli protects macrophages from mitochondria-induced apoptosis.
FIG. 6.
Detection of mitochondrial transmembrane potential in E. coli-infected RAW 264.7 cells by using MitoCapture. RAW cells were seeded in 8-well chamber slides and cultured to confluence. The cells were left untreated (A to C), treated with ST (D to F), or infected with either OmpA+ E. coli (G to I), OmpA+ E. coli plus ST (J to L), or OmpA− E. coli plus ST (M to O). Cells were washed and further incubated with diluted MitoCapture solution for 15 min at 37°C in a CO2 incubator. After further washing, the cells were fixed in 2% paraformaldehyde solution for 10 min at room temperature, washed again, and mounted with a coverslip with antifade solution.
Activity of caspase 3 is inhibited in OmpA+ E. coli-infected macrophages.
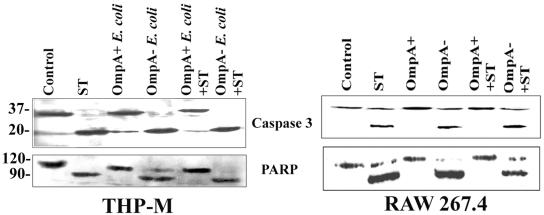
The morphological and cellular changes associated with apoptosis are due to the activation of the caspase cascade, most prominently by caspase 3 (15, 40). Therefore, we first examined whether OmpA+ E. coli infection of macrophages blocks the caspase 3 activation induced by ST. As shown in Fig. 7, caspase 3 was cleaved to a 17-kDa protein in both THP-M and RAW 264.7 cells treated with ST compared to untreated cells. In contrast, OmpA+ E. coli-infected cells showed no such cleavage; OmpA− E. coli-infected cells showed caspase 3 activation similar to that of the ST-treated cells. However, the processing of caspase 3 cleavage induced by ST was completely inhibited in OmpA+ E. coli-infected cells but not in OmpA− E. coli-infected cells.
FIG. 7.
OmpA+ E. coli inhibits the activation of caspase 3 and the cleavage of PARP after induction of apoptosis with ST. Confluent monolayers of either THP-M or RAW 264.7 cells were treated as described in the legend of Fig. 4. Total cell lysates were prepared, separated by SDS-10% polyacrylamide gel electrophoresis, and subjected to immunoblotting with either anti-caspase 3 or anti-PARP antibody (dilution, 1:500). Total cell lysates from untreated cells were used as controls.
The cleavage of PARP, a target of caspase 3, has been used extensively as a marker of caspase 3 activity (40). Therefore, we also examined whether the infection of macrophages with OmpA+ E. coli protects this cellular substrate from caspase 3-mediated cleavage following the induction of apoptosis by ST. Treatment of THP-M and RAW 264.7 cells with ST or infection with OmpA− E. coli induced the cleavage of the 116-kDa full-length PARP into the 85-kDa subunit, whereas infection with OmpA+ E. coli did not (Fig. 7). Again the cleavage of PARP by ST was blocked in OmpA+ E. coli-infected but not in OmpA− E. coli-infected macrophages. These observations suggest that OmpA+ E. coli prevents the activation of caspase 3 by inducing antiapoptotic signals in these macrophage cell lines. However, it is not clear whether E. coli also inhibits caspase 3 directly, besides increasing the expression of BclXL to block cytochrome c release.
OmpA+ E. coli inhibits the activities of both caspase 6 and caspase 9 in macrophages.
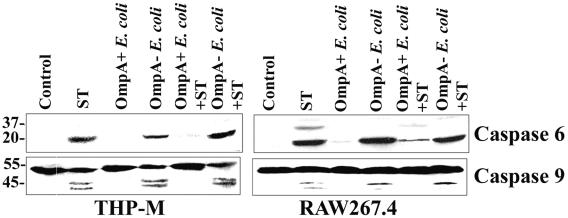
Since caspase 3 activation is suppressed in OmpA+ E. coli-infected cells, we next analyzed the activation of other key caspases in the infected cells by immunoblotting. Gene array analysis of infected THP-M cells suggested that OmpA− E. coli induces the caspase 6 gene significantly, whereas OmpA+ E. coli suppressed the induction by twofold. Therefore, we first examined the activation of caspase 6 in both THP-M and RAW cells stimulated with ST after infection with the bacteria. As shown in Fig. 8, both ST and OmpA− E. coli induced the activation of caspase 6 robustly, whereas OmpA+ E. coli showed no such activation. In addition, OmpA+ E. coli-infected cells were resistant to the stimulus of ST. These results demonstrate that OmpA− E. coli infection induces both the expression of caspase 6 at the RNA level and activation at the protein level.
