Abstract
Enterococci play a dual role in human ecology. They serve as commensal organisms of the gastrointestinal tract and are also leading causes of multiple antibiotic-resistant hospital-acquired infection. Many nosocomial infections result from the ability of microorganisms to form biofilms. The molecular mechanisms involved in enterococcal biofilm formation are only now beginning to be understood. Enterococcal surface protein, Esp, has been reported to contribute to biofilm formation by Enterococcus faecalis. Recent studies have shown that enterococci form biofilms independently of Esp expression. To precisely determine what role Esp plays in E. faecalis biofilm formation, Esp was expressed on the cell surface of genetically well-defined, natively Esp-deficient strains, and isogenic Esp-positive and Esp-deficient strains were compared for their biofilm-forming ability. The results show that Esp expression leads to a significant increase in biofilm formation, irrespective of the strain tested. The contribution of Esp to biofilm formation was found to be most pronounced in the presence of 0.5% (wt/vol) or greater glucose. These results unambiguously define Esp as a key contributor to the ability of E. faecalis to form biofilms.
Biofilms are bacterial communities attached to a biotic or an abiotic substrate and encased in a matrix that may be composed of carbohydrates, DNA, or protein (8, 21, 24, 39). Not only do biofilms play an important role in the pathogenesis of several chronic infections, such as bacterial endocarditis, infectious kidney stones, and cystic fibrosis (25), but they are also central to nosocomial infections related to indwelling medical devices (8). In spite of marked phenotypic differences between biofilm and planktonic cells, such as higher levels of antibiotic resistance and slower growth rate, as shown for the gram-negative pathogen Pseudomonas aeruginosa, only 1% of the genes are differentially regulated between the two modes of growth to a significant extent (40). Thus, biofilm development, though complex, may involve the interplay of only a few crucial factors depending on the milieu.
Enterococci, as members of the commensal human flora, are present in the colon in numbers as high as 108 CFU per g of feces (17). Enterococci are also important opportunistic pathogens and now rank among the most common nosocomial agents infecting the bloodstream, surgical sites, and urinary tract (28). Enterococcal endocarditis, which accounts for 5 to 10% of the total bacterial endocarditis cases reported each year, is becoming increasingly difficult to cure because of growing resistance of enterococci to multiple antibiotics (12). Enterococci have been associated with biofilms on various kinds of indwelling medical devices, such as artificial hip prostheses, intrauterine devices, prosthetic heart valves, central venous catheters, and urinary catheters (7). An understanding of bacterial factors involved in promoting persistence of enterococci in the nosocomial environment or at infection sites is only now emerging.
Enterococcal surface protein, Esp, was initially identified in a virulent gentamicin-resistant Enterococcus faecalis isolate, MMH594 (18), and was recently found to be encoded on a large, 153-kb pathogenicity island (31, 33). An esp homolog has been reported in Enterococcus faecium as well, and infection-derived E. faecalis and E. faecium isolates show enrichment for the esp gene (11, 33). Studies with a mouse model of urinary tract infection have shown that Esp-positive E. faecalis colonizes and persists in urinary bladders in significantly greater numbers than isogenic Esp-deficient strains (32).
A strong correlation between the presence of Esp and the ability of an enterococcal strain to form biofilms in vitro has been reported (36). None of the esp-deficient isolates tested in that study were capable of forming biofilms (36). However, it was also observed that insertional inactivation of esp did not cause a loss of the biofilm phenotype in every mutant tested. These results suggest that, whereas Esp is important in biofilm formation, additional determinants in E. faecalis may also contribute to biofilm formation (36). Moreover, recent studies (20) demonstrated that an esp-negative strain, OG1RF, originally derived from the oral cavity (14), can form biofilms on abiotic surfaces independently of Esp.
To precisely define the role of Esp in enterococcal biofilm formation, we sought to compare Esp-deficient strains FA2-2 (5) and OG1RF (9) to their isogenic Esp-positive derivatives. Using these isogenic strains, we show that Esp expression significantly enhances biofilm formation irrespective of the basal abilities of the tested strains to form biofilms. However, Esp-dependent biofilm formation is influenced by the growth medium and conditions tested.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in the present study are listed in Table 1. Enterococcal strains were cultured in brain heart infusion or Trypticase soy broth (TSB) supplemented with various concentrations of glucose and appropriate antibiotics. Luria-Bertani broth was used for cultivation of Escherichia coli strains. E. coli XL1-Blue was obtained from Stratagene (La Jolla, Calif.), and used as a host for plasmid purifications. The entire esp gene, along with 160-bp region upstream of the putative transcriptional start site, was amplified by using the primers Esp103 (5′-GAGAGAGCTCGGGATGTTCCAGTGACCCC-3′) and Esp104 (5′-GAGAGGATCCGAGGAAGAGACTTCTTCCTCTTGT-3′), which contain recognition sequences (underlined) for SacI and BamHI respectively, at the 5′ ends. The amplified fragment was sequentially restricted with SacI and BamHI (New England Biolabs, Inc., Beverly, Mass.) and purified from a 0.8% low-melting-point agarose gel by using the Wizard Preps DNA purification system (Promega, Madison, Wis.). Plasmid pESPF was generated by ligating the PCR-amplified and -restricted fragment into SacI- and BamHI-cut shuttle vector pAT28 (37). Natively Esp-negative FA2-2 (5) and OG1RF (9) were transformed with pESPF to the generate Esp-positive strains FA2-2(pESPF) and OG1RF(pESPF). FA2-2 and OG1RF were transformed with plasmid pAT28 to generate FA2-2(pAT28) and OG1RF(pAT28), which served as controls. FA2-2(pESPF) was cotransformed with pCUGel (obtained from L. E. Hancock), which harbors the gelE gene cloned downstream of a constitutive aphA promoter in shuttle vector pCU1 (1), to generate the GelE-positive and Esp-positive derivative FA2-2(pESPF, pCUGel). Rifampin at 25 μg/ml and fusidic acid at 10 μg/ml were used for host (FA2-2 and OG1RF) selection; spectinomycin at 500 μg/ml was used for pAT28 and pESPF selection, and chloramphenicol at 10 μg/ml was used for pCUGel selection.