FIG. 8.
Inhibition of ST-induced activation of caspase 6 and caspase 9 by OmpA+ E. coli in macrophages. Both THP-M and RAW cells were treated with ST with or without prior infection with the bacteria as described in Materials and Methods. Total cell lysates were subjected to Western blotting with anti-caspase 6 and anti-caspase 9 antibodies (dilution, 1:500). Total cell lysates from untreated cells were used as controls.
The cleavage of effector caspase 3 may also be mediated by activation of the upstream initiator caspases 8 and 10 induced by TNF-α signaling and caspase 9 induced by mitochondrial disintegration (3, 7). Since we examined caspase 3 activity by 1 h postinfection with bacteria, it is unlikely that TNF-α-mediated death signaling would be involved in the apoptosis induced by OmpA− E. coli. Therefore, we further examined the activity of caspase 9 by Western blotting (Fig. 8). We observed that the amount of active caspase 9 increased after induction of apoptosis by ST in both THP-M and RAW cells; however, this increase was inhibited in cells infected with OmpA+ E. coli prior to ST treatment. By itself, the OmpA+ E. coli did not induce any caspase activation. In contrast, OmpA− E. coli-infected cells could not be protected from ST-induced apoptosis, suggesting that OmpA+ E. coli down-regulates the caspase cascade upstream of caspase 3, probably by maintaining the integrity of mitochondrial membrane permeability.
Complementation of OmpA− E. coli with the ompA gene blocks the activation of caspases in macrophages.
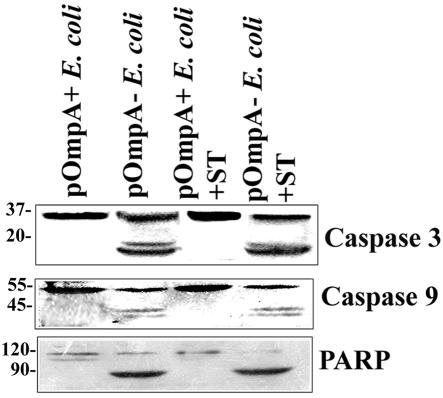
Previous studies (33) coupled with the present data strongly support the hypothesis that expression of OmpA in E. coli K1 is important for the survival of THP-M and RAW cells after infection with E. coli K1. As evidence for this concept, OmpA− E. coli infection of macrophages demonstrably induces apoptosis in these cells 2 to 4 h postinfection. To further confirm the requirement for OmpA expression, the OmpA− E. coli was complemented with a plasmid containing the ompA gene. The expression of OmpA in this strain, designated as pOmpA+ E. coli, was verified by both Western blot analysis and immunofluorescence of the bacteria by using anti-OmpA antibody. The results showed that a significant amount of OmpA was expressed in this strain and that it is surface exposed (data not shown). In contrast, OmpA+ E. coli transformed with the pUC19 plasmid alone (pOmpA− E. coli) showed no reactivity to anti-OmpA antibody. These strains were used to infect the macrophage cell lines with or without ST treatment. As expected, the pOmpA+ E. coli infection of THP-M cells showed no activation of either caspase 3 or caspase 9, whereas pOmpA− E. coli induced the activation of these caspases (Fig. 9). We also examined PARP cleavage in pOmpA+ E. coli-infected cells, and these cells revealed no signs of PARP cleavage. In addition, pOmpA+ E. coli-infected cells were resistant to ST treatment in comparison to pOmpA− E. coli-treated cells. These results, therefore, confirm the requirement for OmpA expression in E. coli for its own survival as well as that of infected macrophages.
FIG. 9.
Complementation of OmpA− E. coli with a plasmid containing the ompA gene suppresses the activation of caspases in THP-M cells. Confluent monolayers of THP-M cells were infected with OmpA− E. coli transformed with either a plasmid containing the ompA gene (pOmpA+ E. coli) or plasmid alone (pOmpA− E. coli) for 1 h. In some experiments the infected cells were treated with ST for an additional 3 h. Total cell lysates were prepared and subjected to Western blotting with anti-caspase 3, anti-caspase 9, or anti-PARP antibodies.
DISCUSSION
Multiplication of E. coli organisms in the blood represents an important event in the pathogenesis of neonatal meningitis, as a certain threshold level of bacteremia is critical for the traversal of bacteria across the blood-brain barrier. Despite this decades-old knowledge, very few studies have focused on the interaction of E. coli K1 with phagocytes. Our recent data have shown that E. coli K1 enters and survives for longer periods of time in both monocytes and macrophages in the newborn rat model of meningitis (33). In vitro studies of both human and murine macrophage-like cell lines demonstrated that E. coli K1 enters these cells with or without opsonization, survives, and multiplies, ultimately leading to rupture of the cells to release progeny into the medium. This entry and survival require the expression of OmpA on the bacterial surface, as it can be demonstrated that OmpA− E. coli, although taken up by the macrophages in small numbers by an OmpA-independent mechanism, is killed in a short period of time. Therefore, we theorize that to multiply the intracellular E. coli somehow induces survival mechanisms in the host monocytes and macrophages. Generally, the infected phagocytes undergo apoptosis to prevent the spreading of the pathogen and also to induce an immune response in the host. However, inhibition of apoptosis appears to be a common defense mechanism in the biology of intracellular pathogens (31). Here, we demonstrated that OmpA+ E. coli-infected macrophages are profoundly resistant to apoptosis including that induced by apoptotic stimuli. Our results indicate that E. coli K1 has evolved mechanisms that may contribute to bacterial antiapoptotic activity via the inhibition of cytochrome c release, which in turn may be due to increased expression of BclXL and/or inhibition of the caspase cascade.
The activation of caspases represents a central step in the apoptosis signaling cascade and transduces regulatory upstream signals into the cell death execution machinery (15). Our results show that E. coli infection of macrophages induces activation of caspase 6 at the RNA level compared to control uninfected cells. However, the amount of pro-caspase 6 at the protein level was not increased, suggesting that the bacteria may be degrading newly synthesized pro-caspase 6 at a faster rate than that of its synthesis. Alternatively, the inhibition of pro-caspase 6 synthesis could be due to the uncoupling of transcription from translation or a feedback inhibition of downstream signaling events by OmpA+ E. coli. An important downstream response of normal macrophage signaling is the production of cytokines. TNF-α is one such critical cytokine produced during the onset of meningitis due to lipopolysaccharide (LPS) released from the bacteria, which induces apoptosis through the TNF-α receptor (38). The binding of TNF-α to its receptors activates caspase 8 and/or caspase 10, which in turn activates other caspases. Despite the presence of LPS on OmpA+ E. coli, the bacteria are able to protect the macrophages from apoptosis, suggesting that the bacteria might interfere with LPS-mediated signaling. Due to this manipulation, OmpA+ E. coli might reduce or block the production of proapoptotic cytokines during the entry process until the bacteria are able to take over the macrophage metabolic machinery. This mechanism provides an advantage to the bacteria in that they are able to derive required nutrition from macrophages. Since we observed the inhibition of caspase 9 and caspase 3 activation when macrophages were infected with E. coli K1, we theorize that the bacteria might be manipulating cytochrome c release into the cytoplasm. Results from both subcellular fractionation analysis and MitoCapture fluorescence studies confirmed that OmpA+ E. coli infection inhibited cytochrome c release. Interestingly, neither heat-killed nor chloramphenicol-treated bacteria could protect the cells from apoptosis (unpublished results), suggesting that newly synthesized E. coli factor(s) might be responsible for the inhibition of apoptosis. In contrast to these data, E. coli K-12 strain DH5-α-infected RAW 264.7 cells undergo apoptosis by 24 h postinfection by activating caspase 3 and caspase 9 (17). These studies further demonstrated that uptake and digestion of bacteria lead to MyD88-independent (Toll-like receptor-independent) apoptosis upon phagocytosis. It remains to be seen whether OmpA− E. coli-induced apoptosis of macrophages as shown here is also a Toll-like receptor-independent mechanism of apoptosis.