TABLE 1.
E. faecalis strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. faecalis strains | ||
| OG1RF | Plasmid-free, esp-negative Rifr Fusr | 9 |
| FA2-2 | Plasmid-free, esp-negative Rifr Fusr | 5 |
| OG1RF(pAT28) | OG1RF transformed with pAT28 | This study |
| OG1RF(pESPF) | OG1RF transformed with pESPF | This study |
| FA2-2(pAT28) | FA2-2 transformed with pAT28 | This study |
| FA2-2(pESPF) | FA2-2 transformed with pESPF | This study |
| FA2-2(pESPF, pCUGel) | FA2-2 cotransformed with pCUGel and pESPF | This study |
| Plasmids | ||
| pAT28 | Shuttle vector | 37 |
| pESPF | esp cloned in pAT28 | This study |
| pCUGel | gelE cloned in pCU1 | This study |
Rifr, rifampin resistant; Fusr, fusidic acid resistant.
Biofilm assay.
The biofilm assay was performed by using microtiter plates as described earlier (36), with slight modifications. Flat-bottom polystyrene (Corning, Inc., Corning, N.Y.), polypropylene, and polyvinyl chloride (Becton Dickinson) microtiter plates were used. Bacterial strains were grown at 37°C for 16 h in TSB containing various amounts of glucose based on the experiment to be performed; the bacterial cells were then pelleted at 6,000 × g for 10 min, and the cell pellet resuspended in 5 ml of fresh medium. The optical densities (ODs) of the bacterial suspensions were measured by using a UV-1201 UV-VIS spectrophotometer (Shimadzu, Kyoto, Japan) and normalized. Bacterial cultures were diluted 1:40 in fresh TSB supplemented with glucose and containing suitable antibiotics. From the diluted culture, 200 μl was dispensed into 12 wells in a single row of a sterile 96-well flat-bottom microtiter plate. After incubation at 37°C for 24 h, the planktonic cells were aspirated, and the wells washed three times with sterile phosphate-buffered saline (PBS). The plates were inverted and allowed to dry for 1 h at room temperature. For biofilm quantification, 200 μl of 0.2% aqueous crystal violet solution was added to each well, and the plates were allowed to stand for 15 min. The wells were subsequently washed thrice with sterile PBS to wash off the excess crystal violet. Crystal violet bound to the biofilm was extracted with 200 μl of an 80:20 (vol/vol) mixture of ethyl alcohol and acetone, and the absorbance of the extracted crystal violet was measured at 595 nm with an ELX-800 (Bio-Tek Instruments, Inc., Winooski, Vt.) automatic microplate reader. As a control, crystal violet binding to wells was measured for wells exposed only to the medium with no bacteria. All biofilm assays were performed in triplicate, with 12 replicates for each strain per assay.
Cell surface hydrophobicity determination.
Bacterial cell surface hydrophobicity was measured as described previously (29). Briefly, bacterial strains were grown overnight at 37°C in TSB containing 0.75% glucose and subsequently diluted 1:50 in 5 ml of fresh medium. The subculture was incubated further at 37°C for 4 h. Log-phase bacteria were harvested by centrifugation, washed twice with PUM buffer (22.2 g of potassium phosphate trihydrate, 7.26 g of monobasic potassium phosphate, 1.8 g of urea, and 0.2 g of magnesium sulfate heptahydrate/liter [pH 7.1]), and resuspended in 5 ml of PUM buffer. Then, 100 μl of n-hexadecane was added to a bacterial cell suspension normalized to an OD at 400 nm (OD400) of 1.0. The mixtures were incubated at 30°C for 10 min, subsequently vortexed vigorously for 2 min, and then allowed to stand for 15 min at room temperature to ensure complete separation of the organic and aqueous phases. The absorbance of the aqueous layer was measured at 400 nm. The percent cell surface hydrophobicity, which is a measure of the percentage of bacterial cells partitioning into the organic phase, was calculated by using the following formula: [1 − (final OD400/initial OD400) × 100]. The assay was performed in triplicate.
Quantitative enzyme-linked immunosorbent assay (ELISA).
Bacterial cultures were grown overnight in TSB supplemented with glucose and antibiotics as appropriate for the experiment performed. Cells were harvested at 6,000 × g for 10 min, and the cell pellet was washed once with sterile PBS. After the wash, the cells were resuspended in 5 ml of carbonate-bicarbonate buffer (1.59 g of sodium carbonate and 2.92 g of sodium bicarbonate/liter [pH 9.6]). The ODs of the bacterial suspensions were measured at 600 nm, and cell densities were normalized by appropriately diluting the cultures in sodium carbonate-bicarbonate buffer. The normalized cultures (100 μl) were added to the wells of a 96-well Immunolon2 HB plate (Dynex Technologies, Inc.), and the plate was incubated at 4°C for 16 h. The wells were then washed three times with PBS supplemented with 0.05% Tween 20 to reduce nonspecific binding. Nonfat dry milk 0.4% (wt/vol; Bio-Rad Laboratories, Hercules, Calif.) in PBS was added to the wells as a blocking agent, and the plate was incubated at 37°C for 2 h. Purified rabbit immunoglobulin G (1:500) specific to the N-terminal region of Esp was added to each well, and the plate was incubated at room temperature for 16 h. After incubation, the wells were washed thrice with sterile PBS containing Tween 20. Goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Sigma-Aldrich Co., St. Louis, Mo.) was used as the secondary antibody, and the plate was incubated at 37°C for 2 h. A 0.1% solution of p-nitrophenylphosphate was added as a substrate for colorimetric detection, and absorbance was measured at 405 nm after 30 min of incubation at 37°C. The experiment was performed in triplicate. Multigroup comparisons were made with analysis of variance by using Bonferroni's test.
Confocal laser scanning microscopy (CLSM).
Bacterial biofilms were grown in six-well polystyrene tissue culture plates (Corning) for 16 h in medium containing various levels of glucose and antibiotics. For imaging, the cell culture supernatant and planktonic cells were removed by gentle vacuum suction, and the biofilms were washed twice with sterile PBS. The biofilms were stained with 100 μl of 1% aqueous acridine orange solution for 10 min and subsequently washed twice with sterile PBS. The biofilms were kept hydrated in 2 ml of PBS during imaging. Confocal images were collected by using a Leica TCS NT microscope, and an argon laser (488 nm) was used for sample excitation. The images were viewed by using the LCS Lite software (ftp://ftp.llt.de/pub/softlib/LCSLite/). The acquired image stacks were quantified by using the COMSTAT program (15). Two parameters were measured by using this program: biovolume and maximum thickness. Biovolume is defined as the ratio of the product of the number of biomass pixels and the voxel size [(pixel size)X × (pixel size)Y × (pixel size)Z] to the substratum area and is expressed in cubic microns per square microns. The maximum thickness is expressed in microns. Statistical significance between the parameters was tested by using the Student t test.
RESULTS
Esp enhances biofilm formation.
In contrast to previously published reports that implied a critical role for Esp in the process of enterococcal biofilm development (36), recent studies demonstrated biofilm formation in an esp-deficient strain (20). Due to these contrasting reports, we sought to compare isogenic Esp+/− strains to determine precisely what role Esp plays in enterococcal biofilm formation. To achieve this, an antibiotic-resistant derivative of the oral isolate OG1 (14), OG1RF (9), shown previously to form biofilms in an Esp-independent manner, was transformed with pESPF, which harbors the esp gene along with its native promoter (unpublished data) cloned in the shuttle vector pAT28 (37). The expression of Esp at the cell surface was confirmed by using an ELISA that used rabbit polyclonal antibodies specific to the N-terminal region of Esp (33).
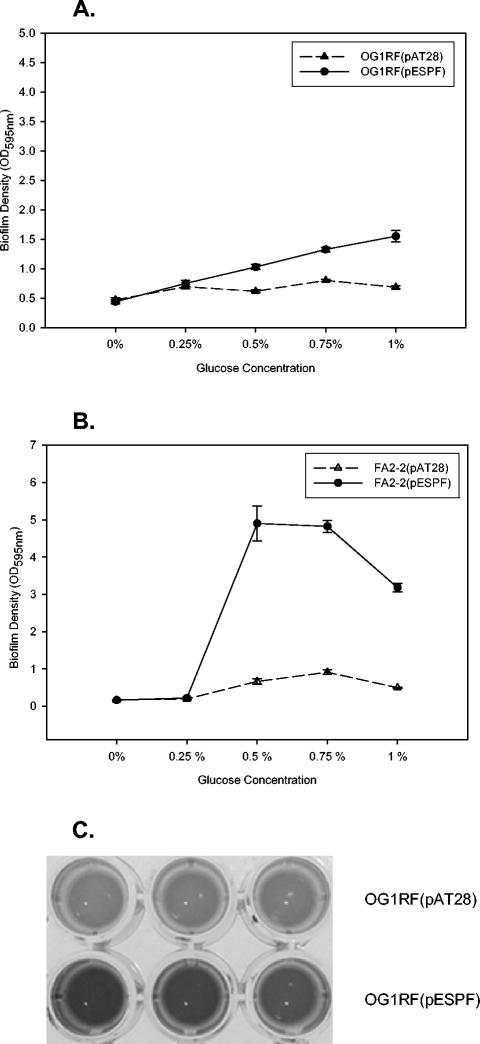
As shown in Fig. 1A, OG1RF(pESPF) showed significantly greater ability to form biofilms on polystyrene in the presence of 0.5, 0.75, or 1% glucose than the Esp-negative control OG1RF(pAT28) (P = 0.001, 0.0002, or 0.0009, respectively). No significant difference was observed when the medium lacked glucose or was supplemented with 0.25% glucose (P = 0.544 or 0.417, respectively).