Proteins of the Bcl-2 family are known to regulate mitochondrial-associated induction of apoptosis (1, 16). The expression of Bcl-2 or Bcl-2-like molecules maintains the membrane potential of mitochondria that prevents the release of cytochrome c for subsequent activation of caspases 9 and 3. E. coli infection of THP-M and RAW cells induces the expression of BclXL both at the gene and protein levels by two- to threefold. A variety of virus-encoded Bcl-2 homologues have been shown to modulate host cell apoptosis after infection (12, 36). Therefore, it is possible that E. coli might secrete a Bcl-2-like protein to control the apoptotic activity induced by proapoptotic cytokines during infection. Interestingly, Toxoplasma gondii infection of U937 cells induces expression the Mcl-1 protein, another antiapoptotic member of the Bcl-2 family (20). Recently, up-regulation of A1, a protein with similarity to Mcl-1, has been described in exudate cells from T. gondii-infected mice (27). Therefore, we cannot rule out the possibility of the involvement of other antiapoptotic proteins in E. coli infection of macrophages. Similar to E. coli K1 survival in macrophages, E. coli isolated from Crohn's disease lesions was able to persist efficiently until 5 days postinfection within J774-A1 macrophages; however, the mechanism by which this organism delays antiapoptosis is not known (13). Of note, we observed that the increase of BclXL expression in E. coli-infected macrophages is more dramatic in the cytosolic extracts than in the mitochondrial fractions. However, when infection with OmpA+ E. coli was followed by ST treatments for 3 h, an increase in BclXL was also observed in the mitochondrial fraction. This suggests that newly synthesized BclXL is stored in the cytoplasm pools and is then recruited to the mitochondria upon apoptotic stimuli. Consequently, the ratio of BclXL to Bax increases at the mitochondrial membrane in favor of survival of the infected cell.
Intriguing results from this study revealed the requirement of OmpA expression on the bacterial surface to induce antiapoptotic mechanisms. Since previous studies showed that OmpA interacts with macrophages directly (33), the interaction of OmpA with a specific receptor on macrophages may induce the expression of a bacterial protein either inside the cell or during invasion, which, in turn, may modulate the expression of BclXL. In the case of OmpA− E. coli infection, the bacteria that phagocytosed in small numbers were killed within a short time, suggesting that the entry of E. coli via an OmpA-independent mechanism renders them susceptible to macrophage killing. In contrast, the binding of OmpA to its receptor may convert the hostile environment of macrophages to a safe haven by modulating the signaling pathways in OmpA+ E. coli infection. These possibilities are currently being examined. Studies by Lakics et al. have demonstrated that BclXL also regulates LPS-inducible gene expression and secretion of proinflammatory and chemotactic cytokines in macrophages, a process that would potentially attenuate inflammation (21). Although the concept of the attenuation of inflammation by E. coli-induced BclXL contradicts the present dogma of E. coli pathophysiology, it could be possible that E. coli down-regulates several signaling mechanisms involved in proinflammatory cytokine release at early stages of dissemination. This situation may provide ample opportunity to E. coli for survival and multiplication to reach threshold levels of bacteremia.
In summary, our data suggest that E. coli K1 induces the expression of the antiapoptotic protein BclXL for its own survival and that of host macrophages. In addition, the bacteria may also block the activation of caspases. Besides up-regulating BclXL, OmpA+ E. coli interaction with macrophages may alter the signaling pathways of the host cell to use it as a protected reservoir for the time required to reach septic levels. Therefore, we feel that the identification of the OmpA receptor on macrophages and a better understanding of how the receptor interacts with OmpA to manipulate the function of host macrophages will provide useful insights for the future development of therapeutic strategies.
Acknowledgments
Our sincere thanks to George McNamara, Childrens Hospital Los Angeles Research Institute Image Core, for assistance with fluorescence imaging and to Martine Torres and Barbara Driscoll for critical reading of the manuscript.
This work was supported by NIH grants AI40567 and HD 41525 (N.V.P).
Editor: A. D. O'Brien
REFERENCES
- 1.Adams, J. M., and S. Cory. 1998. The Bcl-2 protein family: arbiters of cell survival. Science 281:1322-1326. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi, A., and V. M. Dixit. 1998. Death receptors: signaling and modulation. Science 281:1305-1308. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. J., and E. P. Reddy. 1998. Modulation of life and death by the TNF receptor superfamily. Oncogene 17:3261-3270. [DOI] [PubMed] [Google Scholar]
- 4.Belaaouaj, A., K. S. Kim, and S. D. Shapiro. 2000. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 289:1185-1188. [DOI] [PubMed] [Google Scholar]
- 5.Belmokhtar, C. A., J. Hillion, and E. Segal-Bendirdjian. 2001. Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene 20:3354-3362. [DOI] [PubMed] [Google Scholar]
- 6.Bremer, E., S. T. Cole, I. Hindennach, U. Henning, E. Beck, C. Kurz, and H. Schaller. 1982. Export of a protein into the outer membrane of Escherichia coli K12. Stable incorporation of the OmpA protein requires less than 193 amino-terminal amino-acid residues. Eur. J. Biochem. 122:223-231. [DOI] [PubMed] [Google Scholar]
- 7.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 8.Ching, J. C., N. L. Jones, P. J. Ceponis, M. A. Karmali, and P. M. Sherman. 2002. Escherichia coli Shiga-like toxins induce apoptosis and cleavage of poly(ADP-ribose) polymerase via in vitro activation of caspases. Infect. Immun. 70:4669-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fettucciari, K., E. Rosati, L. Scaringi, P. Cornacchione, G. Migliorati, R. Sabatini, I. Fetriconi, R. Rossi, and P. Marconi. 2000. Group B Streptococcus induces apoptosis in macrophages. J. Immunol. 165:3923-3933. [DOI] [PubMed] [Google Scholar]
- 11.Fratazzi, C., R. D. Arbeit, C. Carini, and H. G. Remold. 1997. Programmed cell death of Mycobacterium avium serovar 4-infected human macrophages prevents the mycobacteria from spreading and induces mycobacterial growth inhibition by freshly added, uninfected macrophages. J. Immunol. 158:4320-4327. [PubMed] [Google Scholar]
- 12.Gillet, G., and G. Brun. 1996. Viral inhibition of apoptosis. Trends Microbiol. 4:312-317. [DOI] [PubMed] [Google Scholar]
- 13.Glasser, A. L., J. Boudeau, N. Barnich, M. H. Perruchot, J. F. Colombel, and A. Darfeuille-Michaud. 2001. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69:5529-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goebel, S., U. Gross, and C. G. Luder. 2001. Inhibition of host cell apoptosis by Toxoplasma gondii is accompanied by reduced activation of the caspase cascade and alterations of poly(ADP-ribose) polymerase expression. J. Cell Sci. 114:3495-3505. [DOI] [PubMed] [Google Scholar]
- 15.Green, D. R., and J. C. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 16.Gross, A., J. M. McDonnell, and S. J. Korsmeyer. 1999. Bcl-2 family members and the mitochondria in apoptosis. Genes Dev. 13:1899-1911. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, H., C. Furmann, H. Wagner, and G. Hacker. 2002. Caspase-9/-3 activation and apoptosis are induced in mouse macrophages upon ingestion and digestion of Escherichia coli bacteria. J. Immunol. 169:3172-3179. [DOI] [PubMed] [Google Scholar]
- 18.Kim, M. K., S. Y. Seong, J. Y. Seoh, T. H. Han, H. J. Song, J. E. Lee, J. H. Shin, B. U. Lim, and J. S. Kang. 2002. Orientia tsutsugamushi inhibits apoptosis of macrophages by retarding intracellular calcium release. Infect. Immun. 70:4692-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koedel, U., and H. W. Pfister. 1999. Models of experimental bacterial meningitis: role and limitations. Infect. Dis. Clin. N. Am. 13:549-577. [DOI] [PubMed] [Google Scholar]
- 20.Kozopas, K. M., T. Yang, H. L. Buchan, P. Zhou, and R. W. Craig. 1993. Mcl-1, a gene expressed in programmed myeloid differentiation, has sequence similarity to Blc-2. Proc. Natl. Acad. Sci. USA 90:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakics, V., A. E. Medvedev, S. Okada, and S. N. Vogel. 2000. Inhibition of LPS-induced cytokines by Bcl-xL in a murine macrophage cell line. J. Immunol. 165:2729-2737. [DOI] [PubMed] [Google Scholar]