FIG. 1.
Biofilm formation by Esp-positive and Esp-negative strains at different glucose concentrations. Biofilm formation was quantified by crystal violet staining, followed by back extraction of bound crystal violet into an ethanol-acetone mixture. The y axis represents the ODs of dissolved crystal violet measured at 595 nm. The error bars represent mean ± the standard error. (A) OG1RF(pAT28) (▴) and OG1RF(pESPF) (•); (B) FA2-2(pAT28) (▵) and FA2-2(pESPF) (•). (C) Representative image of biofilm densities of OG1RF(pAT28) and OG1RF(pESPF) strains when grown in the presence of 0.75% glucose, as determined by crystal violet staining.
To determine whether the observed increase in biofilm formation conferred by Esp was independent of the OG1RF strain background, a second Esp-negative strain, FA2-2 (5), which is an antibiotic-resistant derivative of the clinical isolate JH2 (19), was transformed with pESPF, and the expression of Esp at the cell surface was confirmed by using ELISA (33). Similar to the results observed for OG1RF(pESPF), as shown in Fig. 1B, expression of Esp significantly increased the biofilm formation by FA2-2(pESPF) in the presence of 0.5, 0.75, or 1% glucose (P = 0.008, 0.00002, or 0.00002, respectively). This difference also required the presence of glucose, since no difference was observed between Esp-positive and Esp-negative FA2-2 in the complete absence or presence of 0.25% glucose (P = 0.99 or 0.52, respectively) (Fig. 1B). As shown in Fig. 1C, differences in biofilm formation between Esp-positive strain OG1RF(pESPF) and Esp-negative strain OG1RF(pAT28) were easily observed visually after crystal violet staining, when the strains were grown in the presence of 0.75% glucose. Since Esp contributed to biofilm formation in these media in the presence of glucose at levels >0.25%, further biofilm assays were performed with 0.75% glucose.
CLSM.
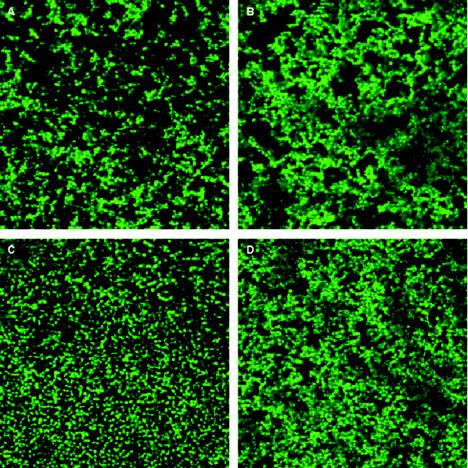
CLSM was used to examine biofilms formed by the Esp-positive and Esp-negative FA2-2 and OG1RF strains (Fig. 2). A 63× Plan APO 1.2 NA water immersion objective was used, which allowed the biofilms to remain hydrated and be observed in situ. Table 2 shows the biovolume and maximum thickness of the biofilms produced by each of the strains. Consistent with the microtiter plate assay, no difference in either biovolume or maximum thickness was observed between the biofilms formed by Esp-positive and Esp-negative strains in the presence of 0.25% glucose. However, at a higher glucose concentration of 0.75%, the biovolume and the maximum thickness of the Esp-positive strain FA2-2(pESPF) biofilms were significantly greater (P < 0.00001) than for FA2-2(pAT28), and those of OG1RF(pESPF) were significantly greater than for OG1RF(pAT28) (P < 0.00001). Importantly, both biovolume and maximum thickness measures for FA2-2(pESPF) and OG1RF(pESPF) were similar, although FA2-2(pESPF) biofilms formed in 0.75% glucose were observed to bind amounts of crystal violet that were more than threefold greater than for OG1RF(pESPF) biofilms. This shows that there is little relationship between biofilm volume and crystal violet binding, raising another important caveat in interpreting and comparing results.
FIG. 2.
CLSM images of Esp-positive and Esp-negative strain pairs grown in TSB containing 0.75% glucose. Each image represents the layer in Z-stack that has the maximum bacterial coverage. The percentage of biomass in each layer of the Z-stack was computed by using the COMSTAT program. (A) FA2-2(pAT28); (B) FA2-2(pESPF); (C) OG1RF(pAT28); (D) OG1RF(pESPF).
TABLE 2.
Quantitative analyses of confocal-microscopy-acquired image stacks from Esp-positive and Esp-negative strains using COMSTAT
| Strain | Mean value ± SEMa with:
|
|||
|---|---|---|---|---|
| 0.25% Glucose
|
0.75% Glucose
|
|||
| Biovolume (μm3/μm2) | Maximum thickness (μm) | Biovolume (μm3/μm2) | Maximum thickness (μm) | |
| FA2-2(pAT28) | 1.55 ± 0.37 | 6.4 ± 0.67 | 2.98 ± 0.79 | 7.4 ± 1.12 |
| FA2-2(pESPF) | 2.17 ± 0.85 | 6.81 ± 1.51 | 8.48 ± 1.79 | 17.29 ± 3.33 |
| OG1RF(pAT28) | 2.33 ± 0.25 | 6.88 ± 1.33 | 2.17 ± 0.45 | 7.14 ± 0.82 |
| OG1RF(pESPF) | 2.53 ± 0.17 | 7.08 ± 1.19 | 7.1 ± 1.18 | 16.04 ± 1.92 |
Biovolume = (number of biomass pixels × voxel size)/ substratum area. Maximum thickness represents the maximum height of the biofilm from the substratum.
Biofilm formation and bacterial cell surface hydrophobicity.
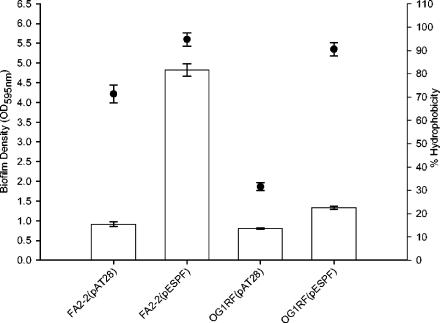
Since the biofilm assays were performed on polystyrene plates, which are inherently hydrophobic surfaces (27), it was of interest to determine the extent to which Esp modulated surface hydrophobicity and the extent to which this may have affected biofilm formation. We determined the relationship between Esp expression and the cell surface hydrophobicity of FA2-2 and OG1RF, by using n-hexadecane extraction, essentially as described previously (29). Esp-positive FA2-2(pESPF) and OG1RF(pESPF) cells were found to be significantly more hydrophobic and partitioned to a greater extent into the organic n-hexadecane phase than did the Esp-negative cells in this assay (Fig. 3), indicating that Esp expression at the cell surface significantly increased the cell surface hydrophobicities of FA2-2 and OG1RF. These results are consistent with earlier findings that demonstrated a significant reduction in the cell surface hydrophobicities of Esp-deficient mutant strains generated from wild-type Esp-positive parent strains (36) and show that the effect observed in that study resulted directly from the loss of Esp expression, as opposed to a polar effect stemming from the mutation. However, as seen in Fig. 3, the relationship between cell surface hydrophobicity and biofilm formation of the tested strains was not proportional supporting the proposition that biofilm formation is a multifactorial process and that Esp-dependent enhanced biofilm formation is not attributable solely to increased cell surface hydrophobicity. A similar conclusion can be drawn by comparing the cell surface hydrophobicity (Fig. 3) with the biovolume measured by using COMSTAT (Table 2).
FIG. 3.
Biofilm densities (□) and cell surface hydrophobicities (•) of Esp-positive and Esp-negative FA2-2 and OG1RF strain pairs. The bacteria were cultured in TSB containing 0.75% glucose, and the cell surface hydrophobicities were measured as described in Materials and Methods. No correlation was observed between cell surface hydrophobicities of the Esp-positive and Esp-negative FA2-2 and OG1RF strains and their ability to form biofilms. The error bars represent the mean ± the standard error.
Biofilm formation on polypropylene and polyvinyl chloride plates.
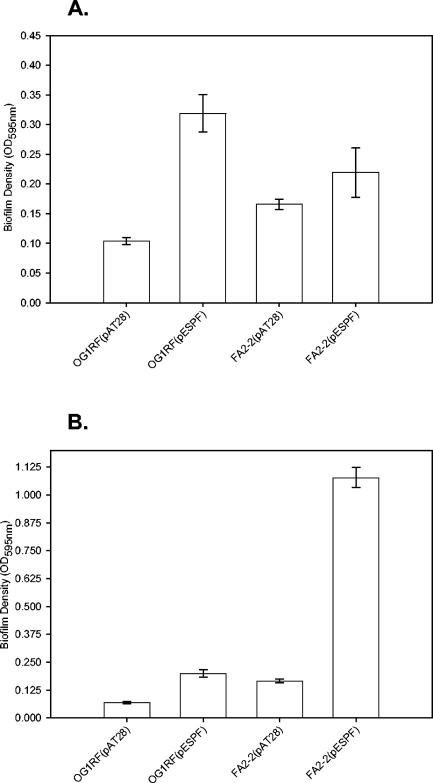
Bacterial adhesion, and consequently biofilm formation, is influenced considerably by the composition of the abiotic substrate (7, 10, 38). Plates made of polyvinyl chloride and polypropylene, which vary significantly in their physical and chemical characteristics (27), were used instead of the conventional polystyrene plates in a 96-well plate biofilm assay to determine how these surfaces affect Esp-mediated biofilm formation by E. faecalis. Similar to that observed with polystyrene plates, both Esp-positive FA2-2(pESPF) and OG1RF(pESPF) formed significantly more biofilms, as measured by crystal violet binding, on the polyvinyl chloride and polypropylene plates, compared to their Esp-negative counterparts (Fig. 4A and B, respectively). However, all of the strains formed correspondingly less biofilm on both the polyvinylchloride and polypropylene substrates compared to polystyrene.