- 22.Leib, S. L., and M. G. Tauber. 1999. Pathogenesis of bacterial meningitis. Infect. Dis. Clin. N. Am. 13:527-547. [DOI] [PubMed] [Google Scholar]
- 23.Maeda, S., H. Yoshida, Y. Mitsuno, Y. Hirata, K. Ogura, Y. Shiratori, and M. Omata. 2002. Analysis of apoptotic and antiapoptotic signalling pathways induced by Helicobacter pylori. Mol. Pathol. 55:286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, S. J., C. P. Reutelingsperger, A. J. McGahon, J. A. Rader, R. C. van Schie, D. M. LaFace, and D. R. Green. 1995. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 182:1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molloy, A., P. Laochumroonvorapong, and G. Kaplan. 1994. Apoptosis, but not necrosis, of infected monocytes is coupled with killing of intracellular bacillus Calmette-Guerin. J. Exp. Med. 180:1499-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagata, S. 1997. Apoptosis by death factor. Cell 88:355-365. [DOI] [PubMed] [Google Scholar]
- 27.Orlfofsky, A., R. D. Somogyi, L. M. Weiss, and M. B. Prystowsky. 1999. The murine antiapoptotic protein A1 is induced in inflammatory macrophages and constitutively expressed in neutrophils. J. Immunol. 163:412-419. [PubMed] [Google Scholar]
- 28.Payne, T. M., R. E. Molestina, and A. P. Sinai. 2003. Inhibition of caspase activation and a requirement for NF-kappaB function in the Toxoplasma gondii-mediated blockade of host apoptosis. J. Cell Sci. 116:4345-4358. [DOI] [PubMed] [Google Scholar]
- 29.Prasadarao, N. V., A. M. Blom, B. O. Villoutreix, and L. C. Linsangan. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169:6352-6360. [DOI] [PubMed] [Google Scholar]
- 30.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberger, C. M., and B. B. Finlay. 2003. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 4:385-396. [DOI] [PubMed] [Google Scholar]
- 32.Santos, R. L., R. M. Tsolis, A. J. Baumler, R. Smith III, and L. G. Adams. 2001. Salmonella enterica serovar Typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect. Immun. 69:2293-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sukumaran, S. K., H. Shimada, and N. V. Prasadarao. 2003. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect. Immun. 71:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tafani, M., D. A. Minchenko, A. Serroni, and J. L. Farber. 2001. Induction of the mitochondrial permeability transition mediates the killing of HeLa cells by staurosporine. Cancer Res. 61:2459-2466. [PubMed] [Google Scholar]
- 35.Tafani, M., J. A. Cohn, and N. O. Karpinich. 2002. Regulation of intracellular pH mediates Bax activation in HeLa cells treated with staurosporine or tumor necrosis factor-alpha. J. Biol. Chem. 277:49569-49576. [DOI] [PubMed] [Google Scholar]
- 36.Teodoro, J. G., and P. E. Branton. 1997. Regulation of apoptosis by viral gene products. J. Virol. 71:1739-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentin-Weigand, P., P. Benkel, M. Rohde, and G. S. Chhatwal. 1996. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 64:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallach, D., M. Boldin, E. Varfolomeev, R. Beyaert, P. Vandenabeele, and W. Fiers. 1997. Cell death induction by receptors of the TNF family: towards a molecular understanding. FEBS Lett. 410:96-106. [DOI] [PubMed] [Google Scholar]
- 39.Williams, G. T. 1994. Programmed cell death: a fundamental protective response to pathogens. Trends Microbiol. 2:463-464. [DOI] [PubMed] [Google Scholar]
- 40.Woo, M., R. Hakem, M. S. Soengas, G. S. Duncan, A. Shahinian, D. Kagi, A. Hakem, M. McCurrach, W. Khoo, S. A. Kaufman, G. Senaldi, T. Howard, S. W. Lowe, and T. W. Mak. 1998. Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 12:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, J. Z., M. Sinha, B. A. Luxon, and X. J. Yu. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72:498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerman, K. C., C. Bonzon, and D. R. Green. 2001. The machinery of programmed cell death. Pharmacol. Ther. 92:57-70. [DOI] [PubMed] [Google Scholar]