FIG. 4.
Esp-mediated biofilm formation on polyvinyl chloride and polypropylene substrates. The ability of Esp-positive and Esp-negative strains to form biofilms on medically relevant surfaces such as polyvinyl chloride and polypropylene was assessed by crystal violet staining. Biofilm formation by FA2-2(pAT28), FA2-2(pESPF), OG1RF(pAT28), and OG1RF(pESPF) was quantified on polyvinyl chloride (A) and polypropylene (B) plates. The error bars represent the mean ± the standard error.
Esp expression and environmental glucose concentration.
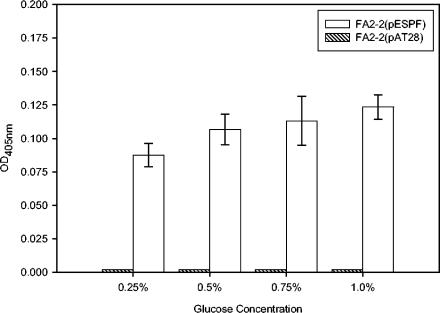
Since we observed that higher glucose concentrations promoted biofilm formation selectively in Esp-positive strains, it was of interest to determine whether glucose upregulates the expression of esp. To examine this, quantitative ELISA with antibodies specific to the N-terminal region of Esp was used to quantify the levels of Esp at the bacterial cell surface. As shown in Fig. 5, although expression tended to be higher, there was no significant difference between the levels of Esp detected on the cell surface of FA2-2(pESPF) grown in the presence of 0.25, 0.5, 0.75, and 1.0% glucose. These results indicate that the significant increase in biofilm formation by Esp-positive E. faecalis, observed in the presence of glucose, did not result from a significant change in the expression of Esp.
FIG. 5.
Quantitative ELISA to ascertain the cell surface levels of Esp in FA2-2(pESPF) grown in the presence of 0.25, 0.5, 0.75, or 1% glucose. The ODs of FA2-2(pESPF) grown in the presence of different glucose concentrations were equalized before they were added to the 96-well plate. The level of Esp on the bacterial cell surface was quantified by ELISA as described in Materials and Methods. Similar levels of Esp were detected on the cell surface of FA2-2(pESPF) when grown in the presence of 0.25, 0.5, 0.75, or 1% glucose. Strain FA2-2(pAT28) was used as negative control.
Gelatinase and Esp in biofilm formation.
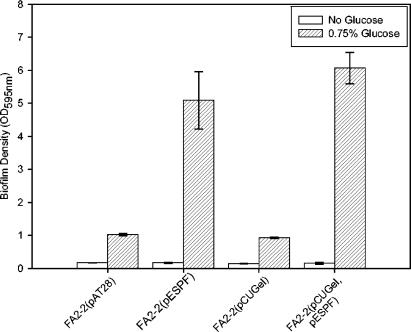
Recently, the secreted enterococcal metalloprotease, gelatinase, was found to play a role in biofilm formation (20). Expression of gelatinase under the influence of a nisin-inducible promoter in a gelE-negative strain, JH2, was found to increase the ability of this strain to form biofilms (20). It was therefore of interest to determine whether the protease, GelE, modulated the ability of Esp to promote biofilm formation. FA2-2, a spontaneous rifampin- and fusidic acid-resistant strain of JH2 (19), although genotypically gelE positive, does not express gelatinase, possibly due to mutation(s) either in gelE and/or one of the regulatory elements of gelE (L. E. Hancock, unpublished data). Therefore, FA2-2 was transformed with pESPF and pCUGel (obtained from L. E. Hancock; gelE cloned in pCU1 under the influence of a constitutive aphA promoter). The phenotypic expression of Esp was confirmed by ELISA as described previously, and that of GelE was confirmed by the observed hydrolysis of casein on 1.5% skimmed milk agar plates. When examined in the standard microtiter plate based biofilm assay by using crystal violet staining, the expression of gelatinase did not enhance or reduce the ability of FA2-2 or FA2-2(pESPF) to form biofilms in the absence or presence of 0.75% glucose (Fig. 6).
FIG. 6.
Biofilm formation by Esp-negative FA2-2(pAT28), Esp-positive FA2-2(pESPF), GelE-positive FA2-2(pCUGel), and Esp- and GelE-positive FA2-2(pCUGel, pESPF) on 96-well polystyrene plates as quantified by crystal violet staining. The error bars represent mean ± the standard error.
DISCUSSION
Infection-derived E. faecalis isolates have been shown to form biofilms in vitro (2, 30), and E. faecalis is isolated from biofilms formed on indwelling medical devices (7, 8). However, an understanding of the process of biofilm formation by E. faecalis is only now beginning to emerge, and the results appear to be contradictory.
In a study using a large number (n = 200) of clinical isolates of E. faecalis, a strong correlation (P < 0.0001) between the presence of Esp and the ability of an enterococcal strain to form biofilms was observed, which suggested a role for Esp in the process of biofilm development in E. faecalis (36). The ability to form biofilms was restricted to only E. faecalis isolates harboring esp, and esp was not detected in any of the E. faecalis isolates unable to produce a biofilm or in E. faecium, Enterococcus avium, or Enterococcus gallinarum. The same study grouped enterococcal isolates into three categories depending on their ability to form biofilms: strong, medium, and weak biofilm producers. It was shown that insertional inactivation of the esp gene in the weak and medium biofilm formers resulted in a significant reduction in the ability of the mutant strains to form biofilms. However, inactivating esp gene expression in the strong biofilm producers had no significant effect on biofilm formation and suggested that other factors may contribute to biofilm formation in these isolates. A more recent report (20) focused on characterizing biofilm formation by a non-infection-derived, esp-negative E. faecalis strain originally isolated from the oral cavity, OG1RF (9), and showed that this strain was able to form a biofilm despite being Esp deficient.
Consistent with the findings of others (36), we found that Esp made a significant contribution to biofilm formation, whether expressed by OG1RF or FA2-2, on three different surfaces tested. However, we found that Esp did not contribute to biofilm formation under all conditions but specifically required glucose at concentrations of ≥0.5% in the media tested. Biofilm formation was quantified by two methods: crystal violet staining and CLSM-acquired image stack analyses by COMSTAT. With both methods of analysis, it was found that Esp significantly enhanced biofilm formation in the presence of glucose at concentrations of ≥0.5%. Moreover, crystal violet binding as a measure of E. faecalis biofilm formation did not reflect biofilm volume or thickness, as measured by image analysis. Thus, whereas crystal violet staining may be a practical method to assess biofilm formation, it does not reflect the biofilm volume quantitatively under the conditions tested.
A number of previous studies have shown that the nutrient content of the growth medium influences biofilm development in different organisms (4, 13, 16, 20, 23, 34). More specifically, environmental factors such as glucose and iron availability, osmolarity, pH, temperature, and anaerobiosis affect biofilm development (22, 23). Since an enhanced ability to form biofilms was observed in Esp-positive strains in a glucose-dependent manner, it was of interest to determine whether Esp expression was regulated by glucose. Quantitative ELISA, however, showed similar levels of Esp on the cell surface of FA2-2(pESPF) grown in the presence of 0.25, 0.5, 0.75, or 1% glucose concentrations, indicating that increasing glucose in the growth medium led to a nominal, but not significant, increase in esp expression. This implies the presence of an as-yet-unidentified glucose-regulated factor(s) that interacts directly or indirectly with Esp to enhance biofilm formation.
Interestingly, an operon, bop, consisting of four open reading frames was identified recently in a strong biofilm-forming E. faecalis strain, T9, and this locus was also found to be present in the strain V583 (16). One of the genes on the bop operon, bopD, which putatively encodes a sugar-binding transcriptional regulator, was found to be critical for the process of biofilm formation (16). Using bopD-specific primers designed from the available V583 sequence information (26), we were able to amplify the entire bop operon from FA2-2, as well as OG1RF genomic DNA. Restriction analysis of the amplified product yielded fragments of expected sizes based on V583 sequence (data not shown). Since our results indicate that glucose does not regulate esp expression, it is possible that bopD regulates the expression of other factor(s), which contribute to biofilm formation synergistically with Esp in these strains. Although this is merely speculative at this stage, a better understanding of the individual factors involved in enterococcal biofilm development will help unravel the regulatory network involved in this process.
Another important parameter that influences biofilm formation is cell surface hydrophobicity (3, 6, 7). Cell surface hydrophobicity has been thought to play a role in the initial stages of biofilm formation by promoting cell-substrate interactions (7). Although Esp expression on the cell surface significantly enhanced the bacterial cell surface hydrophobicity, an increase in hydrophobicity alone did not account for differences in biofilm formation by the E. faecalis strains tested. Interestingly, a previous study (38) examined the contribution of surface proteins to the adherence of E. faecalis to bile drain materials and determined that Esp expression promoted a strong interaction with the substratum, leading to higher numbers of adherent bacteria, whereas another surface protein, aggregation substance, promoted bacterial cell-cell interaction due to positive cooperativity, leading to increased cell numbers at the surface of the substratum. Synergy between these traits may occur and enhance biofilm formation, a prospect that is being investigated further.
Gelatinase, a secreted protease, has recently been implicated in the process of OG1RF biofilm formation (20). In order to examine the possibility of a synergistic effect between gelatinase and Esp on biofilm formation in an isogenic background, we cotransformed FA2-2 with pCUGel and pESPF. In contrast to previous results (20), gelatinase, when constitutively expressed in FA2-2 under the influence of aphA promoter, did not affect the ability of the wild-type Esp-negative FA2-2 or the Esp-positive FA2-2(pESPF) to form biofilms under the conditions tested. The reason for this observed discrepancy is not clear, and whether it is an FA2-2-specific effect remains to be discerned.
The esp gene has been identified to be part of a large pathogenicity island in E. faecalis (31). Infection-derived isolates are enriched for the presence of the pathogenicity island, which is absent or rarely present in commensal isolates (31, 35). Interestingly, strain MMH594 from which the pathogenicity island was first discovered, is a poor biofilm former (20; unpublished observations), leading Kristich et al. (20) to conclude that neither Esp nor the pathogenicity island are essential for biofilm formation. However, the present results show unambiguously that Esp enhances biofilm formation in E. faecalis.
In summary, our results demonstrate that Esp significantly enhances the ability of E. faecalis to form biofilms on three different surfaces in a glucose-dependent manner and that this enhancement is not directly attributable to an increase in cell surface hydrophobicity.
Acknowledgments
This study was supported, in part, by Public Health Service grant AI 059673 (N.S.) from the National Institutes of Health.
We thank Lynn Hancock for providing us with the pCUGel construct and Arne Heydorn for the COMSTAT program. We are indebted to Jim Henthorn for assistance with CLSM. The helpful discussions from members of the University of Oklahoma Health Sciences Center COBRE biofilm group are gratefully acknowledged.
Editor: J. N. Weiser
REFERENCES
- 1.Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K. D. Entian, and F. Gotz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149-1154. [DOI] [PubMed] [Google Scholar]
- 2.Baldassarri, L., R. Cecchini, L. Bertuccini, M. G. Ammendolia, F. Iosi, C. R. Arciola, L. Montanaro, R. Di Rosa, G. Gherardi, G. Dicuonzo, G. Orefici, and R. Creti. 2001. Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Med. Microbiol. Immunol. 190:113-120. [DOI] [PubMed] [Google Scholar]
- 3.Bruinsma, G. M., H. C. van der Mei, and H. J. Busscher. 2001. Bacterial adhesion to surface hydrophilic and hydrophobic contact lenses. Biomaterials 22:3217-3224. [DOI] [PubMed] [Google Scholar]
- 4.Camper, A. K., W. L. Jones, and J. T. Hayes. 1996. Effect of growth conditions and substratum composition on the persistence of coliforms in mixed-population biofilms. Appl. Environ. Microbiol. 62:4014-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewanti, R., and A. C. Wong. 1995. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 26:147-164. [DOI] [PubMed] [Google Scholar]
- 7.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dworniczek, E., K. Kuzko, E. Mroz, L. Wojciech, R. Adamski, B. Sobieszczanska, and A. Seniuk. 2003. Virulence factors and in vitro adherence of Enterococcus strains to urinary catheters. Folia Microbiol. 48:671-678. [DOI] [PubMed] [Google Scholar]
- 11.Eaton, T. J., and M. J. Gasson. 2002. A variant enterococcal surface protein Esp(fm) in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol. Lett. 216:269-275. [DOI] [PubMed] [Google Scholar]
- 12.Giessel, B. E., C. J. Koenig, and R. L. Blake, Jr. 2000. Management of bacterial endocarditis. Am. Fam. Physician 61:1725-1732, 1739. [PubMed] [Google Scholar]
- 13.Gilmore, K. S., P. Srinivas, D. R. Akins, K. L. Hatter, and M. S. Gilmore. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 71:4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 15.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt. 10):2395-2407. [DOI] [PubMed] [Google Scholar]
- 16.Hufnagel, M., S. Koch, R. Creti, L. Baldassarri, and J. Huebner. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189:420-430. [DOI] [PubMed] [Google Scholar]
- 17.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huycke, M. M., C. A. Spiegel, and M. S. Gilmore. 1991. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186:154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 25.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 26.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 27.Peng, M., J. Ping Gong, and Y. Osada. 2003. Substrate effect on the formation of hydrogels with heterogeneous network structure. Chem. Rec. 3:40-50. [DOI] [PubMed] [Google Scholar]
- 28.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg, M., A. Perry, E. A. Bayer, D. L. Gutnick, E. Rosenberg, and I. Ofek. 1981. Adherence of Acinetobacter calcoaceticus RAG-1 to human epithelial cells and to hexadecane. Infect. Immun. 33:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandoe, J. A., I. R. Witherden, J. H. Cove, J. Heritage, and M. H. Wilcox. 2003. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol. 52:547-550. [DOI] [PubMed] [Google Scholar]
- 31.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 32.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 35.Tendolkar, P. M., A. S. Baghdayan, and N. Shankar. 2003. Pathogenic enterococci: new developments in the 21st century. Cell. Mol. Life Sci. 60:2622-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waar, K., H. C. van der Mei, H. J. Harmsen, J. E. Degener, and H. J. Busscher. 2002. Enterococcus faecalis surface proteins determine its adhesion mechanism to bile drain materials. Microbiology 148:1863-1870. [DOI] [PubMed] [Google Scholar]
- 39.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 40.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]